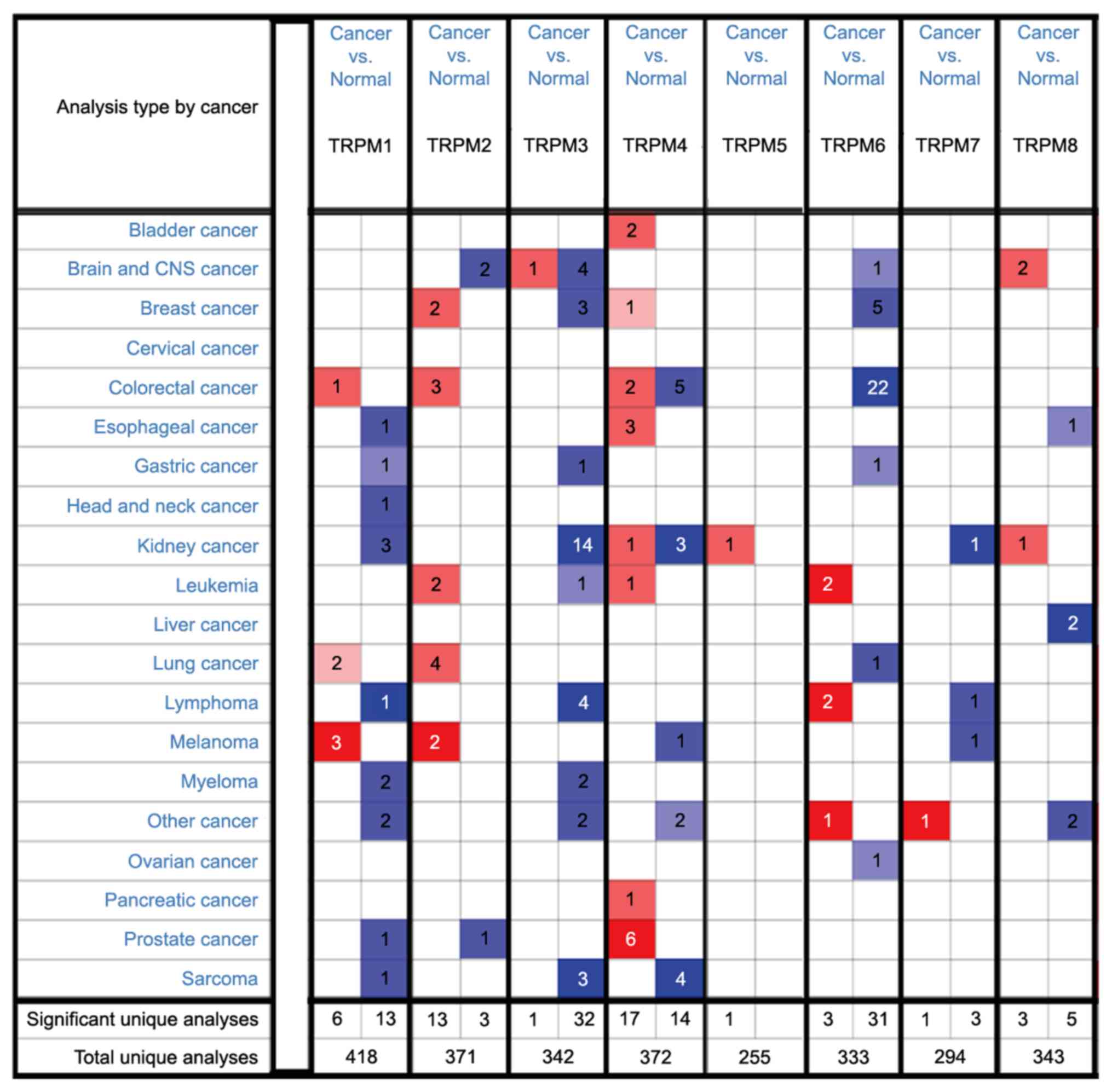
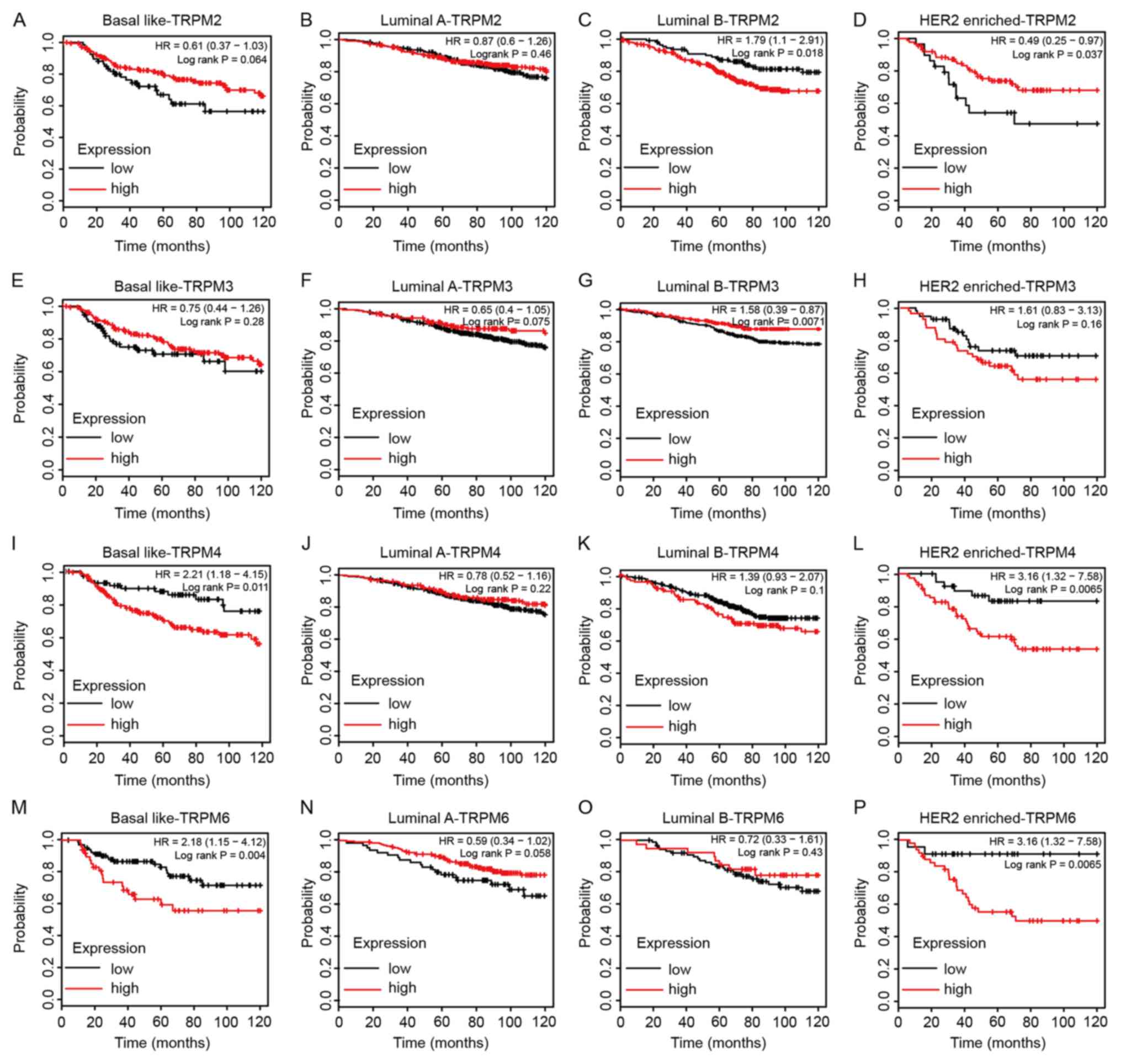
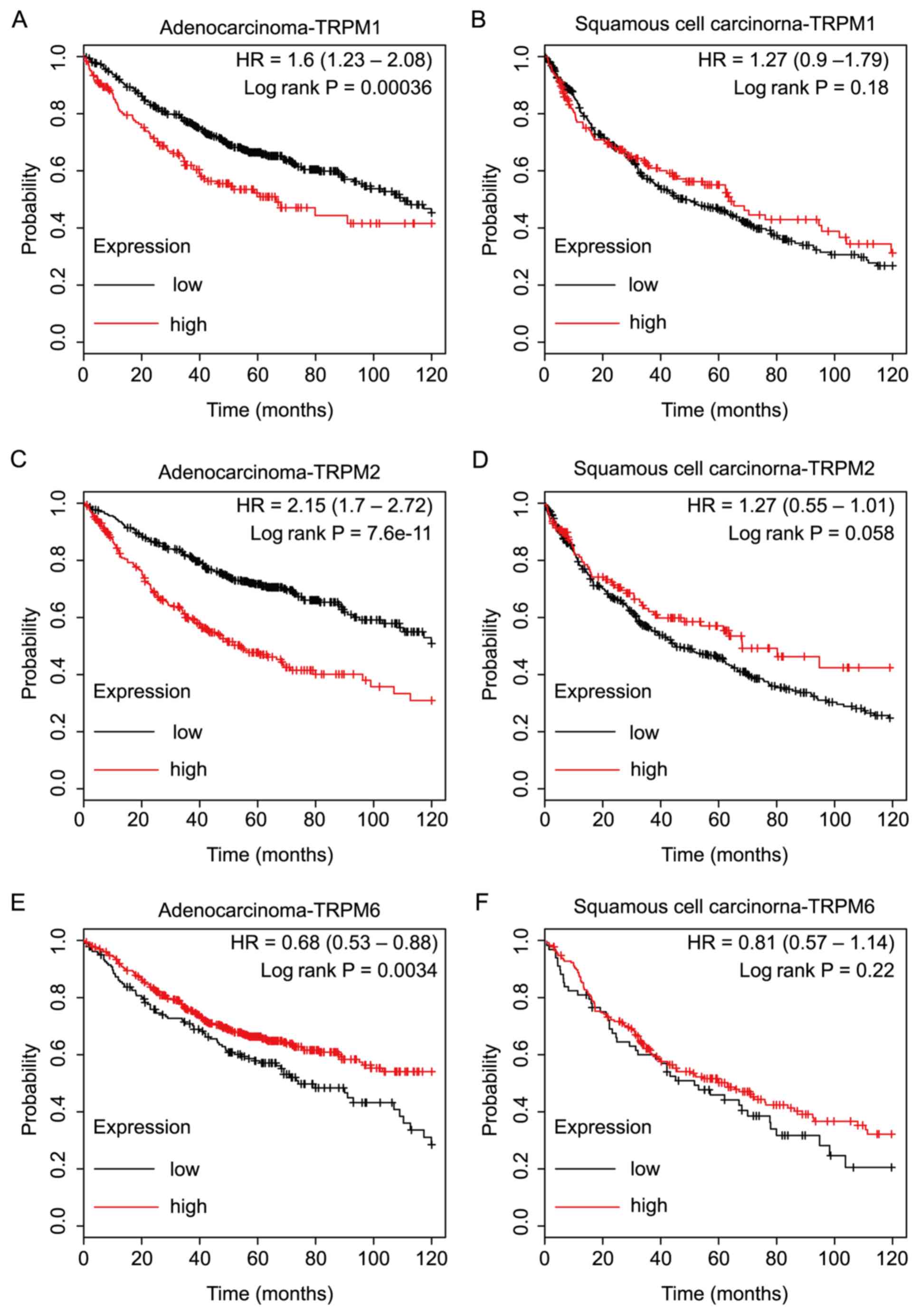
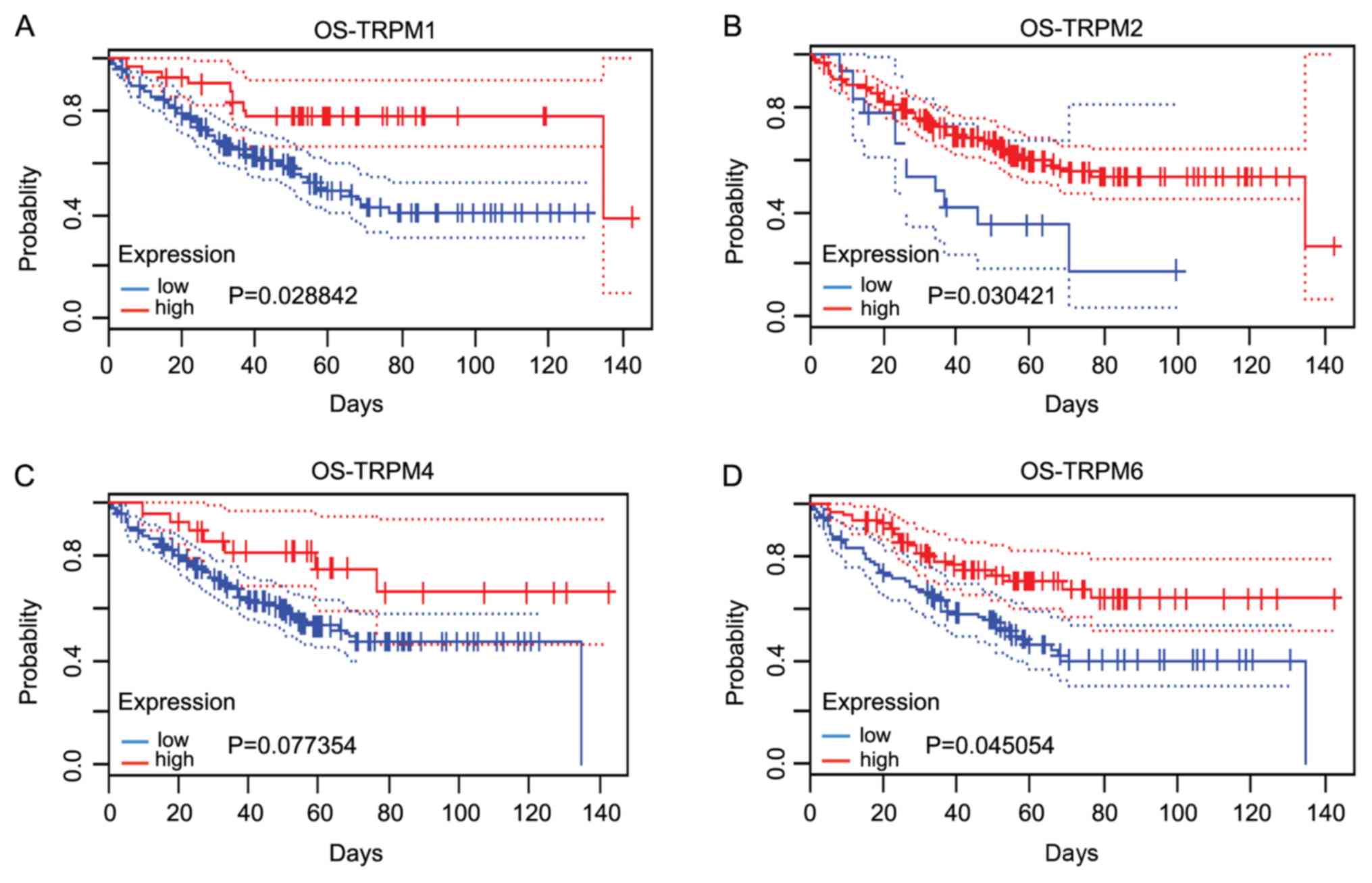
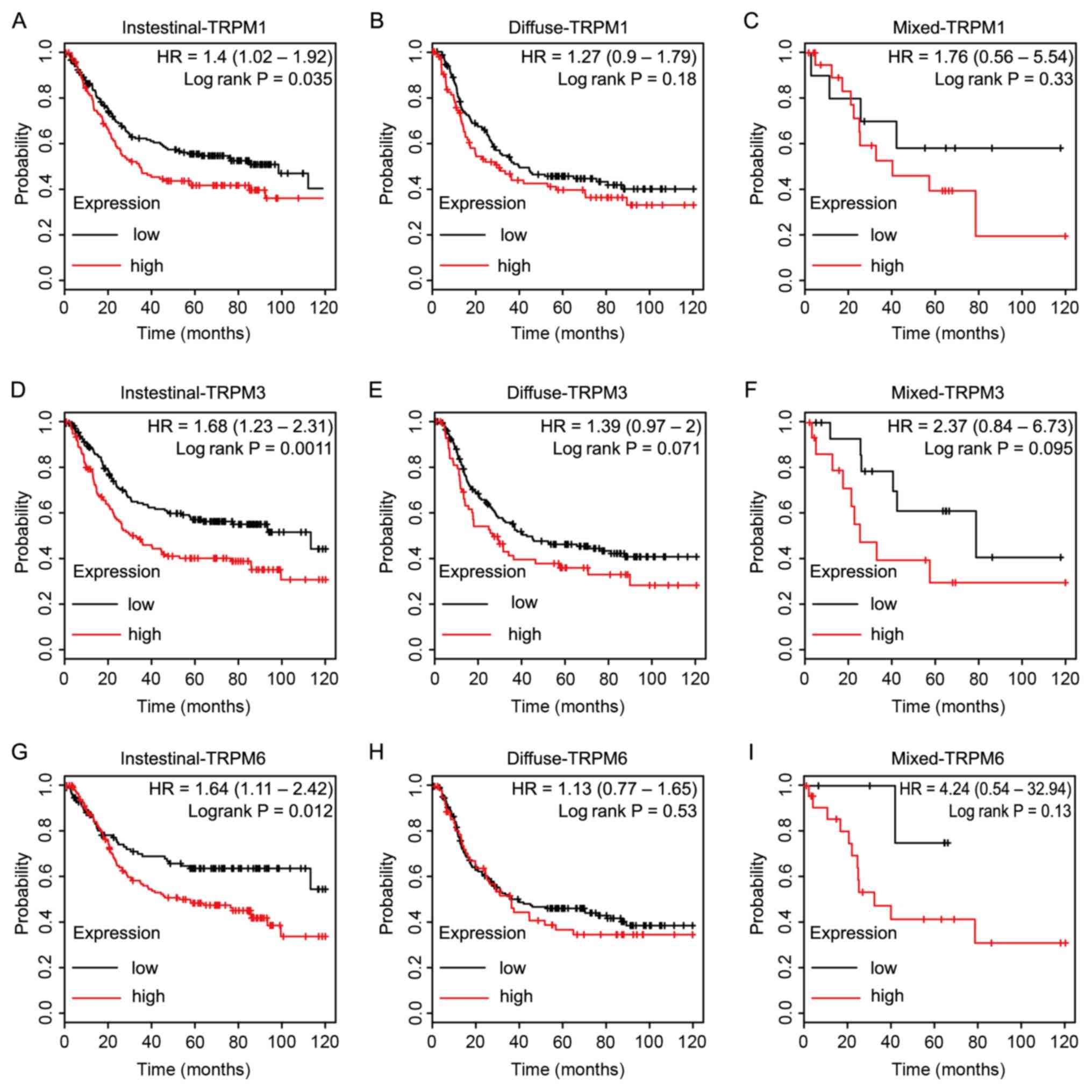
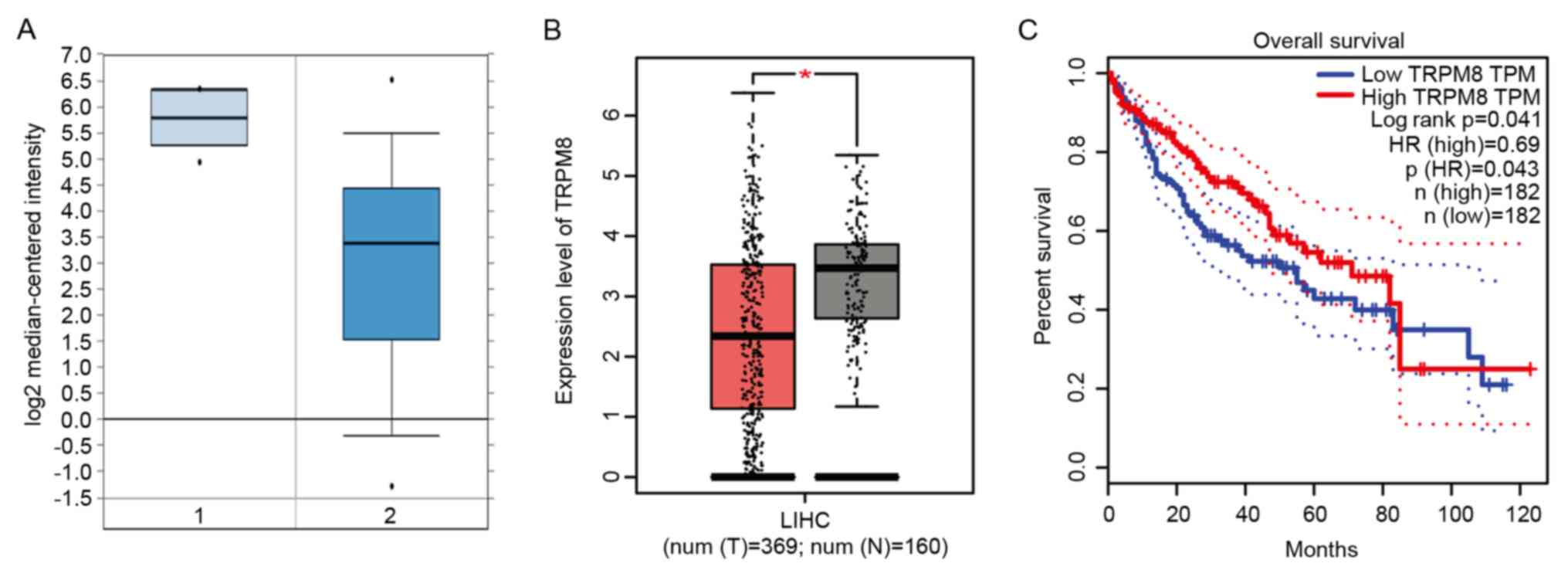
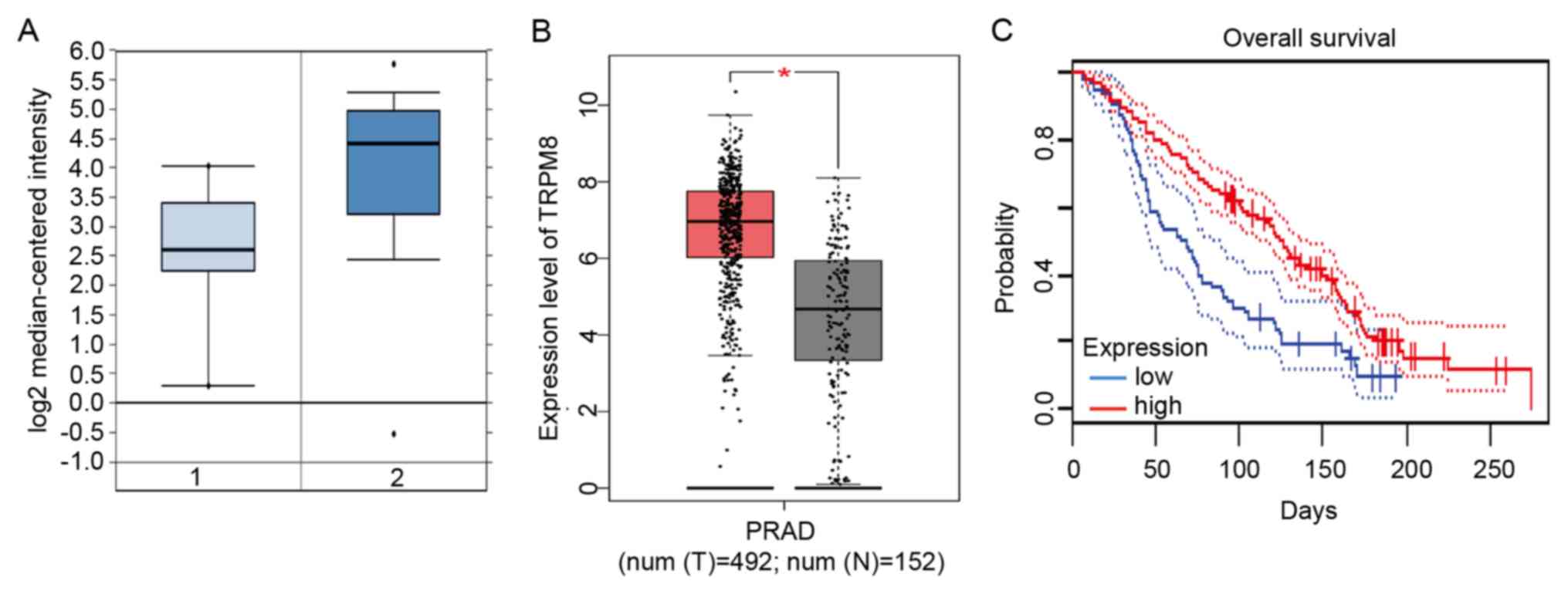
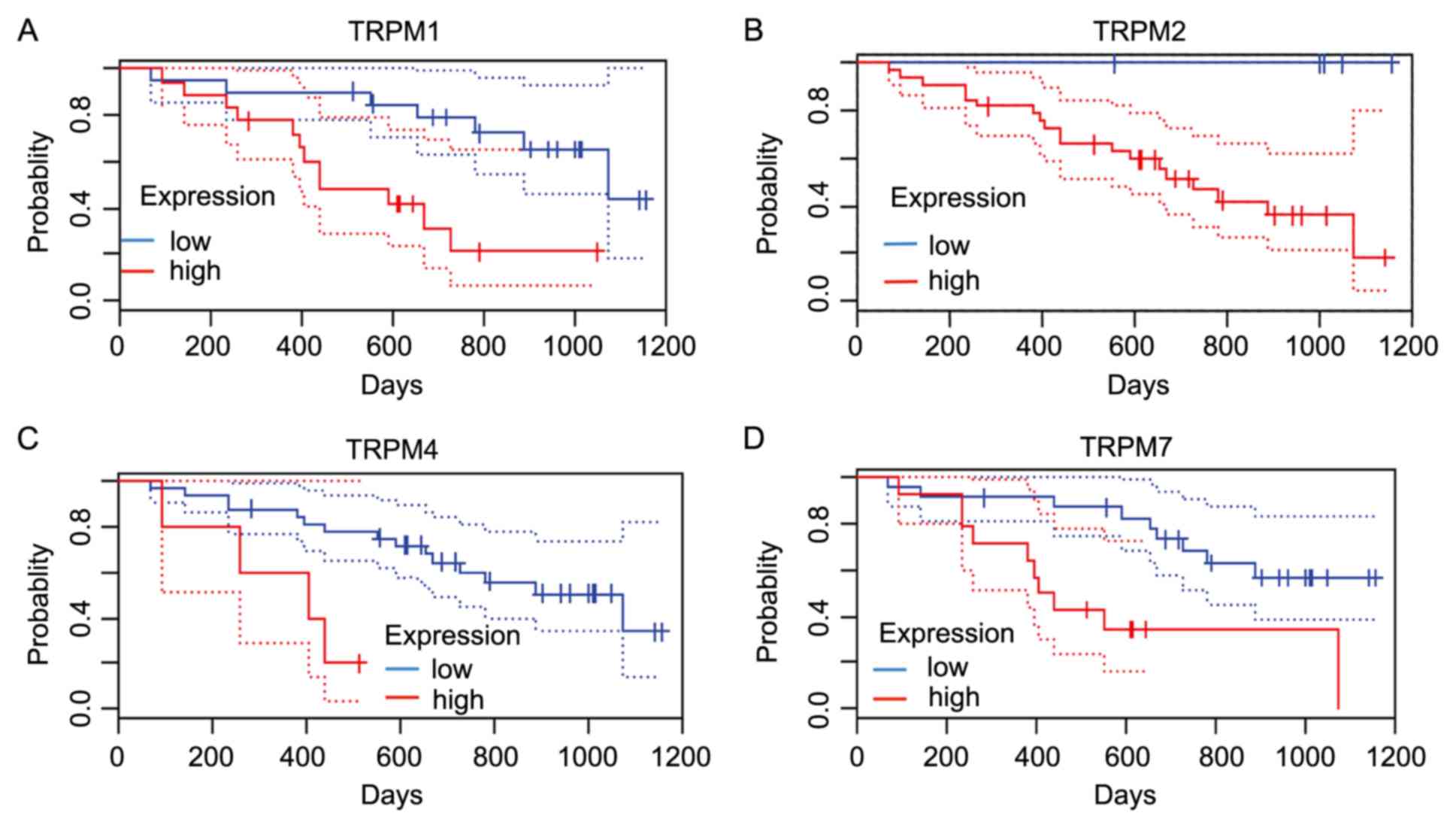
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fidler MM, Soerjomataram I and Bray F: A
global view on cancer incidence and national levels of the human
development index. Int J Cancer. 139:2436–2446. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Montell C and Rubin GM: Molecular
characterization of the Drosophila trp locus: A putative integral
membrane protein required for phototransduction. Neuron.
2:1313–1323. 1989.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Schmitz C and Perraud AL: The TRPM cation
channels in the immune context. Curr Pharm Design. 11:2765–2778.
2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zeng X, Sikka SC, Huang L, Sun C, Xu C,
Jia D, Abdel-Mageed AB, Pottle JE, Taylor JT and Li M: Novel role
for the transient receptor potential channel TRPM2 in prostate
cancer cell proliferation. Prostate Cancer Prostatic Dis.
13:195–201. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Armisen R, Marcelain K, Simon F, Tapia JC,
Toro J, Quest AF and Stutzin A: TRPM4 enhances cell proliferation
through up-regulation of the β-catenin signaling pathway. J Cell
Physiol. 226:103–109. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Meng X, Cai C, Wu J, Cai S, Ye C, Chen H,
Yang Z, Zeng H, Shen Q and Zou F: TRPM7 mediates breast cancer cell
migration and invasion through the MAPK pathway. Cancer Lett.
333:96–102. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hantute-Ghesquier A, Haustrate A,
Prevarskaya N and Lehen'kyi V: TRPM family channels in cancer.
Pharmaceuticals (Basel). 11(2)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wong KK, Banham AH, Yaacob NS and Nur
Husna SM: The oncogenic roles of TRPM ion channels in cancer. J
Cell Physiol, 2019.
|
|
10
|
Nagy Á, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep.
8(9227)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li Q, Birkbak NJ, Gyorffy B, Szallasi Z
and Eklund AC: Jetset: Selecting an optimal microarray probe set to
represent a gene. BMC Bioinformatics. 12(474)2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: A new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2(18)2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gyorffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bhattacharjee A, Richards WG, Staunton J,
Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et
al: Classification of human lung carcinomas by mRNA expression
profiling reveals distinct adenocarcinoma subclasses. Proc Natl
Acad Sci USA. 98:13790–13795. 2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gyorffy B, Surowiak P, Budczies J and
Lanczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8(e82241)2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5(pii: e13091)2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hong Y, Downey T, Eu KW, Koh PK and Cheah
PY: A ‘metastasis-prone’ signature for early-stage mismatch-repair
proficient sporadic colorectal cancer patients and its implications
for possible therapeutics. Clin Exp Metastasis. 27:83–90.
2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sabates-Bellver J, Van der Flier LG, de
Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA,
Bujnicki JM, Menigatti M, et al: Transcriptome profile of human
colorectal adenomas. Mol Cancer Res. 5:1263–1275. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5(pii: e13091)2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gaedcke J, Grade M, Jung K, Camps J, Jo P,
Emons G, Gehoff A, Sax U, Schirmer M, Becker H, et al: Mutated KRAS
results in overexpression of DUSP4, a MAP-kinase phosphatase and
SMYD3, a histone methyltransferase, in rectal carcinomas. Genes
Chromosomes Cancer. 49:1024–1034. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kaiser S, Park YK, Franklin JL, Halberg
RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et
al: Transcriptional recapitulation and subversion of embryonic
colon development by mouse colon tumor models and human colon
cancer. Genome Biol. 8(R131)2007.PubMed/NCBI View Article : Google Scholar
|
|
26
|
D'Errico M, de Rinaldis E, Blasi MF, Viti
V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D,
et al: Genome-wide expression profile of sporadic gastric cancers
with microsatellite instability. Eur J Cancer. 45:461–469.
2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan
DW, Tang HM and Peng ZH: Upregulated INHBA expression is associated
with poor survival in gastric cancer. Med Oncol. 29:77–83.
2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tomlins SA, Mehra R, Rhodes DR, Cao X,
Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA,
Pienta KJ, et al: Integrative molecular concept modeling of
prostate cancer progression. Nat Genet. 39:41–51. 2007.PubMed/NCBI View Article : Google Scholar
|
|
29
|
VVarambally S, Yu J, Laxman B, Rhodes DR,
Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, et
al: Integrative genomic and proteomic analysis of prostate cancer
reveals signatures of metastatic progression. Cancer Cell.
8:393–406. 2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Liu P, Ramachandran S, Ali Seyed M,
Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW,
Jaye DL, et al: Sex-determining region Y box 4 is a transforming
oncogene in human prostate cancer cells. Cancer Res. 66:4011–4019.
2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Vanaja DK, Cheville JC, Iturria SJ and
Young CYF: Transcriptional silencing of zinc finger protein 185
identified by expression profiling is associated with prostate
cancer progression. Cancer Res. 63:3877–3882. 2003.PubMed/NCBI
|
|
32
|
Grasso CS, Wu YM, Robinson DR, Cao X,
Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC,
Asangani IA, et al: The mutational landscape of lethal
castration-resistant prostate cancer. Nature. 487:239–243.
2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Arredouani MS, Lu B, Bhasin M, Eljanne M,
Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA, et al:
Identification of the transcription factor single-minded homologue
2 as a potential biomarker and immunotherapy target in prostate
cancer. Clin Cancer Res. 15:5794–5802. 2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wallace TA, Prueitt RL, Yi M, Howe TM,
Gillespie JW, Yfantis HG, Stephens RM, Caporaso NE, Loffredo CA and
Ambs S: Tumor immunobiological differences in prostate cancer
between African-American and European-American men. Cancer Res.
68:927–936. 2008.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Talantov D, Mazumder A, Yu JX, Briggs T,
Jiang Y, Backus J, Atkins D and Wang Y: Novel genes associated with
malignant melanoma but not benign melanocytic lesions. Clin Cancer
Res. 11:7234–7242. 2005.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Haqq C, Nosrati M, Sudilovsky D, Crothers
J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR III, Allen RE,
Singer MI, et al: The gene expression signatures of melanoma
progression. Proc Natl Acad Sci USA. 102:6092–6097. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Riker AI, Enkemann SA, Fodstad O, Liu S,
Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, et al: The
gene expression profiles of primary and metastatic melanoma yields
a transition point of tumor progression and metastasis. BMC Med
Genomics. 1(13)2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Cutcliffe C, Kersey D, Huang CC, Zeng Y,
Walterhouse D and Perlman EJ: Clear cell sarcoma of the kidney:
up-regulation of neural markers with activation of the sonic
hedgehog and Akt pathways. Clin Cancer Res. 11:7986–7994.
2005.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jones J1, Otu H, Spentzos D, Kolia S, Inan
M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al: Gene
signatures of progression and metastasis in renal cell cancer. Clin
Cancer Res. 11:5730–5739. 2005.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gumz ML1, Zou H, Kreinest PA, Childs AC,
Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et
al: Secreted frizzled-related protein 1 loss contributes to tumor
phenotype of clear cell renal cell carcinoma. Clin Cancer Res.
13:4740–4749. 2007.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Beroukhim R1, Brunet JP, Di Napoli A,
Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin
MA, et al: Patterns of gene expression and copy-number alterations
in von-hippel lindau disease-associated and sporadic clear cell
carcinoma of the kidney. Cancer Res. 69:4674–4681. 2009.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lenburg ME, Liou LS, Gerry NP, Frampton
GM, Cohen HT and Christman MF: Previously unidentified changes in
renal cell carcinoma gene expression identified by parametric
analysis of microarray data. BMC Cancer. 3(31)2003.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yusenko MV, Kuiper RP, Boethe T, Ljungberg
B, van Kessel AG and Kovacs G: High-resolution DNA copy number and
gene expression analyses distinguish chromophobe renal cell
carcinomas and renal oncocytomas. BMC Cancer. 9(152)2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hao Y, Triadafilopoulos G, Sahbaie P,
Young HS, Omary MB and Lowe AW: Gene expression profiling reveals
stromal genes expressed in common between Barrett's esophagus and
adenocarcinoma. Gastroenterology. 131:925–933. 2006.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kimchi ET, Posner MC, Park JO, Darga TE,
Kocherginsky M, Karrison T, Hart J, Smith KD, Mezhir JJ,
Weichselbaum RR, et al: Progression of Barrett's metaplasia to
adenocarcinoma is associated with the suppression of the
transcriptional programs of epidermal differentiation. Cancer Res.
65:3146–3154. 2005.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kim SM, Park YY, Park ES, Cho JY, Izzo JG,
Zhang D, Kim SB, Lee JH, Bhutani MS, Swisher SG, Wu X, Coombes KR,
et al: Prognostic biomarkers for esophageal adenocarcinoma
identified by analysis of tumor transcriptome. PLoS One.
5(e15074)2010.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Liang Y, Diehn M, Watson N, et al: Gene
expression profiling reveals molecularly and clinically distinct
subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A.
102:5814–5819. 2005.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Bredel M1, Bredel C, Juric D, Harsh GR,
Vogel H, Recht LD and Sikic BI: Functional network analysis reveals
extended gliomagenesis pathway maps and three novel MYC-interacting
genes in human gliomas. Cancer Res. 65:8679–8689. 2005.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Lee J, Kotliarova S, Kotliarov Y, Li A, Su
Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al:
Tumor stem cells derived from glioblastomas cultured in bFGF and
EGF more closely mirror the phenotype and genotype of primary
tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403.
2006.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Sun L1, Hui AM, Su Q, Vortmeyer A,
Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey
R, et al: Neuronal and glioma-derived stem cell factor induces
angiogenesis within the brain. Cancer Cell. 9:287–300.
2006.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Murat A, Migliavacca E, Gorlia T, Lambiv
WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven
MC, et al: Stem cell-related ‘self-renewal’ signature and high
epidermal growth factor receptor expression associated with
resistance to concomitant chemoradiotherapy in glioblastoma. J Clin
Oncol. 26:3015–3024. 2008.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ginos MA, Page GP, Michalowicz BS, Patel
KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL and Gaffney PM:
Identification of a gene expression signature associated with
recurrent disease in squamous cell carcinoma of the head and neck.
Cancer Res. 64:55–63. 2004.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Maule M and Merletti F: Cancer transition
and priorities for cancer control. Lancet Oncol. 13:745–746.
2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Bray F, Soerjomataram I, Mery L and Ferlay
J: Improving the quality and coverage of cancer registries
globally. Lancet. 386:1035–1036. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chen J, Luan Y, Yu R, Zhang Z, Zhang J and
Wang W: Transient receptor potential (TRP) channels, promising
potential diagnostic and therapeutic tools for cancer. Biosci
Trends. 8:1–10. 2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Clapham DE: TRP channels as cellular
sensors. Nature. 426:517–524. 2003.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Pedersen SF, Owsianik G and Nilius B: TRP
channels: An overview. Cell Calcium. 38:233–252. 2005.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Gkika D and Prevarskaya N: Molecular
mechanisms of TRP regulation in tumor growth and metastasis.
Biochim Biophys Acta. 1793:953–958. 2009.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Winters S, Martin C, Murphy D and Shokar
NK: Breast cancer epidemiology, prevention and screening. Prog Mol
Biol Transl. 151:1–32. 2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Dai X, Xiang L, Li T and Bai Z: Cancer
hallmarks, biomarkers and breast cancer molecular subtypes. J
Cancer. 7:1281–1294. 2016.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J
and Shi B: Breast cancer intrinsic subtype classification, clinical
use and future trends. Am J Cancer Res. 5:2929–2943.
2015.PubMed/NCBI
|
|
62
|
Spitale A, Mazzola P, Soldini D,
Mazzucchelli L and Bordoni A: Breast cancer classification
according to immunohistochemical markers: Clinicopathologic
features and short-term survival analysis in a population-based
study from the South of Switzerland. Ann Oncol. 20:628–635.
2009.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Sumoza-Toledo A, Espinoza-Gabriel MI and
Montiel-Condado D: Evaluation of the TRPM2 channel as a biomarker
in breast cancer using public databases analysis. Bol Med Hosp
Infant Mex. 73:397–404. 2016.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Stephens PJ, Tarpey PS, Davies H, Van Loo
P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell
GR, et al: The landscape of cancer genes and mutational processes
in breast cancer. Nature. 486:400–404. 2012.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Huang C, Qin Y, Liu H, Liang N, Chen Y, Ma
D, Han Z, Xu X, Zhou X, He J and Li S: Downregulation of a novel
long noncoding RNA TRPM2-AS promotes apoptosis in non-small cell
lung cancer. Tumour Biol. 39(1010428317691191)2017.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691.
2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Bray F, Ren JS, Masuyer E and Ferlay J:
Global estimates of cancer prevalence for 27 sites in the adult
population in 2008. Int J Cancer. 132:1133–1145. 2013.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Kim BJ, Kim SY, Lee S, Jeon JH, Matsui H,
Kwon YK, Kim SJ and So I: The role of transient receptor potential
channel blockers in human gastric cancer cell viability. Can J
Physiol Pharmacol. 90:175–186. 2012.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Almasi S, Kennedy BE, El-Aghil M, Sterea
AM, Gujar S, Partida-Sánchez S and El Hiani Y: TRPM2
channel-mediated regulation of autophagy maintains mitochondrial
function and promotes gastric cancer cell survival via the
JNK-signaling pathway. J Biol Chem. 293:3637–3650. 2018.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Bidaux G, Borowiec AS, Dubois C, Delcourt
P, Schulz C, Vanden Abeele F, Lepage G, Desruelles E, Bokhobza A,
Dewailly E, et al: Targeting of short TRPM8 isoforms induces
4TM-TRPM8-dependent apoptosis in prostate cancer cells. Oncotarget.
7:29063–29080. 2016.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Peng M, Wang Z, Yang Z, Tao L, Liu Q, Yi
LU and Wang X: Overexpression of short TRPM8 variant a promotes
cell migration and invasion and decreases starvation-induced
apoptosis in prostate cancer LNCaP cells. Oncol Lett. 10:1378–1384.
2015.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Deeds J, Cronin F and Duncan LM: Patterns
of melastatin mRNA expression in melanocytic tumors. Hum Pathol.
31:1346–1356. 2000.PubMed/NCBI
|