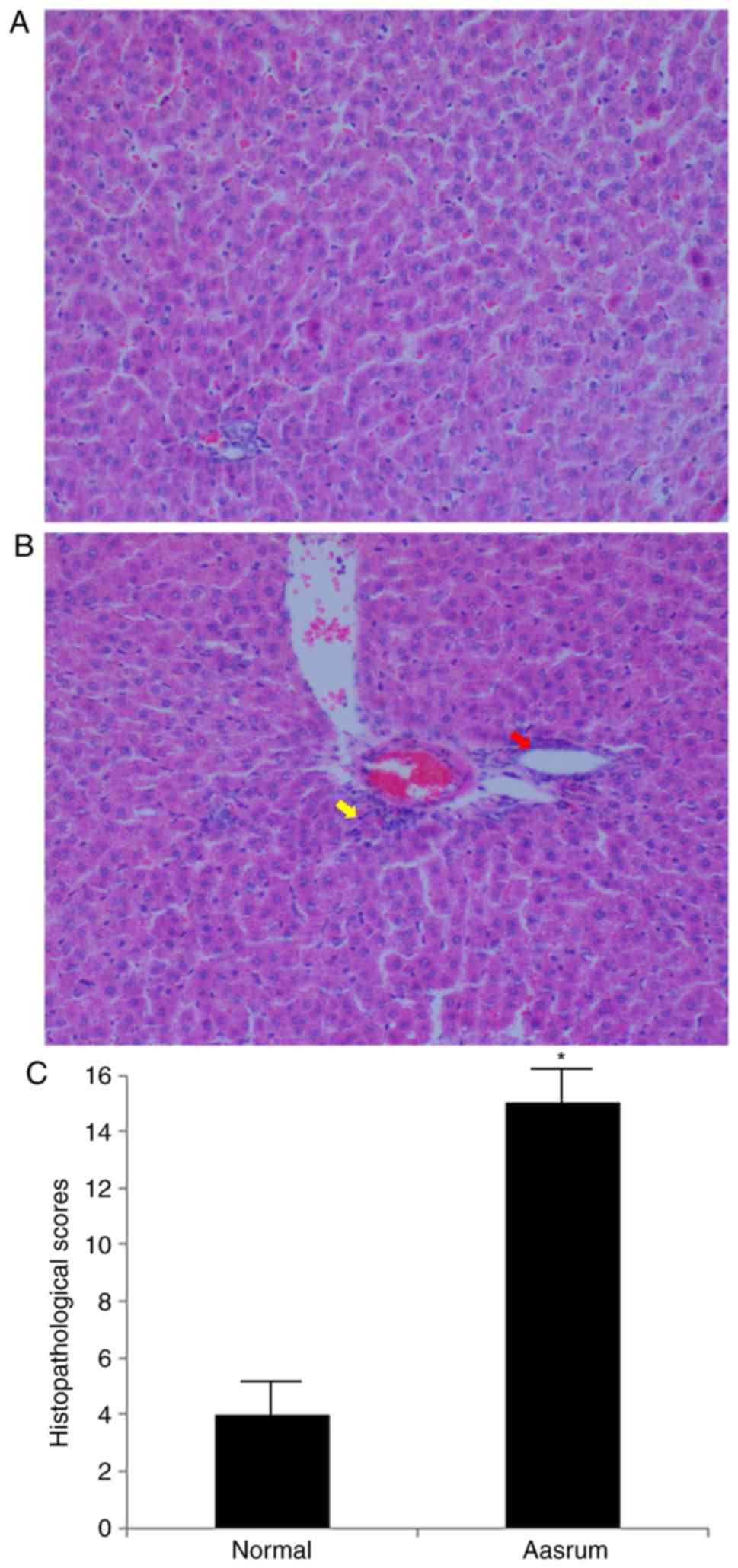
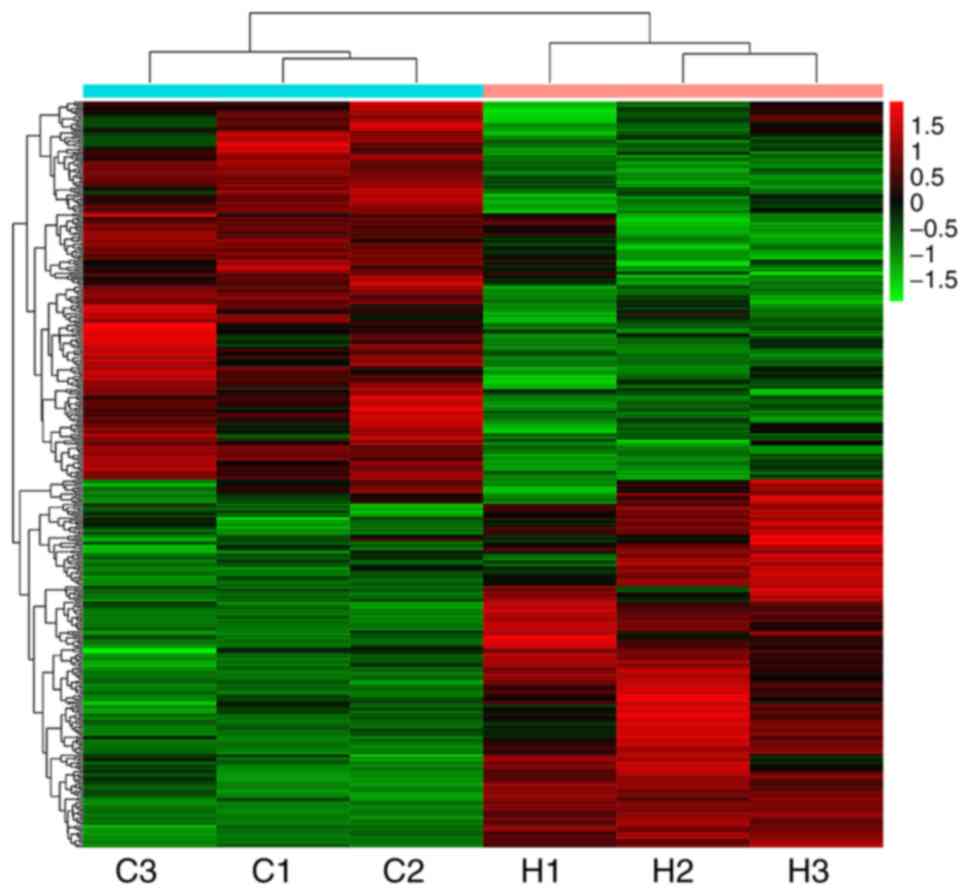
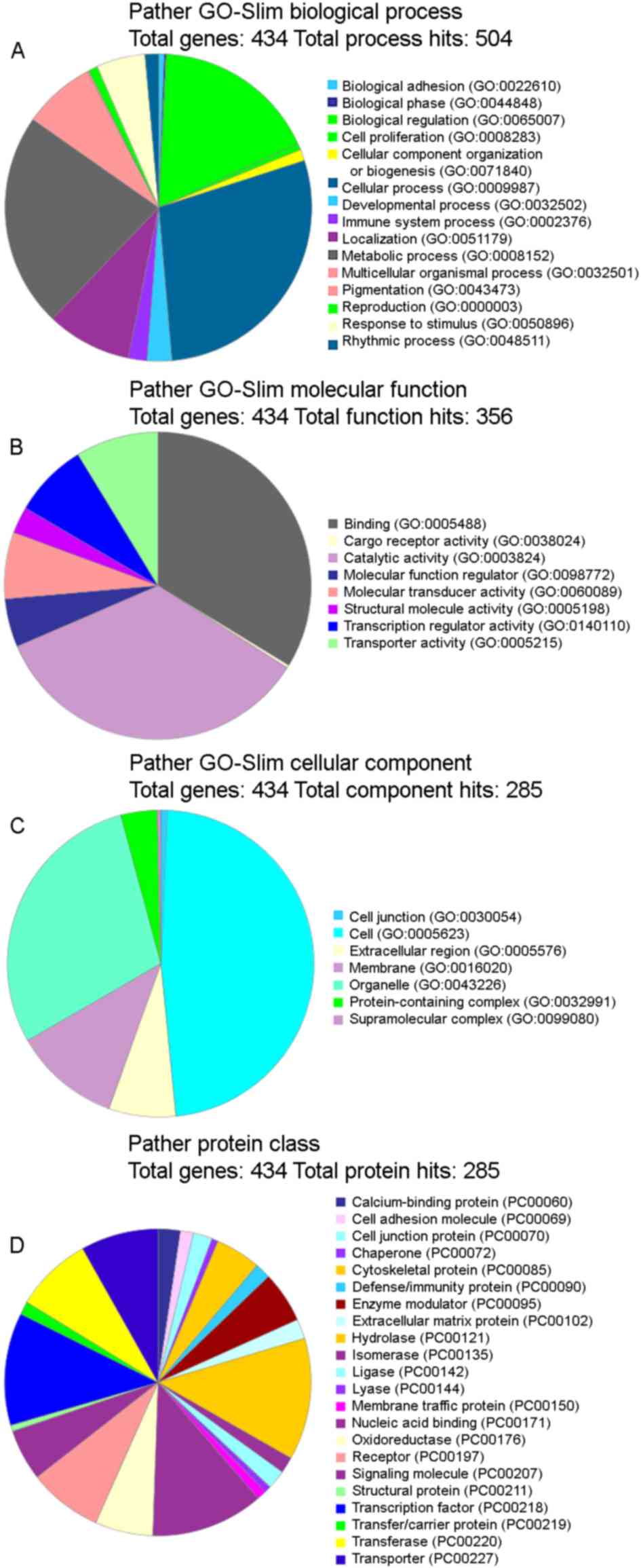
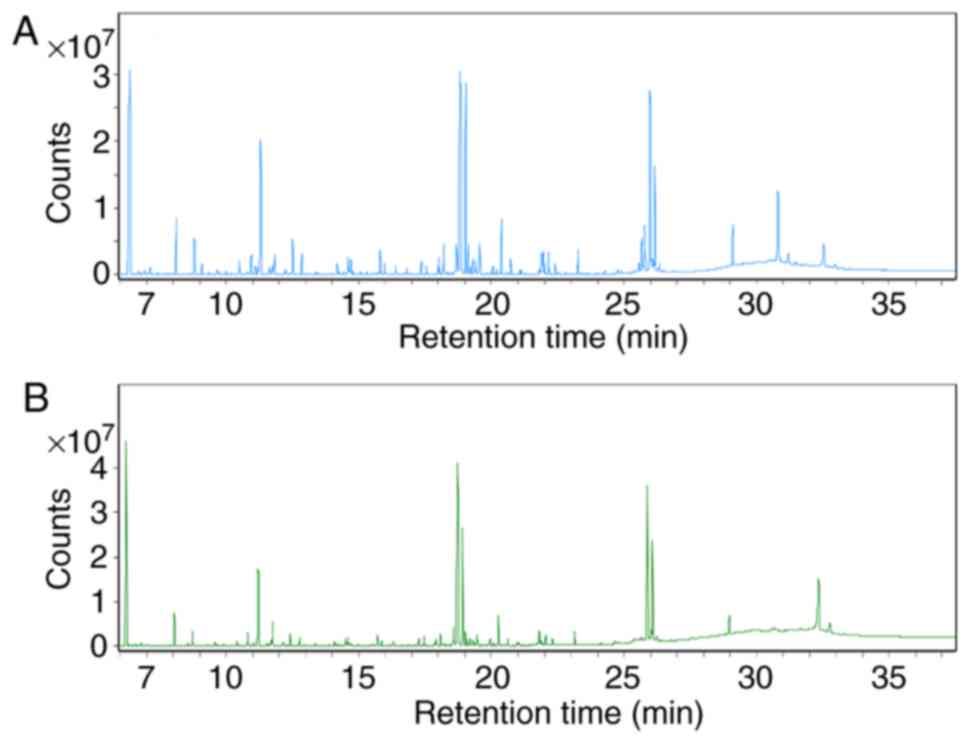
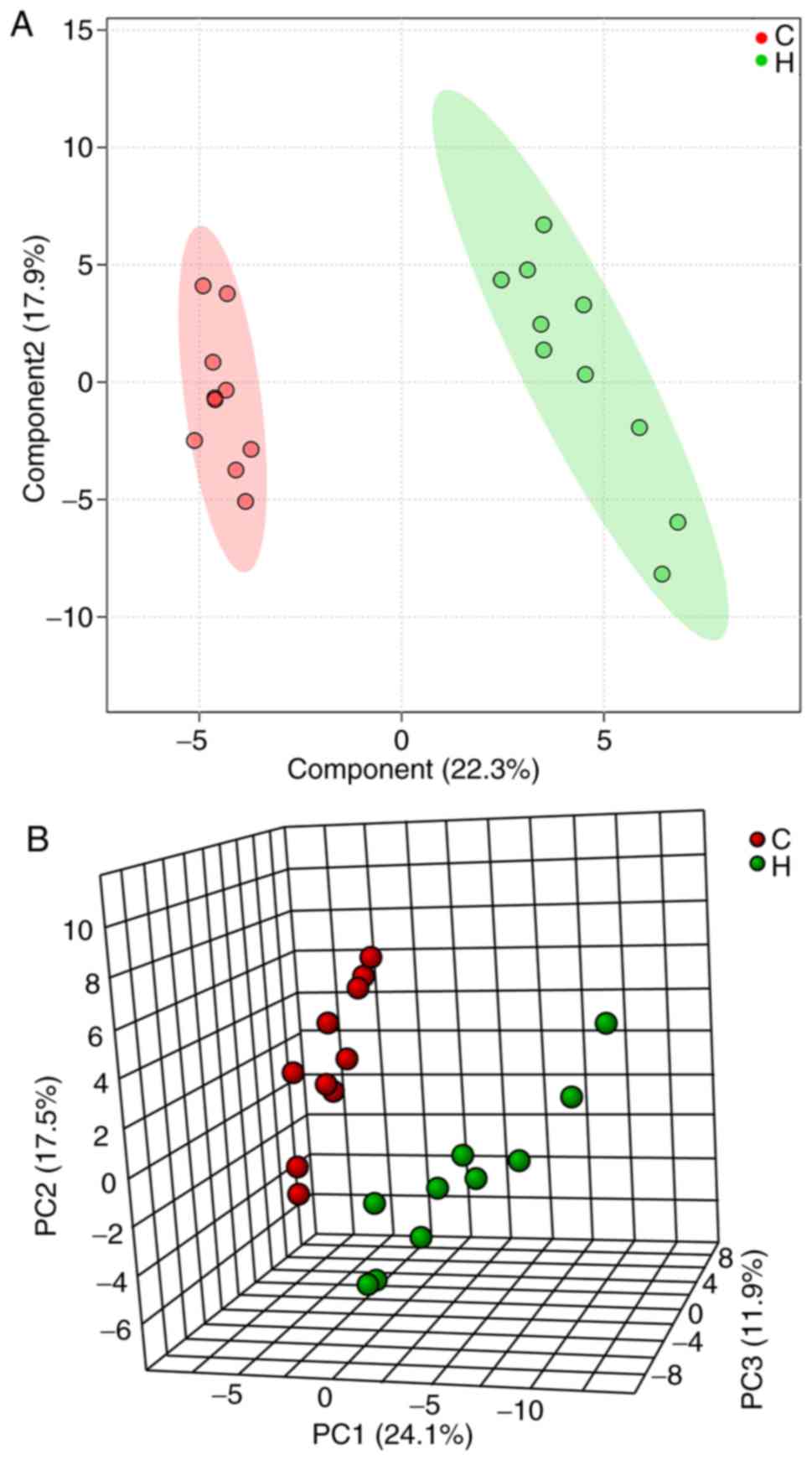
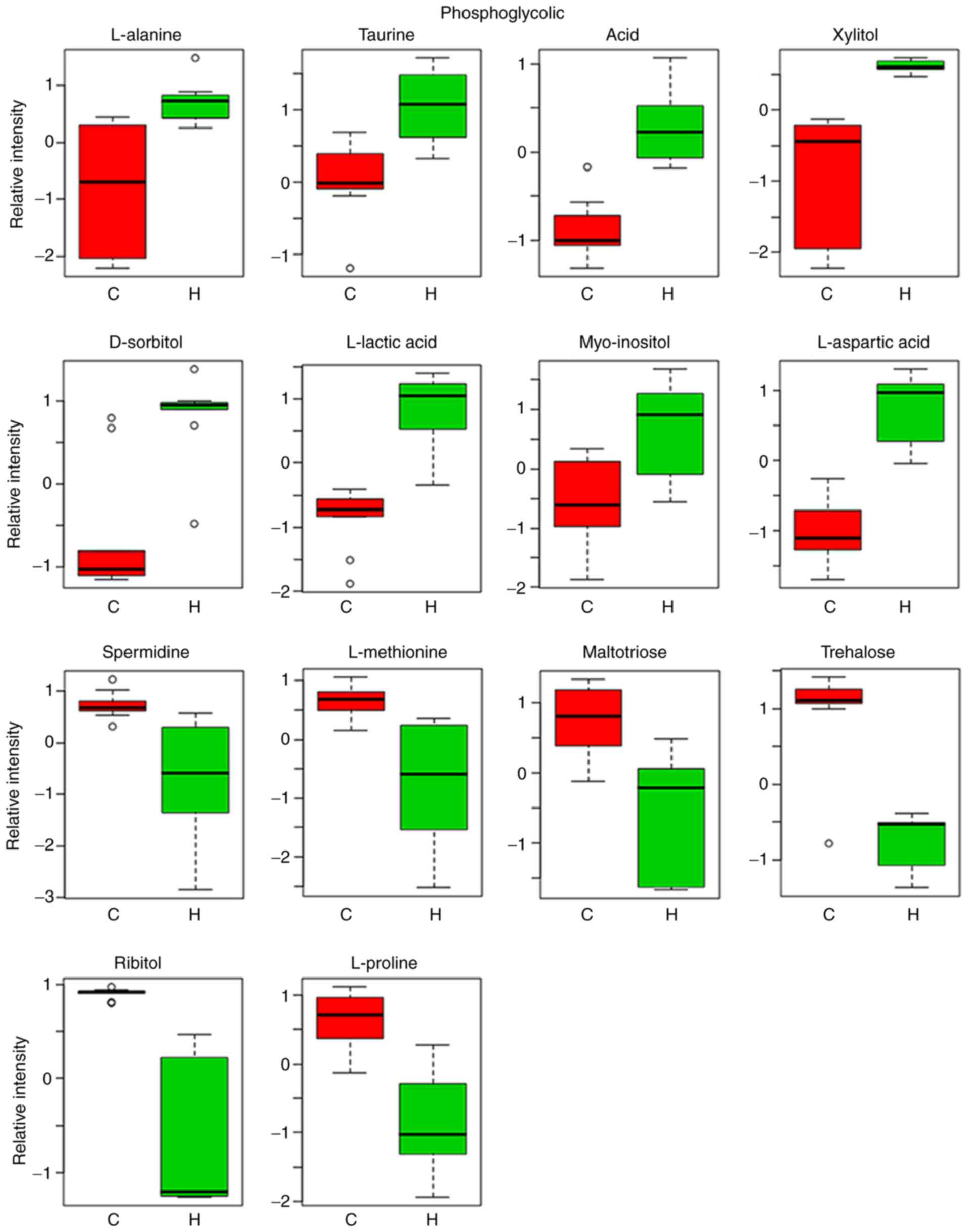
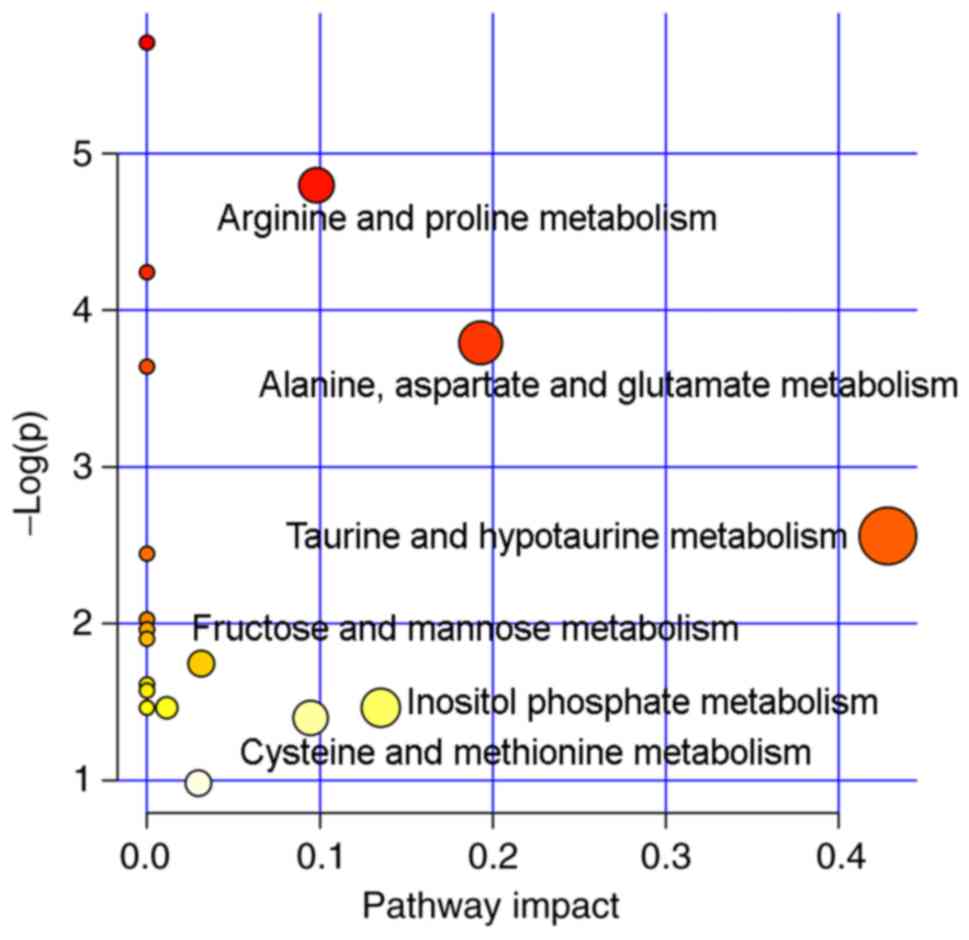
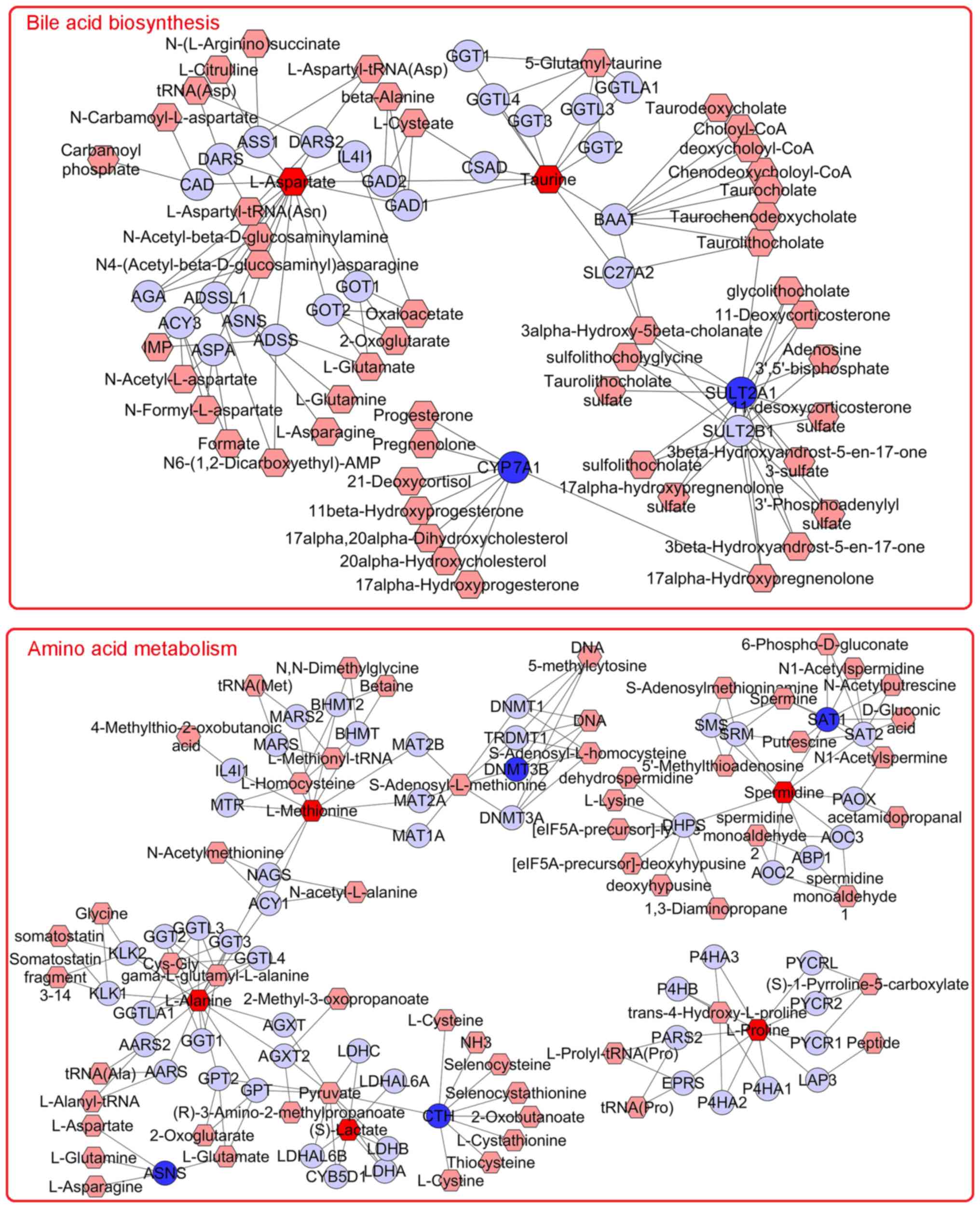
|
1
|
Jing Y, Zhang YF, Shang MY, Liu GX, Li YL,
Wang X and Cai SQ: Chemical constituents from the roots and
rhizomes of Asarum heterotropoides var. mandshuricum
and the in vitro anti-inflammatory activity. Molecules. 22(pii:
E125)2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhang W, Zhang J, Zhang M and Nie L:
Protective effect of Asarum extract in rats with adjuvant
arthritis. Exp Ther Med. 8:1638–1642. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Han S, Huang J, Hou J and Wang S:
Screening epidermal growth factor receptor antagonists from Radix
et Rhizoma Asari by two-dimensional liquid chromatography. J Sep
Sci. 37:1525–1532. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Huang J, Wang HQ, Zhang C, Li GY, Lin RC
and Wang JH: A new tetrahydrofuran-type lignan with
anti-inflammatory activity from Asarum heterotropoides Fr.
Schmidt var. mandshuricum. J Asian Nat Prod Res. 16:387–392.
2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tian HY, Hu J and Wang L: Controlled
observation of non-blister acupoint sticking and electroacupuncture
for bronchial asthma. Zhongguo Zhen Jiu. 33:485–489. 2013.(In
Chinese). PubMed/NCBI
|
|
6
|
Kim JR, Perumalsamy H, Lee JH, Ahn YJ, Lee
YS and Lee SG: Acaricidal activity of Asarum heterotropoides
root-derived compounds and hydrodistillate constitutes toward
Dermanyssus gallinae (Mesostigmata: Dermanyssidae). Exp Appl
Acarol. 68:485–495. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cartus AT, Stegmüller S, Simson N, Wahl A,
Neef S, Kelm H and Schrenk D: Hepatic metabolism of carcinogenic
β-asarone. Chem Res Toxicol. 28:1760–1773. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang Y, Guo S, Cao J, Pang X, Zhang Z,
Chen Z, Zhou Y, Geng Z, Sang Y and Du S: Toxic and Repellent
Effects of Volatile Phenylpropenes from Asarum
heterotropoides on Lasioderma serricorne and Liposcelis
bostrychophila. Molecules. 23(pii: E2131)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yang B, Xie Y, Guo M, Rosner MH, Yang H
and Ronco C: Nephrotoxicity and Chinese herbal medicine. Clin J Am
Soc Nephrol. 13:1605–1611. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Coghlan ML, Haile J, Houston J, Murray DC,
White NE, Moolhuijzen P, Bellgard MI and Bunce M: Deep sequencing
of plant and animal DNA contained within traditional Chinese
medicines reveals legality issues and health safety concerns. PLoS
Genet. 8(e1002657)2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li Y, Han L, Huang C, Dai W, Tian G, Huang
F, Li J, Liu J, Wang Q and Zhou Z: New contributions to asarum
powder on immunology related toxicity effects in lung. Evid Based
Complement Alternat Med. 2018(1054032)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li XW, Morinaga O, Tian M, Uto T, Yu J,
Shang MY, Wang X, Cai SQ and Shoyama Y: Development of an Eastern
blotting technique for the visual detection of aristolochic acids
in Aristolochia and Asarum species by using a monoclonal antibody
against aristolochic acids I and II. Phytochem Anal. 24:645–653.
2013.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Hu L, Shao H, He J, Zhong L, Song Y and Wu
F: Cytotoxicity of safrole in HepaRG cells: Studies on the role of
CYP1A2-mediated ortho-quinone metabolic activation. Xenobiotica.
49:1504–1515. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yang AH, Zhang L, Zhi DX, Liu WL, Gao X
and He X: Identification and analysis of the reactive metabolites
related to the hepatotoxicity of safrole. Xenobiotica.
48:1164–1172. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Patel DN, Ho HK, Tan LL, Tan MM Zhang Q,
Low MY, Chan CL and Koh HL: Hepatotoxic potential of asarones: In
vitro evaluation of hepatotoxicity and quantitative determination
in herbal products. Front Pharmacol. 6(25)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Song J, Zhong RL, Xia Z, Wu H, Zhong QX,
Zhang ZH, Wei YJ, Shi ZQ, Feng L and Jia XB: Research and
application of hepatotoxicity evaluation technique of traditional
Chinese medicine. Zhongguo Zhong Yao Za Zhi. 42:41–48.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ye H, Nelson LJ, Gómez Del Moral M,
Martinez-Naves E and Cubero FJ: Dissecting the molecular
pathophysiology of drug-induced liver injury. World J
Gastroenterol. 24:1373–1385. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xin J, Zhang RC, Wang L and Zhang YQ:
Researches on transcriptome sequencing in the study of traditional
Chinese medicine. Evid Based Complement Alternat Med.
2017(7521363)2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jin J, Kang W, Zhong C, Qin Y, Zhou R, Liu
H, Xie J, Chen L, Qin Y and Zhang S: The pharmacological properties
of Ophiocordyceps xuefengensis revealed by transcriptome analysis.
J Ethnopharmacol. 219:195–201. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Misra BB, Upadhayay RP, Cox LA and Olivier
M: Optimized GC-MS metabolomics for the analysis of kidney tissue
metabolites. Metabolomics. 14(75)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang Y, Li H, Hu T, Li H, Jin G and Zhang
Y: Metabonomic profiling in study hepatoprotective effect of
polysaccharides from Flammulina velutipes on carbon
tetrachloride-induced acute liver injury rats using GC-MS. Int J
Biol Macromol. 110:285–293. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang F, Mao Y, Qiao H, Jiang H, Zhao H,
Chen X, Tong L and Sun X: Protective effects of taurine against
endotoxin-induced acute liver injury after hepatic ischemia
reperfusion. Amino Acids. 38:237–245. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mi H, Muruganujan A, Ebert D, Huang X and
Thomas PD: PANTHER version 14: More genomes, a new PANTHER GO-slim
and improvements in enrichment analysis tools. Nucleic Acids Res.
47 (D1):D419–D426. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fiehn O: Metabolomics by gas
chromatography-mass spectrometry: Combined targeted and untargeted
profiling. Curr Protoc Mol Biol. 114:30.4.1–30.4.32.
2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tavares G, Venturini G, Padilha K, Zatz R,
Pereira AC, Thadhani RI, Rhee EP and Titan SMO: 1,5-Anhydroglucitol
predicts CKD progression in macroalbuminuric diabetic kidney
disease: Results from non-targeted metabolomics. Metabolomics.
14(39)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Chong J, Soufan O, Li C, Caraus I, Li S,
Bourque G, Wishart DS and Xia J: MetaboAnalyst 4.0: Towards more
transparent and integrative metabolomics analysis. Nucleic Acids
Res. 46 (W1):W486–W494. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Su G, Morris JH, Demchak B and Bader GD:
Biological network exploration with Cytoscape 3. Curr Protoc
Bioinformatics. 47:8.13.1–24. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Basu S, Duren W, Evans CR, Burant CF,
Michailidis G and Karnovsky A: Sparse network modeling and
metscape-based visualization methods for the analysis of
large-scale metabolomics data. Bioinformatics. 33:1545–1553.
2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Shou Q, Jin L, Lang J, Shan Q, Ni Z, Cheng
C, Li Q, Fu H and Cao G: Integration of metabolomics and
transcriptomics reveals the therapeutic mechanism underlying
paeoniflorin for the treatment of allergic asthma. Front Pharmacol.
9(1531)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Frenzel C and Teschke R: Herbal
hepatotoxicity: Clinical characteristics and listing compilation.
Int J Mol Sci. 17(pii: E588)2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chiang JYL: Bile acid metabolism and
signaling in liver disease and therapy. Liver Res. 1:3–9.
2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hylemon PB, Takabe K, Dozmorov M,
Nagahashi M and Zhou H: Bile acids as global regulators of hepatic
nutrient metabolism. Liver Res. 1:10–16. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
de Aguiar Vallim TQ, Tarling EJ and
Edwards PA: Pleiotropic roles of bile acids in metabolism. Cell
Metab. 17:657–669. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Shimomura Y and Kitaura Y: Physiological
and pathological roles of branched-chain amino acids in the
regulation of protein and energy metabolism and neurological
functions. Pharmacol Res. 133:215–217. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Nelson C: Less methionine means more
health. Lab Animal. 47(269)2018.
|
|
37
|
Ding HF and Fisher DE: Mechanisms of
p53-mediated apoptosis. Crit Rev Oncog. 9:83–98. 1998.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen P, Li D, Chen Y, Sun J, Fu K, Guan L,
Zhang H, Jiang Y, Li X, Zeng X, et al: p53-mediated regulation of
bile acid disposition attenuates cholic acid-induced cholestasis in
mice. Br J Pharmacol. 174:4345–4361. 2017.PubMed/NCBI View Article : Google Scholar
|