Introduction
In 2018, it was estimated that there were 570,000
new cases and 311,000 CC-associated mortality cases (1). Furthermore, ~90% of cases occur in
low-income and middle-income countries, where there is a lack of
organized screening (2). CC is
preventable to a large extent, and CC at the early stage can be
treated with surgery or radiation (3). Cervical squamous cell carcinoma (CSCC)
accounts for ~70-80% of CC and 20-25% of endocervical
adenocarcinomas (EACs). EAC is commonly associated with worse
clinical outcome and prognosis compared with CSCC (4). Traditionally, clinical stage is
considered the most prominent prognostic parameter, which
determines the modality adopted for treatment to a great extent.
CSCC antigen levels are closely associated with CC during the early
stage (5). Therefore, research on
early-stage CSCC is essential for accurate diagnosis and effective
treatment of CC.
Synthesis of mucin (MUC) is essential for the
formation of the mucous barrier, which helps protect the epithelia
of most organs, such as the stomach, from physical and chemical
damages and infection (6,7). Mucins are generally large
high-molecular-weight glycoproteins that are classified into two
subgroups, the secreted mucins and the membrane-bound mucins
(6,7). MUC2, MUC5AC, MUC5B, MUC6, MUC7, MUC9
and MUC19 are secreted mucins, whereas MUC3A/B, MUC4, MUC12, MUC13,
MUC15, MUC16, MUC17, MUC20 and MUC21 are membrane-bound mucins.
These mucins are considered to play important roles in cellular
interactions, molecular cell signalling and biological processes
(8). Furthermore, it has been
demonstrated that dysregulation of these mucins occurs in different
types of cancer. For example, MUC1 is overexpressed in pancreatic,
lung, breast, colon, ovarian and prostate cancers; MUC4 is
overexpressed in colon adenocarcinoma and pancreatic cancer; and
MUC16 is elevated in ovarian and pancreatic cancers (8).
CA125 was first identified ~38 years ago in a
screening of antibodies against ovarian cancer antigens. After two
decades, it was cloned and characterized as a membrane-bound mucin
named MUC16(9). MUC16 is the largest
membrane-bound mucin and represents an adverse prognostic marker of
human cancer. Previous studies have demonstrated that MUC16 is
overexpressed in several types of cancer, including breast, lung,
pancreatic and colorectal cancers (10,11).
Increasing evidence suggested that MUC16 is associated with immune
responses in human cancer. Gubbels et al (12) reported that ovarian tumour cells with
high levels of MUC16 are unable to be attacked by natural killer
cells and monocytes. Patankar et al (13) demonstrated that tumour-derived MUC16
functions as a suppressor of the immune response that is directed
against ovarian tumours. Furthermore, Fan et al (14) reported that the MUC16 C terminus
promotes forkhead box P3 expression and enrichment of
tumour-associated regulatory cells in tumour tissues, through
tumour-secreted IL-6 activation of the Janus kinase 2/signal
transducer and activator of transcription 3 signalling pathway in
pancreatic cancer. Recent studies have demonstrated that MUC16
mutations are associated with better survival outcomes and immune
responses in gastric and endometrial cancers (15,16).
Furthermore, MUC16 has been indicated to serve as a tumour marker
in different types of gynaecological cancer, including CC (17). Although MUC16 is regarded as one of
the most frequently mutated genes in CC, the associations between
MUC16 mutations, immune responses and clinical prognosis remain
unclear. Subsequently, the present study used mutation, clinical
and RNA-Seq data collected from The Cancer Genome Atlas (TCGA)
database (https://portal.gdc.cancer.gov), in order to
investigate the association between MUC16 mutation and immune
responses, as well as clinical prognosis in CC.
Materials and methods
Raw data
Data associated with mutation, clinical parameters,
copy number variation (CNV), DNA methylation and RNA-Seq of CC
samples were downloaded from the TCGA database. MUC16 RNA-Seq data
from the various types of cancer were downloaded from the TCGA
database (https://portal.gdc.cancer.gov/). The RNA-Seq data were
presented in terms of fragments per kilobase million (FPKM).
Furthermore, the GSE9750 dataset was downloaded from the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE9750)
(18,19). MUC16 expression was assessed in 286
CC and 240 CSCC clinical samples (≤4,000 days of follow-up data)
from the TCGA datasets. Data used in TCGA CNV, DNA methylation and
clinical data analyses were matched with the respective expression
data.
Definitions of clinical survival and
recurrence types
Three types of clinical survival and recurrence
outcomes were selected in the present study: Overall survival (OS),
disease-specific survival (DSS) and progression-free survival
(PFS). The outcomes were defined as follows: OS referred to the
period of time from the date of diagnosis to the date of mortality
from any cause; DSS referred to the period of time from the date of
initial diagnosis to the date of last contact or the date of
mortality from another cause; and PFS referred to the period from
the date of diagnosis to the date of new tumour occurrence
(20).
Patient information and tissue
collection
CC tissues and adjacent normal tissues were obtained
from 9 patients; 3 patients used to detect the MUC16 protein
expression levels between adjacent normal tissue and CC tissue, 3
patients used to detect the MUC16 protein expression levels in
wild-type CC tissues; and 3 patients used to detect the MUC16
protein expression levels in mutant type CC tissues (age range,
44-51 years; median age, 47 years); who underwent radical resection
at The First College of Clinical Medical Science, China Three
Gorges University (Yichang, China) between March 2019 and July
2019. All samples were stored at -80˚C. The inclusion criteria were
as follows: i) All patients were diagnosed with CC, following
colposcopy and cervical tissue biopsy; ii) no chemotherapy or
radiotherapy was performed prior to surgery, and iii) all patients
had complete clinical data. Exclusion criteria: i) Patients with
incomplete clinical data; and ii) patients who refused to
participate in this study. All experimental procedures were
approved by the Ethics Committee of The First College of Clinical
Medical Science, China Three Gorges University (Yichang, China).
Written informed consent was provided by all patients prior to the
study.
Western blotting
Total protein was extracted from CC tissues and
adjacent normal tissues using Cell lysis buffer (cat. no. P0013;
Beyotime Institute of Biotechnology, China), and the protein
concentration was quantified using a BCA Assay kit (cat. no.
P0012S; Beyotime Institute of Biotechnology, China). A total of 50
µg protein was added to each sample well, separated by 5% SDS-PAGE
prior to being transferred onto polyvinylidene fluoride membranes.
Dried, non-fat milk powder was used for blocking, for 1 h at room
temperature. The membranes were incubated with a MUC16 monoclonal
antibody (cat. no. BM5743; 1:2,000; Wuhan Boster Biological
Technology, Ltd.) at 4˚C for 12 h, prior to incubation with a
horseradish peroxidase (HRP)-labelled secondary antibody (cat. no.
BM2006, 1:1,000; Wuhan Boster Biological Technology, Ltd.) at room
temperature for 1 h. ECL luminous liquid A and B (cat. no. P0018S,
Beyotime Institute of Biotechnology) were mixed (1:1) and the
luminescent droplets were dropped onto the film. Protein bands were
visualized using a gel imager (Bio-Rad Laboratories, Inc.). β-actin
(cat. no. BA2305; Wuhan Boster Biological Technology, Ltd.) was
used as the reference protein.
Enrichment analysis
Gene set enrichment analysis (GSEA) was performed to
determine the correlation between the immune response in CC and
MUC16 expression in patients with mutated and wild-type MUC16.
Furthermore, genes were arranged from high to low according to
their correlation with MUC16 expression. Genes and R values were
determined and subsequently assessed via Kyoto Encyclopaedia of
Genes and Genomes and Gene Ontology (GO) analyses within
clusterProfiler v3.10.0 (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
(21). Furthermore, single-sample
(ss)GSEA was performed to calculate the score of immune
response-associated GO terms (22).
The gene set used for the ssGSEA was downloaded from the Molecular
Signatures database (https://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp).
Statistical analysis
Analysis of mutations was performed using R software
version 2.5.0(23). χ2
tests were used to analyze independent of data proportion for the
proportion of MUC16 mutant/wild-type in CSCC/EAC. Student's
unpaired t-test was used to determine differences in expression
levels between two samples. The level of correlation was determined
using Pearson correlation coefficient (R value) analysis.
Kaplan-Meier survival curves and the log-rank test were used to
determine the effects of MUC16 expression on patient survival,
using GraphPad Prism software version 7 (GraphPad Software, Inc.).
FPKM values, from the 20-80th percentiles were considered to
classify the samples, and samples with the lowest log-rank P-values
were selected. All graphs were generated using R software version
2.5.0 (RStudio, Inc.) or GraphPad Prism software version 7
(GraphPad Software, Inc.). Bar graph data are presented as the mean
± SEM.
Results
MUC16 is frequently mutated in CC
The top 10 mutated genes in CC included TTN (32%),
PIK3CA (29%), MUC4 (28%), KMT2C (20%), MUC16 (17%), KMT2D (15%),
FLG (13%), DMD (13%), EP300 (13%) and SYNE1 (13%; Fig. 1A). MUC16 ranked sixth among the most
frequently mutated genes in CC, with a mutational frequency of
17.3%, and most mutations were missense mutations (Fig. 1B). The proportion of MUC16 mutations
in CSCC (19%) was greater than that in EAC (9%; χ2,
P<0.05; Fig. 1C). Furthermore,
C>G and C>T mutations were the major types of single
nucleotide variants (Fig. 1D and
E).
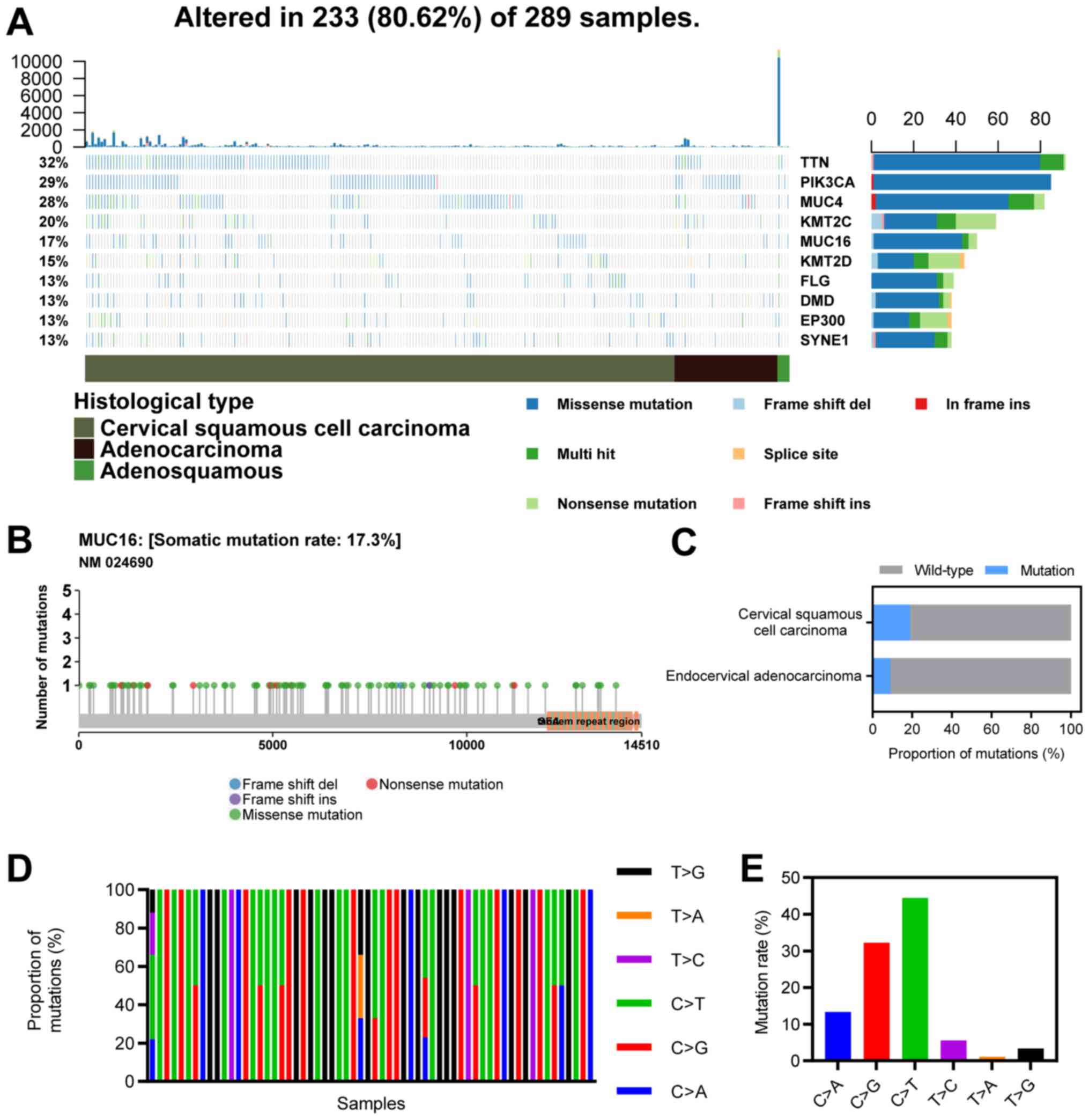 | Figure 1Landscape of MUC16 mutations in CC.
(A) An OncoPlot of the top 10 mutated genes in CC samples from TCGA
database. The upper bar plot indicates the number of genetic
mutations/patient, while the bar plot on the right indicates the
number of genetic mutations/gene. The CC pathology types and
mutation types are represented as annotations at the bottom. (B) A
lollipop plot of MUC16 mutations in CC samples from TCGA database.
Amino acid axis labelled for domain. (C) Proportions of MUC16
mutations between endocervical adenocarcinoma and cervical squamous
cell carcinoma. (D) Proportions of different types of single
nucleotide variants of MUC16 in CC samples from TCGA database. (E)
Numbers of different types of single nucleotide variants of MUC16
in CC samples from TCGA database. CC, cervical cancer; TCGA, The
Cancer Genome Atlas; TTN, Titin; PIK3CA, phosphatidylinositol 3;
MUC4, mucoprotein 4; KMT2C, histone lysine methyltransferase 2C;
MUC16, mucoprotein 16; KMT2D, histone lysine methyltransferase 2D;
FLG, filaggrin; DMD, duchenne muscular dystrophy; EP300, E1A
binding protein p300; SYNE1, spectrin repeat-containing nuclear
envelope protein 1. |
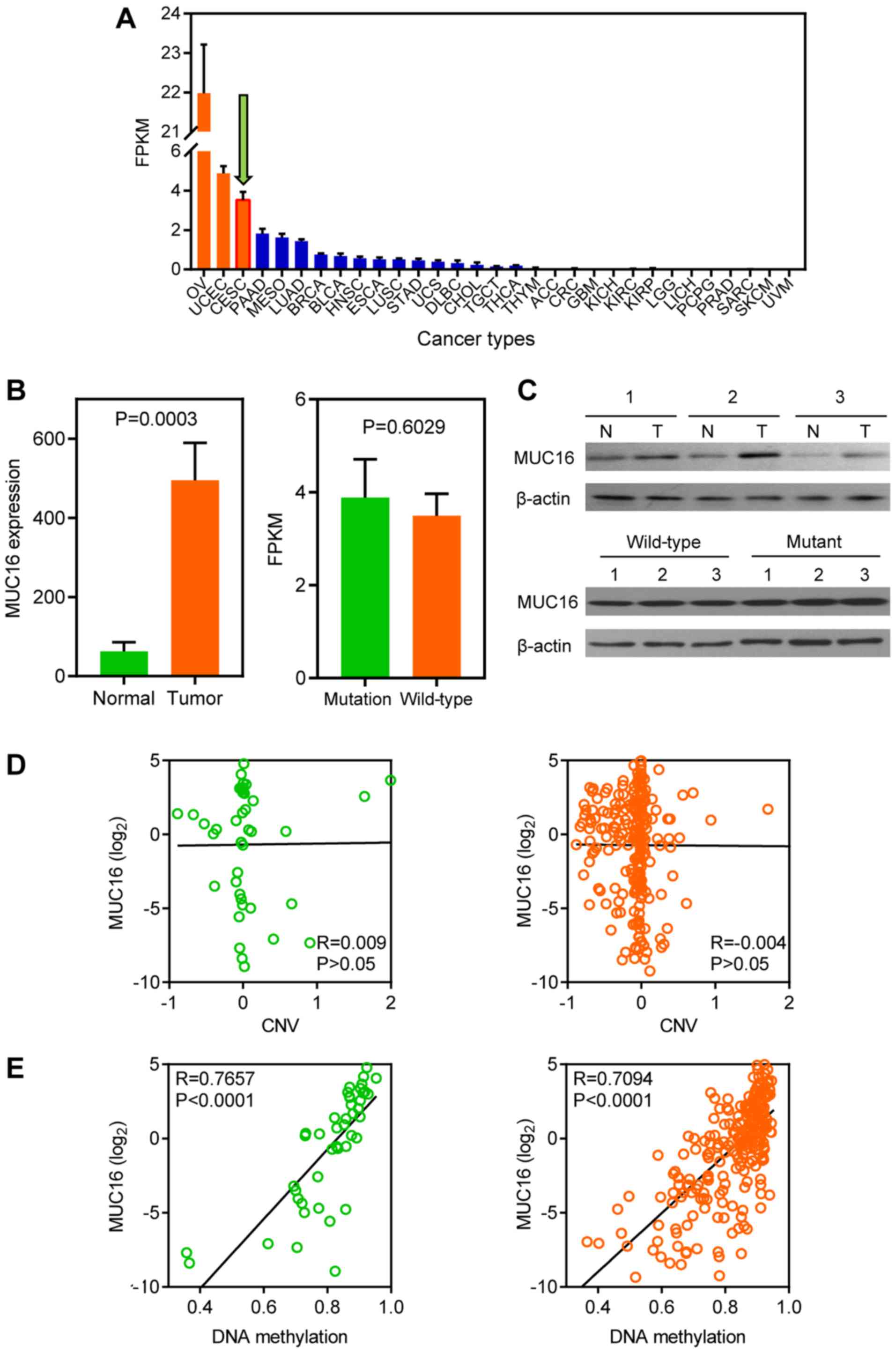
MUC16 expression in CC
MUC16 expression levels in different types of cancer
were analyzed within the TCGA datasets (Fig. 2A). A total of three gynaecologic
cancers, including ovarian, endometrial and CC, ranked as having
the highest MUC16 expression levels. Furthermore, the GEO dataset
was used to assess MUC16 expression in tumour and normal samples,
because TCGA lacks normal sample data for CC. The results
demonstrated that MUC16 was significantly overexpressed in tumour
samples compared with normal samples, according to the GSE9750
dataset (Fig. 2B, left panel).
However, no significant difference was observed between mutated
MUC16 and wild-type MUC16 in TCGA dataset (Fig. 2B, right panel). Subsequently,
clinical human CC and adjacent normal samples were assessed to
further investigate whether MUC16 is associated with CC. The
results demonstrated that MUC16 protein expression was
overexpressed in CC tissues compared with adjacent normal tissues
(Fig. 2C, top panel). However, no
difference was observed between mutant and wild-type CC samples at
the protein level (Fig. 2C, bottom
panel). The CNV and DNA methylation of MUC16 was analyzed, using
TCGA dataset, in order to determine the molecular mechanisms
associated with MUC16 expression in CC. No correlation was observed
between MUC16 expression and CNV in both the MUC16-mutated and the
MUC16-wild-type groups (Fig. 2D).
Conversely, DNA methylation was positively correlated with MUC16
expression in both the MUC16-mutated and the MUC16-wild-type groups
(Fig. 2E). Taken together, the
results demonstrated that MUC16 exhibited no difference between the
MUC16-mutated and the MUC16-wild-type groups, although MUC16 was
overexpressed in CC.
Overexpression of MUC16 is associated
with favourable prognosis in MUC16 mutant patients with CC
For the purpose of evaluating the clinical
significance of MUC16 in the survival of patients with CC and to
determine the association between MUC16 mutational status and
survival, MUC16 expression was assessed in 286 CC clinical samples
(≤4,000 days of follow-up data) from TCGA for OS, DSS and PFS
(24). Although OS is regarded as a
vital endpoint, assessment of OS alone decreases the reliability of
the results of a clinical study. When OS and DSS are used, longer
follow-up is required. Therefore, PFS is used in many clinical
trials as a composite of tumour progression and mortality (24). OS and DSS are associated with
survival, while PFS is associated with recurrence. Overexpression
of MUC16 was significantly associated with a longer OS time [hazard
ratio (HR)=0.156; P=0.0399], a longer DSS time (HR=0.277; P=0.0374)
and a longer PFS time (HR=0.271; P=0.0205) in patients with CC and
mutant MUC16 (Fig. 3A). However,
overexpression of MUC16 was not associated with OS (HR=0.702;
P=0.2122), DSS (HR=0.637; P=0.1522) or PFS (HR=1.570; P=0.0926) in
patients with CC and wild-type MUC16 (Fig. 3B). Furthermore, overexpression of
MUC16 had no significant effect on OS (HR=0.662; P=0.1082), DSS
(HR=0.618; P=0.0765) and PFS (HR=0.701; P=0.1660) in patients with
CC (Fig. 3C). Taken together, these
results suggested that overexpression of MUC16 may specifically
predict a favourable prognosis in patients with CC and mutant
MUC16.
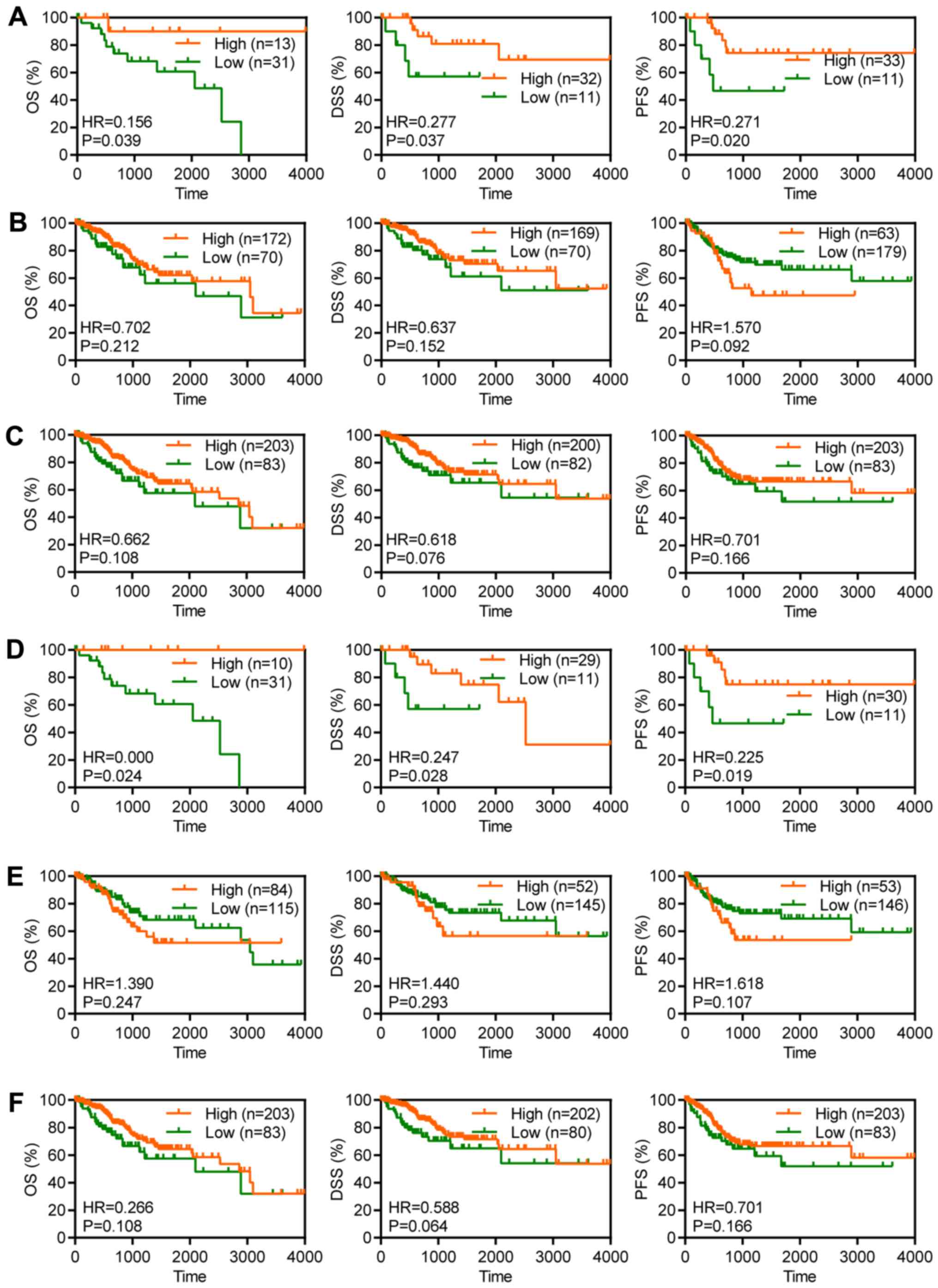 | Figure 3Overexpression of MUC16 was associated
with favourable prognosis of patients with CC and mutant MUC16.
Kaplan-Meier plots depicting the OS, DSS and PFS time in patients
with (A) MUC16-mutant, (B) MUC16-wild-type and (C) all patients
with CC from TCGA database. Kaplan-Meier plots depicting the OS,
DSS and PFS time in patients with (D) MUC16-mutant, (E)
MUC16-wild-type and (F) all patients with cervical squamous cell
carcinoma from TCGA database. CC, cervical cancer; OS, overall
survival; DSS, disease-specific survival; PFS, progression-free
survival; TCGA, The Cancer Genome Atlas; HR, hazard ratio. |
Overexpression of MUC16 is associated
with favourable prognosis in MUC16 mutant patients with CSCC
CSCC accounts for 70-80% of CC and antigen levels
are associated with early-stage CC (4). In order to evaluate the clinical
significance of MUC16 in terms of survival of patients with CSCC
and to determine the association between MUC16 mutational status
and survival, MUC16 expression was assessed in 240 CSCC clinical
samples (≤4,000 days of follow-up) from TCGA for OS, DSS and PFS.
Overexpression of MUC16 was significantly associated with a longer
OS time (HR=0; P=0.0247), a longer DSS time (HR=0.247; P=0.028) and
a longer PFS time (HR=0.225; P=0.019) in patients with mutant MUC16
(Fig. 3D). However, overexpression
of MUC16 was not associated with OS (HR=1.39; P=0.2477), DSS
(HR=1.44; P=0.293) and PFS (HR=1.618; P=0.1079) in patients with
wild-type MUC16 (Fig. 3E).
Furthermore, overexpression of MUC16 had no significant effect on
OS (HR=0.266; P=0.108), DSS (HR=0.588; P=0.0640) and PFS (HR=0.701;
P=0.166) in patients with CSCC (Fig.
3F). Taken together, these results suggested that
overexpression of MUC16 may specifically predict a favourable
prognosis in patients with CSCC and with mutant MUC16.
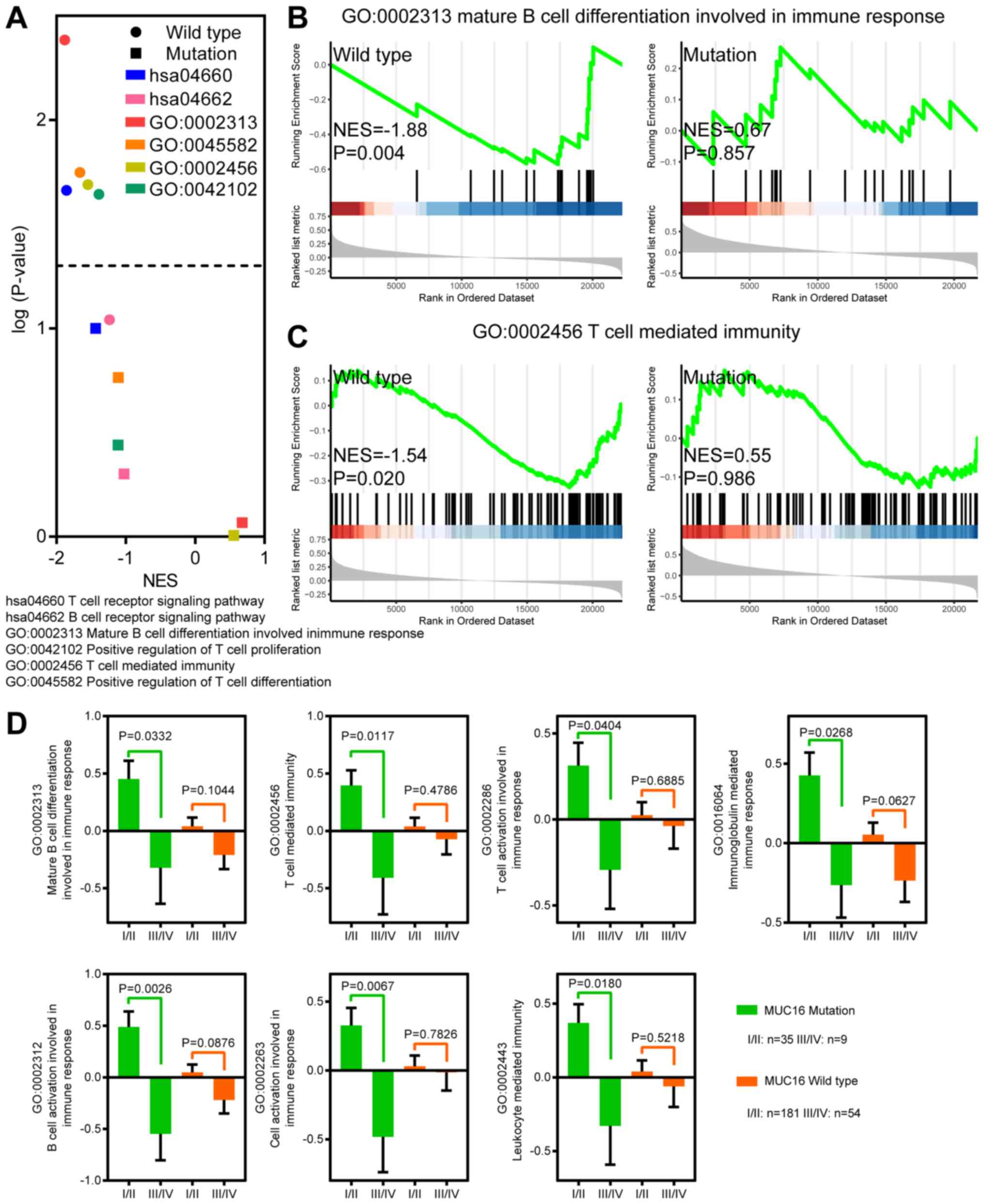
MUC16 mutations are involved in T
cell- and B cell-related immune responses
MUC16 mutant patients with CC with MUC16
overexpression exhibited a longer OS, DSS and PFS time, which
indicates that overexpression of MUC16 may be involved in the
progression and tumorigenesis of MUC16-mutant CC. Previous studies
have reported that MUC16 functions as a suppressor of the immune
response (12-14).
In order to determine whether overexpression of MUC16-mutant could
decrease suppression of the immune response, GSEA was performed to
identify immune response-associated gene sets in MUC16-mutant and
MUC16-wild-type CC samples (Fig.
4A-C). Gene sets associated with T cell and B cell activity
were negatively enriched in CC samples with increased
MUC16-wild-type expression. However, gene sets associated with T
cell and B cell activity were not enriched in CC samples with
increased MUC16-mutant expression. Taken together, these results
suggested that overexpression of MUC16-mutant in patients with CC
may help decrease the suppression of immune responses associated
with T cells and B cells.
Immune responses are upregulated in
early-stage patients with CC and MUC16 mutations
In order to investigate the association between
immune responses, MUC16 mutation and early-stage CC, an analysis of
the differences between early-stage and stages I-IV CC samples was
performed (Fig. 4D). ssGSEA was also
performed to assess the gene sets associated with immune responses.
The results demonstrated that immune responses associated with T
cells and B cells were significantly upregulated in stages I/II
samples with MUC16 mutations. However, no significant difference
was observed between MUC16-wild-type samples of stages I/II and
stages III/IV. Furthermore, leukocyte-mediated immunity was
significantly upregulated in stages I/II samples with MUC16
mutations. Taken together, these results indicated that immune
responses were upregulated in patients with early-stage CC and with
MUC16 mutations.
Discussion
CC is the fourth most common cancer in women
worldwide (1). CC at the early stage
can be treated with surgery or radiation (3). Furthermore, MUC16, which is one of the
most frequently mutated genes in CC, has been proven to serve as a
tumour marker in different types of gynaecologic cancer, including
CC (17). However, the association
between MUC16 mutational status and prognosis of patients with CC
remains unclear. To the best of our knowledge, the present study
was the first to demonstrate the prognosis of patients with MUC16
mutations in CC. A recent study reported that MUC16 is also
frequently mutated in endometrial cancer (16). The results of the present study
demonstrated that the proportion of MUC16 mutations in CSCC is
greater than that in EAC. Furthermore, MUC16 mutations were
associated with early-stage CC. MUC16 expression was subsequently
assessed in CC samples, and the results demonstrated that MUC16 was
overexpressed in gynaecologic cancers. Although MUC16
overexpression was exhibited in CC, there was no difference in
expression between the MUC16-mutated and the MUC16-wild-type
groups. Furthermore, differences in CNV or DNA methylation did not
affect MUC16 expression in either the MUC16-mutated or
MUC16-wild-type samples. These results suggested that the
expression difference in MUC16 between MUC16-mutant and
MUC16-wild-type samples may not have biological significance.
However, it was subsequently observed that overexpression of mutant
MUC16 may have biological significance. Overexpression of mutant
MUC16 specifically predicted favourable OS, DSS and PFS, whereas
these associations were not present for wild-type MUC16 or MUC16
overall. Furthermore, overexpression of MUC16 specifically
predicted favourable OS, DSS and PFS in patients with MUC16-mutant
CSCC. Therefore, the results suggested that overexpression of MUC16
may predict favourable prognosis in both patients with MUC16-mutant
CC and patients with MUC16-mutant CSCC. MUC16 served as an
essential prognostic biomarker in CC, particularly in terms of
clinical survival associated with early-stage disease.
The immune response is of great significance for the
development and progression of CC (25-27).
Increasing evidence demonstrates that gene mutations are associated
with cancer progression, immune response and clinical outcomes in
patients with early-stage disease (28-30).
Furthermore, it has been demonstrated that MUC16 interacts actively
with the immune response (12-14).
Previous studies have reported that MUC16 mutations are closely
associated with superior survival outcomes and the immune response
in gastric and endometrial cancers (12-14,16).
However, very few studies have investigated the effects of MUC16
mutations on the immune response in patients with CC. Previous
studies have demonstrated that MUC16 acts as a suppressor of immune
response (12-14,16).
In the present study, GSEA demonstrated that negative enrichment of
immune response signatures, such as immune responses associated
with T cells and B cells, was observed with overexpression of MUC16
in the MUC16-wild-type CC samples. However, the signatures
associated with T cell and B cell activity revealed no enrichment
with overexpression of MUC16 in the MUC16-mutant CC samples. The
present study demonstrated that MUC16 mutations are associated with
early-stage CC. Thus, the present study investigated the immune
responses associated with T cells and B cells during different
stages of disease in patients with MUC16 mutations. Immune
responses were upregulated in stages I/II of MUC16 patients with CC
patients, but not in MUC16-wild-type patients with CC patients.
In conclusion, the results from the present study
suggested that overexpression of MUC16 may specifically predict
favourable survival and prognosis in patients with MUC16-mutant CC.
Furthermore, MUC16 mutational status was associated with the immune
response in CC, particularly in patients at early stage of disease.
MUC16 mutations are a key marker in immunotherapy for CC patients.
MUC16 overexpression is not only significantly associated with the
prognosis of MUC16-mutant patients with CC, but also lead to the
enrichment of associated immune activity, which can be used to
identify which patients may benefit from immunotherapy in CC. Taken
together, the findings from the present study provided novel and
promising approaches for immunotherapy in CC; however, further
studies are required to investigate these methods.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and analyzed during the
current study are available in the TCGA dataset [https://portal.gdc.cancer.gov/] and GSE9750
dataset [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE9750].
Authors' contributions
HW and CY conceptualized the study, designed the
research and performed the bioinformatics analysis. HW performed
the experiments. HW and HY analyzed and interpreted the data. HW
wrote and edited the manuscript, and HY supervised the project. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
All experimental procedures were approved by the
Ethics Committee of The First College of Clinical Medical Science,
China Three Gorges University (Yichang, China). Written informed
consent was provided by all patients prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that there have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wong FL and Miller JW: Centers for Disease
Control and Prevention's National Breast and Cervical Cancer Early
Detection Program: Increasing Access to Screening. J Womens Health
(Larchmt). 28:427–431. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cancer Genome Atlas Research Network;
Albert Einstein College of Medicine; Analytical Biological
Services; Integrated genomic and molecular characterization of
cervical cancer. Nature 543: 378-384, 2017.
|
|
4
|
Marth C, Landoni F, Mahner S, McCormack M,
Gonzalez-Martin A, Colombo N and Committee EG: Cervical cancer:
ESMO clinical practice guidelines for diagnosis, treatment and
follow-up. Ann Oncol. 28:iv72–iv83. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Reesink-Peters N, van der Velden J, Ten
Hoor KA, Boezen HM, de Vries EG, Schilthuis MS, Mourits MJ, Nijman
HW, Aalders JG, Hollema H, et al: Preoperative serum squamous cell
carcinoma antigen levels in clinical decision making for patients
with early-stage cervical cancer. J Clin Oncol. 23:1455–1462.
2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jonckheere N and Van Seuningen I:
Integrative analysis of the cancer genome atlas and cancer cell
lines encyclopedia large-scale genomic databases: MUC4/MUC16/MUC20
signature is associated with poor survival in human carcinomas. J
Transl Med. 16(259)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hollingsworth MA and Swanson BJ: Mucins in
cancer: Protection and control of the cell surface. Nat Rev Cancer.
4:45–60. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Rao CV, Janakiram NB and Mohammed A:
Molecular pathways: Mucins and drug delivery in cancer. Clin Cancer
Res. 23:1373–1378. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Das S and Batra SK: Understanding the
unique attributes of MUC16 (CA125): Potential implications in
targeted therapy. Cancer Res. 75:4669–4674. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lakshmanan I, Salfity S, Seshacharyulu P,
Rachagani S, Thomas A, Das S, Majhi PD, Nimmakayala RK, Vengoji R,
Lele SM, et al: MUC16 regulates TSPYL5 for lung cancer cell growth
and chemoresistance by suppressing p53. Clin Cancer Res.
23:3906–3917. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Streppel MM, Vincent A, Mukherjee R,
Campbell NR, Chen SH, Konstantopoulos K, Goggins MG, Van Seuningen
I, Maitra A and Montgomery EA: Mucin 16 (cancer antigen 125)
expression in human tissues and cell lines and correlation with
clinical outcome in adenocarcinomas of the pancreas, esophagus,
stomach, and colon. Hum Pathol. 43:1755–1763. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gubbels JA, Felder M, Horibata S, Belisle
JA, Kapur A, Holden H, Petrie S, Migneault M, Rancourt C, Connor
JP, et al: MUC16 provides immune protection by inhibiting synapse
formation between NK and ovarian tumor cells. Mol Cancer.
9(11)2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Patankar MS, Jing Y, Morrison JC, Belisle
JA, Lattanzio FA, Deng Y, Wong NK, Morris HR, Dell A and Clark GF:
Potent suppression of natural killer cell response mediated by the
ovarian tumor marker CA125. Gynecol Oncol. 99:704–713.
2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fan K, Yang C, Fan Z, Huang Q, Zhang Y,
Cheng H, Jin K, Lu Y, Wang Z, Luo G, et al: MUC16 C
terminal-induced secretion of tumor-derived IL-6 contributes to
tumor-associated Treg enrichment in pancreatic cancer. Cancer Lett.
418:167–175. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li X, Pasche B, Zhang W and Chen K:
Association of MUC16 mutation with tumor mutation load and outcomes
in patients with gastric cancer. JAMA Oncol. 4:1691–1698.
2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hu J and Sun J: MUC16 mutations improve
patients' prognosis by enhancing the infiltration and antitumor
immunity of cytotoxic T lymphocytes in the endometrial cancer
microenvironment. Oncoimmunology. 7(e1487914)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Aggarwal P and Kehoe S: Serum tumour
markers in gynaecological cancers. Maturitas. 67:46–53.
2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lee YY, Kim TJ, Kim JY, Choi CH, Do IG,
Song SY, Sohn I, Jung SH, Bae DS, Lee JW, et al: Genetic profiling
to predict recurrence of early cervical cancer. Gynecol Oncol.
131:650–654. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M, et al: Identification of copy number
gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14(7)2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mayakonda A, Lin DC, Assenov Y, Plass C
and Koeffler HP: Maftools: Efficient and comprehensive analysis of
somatic variants in cancer. Genome Res. 28:1747–1756.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Johnson KR, Liauw W and Lassere MN:
Evaluating surrogacy metrics and investigating approval decisions
of progression-free survival (PFS) in metastatic renal cell cancer:
A systematic review. Ann Oncol. 26:485–496. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Punt S, Houwing-Duistermaat JJ, Schulkens
IA, Thijssen VL, Osse EM, de Kroon CD, Griffioen AW, Fleuren GJ,
Gorter A and Jordanova ES: Correlations between immune response and
vascularization qRT-PCR gene expression clusters in squamous
cervical cancer. Mol Cancer. 14(71)2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Heeren AM, Koster BD, Samuels S, Ferns DM,
Chondronasiou D, Kenter GG, Jordanova ES and de Gruijl TD: High and
interrelated rates of PD-L1+CD14+
antigen-presenting cells and regulatory T cells mark the
microenvironment of metastatic lymph nodes from patients with
cervical cancer. Cancer Immunol Res. 3:48–58. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cui JH, Lin KR, Yuan SH, Jin YB, Chen XP,
Su XK, Jiang J, Pan YM, Mao SL, Mao XF, et al: TCR repertoire as a
novel indicator for immune monitoring and prognosis assessment of
patients with cervical cancer. Front Immunol.
9(2729)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Mehnert JM, Panda A, Zhong H, Hirshfield
K, Damare S, Lane K, Sokol L, Stein MN, Rodriguez-Rodriquez L,
Kaufman HL, et al: Immune activation and response to pembrolizumab
in POLE-mutant endometrial cancer. J Clin Invest. 126:2334–2340.
2016.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Choi M, Kadara H, Zhang J, Parra ER,
Rodriguez-Canales J, Gaffney SG, Zhao Z, Behrens C, Fujimoto J,
Chow C, et al: Mutation profiles in early-stage lung squamous cell
carcinoma with clinical follow-up and correlation with markers of
immune function. Ann Oncol. 28:83–89. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
McFarland CD, Yaglom JA, Wojtkowiak JW,
Scott JG, Morse DL, Sherman MY and Mirny LA: The damaging effect of
passenger mutations on cancer progression. Cancer Res.
77:4763–4772. 2017.PubMed/NCBI View Article : Google Scholar
|