Introduction
Influenza A virus (IAV) is a single negative chain
RNA virus belonging to the positive myxovirus family, which is the
main pathogen of seasonal influenza. Seasonal influenza kills
290,000 to 650,000 patients every year (1-3).
Influenza vaccine and antiviral drugs are the main strategies for
the prevention and treatment of influenza virus. However, at
present, the influenza virus vaccine is only aimed at the
corresponding subtypes of strains, and the anti-influenza virus
drugs amantadine and neuraminidase inhibitors have serious drug
resistance (4,5). Therefore, it is urgent to find a
long-term and effective new strategy against influenza virus.
β-defensin 3 is a class of antibacterial peptides
produced by epithelial cells (6,7). It has
been found that β-defensin 3 enhances the resistance ability
against influenza virus (8). MDCK
cells transfected with β-defensin 3 plasmid also showed resistance
to influenza virus (9). Respiratory
epithelial cells are the first line of defense against influenza
virus, so inducing β-defensin 3 in respiratory epithelial cells may
become a new strategy against influenza virus. MAPKs are a key
family of proteins involved in regulating gene expression in
response to extracellular stimuli and activating a variety of
cellular responses, including cell proliferation, differentiation
and apoptosis (10). According to
the literature, the expression and regulation of human β-defensin 3
in human lung epithelial cells are dependent on MAPKs ERK1/2 and
JNK (11). Epigallocatechin gallate
(EGCG) is the most abundant catechin in green tea, which has a
variety of biological functions, including anti-tumor,
anti-inflammatory and immunomodulatory effects (12). Recently, it has been found that EGCG
also has anti-influenza virus effect. Previous studies have shown
that EGCG mainly prevents the virus from entering the cell by
destroying the viral envelope (13).
EGCG can also play an antiviral role by inducing the expression of
antiviral protein (14). However, it
has not been clarified at this stage whether EGCG can play an
antiviral role by inducing the expression of β-defensin 3 in
respiratory epithelial cells and the mechanism of inducing the
expression of β-defensin 3.
Therefore, in the present study, bronchial
epithelial cells (BEAS-2B) were used as a model to explore the
mechanism of β-defensin 3 expression induced by EGCG and the
relationship between β-defensin 3 expression and anti-influenza A
virus. The results confirmed that EGCG pretreatment against
influenza A virus is related to the induction of the expression of
β-defentin 3. EGCG induces the expression of β-defense 3 through
the p38 MAPK, ERK, JNK signaling pathway to inhibit the replication
of influenza A virus H1N1 in BEAS-2B cells. We elucidated part of
the mechanism of EGCG against influenza A virus.
Materials and methods
Cells, virus and reagents
BEAS-2B cells were purchased from Kunming Cell Bank
of the Chinese Academy of Sciences (Kunming, China) and cultured in
KCB medium at 37˚C with 5% CO2. Madin-Darby canine
kidney (MDCK) cells were purchased from Kunming Cell Library of the
Chinese Academy of Sciences (Kunming, China), and cultured in
Dulbecco's modified Eagle's medium (DMEM), with 10% v/v fetal
bovine serum both from (Gibco; Thermo Fisher Scientific, Inc.).
Penicillin (100 U/ml), and 100 μg/ml strepsin both from
(HyClone; GE Healthcare Life Sciences) at 37˚C in 5%
CO2. Influenza A virus (A/PR/8/34 H1N1, PR8) in this
experiment was cultured in the allantoic cavity of chicken embryo
at 9 days of age for 48 h, and the Reed-Muench method was used to
determine the TCID50 of the virus in MDCK cells. The
study was approved by the Ethics Committee of The Affiliated
Hospital of Guizhou Medical university (Guiyang, China).
Effect of EGCG on BEAS-2B cell
activity
Different concentrations of EGCG (Solarbio Life
Science; SE8120) (6.25, 12.5, 25, 50, 100 μg/ml) were added
to the monolayer BEAS-2B cells in 96-well plates. Five wells were
repeated for each concentration and cultured for 48 h. Cell
viability was determined by CCK8 kit (Dojindo Molecular
Technologies Inc.,) and absorbance was detected at 450 nm by BioTek
Instruments, Inc.
Intervention of EGCG on BEAS-2B
cells
Different concentrations of EGCG (0, 6.25, 12.5, 25
and 50 μg/ml) were added to the single-layer BEAS-2B cells
in the 6-well plate and cultured for 24 h. EGCG of 50 μg/ml
was added to the single-layer BEAS-2B cells in the 6-well plate and
cultured for 0, 2, 4, 12, 24 and 48 h. One hour before adding 50
μg/ml EGCG, 20 μM SB203580, 20 μM U1026 or 10
μM SP600125 dissolved in dimethyl sulfone was added to
inhibit the MAPK signaling pathway.
Transfection of silenced HBD3 in
BEAS-2B cells
When 50% of the bottom area of the confocal dish was
covered by cells, the culture solution was discarded. After rinsing
3 times with 1X PBS, serum-free DMEM basal medium was added at 2
ml/well. After culture for 12 h, 120 μl ribo FECTCP buffer,
5 μl, 20 μM siRNA and 12 μl ribo FECTCP
reagent was taken. HBD3, sequence 5'-TGTGGAAATGCCTTCTTAA-3' and
control siRNA (NC), article number: N Control_05815, transfection
reagent and primer design were provided by Guangzhou RiboBio Co.,
Ltd. The mixture was placed at room temperature for 15 min to form
a transfection complex, then mixed with 1,863 μl of
serum-free DMEM medium (confocal dish 1 well system). Then cells
were added and incubated at 37˚C for 24 h.
Virus infection of BEAS-2B cells
At 24 h after transfection, the original culture
solution was removed and rinsed 3 times with 1X PBS. The
experimental groups were control, IAV, EGCG + IAV, EGCG + siNC and
EGCG + IAV + siHBD3 group. In IAV, EGCG + IAV and EGCG + IAV +
siHBD3 group, 100X TCID50 H1N1 collected after chicken
embryo culture was added, and 50 μg/ml EGGC was added to
EGCG + IAV group, EGCG + siNC group and EGCG + IAV + siHBD3 group
for 12 h culture. After washing twice with PBS, the medium was
changed to KCB and cultured for 12 h.
RNA extraction and real-time
fluorescence quantitative PCR
Total RNA was extracted using total RNA extraction
kit (Tiangen Biotech Co., Ltd.,) and tested on NanoDrop 2000
(Thermo Fisher Scientific, Inc.) UV spectrophotometer for RNA
purity and concentration. cDNA was synthesized using
PrimeScriptTM RT kit with gDNA Eraser (Takara Bio,
Inc.). Quantitative PCR was performed on CFX96 real-time
fluorescent quantitative PCR instrument (Bio-Rad Laboratories,
Inc.). Primer sequence-β-defensin 3 forward: 5'-TTATTGCAGAGT
CAGAGGCGG-3', and reverse: 5'-CGAGCACTTGCCGAT CTGTT-3'. GAPDH
forward, 5'-AGAAGGCTGGGGCTCAT TTG-3' and reverse
5'-AGGGGCCATCCACAGTCTTC-3'. The expression of β-defensin 3 mRNA was
calculated using the 2-ΔΔCq method.
Western blot analysis
Cells were washed with PBS at 37˚C and RIPA lysate
buffer was added into the cells and centrifuged at 4˚C for 5 min at
14,800 x g to obtain complete cell lysates. Protein samples (20 μg)
were electrophoresed using SDS polyacrylamide gel (Bio-Rad
Laboratories, Inc.) and the target protein molecules were
transferred to a polyvinylidene fluoride membrane. Polyvinylidene
fluoride membrane was sealed for 2 h. Then the corresponding
resistance p-ERK (cat. no. 4370; 1:1,000), and ERK (cat. no. 4695;
1:1,000) (both from Cell Signaling Technology, Inc.), p-JNK (Abcam;
1:1,000), JNK (Abcam; cat. no. ab179461; 1:1,000), p-p38 lightning
MAPK (Cell Signaling Technology, Inc.; cat. no. 9211; 1:1,000), p38
lightning MAPK (Cell Signaling Technology, Inc.; cat. no. 9212;
1:1,000), NP (BIOSS; cat. no. bs-4976R; 1:1,000) and GAPDH (Boster
Biological Technology; cat. no. M00227-5; 1:700) were added and
incubated overnight at 4˚C. After washing, horseradish
peroxidase-labeled secondary antibodies (Boster Biological
Technology; cat. no. BA1054; 1:5,000) were added for incubation and
exposure in the gel imaging system.
Cytokine detection
The supernatant of cell culture was taken and the
content of β-defensin 3 was detected by human β-defensin 3 ELISA
kit (Wuhan Gene-beauty Organism).
Immunofluorescence detection of
influenza virus replication in BEAS-2B cells
When the BEAS-2B cells were fused to 80%, the cells
were washed with PBS twice, fixed with 4% paraformaldehyde for 30
min, and then treated with 0.2% triton for 2 min. After blocking
with 5% BSA, rabbit anti-NP antibody (Boster Biological Technology)
and rabbit anti-HA antibody (Cell Signaling Technology, Inc.) were
added at 4˚C overnight. After washing with PBS, fluorescence
labeled secondary antibody (Boster Biological Technology) was added
avoiding light for 30 min and photographed under a confocal
microscope.
Statistical analysis
Data are expressed as the mean ± SEM. Statistical
analysis was performed using one-way analysis of variance (ANOVA),
with Bonferroni multiple comparison post hoc test, or Student's
t-test. Statistical calculations were carried out using the
GraphPad InStat3 software (GraphPad Software, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
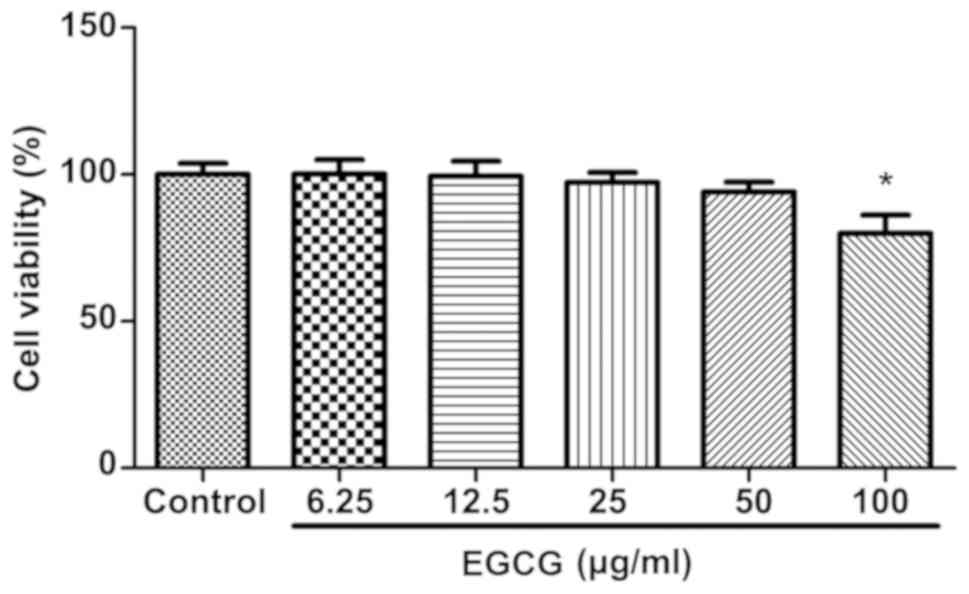
Effects of EGCG on cell activity
Different concentrations of EGCG (0, 6.25, 12.5, 25,
50, 100 μg/ml) were applied to BEAS-2B cells for 48 h, and
CCK8 kit was used to detect cell activity. The results showed that
the activity of BEAS-2B cells was significantly inhibited only by
100 μg/ml of EGCG compared with the control group, and the
remaining concentrations were not significantly different from
those in the control group (Fig. 1).
Therefore, EGCG of up to 50 μg/ml was selected for the next
experiment.
Expression of β-defensin 3 in BEAS-2B
cells induced by EGCG
After confirming that EGCG at concentrations of up
to 50 μg/ml had no effect on cell activity, the time-dose
relationship was studied between EGCG-induced expression of
β-defensin 3 in BEAS-2B cells. After the intervention of different
concentrations of EGCG in BEAS-2B cells, the expression level of
β-defensin 3 mRNA was detected by real-time fluorescence
quantitative PCR and the protein expression level of β-defensin 3
was detected by ELISA. The results showed that the expression of
β-defensin 3 in BEAS-2B cells was up-regulated in a dose-dependent
manner, with the highest concentration of 50 μg/ml (Fig. 2A and B). When EGCG was applied to BEAS-2B cells
at a concentration of 50 μg/ml, the expression of β-defense
3 mRNA and protein in BEAS-2B cells at different time periods was
up-regulated in a time-dependent manner and reached its peak at 24
h (Fig. 2C and D).
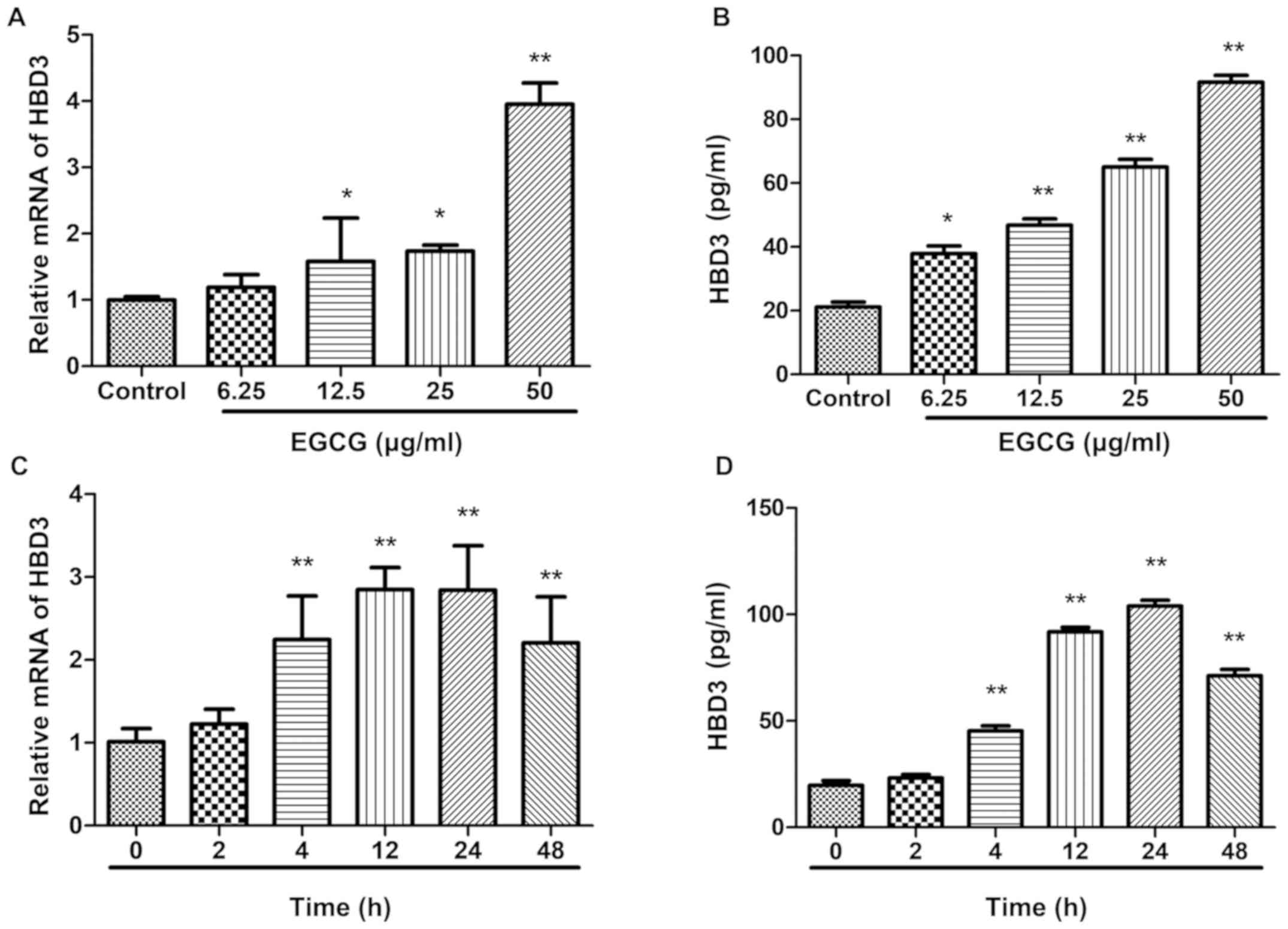 | Figure 2Effects of EGCG on the expression of
HBD3 in BEAS-2B cells. Real-time RT-PCR assay for HBD3 mRNA
stimulated and ELISA assay for protein stimulated with EGCG.
BEAS-2B cells were incubated with 0, 6.25, 12.5, 25, and 50
μg/ml EGCG for 24 h. The expression of HBD3 was quantified
by SYBR-Green Real-time RT-PCR. GAPDH mRNA was used as an internal
control. Data are the means ± SEM of three separate experiments.
mRNA expression of HBD3 is significantly higher at 50 μg/ml
EGCG than the control without EGCG. Detection of HBD3 mRNA
expression level by real-time RT-PCR and protein expression level
by ELISA in BEAS-2B cells after 50 μg/ml EGCG stimulation
for different times (0, 2, 4, 12, 24 and 48 h). Data are shown as
means ± SEM of three separate experiments. The expression of HBD3
mRNA is significantly higher at 24 h than the control at 0 h
(*P<0.05, **P<0.01). The data are
representative of three independent experiments: (A) Effects of
different concentrations of EGCG on HBD3 mRNA expression; (B)
Effects of different concentrations of EGCG on HBD3 protein
expression; (C) HBD3 mRNA expression at different time points; (D)
HBD3 protein expression at different time points. EGCG,
Epigallocatechin gallate; SEM, standard error of the mean; GAPDH,
glyceraldehyde-3-phosphate dehydrogenase; ELISA, enzyme-linked
immunosorbent assay. |
Expression of phosphorylation protein
in MAPK signaling pathway induced by EGCG in BEAS-2B cells
It was confirmed that β-defensin 3 was most strongly
induced by 50 μg/ml EGCG, so the signal pathway of
β-defensin 3 expression in BEAS-2B cells was determined by 50
μg/ml EGCG. BEAS-2B cells were treated with EGCG for 0, 2,
4, 8, 12 and 24 h. The effects of EGCG on the activation of p38
MAPK, ERK and JNK in BEAS-2B cells were determined. The results
showed that EGCG did not affect the levels of total p38 MAPK, ERK
and JNK protein over time, but significantly increased the levels
of phosphorylated p38 MAPK, p-ERK and p-JNK (Fig. 3).
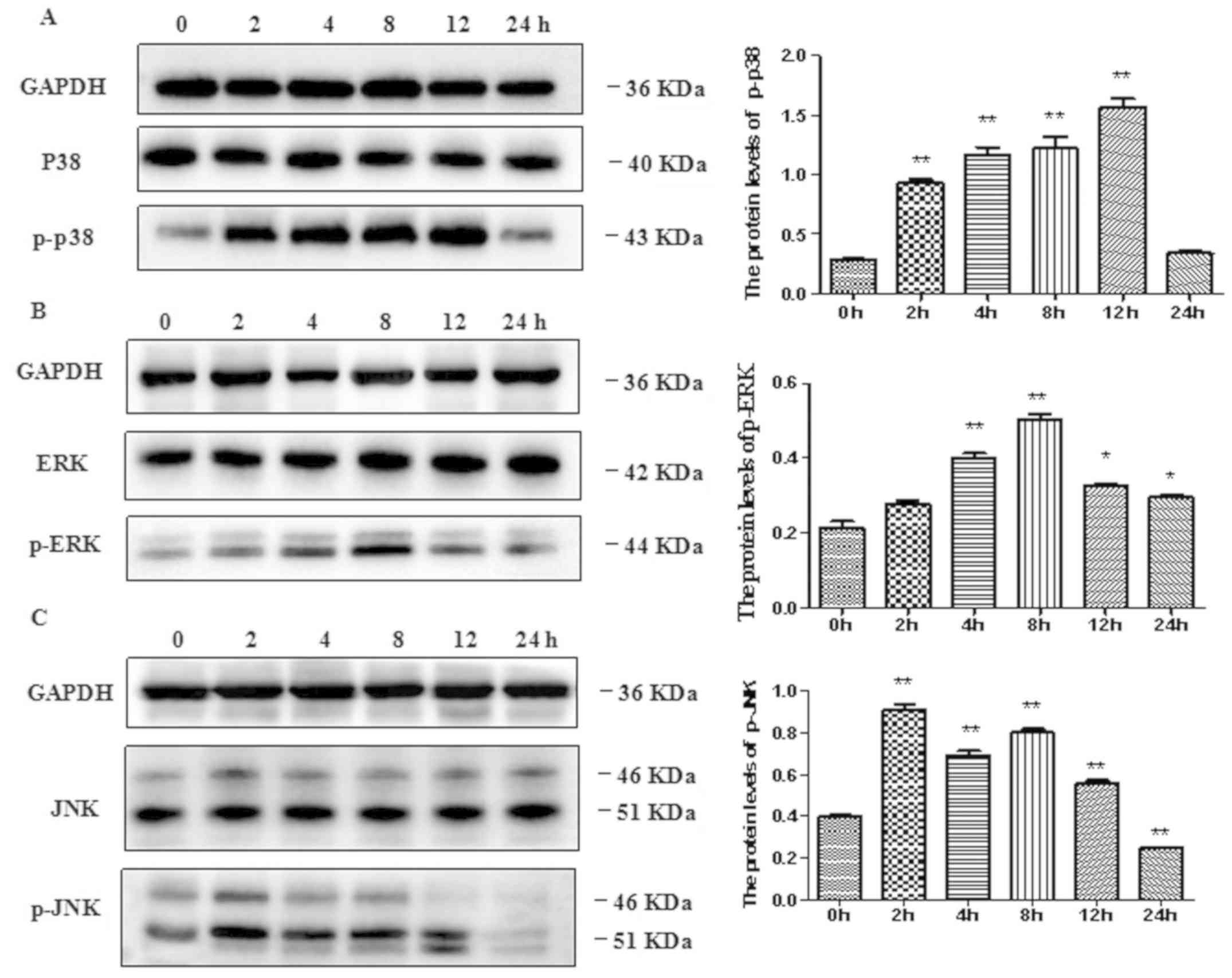 | Figure 3Effects of EGCG on the expressions of
(A) p-p38, (B) p-ERK and (C) p-JNK proteins in BEAS-2B cells.
BEAS-2B cells were treated with 50 µg/ml EGCG for the indicated
times. At each time, whole-cell lysates were prepared and used for
p-p38 MAPK, p38 MAPK, p-ERK, ERK, p-JNK, JNK, or GAPDH western with
respective antibodies. Data are shown as means ± SEM of three
separate experiments. The treatment with 50 μg/ml EGCG
triggered the phosphorylation (activation) of ERK, p38 MAPK, and
JNK (*P<0.05, **P<0.01). The data are
representative of three independent experiments. EGCG,
Epigallocatechin gallate; GAPDH, glyceraldehyde-3-phosphate
dehydrogenase; SEM, standard error of the mean. |
Inhibition of MAPK signaling pathway
induces expression of β-defensin 3 mRNA and protein in BEAS-2B
cells induced by EGCG
The expression of β-defensin 3 induced by p38 MAPK,
ERK and JNK signaling pathway in EGCG was further detected using 20
μM SB203580, 20 μM U1026 or 10 μM SP600125 in
the pretreatment of the BEAS-2B cells for 1 h and then 50
μg/ml EGCG was added to the cells. Pretreatment with 20
μM SB203580, 20 μM U1026 and 10 μM SP600125
significantly inhibited the expression of β-defensin 3 mRNA and
protein induced by EGCG (Fig.
4).
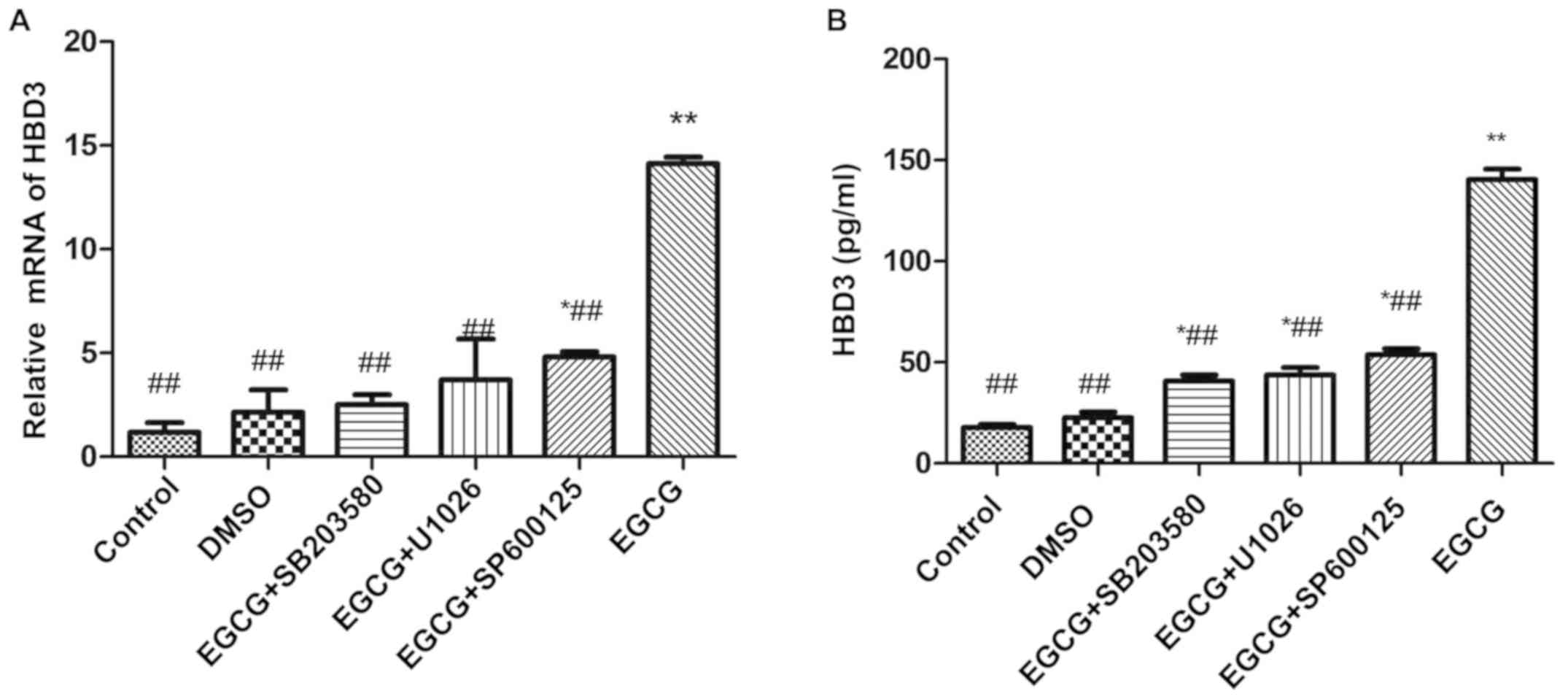 | Figure 4Effects of EGCG on the expression of
HBD3 in BEAS-2B cells. (A) Real-time RT-PCR assay for the
upregulated expression of HBD3 mRNA treated with inhibitors for
intracellular pathways. The cells were treated with inhibitors for
intracellular pathways in a series of experiments. BEAS-2B cells
were pre-incubated with SB203580 (an inhibitor of p38 MAPK), U1026
(an inhibitor of ERK), and SP600125 (an inhibitor of JNK) for 1 h
and then treated with 50 µg/ml EGCG for 12 h. SB203580 (an
inhibitor of p38 MAPK), U1026 (an inhibitor of ERK), and SP600125
(an inhibitor of JNK) completely inhibited the upregulated
expression of HBD3 by EGCG (*P<0.05,
**P<0.01, compared with the control group;
#P<0.05, ##P<0.01 compared with the
EGCG group). The data are representative of three independent
experiments. (B) Competitive ELISA for the quantification of HBD3
secretion by cultured BEAS-2B cells after EGCG and inhibitor
treatments. The cell were preincubated with SB203580, U1026, and
SP600125 for 1 h and then treated with 50 μg/ml EGCG for 12
h. Serial dilutions of synthetic mature HBD3 peptide were used to
generate a standard curve, and the HBD3 peptide concentration of
each experimental group was considered. In accordance with the
RT-PCR result, SB203580, U1026, and SP600125 completely inhibited
the secretion of HBD3 by EGCG (*P<0.05,
**P<0.01, compared with the control group;
##P<0.01, compared with the EGCG group). The data are
representative of three independent experiments. EGCG,
Epigallocatechin gallate. |
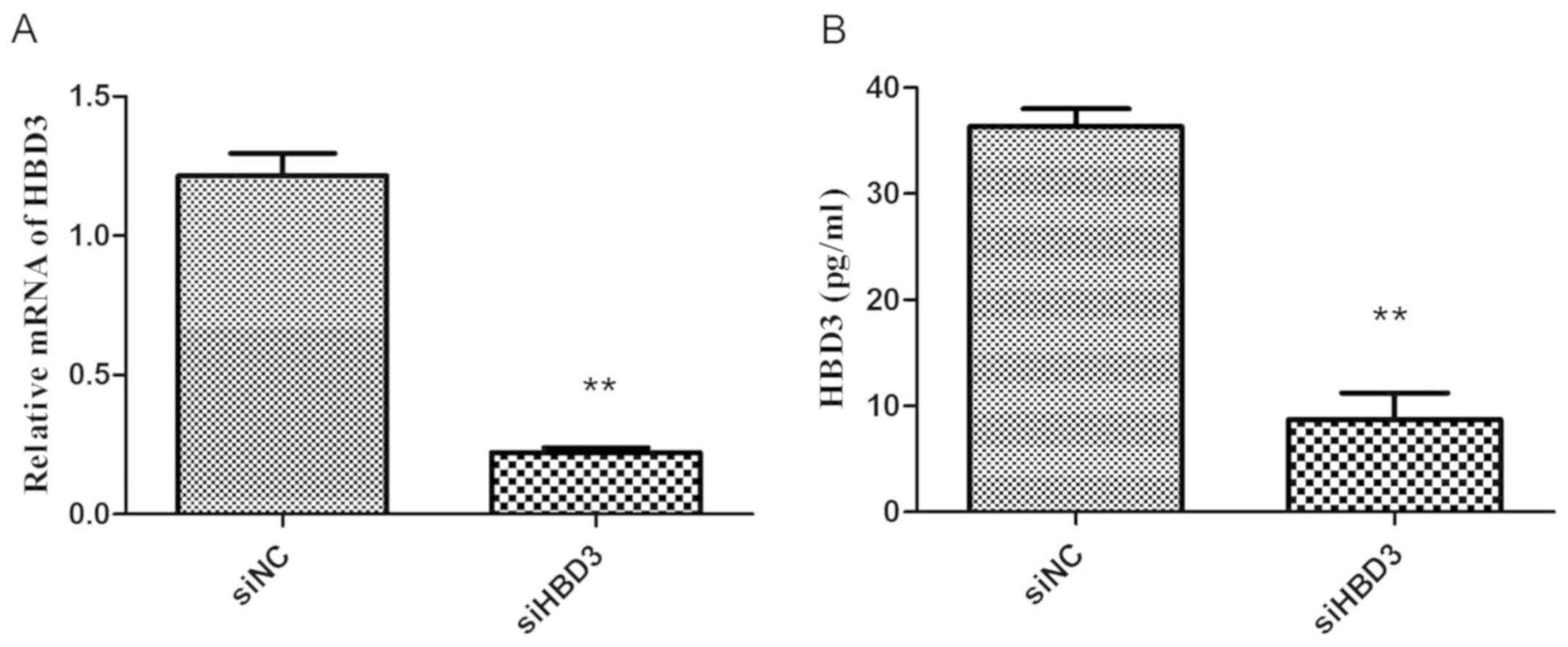
Expression of HBD3 mRNA and protein in BEAS-2B cells
after silent transfection qPCR and ELISA were used to detect the
expression of HBD3 mRNA and protein in BEAS-2B cells of siNC and
siHBD3. The results showed that compared with the siNC group, the
expression of HBD3 mRNA and protein in the siHBD3 group was
significantly reduced (P<0.01) (Fig.
5). This suggested the successful silencing of the HBD3 gene of
BEAS-2B cells.
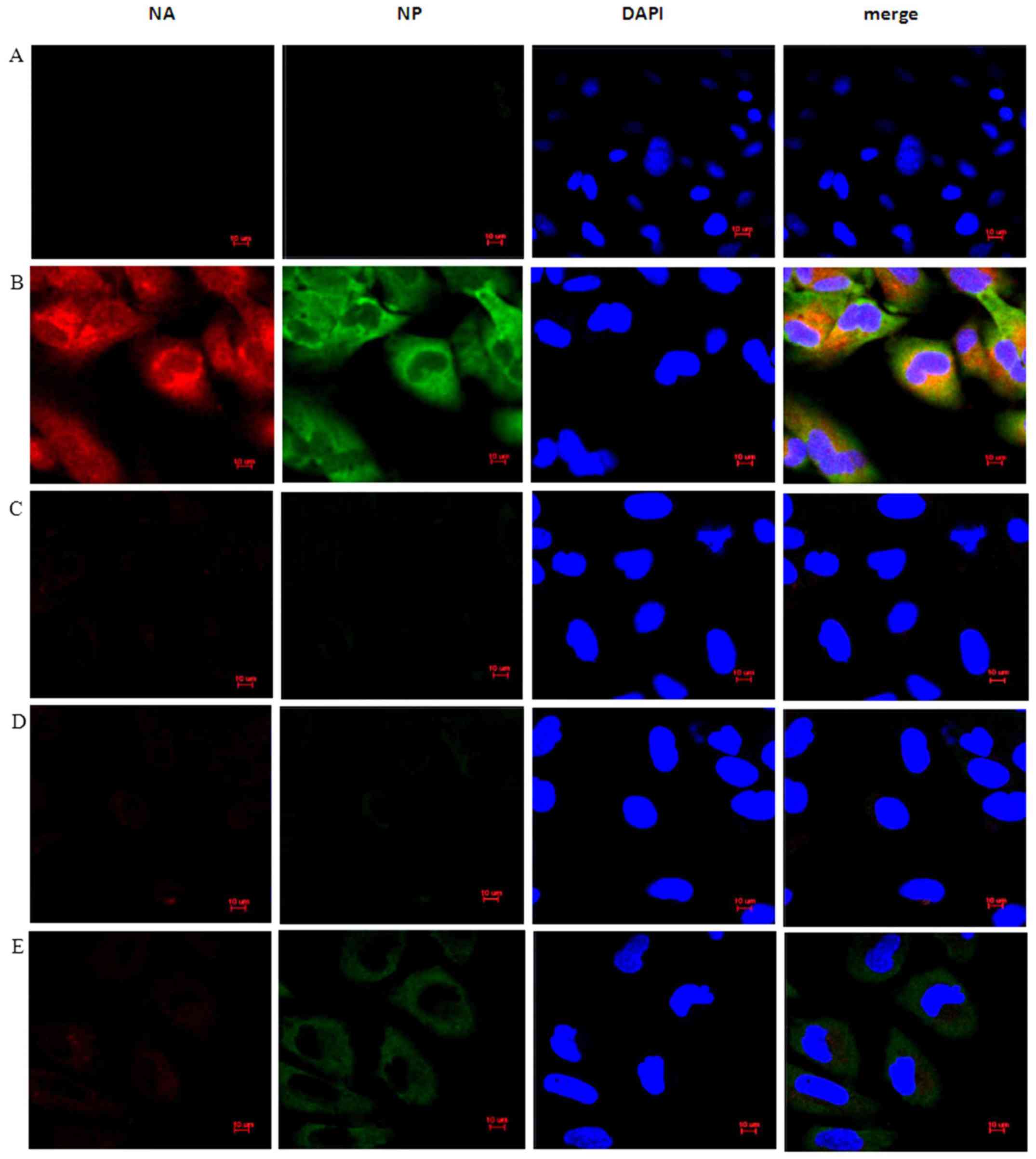
EGCG induces BEAS-2B cells against
influenza A virus H1N1
The above study confirmed that EGCG can induce the
expression of β-defensin 3 in BEAS-2B cells. Whether EGCG
anti-influenza virus H1N1 was related to the induction of
β-defensin 3 expression was explored. Immunofluorescence was used
to detect the expression of HA and NP proteins in BEAS-2B cells.
The results showed that compared with the IAV group (Fig. 6B), the expression of HA and NP in the
EGCG + IAV group (Fig. 6C) was
significantly inhibited, but the inhibition of the expression of HA
and NP in the EGCG + IAV + siHBD3 group (Fig. 6E) was significantly lower than that
in the EGCG + IAV group (Fig. 6C),
indicating that silencing HBD3 inhibited the antiviral effect of
EGCG (Fig. 6).
Discussion
In this study, EGCG was used to intervene in BEAS-2B
cells, and the results showed that EGCG significantly up-regulated
the expression levels of β-defensin 3 mRNA and protein in BEAS-2B
cells. Therefore, it was confirmed that EGCG can induce the
production of HBD3 in BEAS-2B cells, and EGCG stimulates the
up-regulation of HBD3 mRNA and protein expression in a
concentration- and time-dependent manner. Gan et al
(14) have shown that the expression
levels of β-defensin 2 mRNA and protein in 16HBECs cells are in a
concentration- and time-dependent relationship with paeoniflorin
(PF) stimulation, which is similar to the trend in this study, but
the concentration and the length of time were different, which
related to the experimental subjects and the different types of
drugs and defensins. β-defensin 3 has strong antiviral (9) and antibacterial activity (15). HBD3 can be induced by viruses,
bacteria, microbial products, toll-like receptors, epidermal cell
growth factors, and pro-inflammatory factors (16). Although the exact mechanism of action
is not yet clear, it is currently believed that HBD3 works through
ion channels (17).
The mechanism of EGCG-induced β-defensin 3
expression in BEAS-2B cells was explored. MAPKs in mammals include
c-Jun NH2 terminal kinase (JNK), p38 MAPK and extracellular
signal-regulated kinase (ERK). These enzymes are serine-threonine
protein kinases that can regulate various cellular activities,
including proliferation, differentiation, apoptosis, survival,
inflammation and innate immunity (18-20).
It has been confirmed that EGCG can regulate the activation of p38
MAPK, ERK, and JNK signaling pathways in colon cancer cells
(21). ERK and JNK-dependent HBD3
expression has been confirmed in Helicobacter pylori
infected gastric epithelial cells (22). In this study, it was found that EGCG
can significantly increase the level of phosphorylated (p)-p38
MAPK, ERK and JNK, and the results show that EGCG can activate the
p38 MAPK, ERK and JNK signaling pathways. On the other hand, p38
MAPK, ERK, or JNK inhibitors were used to inhibit the signaling
pathway. p38 MAPK, ERK, or JNK inhibitors significantly reduced the
expression level of β-defensin 3 in BEAS-2B cells induced by EGCG.
The results in this study were similar to other studies which
suggested that the expression of β-defensin 3 is also regulated by
p38 MAPK, ERK and JNK signaling pathways (14,23,24).
Therefore, it is suggested in this study that EGCG can induce
β-defensin 3 expression in BEAS-2B cells through p38 MAPK, ERK and
JNK signaling pathways. In addition, the expression of β-defensin 3
mRNA and protein peaked at 12 or 24 h under EGCG stimulation, while
the activation peak of MAPK pathway was 8 or 12 h, but the
expression at 24 h was significantly reduced. The reason may be
that since β-defensin 3 was downstream of MAPK pathway, when MAPKs
start to be highly expressed at 8 h, the downstream β-defensin 3
was gradually activated to express. At 12 h, β-defensin 3
expression began to increase sharply, and although the expression
of MAPKs was reduced at 24 h, the gradual reactions persisted.
β-defensin 3 continued to maintain an expression peak, and the
expression level was slightly higher than at 12 h.
EGCG has been proven to be resistant to influenza
virus (13), but the role of EGCG in
immune modulation against influenza virus remains to be further
studied. Our experiments found that EGCG pretreatment can
significantly inhibit the expression of HA and NP proteins of
influenza virus in BEAS-2B cells. However, silencing HBD3
expression in cells will reduce the ability of EGCG to inhibit the
expression of HA and NP of influenza virus. In the report of EGCG
against influenza A virus, Colpitts and Schang pointed out that
EGCG competitively binds to the sialic acid receptor on the surface
of the virus, thereby inhibiting influenza disease (25). Kesic et al found that EGCG can
upregulate the expression of Nrf-2 gene, the most important
transcriptional regulator in the second phase of the cellular
antioxidant pathway, to inhibit influenza virus (26). In a recent study, it was pointed out
that EGCG can significantly down-regulate TLR4 and NF-κB protein
levels and treat acute lung injury caused by swine influenza virus
H9N2(27). These findings indicate
that EGCG can fight influenza virus through a variety of
mechanisms, but there are few reports on EGCG inhibiting influenza
virus by regulating HBD3 secretion. However, our results confirm
that EGCG can induce HBD3 expression in BEAS-2B cells and inhibit
the expression of HA and NP in influenza A virus. Because HA plays
an important role in the process of virus introduction into host
cells, NP is a key protein for viral transcription and replication,
it is speculated that EGCG can regulate the concentration of HBD3
produced by cells to inhibit influenza virus adsorption, invasion
and replication, and then produce anti-influenza virus effects.
In conclusion, it has been shown in this study that
EGCG can induce the expression of β-defensin 3 through p38 MAPK,
ERK and JNK signaling pathways in BEAS-2B cells to inhibit
influenza A virus H1N1, which indicates that EGCG may be an
effective drug against influenza A virus.
Acknowledgements
Not applicable.
Funding
The study was supported by Baiyun Science and
Technology Plan Project of Guiyang (grant no. [2019]33; ‘Research
on the Mechanism of Epigallocatechin Gallate Inhibiting Influenza A
Virus Based on NF-κB Signaling Pathway’) and the National Natural
Science Foundation of China (grant no. 81260249; ‘Research on the
Mechanism of Influenza A Virus Induces β-defensins Production in
Respiratory Rpithelial Cells Based on NF-κB and p38 MAPK Signaling
Pathways’).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QM conceived the study and wrote the manuscript. YJ
interpreted and analyzed the data. LZ and ZZ designed the study and
performed the experiment. TR was responsible for the analysis and
discussion of the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The Affiliated Hospital of Guizhou Medical university (Guiyang,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Horimoto T and Kawaoka Y: Influenza:
Lessons from past pandemics, warnings from current incidents. Nat
Rev Microbiol. 3:591–600. 2005.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hause BM, Collin EA, Liu R, Huang B, Sheng
Z, Lu W, Wang D, Nelson EA and Li F: Characterization of a novel
influenza virus in cattle and Swine: Proposal for a new genus in
the Orthomyxoviridae family. MBio. 5:e00031–e14.
2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lee VJ, Ho ZJM, Goh EH, Campbell H, Cohen
C, Cozza V, Fitzner J, Jara J, Krishnan A and Bresee J: WHO Working
Group on Influenza Burden of Disease. Advances in measuring
influenza burden of disease. Influenza Other Respir Viruses.
12:3–9. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mungall BA, Xu X and Klimov A:
Surveillance of influenza isolates for susceptibility to
neuraminidase inhibitors during the 2000-2002 influenza seasons.
Virus Res. 103:195–197. 2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Nguyen JT, Smee DF, Barnard DL, Julander
JG, Gross M, de Jong MD and Went GT: Efficacy of combined therapy
with amantadine, oseltamivir, and ribavirin in vivo against
susceptible and amantadine-resistant influenza A viruses. PLoS One.
7(e31006)2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Pazgier M, Li X, Lu W and Lubkowski J:
Human defensins: Synthe- sis and structural properties. Curr Pharm
Des. 13:3096–3118. 2007.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Shah R and Chang TL: Defensins in viral
infection. In: Small Wonders: Peptides for Disease Control.
Rajasekaran K, Cary JW, Jaynes JM, Montesinos E (eds). Vol 1095.
ACS Symposium Series, pp137-171, 2012. https://doi.org/10.1021/bk-2012-1095.ch007.
|
|
8
|
Jiang Y, Yang D, Li W, Wang B, Jiang Z and
Li M: Antiviral activity of recombinant mouse β-defensin 3 against
influenza A virus in vitro and in vivo. Antivir Chem Chemother.
22:255–262. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Jiang Y, Wang Y, Kuang Y, Wang B, Li W,
Gong T, Jiang Z, Yang D and Li M: Expression of mouse β-defensin-3
in MDCK cells and its anti-influenza-virus activity. Arch Virol.
154:639–647. 2009.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yang SH, Sharrocks AD and Whitmarsh AJ:
MAP kinase signalling cascades and transcriptional regulation.
Gene. 513:1–13. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Haarmann H, Steiner T, Schreiber F,
Heinrich A, Zweigner J, N’Guessan PD and Slevogt H: The role and
regulation of Moraxella catarrhalis-induced human
beta-defensin 3 expression in human pulmonary epithelial cells.
Biochem Biophys Res Commun. 467:46–52. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chacko SM, Thambi PT, Kuttan R and
Nishigaki I: Beneficial effects of green tea: A literature review.
Chin Med. 5(13)2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kim M, Kim SY, Lee HW, Shin JS, Kim P,
Jung YS, Jeong HS, Hyun JK and Lee CK: Inhibition of influenza
virus internalization by (-)-epigallocatechin-3-gallate. Antiviral
Res. 100:460–472. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Gan Y, Cui X, Ma T, Liu Y, Li A and Huang
M: Paeoniflorin upregulates β-defensin-2 expression in human
bronchial epithelial cell through the p38 MAPK, ERK, and NF-κB
signaling pathways. Inflammation. 37:1468–1475. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Fusco A, Savio V, Cammarota M, Alfano A,
Schiraldi C and Donnarumma G: Schiraldi C and Donnarumma G: Beta-
defensin-2 and beta-defensin-3 reduce intestinal damage caused by
Salmonella typhimurium modulating the expression of
cytokines and enhancing the probiotic activity of Enterococcus
faecium. J Immunol Res. 2017(6976935)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Valore EV, Park CH, Quayle AJ, Wiles KR,
McCray PB Jr and Ganz T: Human beta-defensin-1: An antimicrobial
peptide of urogenital tissues. J Clin Invest. 101:1633–1642.
1998.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Reichel M, Heisig A, Heisig P and Kampf G:
Skin bacteria after chlorhexidine exposure-is there a difference in
response to human beta-Defensin-3? Eur J Clin Microbiol Infect Dis.
29:623–632. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bezniakow N, Gos M and Obersztyn E: The
RASopathies as an example of RAS/MAPK pathway disturbances -
clinical presentation and molecular pathogenesis of selected
syndromes. Dev Period Med. 18:285–296. 2014.PubMed/NCBI
|
|
19
|
Kono M, Tatsumi K, Imai AM, Saito K,
Kuriyama T and Shirasawa H: Kuriyama T and Shirasawa H: Inhibition
of human coronavirus 229E infection in human epithelial lung cells
(L132) by chloroquine: Involvement of p38 MAPK and ERK. Antiviral
Res. 77:150–152. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sui X, Kong N, Ye L, Han W, Zhou J, Zhang
Q, He C and Pan H: p38 and JNK MAPK pathways control the balance of
apoptosis and autophagy in response to chemotherapeutic agents.
Cancer Lett. 344:174–179. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Cerezo-Guisado MI, Zur R, Lorenzo MJ,
Risco A, Martín-Serrano MA, Alvarez-Barrientos A, Cuenda A and
Centeno F: Implication of Akt, ERK1/2 and alternative p38MAPK
signalling pathways in human colon cancer cell apoptosis induced by
green tea EGCG. Food Chem Toxicol. 84:125–132. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Boughan PK, Argent RH, Body-Malapel M,
Park JH, Ewings KE, Bowie AG, Ong SJ, Cook SJ, Sorensen OE, Manzo
BA, et al: Nucleotide-binding oligomerization domain-1 and
epidermal growth factor receptor: Critical regulators of
beta-defensins during Helicobacter pylori infection. J Biol
Chem. 281:11637–11648. 2006.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu J, Du X, Chen J, Hu L and Chen L: The
induction expression of human β-defensins in gingival epithelial
cells and fibroblasts. Arch Oral Biol. 58:1415–1421.
2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Paoletti I, Buommino E, Fusco A, Baudouin
C, Msika P, Tufano MA, Baroni A and Donnarumma G: Patented natural
avocado sugar modulates the HBD-2 and HBD-3 expression in human
keratinocytes through toll-like receptor-2 and ERK/MAPK activation.
Arch Dermatol Res. 304:619–625. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Colpitts CC and Schang LM: A small
molecule inhibits virion attachment to heparan sulfate- or sialic
acid-containing glycans. J Virol. 88:7806–7817. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kesic MJ, Simmons SO, Bauer R and Jaspers
I: Nrf2 expression modifies influenza A entry and replication in
nasal epithelial cells. Free Radic Biol Med. 51:444–453.
2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Xu MJ, Liu BJ, Wang CL, Wang GH, Tian Y,
Wang SH, Li J, Li PY, Zhang RH, Wei D, et al:
Epigallocatechin-3-gallate inhibits TLR4 signaling through the
67-kDa laminin receptor and effectively alleviates acute lung
injury induced by H9N2 swine influenza virus. Int Immunopharmacol.
52:24–33. 2017.PubMed/NCBI View Article : Google Scholar
|