|
1
|
Marsh JA and Forman-Kay JD: Sequence
determinants of compaction in intrinsically disordered proteins.
Biophysical J. 98:2383–2390. 2010.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bah A and Forman-Kay JD: Modulation of
intrinsically disordered protein function by post-translational
modifications. J Biol Chem. 291:6696–6705. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Adaniya SM, O-Uchi J, Cypress MW, Kusakari
Y and Jhun BS: Posttranslational modifications of mitochondrial
fission and fusion proteins in cardiac physiology and
pathophysiology. Am J Physiol Cell Physiol. 316:C583–C604.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ametzazurra A, Larrea E, Civeira MP,
Prieto J and Aldabe R: Implication of human
N-alpha-acetyltransferase 5 in cellular proliferation and
carcinogenesis. Oncogene. 27:7296–7306. 2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Christensen DG, Baumgartner JT, Xie X, Jew
KM, Basisty N, Schilling B, Kuhn ML and Wolfe AJ: Mechanisms,
detection, and relevance of protein acetylation in prokaryotes.
mBio. 10:e02708–18. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Drazic A, Myklebust LM, Ree R and Arnesen
T: The world of protein acetylation. Biochim Biophys Acta.
1864:1372–1401. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Verdin E and Ott M: 50 years of protein
acetylation: From gene regulation to epigenetics, metabolism and
beyond. Nat Rev Mol Cell Biol. 16:258–264. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lee TY, Hsu JB, Lin FM, Chang WC, Hsu PC
and Huang HD: N-Ace: Using solvent accessibility and
physicochemical properties to identify protein N-acetylation sites.
J Comput Chem. 31:2759–2771. 2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hollebeke J, Van Damme P and Gevaert K:
N-terminal acetylation and other functions of
Nalpha-acetyltransferases. Biol Chem. 393:291–298. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Thao S, Chen CS, Zhu H and
Escalante-Semerena JC: Nepsilon-lysine acetylation of a bacterial
transcription factor inhibits Its DNA-binding activity. PLoS One.
5(e15123)2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yang XJ and Gregoire S: Metabolism,
cytoskeleton and cellular signalling in the grip of protein
Nepsilon- and O-acetylation. EMBO Rep. 8:556–562. 2007.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Allfrey VG, Faulkner R and Mirsky AE:
Acetylation and methylation of histones and their possible role in
the regulation of RNA synthesis. Proc Natl Acad Sci USA.
51:786–794. 1964.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Verdone L, Caserta M and Di Mauro E: Role
of histone acetylation in the control of gene expression. Biochem
Cell Biol. 83:344–353. 2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Choudhary C, Kumar C, Gnad F, Nielsen ML,
Rehman M, Walther TC, Olsen JV and Mann M: Lysine acetylation
targets protein complexes and co-regulates major cellular
functions. Science. 325:834–840. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
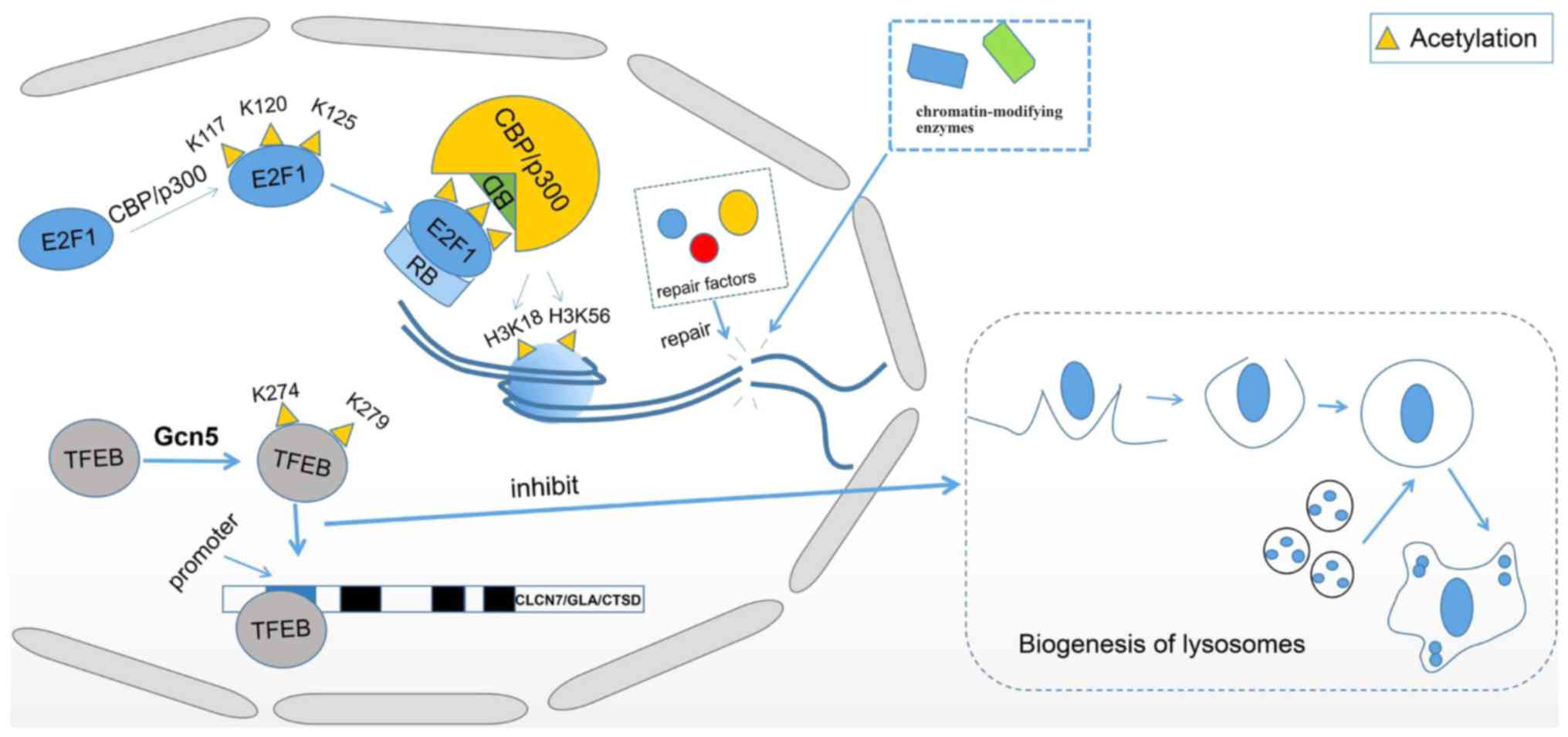
Allis CD, Berger SL, Cote J, Dent S,
Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar
R, et al: New nomenclature for chromatin-modifying enzymes. Cell.
131:633–636. 2007.PubMed/NCBI View Article : Google Scholar
|
|
16
|
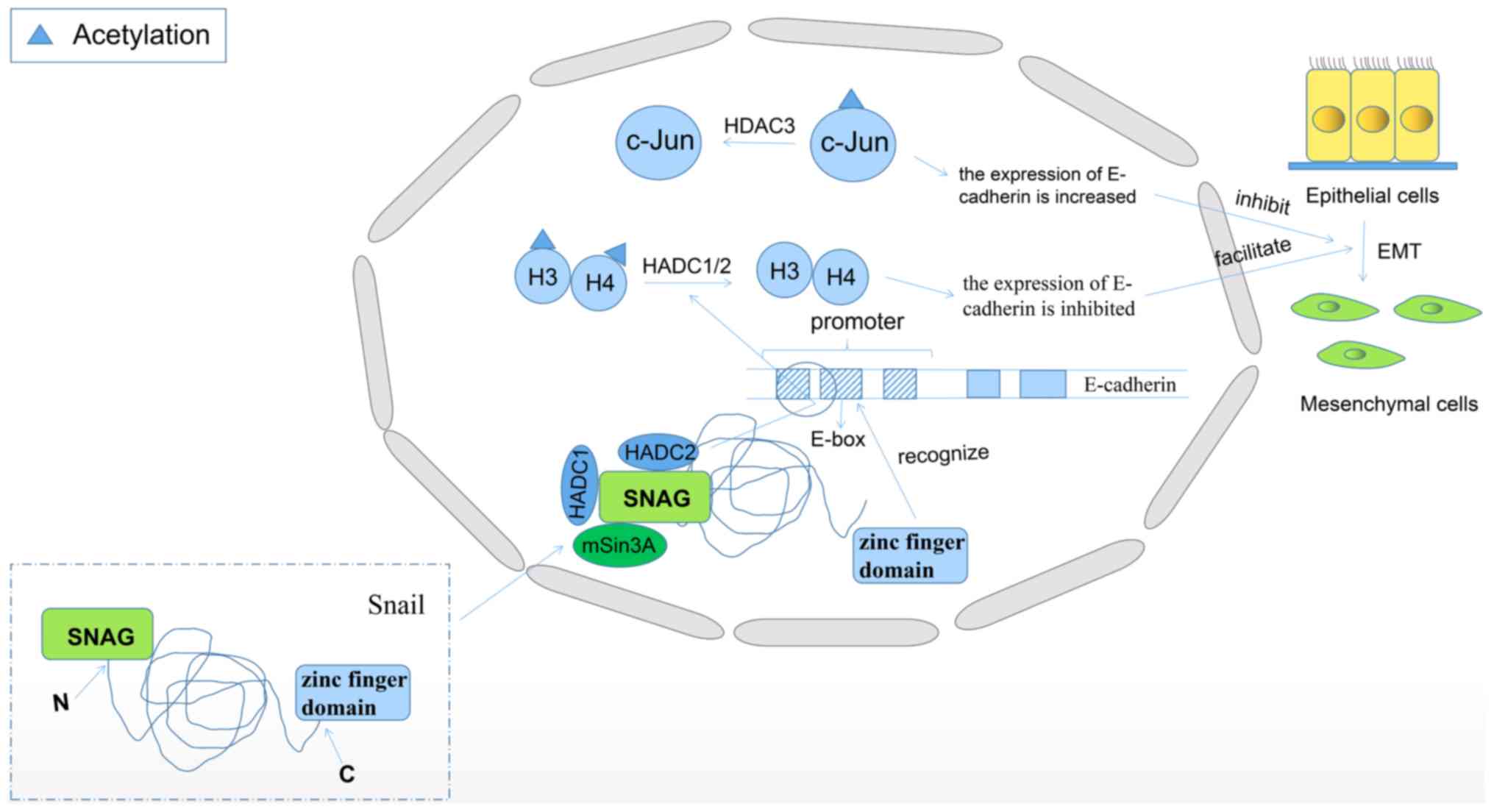
Li P, Ge J and Li H: Lysine
acetyltransferases and lysine deacetylases as targets for
cardiovascular disease. Nat Rev Cardiol. 17:96–115. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Song L, Wang G, Malhotra A, Deutscher MP
and Liang W: Reversible acetylation on Lys501 regulates the
activity of RNase II. Nucleic Acids Res. 44:1979–1988.
2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sterner DE and Berger SL: Acetylation of
histones and transcription-related factors. Microbiol Mol Biol Rev.
64:435–459. 2000.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ruhlmann F, Windhof-Jaidhauser IM, Menze
C, Beißbarth T, Bohnenberger H, Ghadimi M and Dango S: The
prognostic capacities of CBP and p300 in locally advanced rectal
cancer. World J Surg Oncol. 17(224)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Narita T, Weinert BT and Choudhary C:
Functions and mechanisms of non-histone protein acetylation. Nat
Rev Mol Cell Biol. 20:156–174. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hwang CS, Shemorry A and Varshavsky A:
N-terminal acetylation of cellular proteins creates specific
degradation signals. Science. 327:973–977. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Vetting MW, S de Carvalho LP, Yu M, Hegde
SS, Magnet S, Roderick SL and Blanchard JS: Structure and functions
of the GNAT superfamily of acetyltransferases. Arch Biochem
Biophys. 433:212–226. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ruiz-Garcia AB, Sendra R, Galiana M,
Pamblanco M, Perez-Ortin JE and Tordera V: HAT1 and HAT2 proteins
are components of a yeast nuclear histone acetyltransferase enzyme
specific for free histone H4. J Biol Chem. 273:12599–12605.
1998.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Miskiewicz K, Jose LE, Bento-Abreu A,
Fislage M, Taes I, Kasprowicz J, Swerts J, Sigrist S, Versées W,
Robberecht W and Verstreken P: ELP3 controls active zone morphology
by acetylating the ELKS family member Bruchpilot. Neuron.
72:776–788. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sampath V, Liu B, Tafrov S, Srinivasan M,
Rieger R, Chen EI and Sternglanz R: Biochemical characterization of
Hpa2 and Hpa3, two small closely related acetyltransferases from
Saccharomyces cerevisiae. J Biol Chem. 288:21506–21513.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sapountzi V and Cote J: MYST-family
histone acetyltransferases: Beyond chromatin. Cell Mol Life Sci.
68:1147–1156. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
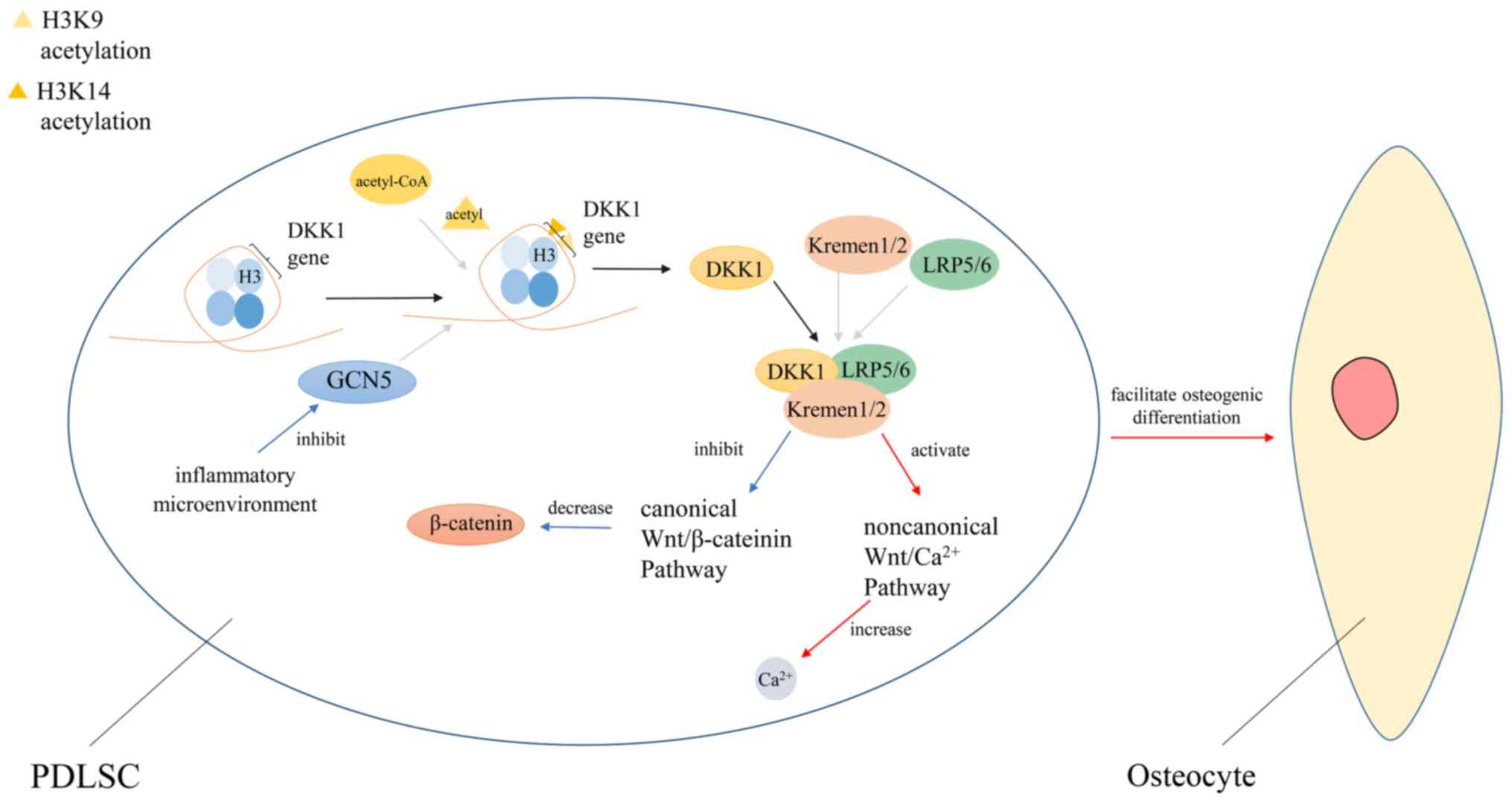
|
Reiter C, Heise F, Chung HR and
Ehrenhofer-Murray AE: A link between Sas2-mediated H4 K16
acetylation, chromatin assembly in S-phase by CAF-I and Asf1, and
nucleosome assembly by Spt6 during transcription. FEMS Yeast Res.
15(fov073)2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Church M, Smith KC, Alhussain MM, Pennings
S and Fleming AB: Sas3 and Ada2(Gcn5)-dependent histone H3
acetylation is required for transcription elongation at the
de-repressed FLO1 gene. Nucleic Acids Res. 45:4413–4430.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yan Y, Barlev NA, Haley RH, Berger SL and
Marmorstein R: Crystal structure of yeast Esa1 suggests a unified
mechanism for catalysis and substrate binding by histone
acetyltransferases. Mol Cell. 6:1195–1205. 2000.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang F, Marshall CB and Ikura M:
Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis:
Structural and functional versatility in target recognition. Cell
Mol Life Sci. 70:3989–4008. 2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hu LI, Lima BP and Wolfe AJ: Bacterial
protein acetylation: The dawning of a new age. Mol Microbiol.
77:15–21. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Pelletier N, Champagne N, Stifani S and
Yang XJ: MOZ and MORF histone acetyltransferases interact with the
Runt-domain transcription factor Runx2. Oncogene. 21:2729–2740.
2002.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Rokudai S, Laptenko O, Arnal SM, Taya Y,
Kitabayashi I and Prives C: MOZ increases p53 acetylation and
premature senescence through its complex formation with PML. Proc
Natl Acad Sci USA. 110:3895–3900. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fournier M, Orpinell M, Grauffel C, Scheer
E, Garnier JM, Ye T, Chavant V, Joint M, Esashi F, Dejaegere A, et
al: KAT2A/KAT2B-targeted acetylome reveals a role for PLK4
acetylation in preventing centrosome amplification. Nat Commun.
7(13227)2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bao X, Liu H, Liu X, Ruan K, Zhang Y,
Zhang Z, Hu Q, Liu Y, Akram S, Zhang J, et al: Mitosis-specific
acetylation tunes Ran effector binding for chromosome segregation.
J Mol Cell Biol. 10:18–32. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ghosh TK, Aparicio-Sanchez JJ, Buxton S,
Ketley A, Mohamed T, Rutland CS, Loughna S and Brook JD:
Acetylation of TBX5 by KAT2B and KAT2A regulates heart and limb
development. J Mol Cell Cardiol. 114:185–198. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cheng X, Ma X, Zhu Q, Song D, Ding X, Li
L, Jiang X, Wang X, Tian R, Su H, et al: Pacer is a mediator of
mTORC1 and GSK3-TIP60 signaling in regulation of autophagosome
maturation and lipid metabolism. Mol Cell. 73:788–802.e7.
2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Miotto B and Struhl K: HBO1 histone
acetylase is a coactivator of the replication licensing factor
Cdt1. Genes Dev. 22:2633–2638. 2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yuan H, Rossetto D, Mellert H, Dang W,
Srinivasan M, Johnson J, Hodawadekar S, Ding EC, Speicher K,
Abshiru N, et al: MYST protein acetyltransferase activity requires
active site lysine autoacetylation. EMBO J. 31:58–70.
2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chang R, Zhang Y, Zhang P and Zhou Q:
Snail acetylation by histone acetyltransferase p300 in lung cancer.
Thoracic Cancer. 8:131–137. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Yang Y, Cui J, Xue F, Zhang C, Mei Z, Wang
Y, Bi M, Shan D, Meredith A, Li H and Xu ZQ: Pokemon (FBI-1)
interacts with Smad4 to repress TGF-β-induced transcriptional
responses. Biochim Biophys Acta. 1849:270–281. 2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cazzalini O, Sommatis S, Tillhon M, Dutto
I, Bachi A, Rapp A, Nardo T, Scovassi AI, Necchi D, Cardoso MC, et
al: CBP and p300 acetylate PCNA to link its degradation with
nucleotide excision repair synthesis. Nucleic Acids Res.
42:8433–8448. 2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Senf SM, Sandesara PB, Reed SA and Judge
AR: p300 Acetyltransferase activity differentially regulates the
localization and activity of the FOXO homologues in skeletal
muscle. Am J Physiol Cell Physiol. 300:C1490–C1501. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lee K and Seo PJ: The HAF2 protein shapes
histone acetylation levels of PRR5 and LUX loci in Arabidopsis.
Planta. 248:513–518. 2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Nakakura T, Nemoto T, Suzuki T,
Asano-Hoshino A, Tanaka H, Arisawa K, Nishijima Y, Kiuchi Y and
Hagiwara H: Adrenalectomy facilitates ATAT1 expression and
alpha-tubulin acetylation in ACTH-producing corticotrophs. Cell
Tissue Res. 366:363–370. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang
Z, Yang T, Fu X, Jung SY, Wang Y, et al: Acetylation of Smc3 by
Eco1 is required for S phase sister chromatid cohesion in both
human and yeast. Mol Cell. 31:143–151. 2008.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wu H, Moshkina N, Min J, Zeng H, Joshua J,
Zhou MM and Plotnikov AN: Structural basis for substrate
specificity and catalysis of human histone acetyltransferase 1.
Proc Natl Acad Sci USA. 109:8925–8930. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
de Ruijter AJ, van Gennip AH, Caron HN,
Kemp S and van Kuilenburg AB: Histone deacetylases (HDACs):
Characterization of the classical HDAC family. Biochem J.
370:737–749. 2003.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Banik D, Moufarrij S and Villagra A:
Immunoepigenetics combination therapies: An overview of the role of
HDACs in cancer immunotherapy. Int J Mol Sci.
20(2241)2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Parra M: Class IIa HDACs-new insights into
their functions in physiology and pathology. FEBS J. 282:1736–1744.
2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Muller BM, Jana L, Kasajima A, Lehmann A,
Prinzler J, Budczies J, Winzer KJ, Dietel M, Weichert W and Denkert
C: Differential expression of histone deacetylases HDAC1, 2 and 3
in human breast cancer-overexpression of HDAC2 and HDAC3 is
associated with clinicopathological indicators of disease
progression. BMC Cancer. 13(215)2013.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Miller KM, Tjeertes JV, Coates J, Legube
G, Polo SE, Britton S and Jackson SP: Human HDAC1 and HDAC2
function in the DNA-damage response to promote DNA nonhomologous
end-joining. Nat Struct Mol Biol. 17:1144–1151. 2010.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Saito S, Zhuang Y, Suzuki T, Ota Y,
Bateman ME, Alkhatib AL, Morris GF and Lasky JA: HDAC8 inhibition
ameliorates pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol.
316:L175–L186. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Winbanks CE, Wang B, Beyer C, Koh P, White
L, Kantharidis P and Gregorevic P: TGF-beta regulates miR-206 and
miR-29 to control myogenic differentiation through regulation of
HDAC4. J Biol Chem. 286:13805–13814. 2011.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Cho Y, Sloutsky R, Naegle KM and Cavalli
V: Injury-induced HDAC5 nuclear export is essential for axon
regeneration. Cell. 155:894–908. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Bradley EW, Carpio LR, Olson EN and
Westendorf JJ: Histone deacetylase 7 (Hdac7) suppresses chondrocyte
proliferation and β-catenin activity during endochondral
ossification. J Biol Chem. 290:118–126. 2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Hu Y, Sun L, Tao S, Dai M, Wang Y, Li Y
and Wu J: Clinical significance of HDAC9 in hepatocellular
carcinoma. Cell Mol Biol (Noisy-le-Grand). 65:23–28.
2019.PubMed/NCBI
|
|
58
|
Bitler BG, Wu S, Park PH, Hai Y, Aird KM,
Wang Y, Zhai Y, Kossenkov AV, Vara-Ailor A, Rauscher FJ III, et al:
ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell
Biol. 19:962–973. 2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Radhakrishnan R, Li Y, Xiang S, Yuan F,
Yuan Z, Telles E, Fang J, Coppola D, Shibata D, Lane WS, et al:
Histone deacetylase 10 regulates DNA mismatch repair and may
involve the deacetylation of MutS homolog 2. J Biol Chem.
290:22795–22804. 2015.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zerr P, Palumbo-Zerr K, Huang J, Tomcik M,
Sumova B, Distler O, Schett G and Distler JH: Sirt1 regulates
canonical TGF-β signalling to control fibroblast activation and
tissue fibrosis. Ann Rheum Dis. 75:226–233. 2016.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Yuan Q, Zhan L, Zhou QY, Zhang LL, Chen
XM, Hu XM and Yuan XC: SIRT2 regulates microtubule stabilization in
diabetic cardiomyopathy. Eur J Pharmacol. 764:554–561.
2015.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Ahn BH, Kim HS, Song S, Lee IH, Liu J,
Vassilopoulos A, Deng CX and Finkel T: A role for the mitochondrial
deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad
Sci USA. 105:14447–14452. 2008.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Jeong SM, Xiao C, Finley LW, Lahusen T,
Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al:
SIRT4 has tumor-suppressive activity and regulates the cellular
metabolic response to DNA damage by inhibiting mitochondrial
glutamine metabolism. Cancer Cell. 23:450–463. 2013.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Rardin MJ, He W, Nishida Y, Newman JC,
Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, et al: SIRT5
regulates the mitochondrial lysine succinylome and metabolic
networks. Cell Metab. 18:920–933. 2013.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Kaluski S, Portillo M, Besnard A, Stein D,
Einav M, Zhong L, Ueberham U, Arendt T, Mostoslavsky R, Sahay A and
Toiber D: Neuroprotective functions for the histone Deacetylase
SIRT6. Cell Rep. 18:3052–3062. 2017.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Barber MF, Michishita-Kioi E, Xi Y,
Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL,
Chen K, et al: SIRT7 links H3K18 deacetylation to maintenance of
oncogenic transformation. Nature. 487:114–118. 2012.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Yuan L, Chen X, Cheng L, Rao M, Chen K,
Zhang N, Meng J, Li M, Yang LT, Yang PC, et al: HDAC11 regulates
interleukin-13 expression in CD4+T cells in the heart. J Mol Cell
Cardiol. 122:1–10. 2018.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Sahakian E, Chen J, Powers JJ, Chen X,
Maharaj K, Deng SL, Achille AN, Lienlaf M, Wang HW, Cheng F, et al:
Essential role for histone deacetylase 11 (HDAC11) in neutrophil
biology. J Leukoc Biol. 102:475–486. 2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Chatterjee SS, Saj A, Gocha T, Murphy M,
Gonsalves FC, Zhang X, Hayward P, Akgöl Oksuz B, Shen SS, Madar A,
et al: Inhibition of β-catenin-TCF1 interaction delays
differentiation of mouse embryonic stem cells. J Cell Biol.
211:39–51. 2015.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Abu-Elmagd M, Robson L, Sweetman D, Hadley
J, Francis-West P and Munsterberg A: Wnt/Lef1 signaling acts via
Pitx2 to regulate somite myogenesis. Dev Biol. 337:211–219.
2010.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Wapenaar H and Dekker FJ: Histone
acetyltransferases: Challenges in targeting bi-substrate enzymes.
Clin Epigenetics. 8(59)2016.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Chen GD, Yu WD and Chen XP: SirT1
activator represses the transcription of TNFα in THP1 cells of a
sepsis model via deacetylation of H4K16. Mol Med Rep. 14:5544–5550.
2016.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Michishita E, McCord RA, Berber E, Kioi M,
Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL,
Barrett JC, et al: SIRT6 is a histone H3 lysine 9 deacetylase that
modulates telomeric chromatin. Nature. 452:492–496. 2008.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Kim TK and Shiekhattar R: Architectural
and functional commonalities between enhancers and promoters. Cell.
162:948–959. 2015.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Rada-Iglesias A, Bajpai R, Swigut T,
Brugmann SA, Flynn RA and Wysocka J: A unique chromatin signature
uncovers early developmental enhancers in humans. Nature.
470:279–283. 2011.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Pradeepa MM, Grimes GR, Kumar Y, Olley G,
Taylor GC, Schneider R and Bickmore WA: Histone H3 globular domain
acetylation identifies a new class of enhancers. Nat Genet.
48:681–686. 2016.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Pradeepa MM: Causal role of histone
acetylations in enhancer function. Transcription. 8:40–47.
2017.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Dhar S, Gursoy-Yuzugullu O, Parasuram R
and Price BD: The tale of a tail: Histone H4 acetylation and the
repair of DNA breaks. Philos Trans R Soc Lond B Biol Sci.
372(20160284)2017.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Noguchi C, Singh T, Ziegler MA, Peake JD,
Khair L, Aza A, Nakamura TM and Noguchi E: The NuA4
acetyltransferase and histone H4 acetylation promote replication
recovery after topoisomerase I-poisoning. Epigenetics Chromatin.
12(24)2019.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Vadla R, Chatterjee N and Haldar D:
Cellular environment controls the dynamics of histone H3 lysine 56
acetylation in response to DNA damage in mammalian cells. J Biosci.
45(19)2020.PubMed/NCBI
|
|
81
|
Koprinarova M, Schnekenburger M and
Diederich M: Role of histone acetylation in cell cycle regulation.
Curr Top Med Chem. 16:732–744. 2016.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Gao FH, Hu XH, Li W, Liu H, Zhang YJ, Guo
ZY, Xu MH, Wang ST, Jiang B, Liu F, et al: Oridonin induces
apoptosis and senescence in colorectal cancer cells by increasing
histone hyperacetylation and regulation of p16, p21, p27 and c-myc.
BMC Cancer. 10(610)2010.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Kim E, Bisson WH, Löhr CV, Williams DE, Ho
E, Dashwood RH and Rajendran P: Histone and non-histone targets of
dietary Deacetylase inhibitors. Curr Top Med Chem. 16:714–731.
2016.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Glozak MA, Sengupta N, Zhang X and Seto E:
Acetylation and deacetylation of non-histone proteins. Gene.
363:15–23. 2005.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Suzuki M, Ikeda A and Bartlett JD: Sirt1
overexpression suppresses fluoride-induced p53 acetylation to
alleviate fluoride toxicity in ameloblasts responsible for enamel
formation. Arch Toxicol. 92:1283–1293. 2018.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Ito A, Lai CH, Zhao X, Saito S, Hamilton
MH, Appella E and Yao TP: p300/CBP-mediated p53 acetylation is
commonly induced by p53-activating agents and inhibited by MDM2.
EMBO J. 20:1331–1340. 2001.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Iyer NG, Xian J, Chin SF, Bannister AJ,
Daigo Y, Aparicio S, Kouzarides T and Caldas C: p300 is required
for orderly G1/S transition in human cancer cells. Oncogene.
26:21–29. 2007.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Shi D, Dai C, Qin J and Gu W: Negative
regulation of the p300-p53 interplay by DDX24. Oncogene.
35:528–536. 2016.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Morton DJ, Patel D, Joshi J, Hunt A,
Knowell AE and Chaudhary J: ID4 regulates transcriptional activity
of wild type and mutant p53 via K373 acetylation. Oncotarget.
8:2536–2549. 2017.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Ou HL and Schumacher B: DNA damage
responses and p53 in the aging process. Blood. 131:488–495.
2018.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Miyajima C, Kawarada Y, Inoue Y, Suzuki C,
Mitamura K, Morishita D, Ohoka N, Imamura T and Hayashi H:
Transcriptional Coactivator TAZ negatively regulates tumor
suppressor p53 activity and cellular senescence. Cells.
9(171)2020.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Reed SM and Quelle DE: p53 Acetylation:
Regulation and consequences. Cancers. 7:30–69. 2014.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Shan W, Jiang Y, Yu H, Huang Q, Liu L, Guo
X, Li L, Mi Q, Zhang K and Yang Z: HDAC2 overexpression correlates
with aggressive clinicopathological features and DNA-damage
response pathway of breast cancer. Am J Cancer Res. 7:1213–1226.
2017.PubMed/NCBI
|
|
94
|
Zhang L, Kang W, Lu X, Ma S, Dong L and
Zou B: Weighted gene co-expression network analysis and
connectivity map identifies lovastatin as a treatment option of
gastric cancer by inhibiting HDAC2. Gene. 681:15–25.
2019.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Brandl A, Wagner T, Uhlig KM, Knauer SK,
Stauber RH, Melchior F, Schneider G, Heinzel T and Krämer OH:
Dynamically regulated sumoylation of HDAC2 controls p53
deacetylation and restricts apoptosis following genotoxic stress. J
Mol Cell Biol. 4:284–293. 2012.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Li D, Marchenko ND and Moll UM: SAHA shows
preferential cytotoxicity in mutant p53 cancer cells by
destabilizing mutant p53 through inhibition of the HDAC6-Hsp90
chaperone axis. Cell Death Differ. 18:1904–1913. 2011.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Marrogi AJ, Khan MA, van Gijssel HE, Welsh
JA, Rahim H, Demetris AJ, Kowdley KV, Hussain SP, Nair J, Bartsch
H, et al: Oxidative stress and p53 mutations in the carcinogenesis
of iron overload-associated hepatocellular carcinoma. J Natl Cancer
Inst. 93:1652–1655. 2001.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Zhou Y, Que KT, Zhang Z, Yi ZJ, Zhao PX,
You Y, Gong JP and Liu ZJ: Iron overloaded polarizes macrophage to
proinflammation phenotype through ROS/acetyl-p53 pathway. Cancer
Med. 7:4012–4022. 2018.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Tu W, Zhang Q, Liu Y, Han L, Wang Q, Chen
P, Zhang S, Wang A and Zhou X: Fluoride induces apoptosis via
inhibiting SIRT1 activity to activate mitochondrial p53 pathway in
human neuroblastoma SH-SY5Y cells. Toxicol Appl Pharmacol.
347:60–69. 2018.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Wingelhofer B, Neubauer HA, Valent P, Han
X, Constantinescu SN, Gunning PT, Müller M and Moriggl R:
Implications of STAT3 and STAT5 signaling on gene regulation and
chromatin remodeling in hematopoietic cancer. Leukemia.
32:1713–1726. 2018.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Chen Y, Wu R, Chen HZ, Xiao Q, Wang WJ, He
JP, Li XX, Yu XW, Li L, Wang P, et al: Enhancement of hypothalamic
STAT3 acetylation by nuclear receptor Nur77 dictates leptin
sensitivity. Diabetes. 64:2069–2081. 2015.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Zheng J, van de Veerdonk FL, Crossland KL,
Smeekens SP, Chan CM, Al Shehri T, Abinun M, Gennery AR, Mann J,
Lendrem DW, et al: Gain-of-function STAT1 mutations impair STAT3
activity in patients with chronic mucocutaneous candidiasis (CMC).
Eur J Immunol. 45:2834–2846. 2015.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Ma L, Huang C, Wang XJ, Xin DE, Wang LS,
Zou QC, Zhang YS, Tan MD, Wang YM, Zhao TC, et al: Lysyl oxidase 3
is a dual-specificity enzyme involved in STAT3 Deacetylation and
Deacetylimination modulation. Mol Cell. 65:296–309. 2017.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Hansen DV, Hanson JE and Sheng M:
Microglia in Alzheimer's disease. J Cell Biol. 217:459–472.
2018.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Dani M, Wood M, Mizoguchi R, Fan Z, Walker
Z, Morgan R, Hinz R, Biju M, Kuruvilla T, Brooks DJ and Edison P:
Microglial activation correlates in vivo with both tau and amyloid
in Alzheimer's disease. Brain. 141:2740–2754. 2018.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Eufemi M, Cocchiola R, Romaniello D,
Correani V, Di Francesco L, Fabrizi C, Maras B and Schininà ME:
Acetylation and phosphorylation of STAT3 are involved in the
responsiveness of microglia to beta amyloid. Neurochem Int.
81:48–56. 2015.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Xu YS, Liang JJ, Wang Y, Zhao XJ, Xu L, Xu
YY, Zou QC, Zhang JM, Tu CE, Cui YG, et al: STAT3 undergoes
acetylation-dependent mitochondrial translocation to regulate
pyruvate metabolism. Sci Rep. 6(39517)2016.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Gough DJ, Corlett A, Schlessinger K,
Wegrzyn J, Larner AC and Levy DE: Mitochondrial STAT3 supports
Ras-dependent oncogenic transformation. Science. 324:1713–1716.
2009.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Wang Y, Huang Y, Liu J, Zhang J, Xu M, You
Z, Peng C, Gong Z and Liu W: Acetyltransferase GCN5 regulates
autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep.
21(e48335)2020.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Bao J, Zheng L, Zhang Q, Li X, Zhang X, Li
Z, Bai X, Zhang Z, Huo W, Zhao X, et al: Deacetylation of TFEB
promotes fibrillar Aβ degradation by upregulating lysosomal
biogenesis in microglia. Protein Cell. 7:417–433. 2016.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Zhang J, Wang J, Zhou Z, Park JE, Wang L,
Wu S, Sun X, Lu L, Wang T, Lin Q, et al: Importance of TFEB
acetylation in control of its transcriptional activity and
lysosomal function in response to histone deacetylase inhibitors.
Autophagy. 14:1043–1059. 2018.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Brijmohan AS, Batchu SN, Majumder S,
Alghamdi TA, Thieme K, McGaugh S, Liu Y, Advani SL, Bowskill BB,
Kabir MG, et al: HDAC6 inhibition promotes transcription factor EB
activation and is protective in experimental kidney disease. Front
Pharmacol. 9(34)2018.PubMed/NCBI View Article : Google Scholar
|
|
113
|
She A, Kurtser I, Reis SA, Hennig K, Lai
J, Lang A, Zhao WN, Mazitschek R, Dickerson BC, Herz J and Haggarty
SJ: Selectivity and kinetic requirements of HDAC inhibitors as
progranulin enhancers for treating frontotemporal dementia. Cell
Chem Biol. 24:892–906.e5. 2017.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Manickavinayaham S, Velez-Cruz R, Biswas
AK, Bedford E, Klein BJ, Kutateladze TG, Liu B, Bedford MT and
Johnson DG: E2F1 acetylation directs p300/CBP-mediated histone
acetylation at DNA double-strand breaks to facilitate repair. Nat
Commun. 10(4951)2019.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Chen L, Wei T, Si X, Wang Q, Li Y, Leng Y,
Deng A, Chen J, Wang G, Zhu S and Kang J: Lysine acetyltransferase
GCN5 potentiates the growth of non-small cell lung cancer via
promotion of E2F1, cyclin D1, and cyclin E1 expression. J Biol
Chem. 288:14510–14521. 2013.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Lin Y, Dong C and Zhou BP: Epigenetic
regulation of EMT: The snail story. Curr Pharm Des. 20:1698–1705.
2014.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Hsu DS, Wang HJ, Tai SK, Chou CH, Hsieh
CH, Chiu PH, Chen NJ and Yang MH: Acetylation of snail modulates
the cytokinome of cancer cells to enhance the recruitment of
macrophages. Cancer Cell. 26:534–548. 2014.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Xu W, Liu H, Liu ZG, Wang HS, Zhang F,
Wang H, Zhang J, Chen JJ, Huang HJ, Tan Y, et al: Histone
deacetylase inhibitors upregulate Snail via Smad2/3 phosphorylation
and stabilization of Snail to promote metastasis of hepatoma cells.
Cancer Lett. 420:1–13. 2018.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Zhang L, Shan X, Chen Q, Xu D, Fan X, Yu
M, Yan Q and Liu J: Downregulation of HDAC3 by ginsenoside Rg3
inhibits epithelial-mesenchymal transition of cutaneous squamous
cell carcinoma through c-Jun acetylation. J Cell Physiol.
234:22207–22219. 2019.PubMed/NCBI View Article : Google Scholar
|
|
121
|
McMahon SB: MYC and the control of
apoptosis. Cold Spring Harb Perspect Med. 4(a014407)2014.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Kenneth NS, Ramsbottom BA, Gomez-Roman N,
Marshall L, Cole PA and White RJ: TRRAP and GCN5 are used by c-Myc
to activate RNA polymerase III transcription. Proc Natl Acad Sci
USA. 104:14917–14922. 2007.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Mao B, Zhao G, Lv X, Chen HZ, Xue Z, Yang
B, Liu DP and Liang CC: Sirt1 deacetylates c-Myc and promotes
c-Myc/Max association. Int J Biochem Cell Biol. 43:1573–1581.
2011.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Bhadury J, Nilsson LM, Muralidharan SV,
Green LC, Li Z, Gesner EM, Hansen HC, Keller UB, McLure KG and
Nilsson JA: BET and HDAC inhibitors induce similar genes and
biological effects and synergize to kill in Myc-induced murine
lymphoma. Proc Natl Acad Sci USA. 111:E2721–E2730. 2014.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Nebbioso A, Carafa V, Conte M, Tambaro FP,
Abbondanza C, Martens J, Nees M, Benedetti R, Pallavicini I,
Minucci S, et al: c-Myc modulation and acetylation is a key HDAC
inhibitor target in cancer. Clin Cancer Res. 23:2542–2555.
2017.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Wang F, Wang Z, Li D and Chen Q:
Identification and characterization of a Bursaphelenchus xylophilus
(Aphelenchida: Aphelenchoididae) Thermotolerance-related gene:
Bx-HSP90. Int J Mol Sci. 13:8819–8833. 2012.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Kovacs JJ, Murphy PJ, Gaillard S, Zhao X,
Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB and Yao TP: HDAC6
regulates Hsp90 acetylation and chaperone-dependent activation of
glucocorticoid receptor. Mol Cell. 18:601–607. 2005.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Scroggins BT, Robzyk K, Wang D, Marcu MG,
Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, et al:
An acetylation site in the middle domain of Hsp90 regulates
chaperone function. Mol Cell. 25:151–159. 2007.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Meng Q, Chen X, Sun L, Zhao C, Sui G and
Cai L: Carbamazepine promotes Her-2 protein degradation in breast
cancer cells by modulating HDAC6 activity and acetylation of Hsp90.
Mol Cell Biochem. 348:165–171. 2011.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Rahimi N and Costello CE: Emerging roles
of post-translational modifications in signal transduction and
angiogenesis. Proteomics. 15:300–309. 2015.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Yan MS, Turgeon PJ, Man HJ, Dubinsky MK,
Ho JJD, El-Rass S, Wang YD, Wen XY and Marsden PA:
Acetyltransferase 7 (KAT7)-dependent intragenic histone acetylation
regulates endothelial Histone cell gene regulation. J Biol Chem.
293:4381–4402. 2018.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Alomer RM, da Silva EML, Chen J, Piekarz
KM, McDonald K, Sansam CG, Sansam CL and Rankin S: Esco1 and Esco2
regulate distinct cohesin functions during cell cycle progression.
Proc Natl Acad Sci USA. 114:9906–9911. 2017.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Rowland BD, Roig MB, Nishino T, Kurze A,
Uluocak P, Mishra A, Beckouët F, Underwood P, Metson J, Imre R, et
al: Building sister chromatid cohesion: Smc3 acetylation
counteracts an antiestablishment activity. Mol Cell. 33:763–774.
2009.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Gallinari P, Di Marco S, Jones P, Pallaoro
M and Steinkuhler C: HDACs, histone deacetylation and gene
transcription: From molecular biology to cancer therapeutics. Cell
Res. 17:195–211. 2007.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Schlesinger J, Schueler M, Grunert M,
Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I and
Sperling SR: The cardiac transcription network modulated by Gata4,
Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS
Genet. 7(e1001313)2011.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Leszczynska KB, Foskolou IP, Abraham AG,
Anbalagan S, Tellier C, Haider S, Span PN, O'Neill EE, Buffa FM and
Hammond EM: Hypoxia-induced p53 modulates both apoptosis and
radiosensitivity via AKT. J Clin Invest. 125:2385–2398.
2015.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Vadvalkar SS, Matsuzaki S, Eyster CA,
Giorgione JR, Bockus LB, Kinter CS, Kinter M and Humphries KM:
Decreased mitochondrial pyruvate transport activity in the diabetic
heart: Role of mitochondrial pyruvate carrier 2 (MPC2) Acetylation.
J Biol Chem. 292:4423–4433. 2017.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Hu H, Zhu W, Qin J, Chen M, Gong L, Li L,
Liu X, Tao Y, Yin H, Zhou H, et al: Acetylation of PGK1 promotes
liver cancer cell proliferation and tumorigenesis. Hepatology.
65:515–528. 2017.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Wang Y, Wang F, Bao X and Fu L: Systematic
analysis of lysine acetylome reveals potential functions of lysine
acetylation in Shewanella baltica, the specific spoilage organism
of aquatic products. J Proteomics. 205(103419)2019.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Chen J, Rong X, Fan G, Li S and Li Q:
Effects of different concentrations of putrescine on proliferation,
migration and apoptosis of human skin fibroblasts. Nan Fang Yi Ke
Da Xue Xue Bao. 35:758–762. 2015.PubMed/NCBI(In Chinese).
|
|
141
|
Mo R, Yang M, Chen Z, Cheng Z, Yi X, Li C,
He C, Xiong Q, Chen H, Wang Q and Ge F: Acetylome analysis reveals
the involvement of lysine acetylation in photosynthesis and carbon
metabolism in the model cyanobacterium Synechocystis sp. PCC 6803.
J Proteome Res. 14:1275–1286. 2015.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Yu BJ, Kim JA, Moon JH, Ryu SE and Pan JG:
The diversity of lysine-acetylated proteins in Escherichia coli. J
Microbiol Biotechnol. 18:1529–1536. 2008.PubMed/NCBI
|
|
143
|
Savoia M, Cencioni C, Mori M, Atlante S,
Zaccagnini G, Devanna P, Di Marcotullio L, Botta B, Martelli F,
Zeiher AM, et al: P300/CBP-associated factor regulates
transcription and function of isocitrate dehydrogenase 2 during
muscle differentiation. FASEB J. 33:4107–4123. 2019.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Baeza J, Smallegan MJ and Denu JM:
Mechanisms and dynamics of protein acetylation in mitochondria.
Trends Biochem Sci. 41:231–244. 2016.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Wagner GR and Hirschey MD: Nonenzymatic
protein acylation as a carbon stress regulated by sirtuin
deacylases. Mol Cell. 54:5–16. 2014.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Ronowska A, Szutowicz A, Bielarczyk H,
Gul-Hinc S, Klimaszewska-Łata J, Dyś A, Zyśk M and Jankowska-Kulawy
A: The regulatory effects of Acetyl-CoA distribution in the healthy
and diseased brain. Front Cell Neurosci. 12(169)2018.PubMed/NCBI View Article : Google Scholar
|
|
147
|
Zaini MA, Muller C, de Jong TV, Ackermann
T, Hartleben G, Kortman G, Gührs KH, Fusetti F, Krämer OH, Guryev V
and Calkhoven CF: A p300 and SIRT1 regulated acetylation switch of
C/EBPα controls mitochondrial function. Cell Rep. 22:497–511.
2018.PubMed/NCBI View Article : Google Scholar
|
|
148
|
Priante G, Gianesello L, Ceol M, Del Prete
D and Anglani F: Cell death in the kidney. Int J Mol Sci.
20(3598)2019.PubMed/NCBI View Article : Google Scholar
|
|
149
|
De Rasmo D, Signorile A, De Leo E,
Polishchuk EV, Ferretta A, Raso R, Russo S, Polishchuk R, Emma F
and Bellomo F: Mitochondrial dynamics of proximal tubular
epithelial cells in Nephropathic Cystinosis. Int J Mol Sci.
21(192)2019.PubMed/NCBI View Article : Google Scholar
|
|
150
|
Silva Junior G, Liborio AB, Mota RM, Abreu
KLS, Silva AEB, Silva SMHA and Daher EF: Acute kidney injury in
AIDS: Frequency, RIFLE classification and outcome. Brazilian J Med
Biol Res. 43:1102–1108. 2010.PubMed/NCBI View Article : Google Scholar
|
|
151
|
Kellum JA, Sileanu FE, Murugan R, Lucko N,
Shaw AD and Clermont G: Classifying AKI by urine output versus
serum creatinine level. J Am Soc Nephrol. 26:2231–2238.
2015.PubMed/NCBI View Article : Google Scholar
|
|
152
|
Cianciolo Cosentino C, Skrypnyk NI, Brilli
LL, Chiba T, Novitskaya T, Woods C, West J, Korotchenko VN,
McDermott L, Day BW, et al: Histone deacetylase inhibitor enhances
recovery after AKI. J Am Soc Nephrol. 24:943–953. 2013.PubMed/NCBI View Article : Google Scholar
|
|
153
|
Ranganathan P, Hamad R, Mohamed R,
Jayakumar C, Muthusamy T and Ramesh G: Histone deacetylase-mediated
silencing of AMWAP expression contributes to cisplatin
nephrotoxicity. Kidney Int. 89:317–326. 2016.PubMed/NCBI View Article : Google Scholar
|
|
154
|
Fan H, Yang HC, You L, Wang YY, He WJ and
Hao CM: The histone deacetylase, SIRT1, contributes to the
resistance of young mice to ischemia/reperfusion-induced acute
kidney injury. Kidney Int. 83:404–413. 2013.PubMed/NCBI View Article : Google Scholar
|
|
155
|
Xu S, Gao Y, Zhang Q, Wei S, Chen Z, Dai
X, Zeng Z and Zhao KS: SIRT1/3 activation by resveratrol attenuates
acute kidney injury in a septic rat model. Oxid Med Cell Longev.
2016(7296092)2016.PubMed/NCBI View Article : Google Scholar
|
|
156
|
Ozkok A, Ravichandran K, Wang Q,
Ljubanovic D and Edelstein CL: NF-κB transcriptional inhibition
ameliorates cisplatin-induced acute kidney injury (AKI). Toxicol
Lett. 240:105–113. 2016.PubMed/NCBI View Article : Google Scholar
|
|
157
|
Morigi M, Perico L, Rota C, Longaretti L,
Conti S, Rottoli D, Novelli R, Remuzzi G and Benigni A: Sirtuin
3-dependent mitochondrial dynamic improvements protect against
acute kidney injury. J Clin Invest. 125:715–726. 2015.PubMed/NCBI View Article : Google Scholar
|
|
158
|
Zhang Q, Liu X, Li N, Zhang J, Yang J and
Bu P: Sirtuin 3 deficiency aggravates contrast-induced acute kidney
injury. J Transl Med. 16(313)2018.PubMed/NCBI View Article : Google Scholar
|
|
159
|
Ning YC, Cai GY, Zhuo L, Gao JJ, Dong D,
Cui SY, Shi SZ, Feng Z, Zhang L, Sun XF and Chen XM: Beneficial
effects of short-term calorie restriction against cisplatin-induced
acute renal injury in aged rats. Nephron Exp Nephrol. 124:19–27.
2013.PubMed/NCBI View Article : Google Scholar
|
|
160
|
Scharman EJ and Troutman WG: Prevention of
kidney injury following rhabdomyolysis: A systematic review. Ann
Pharmacother. 47:90–105. 2013.PubMed/NCBI View Article : Google Scholar
|
|
161
|
Jian B, Yang S, Chaudry IH and Raju R:
Resveratrol restores sirtuin 1 (SIRT1) activity and pyruvate
dehydrogenase kinase 1 (PDK1) expression after hemorrhagic injury
in a rat model. Mol Med. 20:10–16. 2014.PubMed/NCBI View Article : Google Scholar
|
|
162
|
Si Y, Bao H, Han L, Chen L, Zeng L, Jing
L, Xing Y and Geng Y: Dexmedetomidine attenuation of renal
ischaemia-reperfusion injury requires sirtuin 3 activation. Br J
Anaesth. 121:1260–1271. 2018.PubMed/NCBI View Article : Google Scholar
|
|
163
|
Chaix MA, Andelfinger G and Khairy P:
Genetic testing in congenital heart disease: A clinical approach.
World J Cardiol. 8:180–191. 2016.PubMed/NCBI View Article : Google Scholar
|
|
164
|
Yuan S, Zaidi S and Brueckner M:
Congenital heart disease: Emerging themes linking genetics and
development. Curr Opin Genet Dev. 23:352–359. 2013.PubMed/NCBI View Article : Google Scholar
|
|
165
|
Wu G, Nan C, Rollo JC, Huang X and Tian J:
Sodium valproate-induced congenital cardiac abnormalities in mice
are associated with the inhibition of histone deacetylase. J Biomed
Sci. 17(16)2010.PubMed/NCBI View Article : Google Scholar
|
|
166
|
Zhong L, Zhu J, Lv T, Chen G, Sun H, Yang
X, Huang X and Tian J: Ethanol and its metabolites induce histone
lysine 9 acetylation and an alteration of the expression of heart
development-related genes in cardiac progenitor cells. Cardiovasc
Toxicol. 10:268–274. 2010.PubMed/NCBI View Article : Google Scholar
|
|
167
|
Li L, Zhu J, Tian J, Liu X and Feng C: A
role for Gcn5 in cardiomyocyte differentiation of rat mesenchymal
stem cells. Mol Cell Biochem. 345:309–316. 2010.PubMed/NCBI View Article : Google Scholar
|
|
168
|
Pillai VB, Sundaresan NR, Jeevanandam V
and Gupta MP: Mitochondrial SIRT3 and heart disease. Cardiovasc
Res. 88:250–256. 2010.PubMed/NCBI View Article : Google Scholar
|
|
169
|
Sundaresan NR, Samant SA, Pillai VB,
Rajamohan SB and Gupta MP: SIRT3 is a stress-responsive deacetylase
in cardiomyocytes that protects cells from stress-mediated cell
death by deacetylation of Ku70. Mol Cell Biol. 28:6384–6401.
2008.PubMed/NCBI View Article : Google Scholar
|
|
170
|
Zhang L, Wang H, Zhao Y, Wang J,
Dubielecka PM, Zhuang S, Qin G, Chin YE, Kao RL and Zhao TC:
Myocyte-specific overexpressing HDAC4 promotes myocardial
ischemia/reperfusion injury. Mol Med. 24(37)2018.PubMed/NCBI View Article : Google Scholar
|
|
171
|
Möller T and de Lange C: Myocardial
fibrosis in congenital heart disease. Tidsskr Nor Laegeforen: 138:
2018 (In Norwegian) doi: 10.4045/tidsskr.18.0864.
|
|
172
|
Pang M and Zhuang S: Histone deacetylase:
A potential therapeutic target for fibrotic disorders. J Pharmacol
Exp Ther. 335:266–272. 2010.PubMed/NCBI View Article : Google Scholar
|
|
173
|
Morimoto T, Sunagawa Y, Fujita M and
Hasegawa K: Novel heart failure therapy targeting transcriptional
pathway in cardiomyocytes by a natural compound, curcumin. Circ J.
74:1059–1066. 2010.PubMed/NCBI View Article : Google Scholar
|
|
174
|
Hu M, Zhang Q, Tian XH, Wang JL, Niu YX
and Li G: lncRNA CCAT1 is a biomarker for the proliferation and
drug resistance of esophageal cancer via the miR-143/PLK1/BUBR1
axis. Mol Carcinog. 58:2207–2217. 2019.PubMed/NCBI View Article : Google Scholar
|
|
175
|
Li J and Qi Y: Ginsenoside Rg3 inhibits
cell growth, migration and invasion in Caco-2 cells by
downregulation of lncRNA CCAT1. Exp Mol Pathol. 106:131–138.
2019.PubMed/NCBI View Article : Google Scholar
|
|
176
|
Zhang E, Han L, Yin D, He X, Hong L, Si X,
Qiu M, Xu T, De W, Xu L, et al: H3K27 acetylation activated-long
non-coding RNA CCAT1 affects cell proliferation and migration by
regulating SPRY4 and HOXB13 expression in esophageal squamous cell
carcinoma. Nucleic Acids Res. 45:3086–3101. 2017.PubMed/NCBI View Article : Google Scholar
|
|
177
|
Zhang H, Xie C, Yue J, Jiang Z, Zhou R,
Xie R, Wang Y and Wu S: Cancer-associated fibroblasts mediated
chemoresistance by a FOXO1/TGFβ1 signaling loop in esophageal
squamous cell carcinoma. Mol Carcinog. 56:1150–1163.
2017.PubMed/NCBI View Article : Google Scholar
|
|
178
|
Zhao Y, Yang J, Liao W, Liu X, Zhang H,
Wang S, Wang D, Feng J, Yu L and Zhu WG: Cytosolic FoxO1 is
essential for the induction of autophagy and tumour suppressor
activity. Nat Cell Biol. 12:665–675. 2010.PubMed/NCBI View Article : Google Scholar
|
|
179
|
Yu H, Ye W, Wu J, Meng X, Liu RY, Ying X,
Zhou Y, Wang H, Pan C and Huang W: Overexpression of sirt7 exhibits
oncogenic property and serves as a prognostic factor in colorectal
cancer. Clin Cancer Res. 20:3434–3445. 2014.PubMed/NCBI View Article : Google Scholar
|
|
180
|
Li D, Sun X, Zhang L, Yan B, Xie S, Liu R,
Liu M and Zhou J: Histone deacetylase 6 and cytoplasmic linker
protein 170 function together to regulate the motility of
pancreatic cancer cells. Protein Cell. 5:214–223. 2014.PubMed/NCBI View Article : Google Scholar
|
|
181
|
Sharma S and Taliyan R: Histone
deacetylase inhibitors: Future therapeutics for insulin resistance
and type 2 diabetes. Pharmacol Res. 113:320–326. 2016.PubMed/NCBI View Article : Google Scholar
|
|
182
|
Berry JM, Cao DJ, Rothermel BA and Hill
JA: Histone deacetylase inhibition in the treatment of heart
disease. Expert Opin Drug Saf. 7:53–67. 2008.PubMed/NCBI View Article : Google Scholar
|
|
183
|
Yoon S, Kang G and Eom GH: HDAC
Inhibitors: Therapeutic potential in fibrosis-associated human
diseases. Int J Mol Sci. 20(1329)2019.PubMed/NCBI View Article : Google Scholar
|
|
184
|
Clawson GA: Histone deacetylase inhibitors
as cancer therapeutics. Ann Transl Med. 4(287)2016.PubMed/NCBI View Article : Google Scholar
|
|
185
|
Peserico A and Simone C: Physical and
functional HAT/HDAC interplay regulates protein acetylation
balance. J Biomed Biotechnol. 2011(371832)2011.PubMed/NCBI View Article : Google Scholar
|
|
186
|
Qiu X, Xiao X, Li N and Li Y: Histone
deacetylases inhibitors (HDACis) as novel therapeutic application
in various clinical diseases. Prog Neuropsychopharmacol Biol
Psychiatry. 72:60–72. 2017.PubMed/NCBI View Article : Google Scholar
|
|
187
|
Xu W, Ngo L, Perez G, Dokmanovic M and
Marks PA: Intrinsic apoptotic and thioredoxin pathways in human
prostate cancer cell response to histone deacetylase inhibitor.
Proc Natl Acad Sci USA. 103:15540–15545. 2006.PubMed/NCBI View Article : Google Scholar
|
|
188
|
Rosato RR and Grant S: Histone deacetylase
inhibitors: Insights into mechanisms of lethality. Expert Opin Ther
Targets. 9:809–824. 2005.PubMed/NCBI View Article : Google Scholar
|
|
189
|
Minucci S and Pelicci PG: Histone
deacetylase inhibitors and the promise of epigenetic (and more)
treatments for cancer. Nat Rev Cancer. 6:38–51. 2006.PubMed/NCBI View Article : Google Scholar
|
|
190
|
Lane AA and Chabner BA: Histone
deacetylase inhibitors in cancer therapy. J Clin Oncol.
27:5459–5468. 2009.PubMed/NCBI View Article : Google Scholar
|
|
191
|
Eckschlager T, Plch J, Stiborova M and
Hrabeta J: Histone deacetylase inhibitors as anticancer drugs. Int
J Mol Sci. 18(1414)2017.PubMed/NCBI View Article : Google Scholar
|
|
192
|
Di Pompo G, Salerno M, Rotili D, Valente
S, Zwergel C, Avnet S, Lattanzi G, Baldini N and Mai A: Novel
histone deacetylase inhibitors induce growth arrest, apoptosis, and
differentiation in sarcoma cancer stem cells. J Med Chem.
58:4073–4079. 2015.PubMed/NCBI View Article : Google Scholar
|
|
193
|
Debeb BG, Lacerda L, Larson R, Wolfe AR,
Krishnamurthy S, Reuben JM, Ueno NT, Gilcrease M and Woodward WA:
Histone deacetylase inhibitor-induced cancer stem cells exhibit
high pentose phosphate pathway metabolism. Oncotarget.
7:28329–28339. 2016.PubMed/NCBI View Article : Google Scholar
|
|
194
|
Cao D, Wang M, Qiu X, Liu D, Jiang H, Yang
N and Xu RM: Structural basis for allosteric, substrate-dependent
stimulation of SIRT1 activity by resveratrol. Genes Dev.
29:1316–1325. 2015.PubMed/NCBI View Article : Google Scholar
|
|
195
|
Li J, Chong T, Wang Z, Chen H and Li H,
Cao J, Zhang P and Li H: A novel anticancer effect of resveratrol:
Reversal of epithelial-mesenchymal transition in prostate cancer
cells. Mol Med Rep. 10:1717–1724. 2014.PubMed/NCBI View Article : Google Scholar
|
|
196
|
Moussaieff A, Rouleau M, Kitsberg D, Cohen
M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M,
et al: Glycolysis-mediated changes in acetyl-CoA and histone
acetylation control the early differentiation of embryonic stem
cells. Cell Metab. 21:392–402. 2015.PubMed/NCBI View Article : Google Scholar
|
|
197
|
Hezroni H, Tzchori I, Davidi A, Mattout A,
Biran A, Nissim-Rafinia M, Westphal H and Meshorer E: H3K9 histone
acetylation predicts pluripotency and reprogramming capacity of ES
cells. Nucleus. 2:300–309. 2011.PubMed/NCBI View Article : Google Scholar
|
|
198
|
Zhang Y, Cui P, Li Y, Feng G, Tong M, Guo
L, Li T, Liu L, Li W and Zhou Q: Mitochondrially produced ATP
affects stem cell pluripotency via Actl6a-mediated histone
acetylation. FASEB J. 32:1891–1902. 2018.PubMed/NCBI View Article : Google Scholar
|
|
199
|
Qiao Y, Wang R, Yang X, Tang K and Jing N:
Dual roles of histone H3 lysine 9 acetylation in human embryonic
stem cell pluripotency and neural differentiation. J Biol Chem.
290:2508–2520. 2015.PubMed/NCBI View Article : Google Scholar
|
|
200
|
Jacobs KM, Misri S, Meyer B, Raj S, Zobel
CL, Sleckman BP, Hallahan DE and Sharma GG: Unique epigenetic
influence of H2AX phosphorylation and H3K56 acetylation on normal
stem cell radioresponses. Mol Biol Cell. 27:1332–1345.
2016.PubMed/NCBI View Article : Google Scholar
|
|
201
|
Wen Y, Yang H, Wu J, Wang A, Chen X, Hu S,
Zhang Y, Bai D and Jin Z: COL4A2 in the tissue-specific
extracellular matrix plays important role on osteogenic
differentiation of periodontal ligament stem cells. Theranostics.
9:4265–4286. 2019.PubMed/NCBI View Article : Google Scholar
|
|
202
|
Li B, Sun J, Dong Z, Xue P, He X, Liao L,
Yuan L and Jin Y: GCN5 modulates osteogenic differentiation of
periodontal ligament stem cells through DKK1 acetylation in
inflammatory microenvironment. Sci Rep. 6(26542)2016.PubMed/NCBI View Article : Google Scholar
|
|
203
|
Liu N, Shi S, Deng M, Tang L, Zhang G, Liu
N, Ding B, Liu W, Liu Y, Shi H, et al: High levels of β-catenin
signaling reduce osteogenic differentiation of stem cells in
inflammatory microenvironments through inhibition of the
noncanonical Wnt pathway. J Bone Miner Res. 26:2082–2095.
2011.PubMed/NCBI View Article : Google Scholar
|
|
204
|
Ishitani T, Kishida S, Hyodo-Miura J, Ueno
N, Yasuda J, Waterman M, Shibuya H, Moon RT, Ninomiya-Tsuji J and
Matsumoto K: The TAK1-NLK mitogen-activated protein kinase cascade
functions in the Wnt-5a/Ca(2+) pathway to antagonize
Wnt/beta-catenin signaling. Mol Cell Biol. 23:131–139.
2003.PubMed/NCBI View Article : Google Scholar
|
|
205
|
Wang H, Diao D, Shi Z, Zhu X, Gao Y, Gao
S, Liu X, Wu Y, Rudolph KL, Liu G, et al: SIRT6 controls
hematopoietic stem cell homeostasis through epigenetic regulation
of Wnt signaling. Cell Stem Cell. 18:495–507. 2016.PubMed/NCBI View Article : Google Scholar
|
|
206
|
Wagner VP, Martins MD and Castilho RM:
Histones acetylation and cancer stem cells (CSCs). Methods Mol
Biol. 1692:179–193. 2018.PubMed/NCBI View Article : Google Scholar
|
|
207
|
Liu C, Liu L, Shan J, Shen J, Xu Y, Zhang
Q, Yang Z, Wu L, Xia F, Bie P, et al: Histone deacetylase 3
participates in self-renewal of liver cancer stem cells through
histone modification. Cancer Lett. 339:60–69. 2013.PubMed/NCBI View Article : Google Scholar
|
|
208
|
Liu D, Zhou P, Zhang L, Gong W, Huang G,
Zheng Y and He F: HDAC1/DNMT3A-containing complex is associated
with suppression of Oct4 in cervical cancer cells. Biochemistry
(Mosc). 77:934–940. 2012.PubMed/NCBI View Article : Google Scholar
|
|
209
|
Choi HJ, Park JH, Park M, Won HY, Joo HS,
Lee CH, Lee JY and Kong G: UTX inhibits EMT-induced breast CSC
properties by epigenetic repression of EMT genes in cooperation
with LSD1 and HDAC1. EMBO Rep. 16:1288–1298. 2015.PubMed/NCBI View Article : Google Scholar
|
|
210
|
Salvador MA, Wicinski J, Cabaud O, Toiron
Y, Finetti P, Josselin E, Lelièvre H, Kraus-Berthier L, Depil S,
Bertucci F, et al: The histone deacetylase inhibitor abexinostat
induces cancer stem cells differentiation in breast cancer with low
Xist expression. Clin Cancer Res. 19:6520–6531. 2013.PubMed/NCBI View Article : Google Scholar
|
|
211
|
Corbin AS, Agarwal A, Loriaux M, Cortes J,
Deininger MW and Druker BJ: Human chronic myeloid leukemia stem
cells are insensitive to imatinib despite inhibition of BCR-ABL
activity. J Clin Invest. 121:396–409. 2011.PubMed/NCBI View Article : Google Scholar
|
|
212
|
Li L, Wang L, Li L, Wang Z, Ho Y, McDonald
T, Holyoake TL, Chen W and Bhatia R: Activation of p53 by SIRT1
inhibition enhances elimination of CML leukemia stem cells in
combination with imatinib. Cancer Cell. 21:266–281. 2012.PubMed/NCBI View Article : Google Scholar
|
|
213
|
Hsu YC, Wu YT, Tsai CL and Wei YH: Current
understanding and future perspectives of the roles of sirtuins in
the reprogramming and differentiation of pluripotent stem cells.
Exp Biol Med (Maywood). 243:563–575. 2018.PubMed/NCBI View Article : Google Scholar
|
|
214
|
Sun J, Wei HM, Xu J, Chang JF, Yang Z, Ren
X, Lv WW, Liu LP, Pan LX, Wang X, et al: Histone H1-mediated
epigenetic regulation controls germline stem cell self-renewal by
modulating H4K16 acetylation. Nat Commun. 6(8856)2015.PubMed/NCBI View Article : Google Scholar
|
|
215
|
Hou T, Zheng G, Zhang P, Jia J, Li J, Xie
L, Wei C and Li Y: LAceP: Lysine acetylation site prediction using
logistic regression classifiers. PLoS One. 9(e89575)2014.PubMed/NCBI View Article : Google Scholar
|
|
216
|
Chen C, Huang H and Wu CH: Protein
bioinformatics databases and resources. Methods Mol Biol.
1558:3–39. 2017.PubMed/NCBI View Article : Google Scholar
|
|
217
|
Wang L, Du Y, Lu M and Li T: ASEB: A web
server for KAT-specific acetylation site prediction. Nucleic Acids
Res. 40:W376–379. 2012.PubMed/NCBI View Article : Google Scholar
|
|
218
|
Deng W, Wang C, Zhang Y, Xu Y, Zhang S,
Liu Z and Xue Y: GPS-PAIL: Prediction of lysine
acetyltransferase-specific modification sites from protein
sequences. Sci Rep. 6(39787)2016.PubMed/NCBI View Article : Google Scholar
|
|
219
|
Shi SP, Qiu JD, Sun XY, Suo SB, Huang SY
and Liang RP: PLMLA: Prediction of lysine methylation and lysine
acetylation by combining multiple features. Mol Biosyst.
8:1520–1527. 2012.PubMed/NCBI View Article : Google Scholar
|