The functions of biological systems are complicated
and diverse, and are largely dependent on the regulation of protein
function. After translation is complete, the proteins need to
undergo different degrees of chemical modifications, known as
post-translational modifications (PTMs), which can change
structures by altering the physicochemical properties of the
primary sequences and adjusting protein compactness by changing
their charge (1). In addition, PTMs
can interfere with the shift of protein state (2). The main types of PTM include
methylation, acetylation, glycosylation, ubiquitination and
phosphorylation (3). Enzymatic
acetylation modifies ~50% of yeast proteins and up to 90% of higher
eukaryotic proteins (4). Fewer
acetylated proteins have been identified in prokaryotes (5). The main mechanism of protein
acetylation, which is one of the most advanced topics in PTM
studies, is that acetyl donors (such as acetyl-CoA) transfer acetyl
groups to the proteins under the catalysis of acetyltransferase
(6). Acetylation occurs mainly on
lysine and can be divided into histone acetylation and non-histone
protein acetylation (7). There are
currently three well-known forms of acetylation: Nα-acetylation,
Nε-acetylation and O-acetylation (8). Nα-acetylation refers to the addition of
an acetyl group to the α-amino group of the N-terminal amino acid,
which is an irreversible process; ~85% of human protein is modified
by Nα-acetylation (9).
Nε-acetylation refers to the addition of an acetyl group to the
ε-amino group of the lysine residue, which is a reversible process
(10). O-acetylation refers to the
addition of an acetyl group to the tyrosine/serine/threonine
hydroxyl group (11).
The research history of protein acetylation has
spanned >50 years. Lysine acetylation in histones was first
discovered and proposed by Vincent Allfrey in 1964(12), which was considered to be related to
the regulation of gene transcription (13). Histones contain a large number of two
basic amino acids, lysine and arginine, and therefore have a
positive charge. If lysine is acetylated, it will no longer be
positively charged, so the binding of DNA to the histone is
relaxed, which facilitates gene transcription (12). In 2009, >1,000 types of acetylated
non-histone proteins were historically discovered by studying
metabolic pathways of different species (14). In accordance with the large amount of
non-histone protein acetylation, histone acetyltransferases (HATs)
and histone deacetylases (HDACs) were renamed to lysine
acetyltransferases (KATs) and lysine deacetylases (KDACs),
respectively (15,16). It was discovered that acetylation
could affect the enzyme activity of nucleases, thereby regulating
the level of substrate RNA (17).
This discovery indicates that organisms can achieve self-regulation
of cells through nucleases.
It has been reported that there is a wide range of
protein acetylation, which could mean that they have crucial
physiological functions in various biological activities. Protein
acetylation is one of the major regulators of gene transcription
(18). Most HATs are localized in
the nucleus and function as transcriptional co-activators (19). Acetylation involves the regulation of
>100 non-histone proteins, including transcription factors
(TFs), transcriptional coactivators and nuclear receptors (20). Protein acetylation is also associated
with protein degradation. Early studies demonstrated that proteins
with free α-amino groups can be degraded by ATP-dependent ubiquitin
degradation, and that ubiquitin-mediated protein degradation can be
prevented when the N-terminal α-amino group is acetylated (21). Besides, protein acetylation can
regulate a variety of signaling pathways and affect the cell cycle.
In this review, the latest advances in protein acetylation of both
histone and non-histone proteins will be presented, and the
interaction between diseases and protein acetylation will be
discussed.
Acetylation is catalyzed by KATs, which are
distributed in both the nucleus and the cytoplasm. Previous studies
have identified >20 types of KATs, they can be primarily divided
into five groups on the basis of their similarity in several
homology regions and acetylation-related motifs: i) General control
of amino acid synthesis protein 5 (GCN5) family, including GCN5
(KAT2A), p300/CBP-associated factor (PCAF; KAT2B), histone
acetyltransferase 1 (HAT1), elongator acetyltransferase complex
subunit 3, histone acetyltransferase HPA2 (HPA2) and HPA3, which is
currently the most typical family of KATs (22-25).
ii) MYST family, primarily including 60 kDa tat-protein (KAT5),
monocytic leukemia zinc finger protein (KAT6A), MOZ-related factor
(KAT6B), histone acetyltransferase binding to ORC1 (KAT7), ortholog
of Drosophila males-absent on the first protein (MOF; KAT8),
histone acetyltransferase SAS2 (SAS2), SAS3 and ESA1. The
classification of this family is due to the presence of the highly
conserved MYST domain that consists of an acetyl-CoA binding motif
and a zinc finger. According to additional structural features,
members of this family can be classified (26-29).
iii) p300/cAMP response element-binding protein (CBP or CREB)
family (KAT3A/KAT3B), which is closely related to cell
differentiation and apoptosis, and has >75 non-histone
substrates. Members of this family have four separate
trans-activation domains, including the cysteine-histidine-rich
region 1, the CREB-interacting kinase-inducible domain interacting
domain, another cysteine-histidine-rich region and the nuclear
receptor co-activator binding domain. p300/CBP is also a
coactivator of various TFs, and it can link chromatin remodeling
and transcriptional processes to coordinate physiological
activities, such as signal transduction, in vivo (30). iv) Transcription initiation factor
TFIID 230/250 kDa subunit (TAFII230/250) family. This family in
humans is TAFII250, and it is a component of the TF complex TAFIID
(31). v) Others, including
α-tubulin N-acetyltransferase 1, establishment of sister chromatid
cohesion N-acetyltransferase (ESCO)1, ESCO2 and HAT1, among which
ESCO1 and ESCO2 are two N-acetyltransferases. Different types of
KATs play different roles in cells, and stable expression of
various KATs is vital for maintaining the physiological activities
of cells (Table I) (23-25,27-29,32-47).
Previous research has demonstrated that KDACs can be
primarily divided into four categories (48). Class I includes HDAC1, HDAC2, HDAC3
and HDAC8, which can be found in the nucleus (49). Class II can be found in both the
nucleus and the cytoplasm. According to the different catalytic
sites, it can be further divided into class IIA and class IIB.
Class IIA, including HDAC4, HDAC5, HDAC7 and HDAC9, have a
catalytic site and they can perform nuclear transport under cell
signal stimulation. Class IIB, including HDAC6 and HDAC10, have two
catalytic sites and they are mainly located in the cytoplasm
(49). Both class I and class II are
zinc-dependent enzymes. Class III, including sirtuin 1-7 (SIRT1-7),
are NAD+-dependent enzymes. This class of enzymes have a
wide range of subcellular localization (49). Class IV includes only one member,
HDAC11, which is a zinc-dependent enzyme. HDAC11 mediates
deacetylation of lysine on the N-terminal tail of the core histones
(50). In addition to these four
categories, T cell transcription factor 1 and lymphoid
enhancer-binding factor 1 are also two KDACs located in the nucleus
(Table II) (51-70).
Typically, acetyltransferases and deacetylases work together to
regulate lysine acetylation and other various physiological
processes in the organism. If the balance of acetylation and
deacetylation becomes dysregulated, it is likely to cause tumor
growth and a number of non-neoplastic diseases such as inflammatory
diseases and neurological disorders (71).
It has been reported that combinations of
monomethylation of histone H3 at lysine 4 (H3K4me1) and histone 3
lysine 27 acetylation (H3K27ac) or H3K27me3 are often used as a
basis to differentiate active enhancers from inactive enhancers and
poised enhancers (74,75). However, this method of identification
does not completely distinguish between other types of enhancers,
such as super-enhancer (76). It has
been found that H3K122ac is also enriched with H3K27ac on the
active enhancer. H3K122ac can be used as a marker to identify some
novel enhancers, but some of these novel enhancers will also be
enriched in H3K27ac. This characteristic provides new ideas for
comprehensive identification enhancers (77). Histone acetylation also plays a role
in the repair of DNA replication forks. Nucleosome
acetyltransferase of H4 (NuA4) is involved in acetylation of H4 on
four lysine residues at position 5, 8, 12 and 16, which is
Nε-acetylation. This modification changes the structure of
chromatin, facilitating the repair of broken DNA replication forks
(78). SWI1 promotes histone H4
acetylation by stabilizing the expression of NuA4. Loss of SWI1
leads to the instability of chromatin modification-related protein
vid21, a regulatory subunit of NuA4, leading to a reduction in
histone H4 acetylation (79). It is
reported that the level of H3K56ac increases from low to high cell
density and H3K56ac was observed to increase when lactic acid
levels rose. This phenomenon may be attributed to changes in the
levels of SIRT6. Furthermore, the level of H3K56ac was increased in
cells with low acetylation immediately after DNA damage, and the
level was decreased in cells with high acetylation immediately
after DNA damage, which indicates the association between
acetylation and repair after DNA damage (80). Moreover, histone acetyltransferase
Gcn5p is a catalytic subunit of a nuclear HAT. Gcn5p catalyzes the
acetylation of histone H3 and H4 at specific lysines, which is N-ε
acetylation at specific lysines in the amino-terminal domains,
promoting cell growth. These results suggest that the acetylation
of specific lysines at H3 and H4 is essential for normal cell cycle
progression (81). Oridonin is a
tetracycline diterpenoid compound that is an important traditional
Chinese herb. It has been reported that oridonin inhibits tumor
cell proliferation and induces apoptosis, possibly by inducing the
hyperacetylation of histone H3(82).
As studies of histone acetylation have gradually
deepened, researchers proposed the idea that non-histone proteins,
such as p53, could also be acetylated. Although non-histone protein
acetylation has been studied for a shorter period of time compared
with histone acetylation, non-histone protein acetylation has been
highlighted recently due to its extensive regulatory functions.
There are numerous types of non-histone proteins that can be
acetylated, among which TFs are the main members (83). These non-histone proteins are widely
involved in a variety of physiological processes in different ways,
including gene transcription and protein folding (71).
As a tumor suppressor, p53 actively participates in
the regulation of tumor formation and can be acetylated by the
p300/CBP family in the way of Nε-acetylation. The p300/CBP family
can acetylate the C-terminal lysine of p53 and further activate
specific DNA binding sites on p53(84). When DNA is damaged, p300/CBP family
members binds to the promoter of p53 to increase the
transcriptional activity of its gene in order to enhance p53
stability (85). p300/CBP not only
regulates p53 activity in cells by acetylating p53, but also causes
the inactivation of E3 ubiquitin-protein ligase murine double
minute 2 (MDM2) to regulate p53. MDM2 also inhibits
p300/CBP-mediated p53 acetylation, and p53 can be effectively
degraded by MDM2 after being deacetylated (86). p53 acetylation is also associated
with the regulation of apoptosis. p300 is a key TF that promotes
cell transformation from G1 to S phase and regulates p53
transcriptional activity via acetylation (87,88). It
has been demonstrated that DEAD-box RNA helicase 24 (DDX24)
interacts with p300 to inhibit p300-mediated p53 acetylation. When
DDX24 was knocked out in human lung cancer cells, it was found that
the level of p53 acetylation was significantly increased, and
G1/S arrest was observed in these cells. Following
which, cells showed apoptosis (88).
Inhibitor of DNA binding 4 is a differentiation inhibitory protein
that promotes p53-dependent apoptosis by increasing the level of
p53 acetylation (89). Moreover, it
has been reported that the acetylation of p53 is associated with
aging (90). Transcriptional
coactivator with a PDZ-binding motif (TAZ) inhibits p300-mediated
p53 acetylation by suppressing the binding of p53 and p300(91). Furthermore, experiments have revealed
that TAZ-knockout causes p53-dependent cellular senescence in
normal human fibroblasts, which may contribute to tumorigenesis by
suppressing p53-mediated cellular senescence (91). The association of p53 acetylation
with apoptosis and senescence suggests that p53 is related to
cancer (92). Overexpression of
HDAC2 is found in a variety of cancer cells, such as breast cancer
and gastric cancer cells (93,94).
HDAC2 causes the deacetylation of the C-terminal lysine on p53, and
functions as a corepressor involved in the regulation of target
genes. If KDAC inhibitors (KDACIs) are used to keep some key lysine
residues highly acetylated on p53, the stability of p53 can be
enhanced (95). KDACIs also inhibit
HDAC6 and promote the degradation of mutant p53 via MDM2 and CHIP
ligase, which is hypothesized to be a mechanism for eliminating
mutant p53(96). Iron overload and
fluoride may also be associated with p53 acetylation. Some studies
have demonstrated that iron overload in macrophages may promote the
production of reactive oxygen species, increase p53
acetyltransferase activity, and ultimately lead to macrophage M1
polarization by inducing the expression and acetylation of p53
(97,98). Fluoride can induce the acetylation of
K379 on p53, which can be recovered by SIRT1 (85,99).
Further study has indicated that SIRT1 deacetylates
fluoride-induced p53 acetylation to attenuate fluorescence-induced
cell growth inhibition, mitochondrial damage, DNA damage and
apoptosis (85). As one of the
typical representatives of non-histone protein acetylation, p53
acetylation broadens the scope of acetylation modification,
providing an important basis for further studies on protein
acetylation.
Signal transducer and activator of transcription 3
(STAT3) serves a dual role in transmitting signals and initiating
gene transcription (100). The
nuclear receptor Nur77 can recruit p300 and reduce HDAC1
expression, which promotes STAT3 acetylation in the way of
Nε-acetylation and enhance the transcriptional activity of
STAT3(101). Decreased
transcriptional capacity of STAT3-dependent genes can be reversed
using KDACI trichostatin A or ITF2357 to inhibit acetylation of
STAT1 or STAT3. This may be useful in the treatment of chronic
mucocutaneous candidiasis (102).
In the nucleus, lysyl oxidase like 3 associates with STAT3 to
deacetylate and deacetyliminate STAT3 on multiple acetyl-lysine
sites. As the result of this, STAT3 dimerization is disrupted and
STAT3 transcription activity is inhibited (103). The acetylation of K685 on STAT3 can
be regulated by p300/CBP, which enhances sequence-specific DNA
binding ability and transcriptional activity of STAT3. Microglia
are the innate immune cells of the central nervous system, which
can inhibit the toxic toxic accumulation of β-amyloid. However,
activated microglia can engulf synapses and activate inflammatory
cytokines, causing the loss of synapses and the damage of neurons
(104). It is reported that the
activation of microglia is related to the deposition of amyloid
protein (105). The synthetic form
of Aβ peptides was used to treat primary and immortalized
microglial cells, and then the relative abundance of acetylated and
phosphorylated STAT3 was measured at different stages. In the early
stage, the level of STAT3 acetylation on K685 was increased. Then,
in the delayed one, its isoform will be phosphorylated on Y705
residue (106). Although the
association between acetylation and phosphorylation is still
unknown in this event, the acetylation of STAT3 is associated with
the nervous system (106). Studies
have also found that acetylation and deacetylation of STAT3 can
regulate the tricarboxylic acid cycle (107,108).
Serum starvation and reintroduction or insulin stimulation can
induce STAT3 CBP acetylation in serum starved cells and result in
the transfer of STAT3 into mitochondria (107). If STAT3 is deacetylated by SIRT5,
STAT3 will reduce the association with the pyruvate dehydrogenase
complex E1 and slow the conversion of pyruvate to acetyl-CoA
(107).
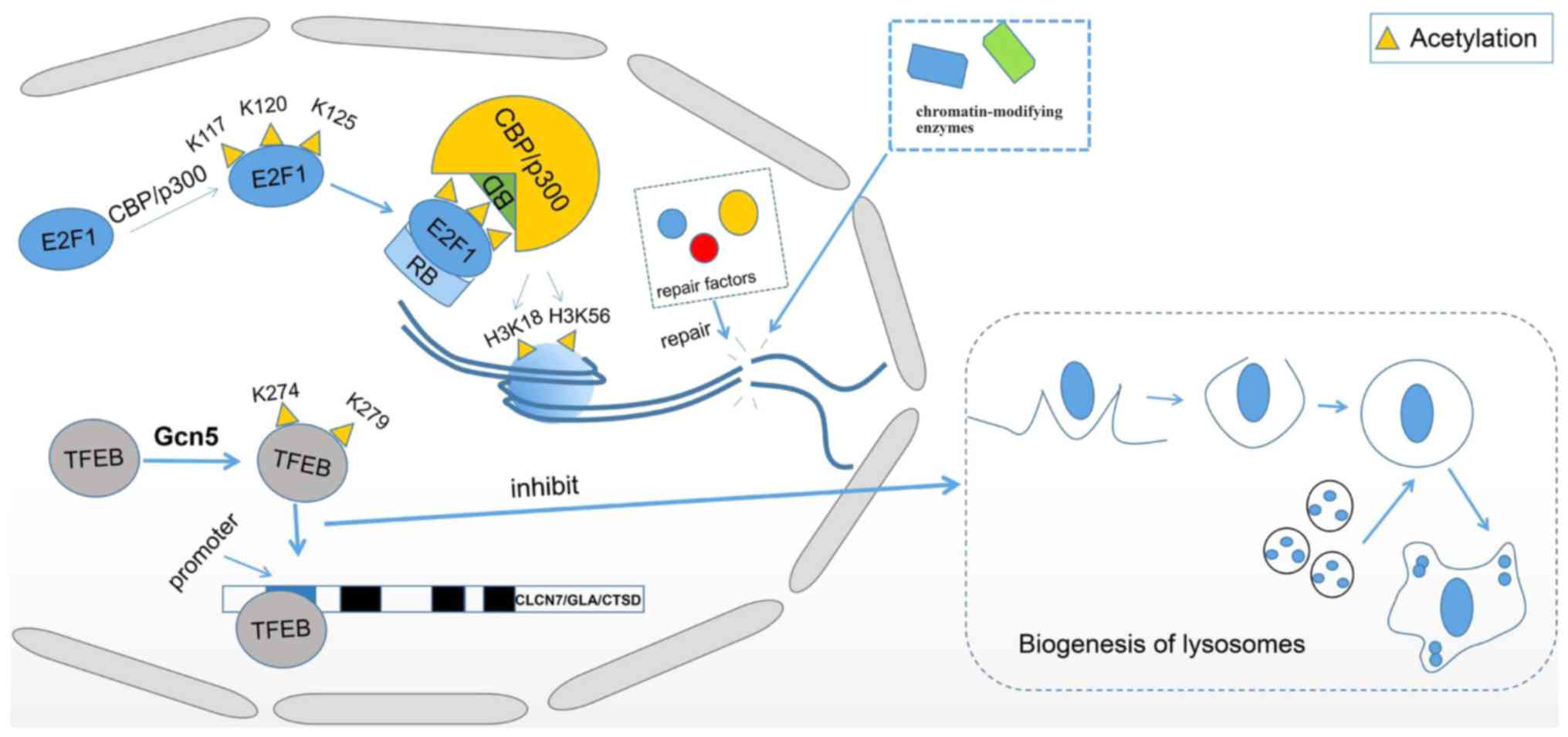
Transcription factor EB (TFEB), a primary TF for
autophagy and lysosome-related gene expression, can be acetylated
by GCN5 at K274 and K279 in the way of Nε-acetylation, which
inhibits the binding of TFEB to promoters of its target genes
chloride voltage-gated channel 7, galactosidase α and cathepsin D
by interfering with TFEB dimerization (109). Interfered TFEB dimerization results
in a decreased binding affinity to DNA, which inhibits its
transcriptional activity and ultimately suppresses the biogenesis
of lysosomes and aggregation of autophagosomes (Fig. 1) (109). NAD+-dependent
deacetylase SIRT1 deacetylates TFEB at K116, enhancing lysosomal
function by upregulating transcriptional levels of TFEB downstream
targets (110). HDAC inhibitor
suberoylanilide hydroxamic acid (SAHA) upregulates TFEB acetylation
by recruiting increased levels of acetyl-Coenzyme A
acetyltransferase 1 to TFEB; the acetylation sites on TFEB induced
by SAHA are K91, K103, K116 and K430(111). It has been reported that SIRT1
deacetylates TFEB at lysine residue 116 and the deacetylation of
TFEB in microglia induces the regulatory ability of microglia to
degrade fibrillar Aβ, the deacetylation process further reduces the
number of deposited amyloid plaques by facilitating lysosomal
biogenesis, which could be helpful in the treatment of Alzheimer's
disease (110). In human diabetic
kidney disease, deacetylation of TFEB by HDAC6 has been observed,
which promotes its activation (112). Tubastatin A, an inhibitor of HDAC6,
increases the acetylation level of TFEB accordingly (113). In addition, experiments have
demonstrated that the use of tubastatin A can attenuate renal
injury, which indicates that TFEB may be a promising target for
renal diseases treatment (112).
Recently, it has been demonstrated that acetylation of E2F
transcription factor 1 (E2F1) is associated with the repair of DNA
double-strand breaks (DSBs). The acetylation of E2F1 at K117, K120
and K125 by acetyltransferase p300/CBP creates a binding motif for
the bromodomains of p300/CBP, which results in the recruitment of
p300/CBP to DSBs with the help of retinoblastoma tumor-suppressor
protein, an important regulator of E2F1. Subsequently, p300/CBP
acetylates H3K18 and H3K56, and then facilitates the recruitment of
chromatin-modifying enzymes and repair factors for DSBs (Fig. 1) (114). GCN5 catalyzes the histone
acetylation at the promoter regions of E2F1, enhancing the
transcription of target genes cyclin D1 and cyclin E1. The
expression of GCN5 promotes cell growth and the G1/S phase
transition in lung cancer cells, which is aided by E2F1 to control
the transcription of cyclin E1 and cyclin D1. These data suggest
that the interaction between GCN5 and E2F1 may be a potential
target for lung cancer treatment (115).
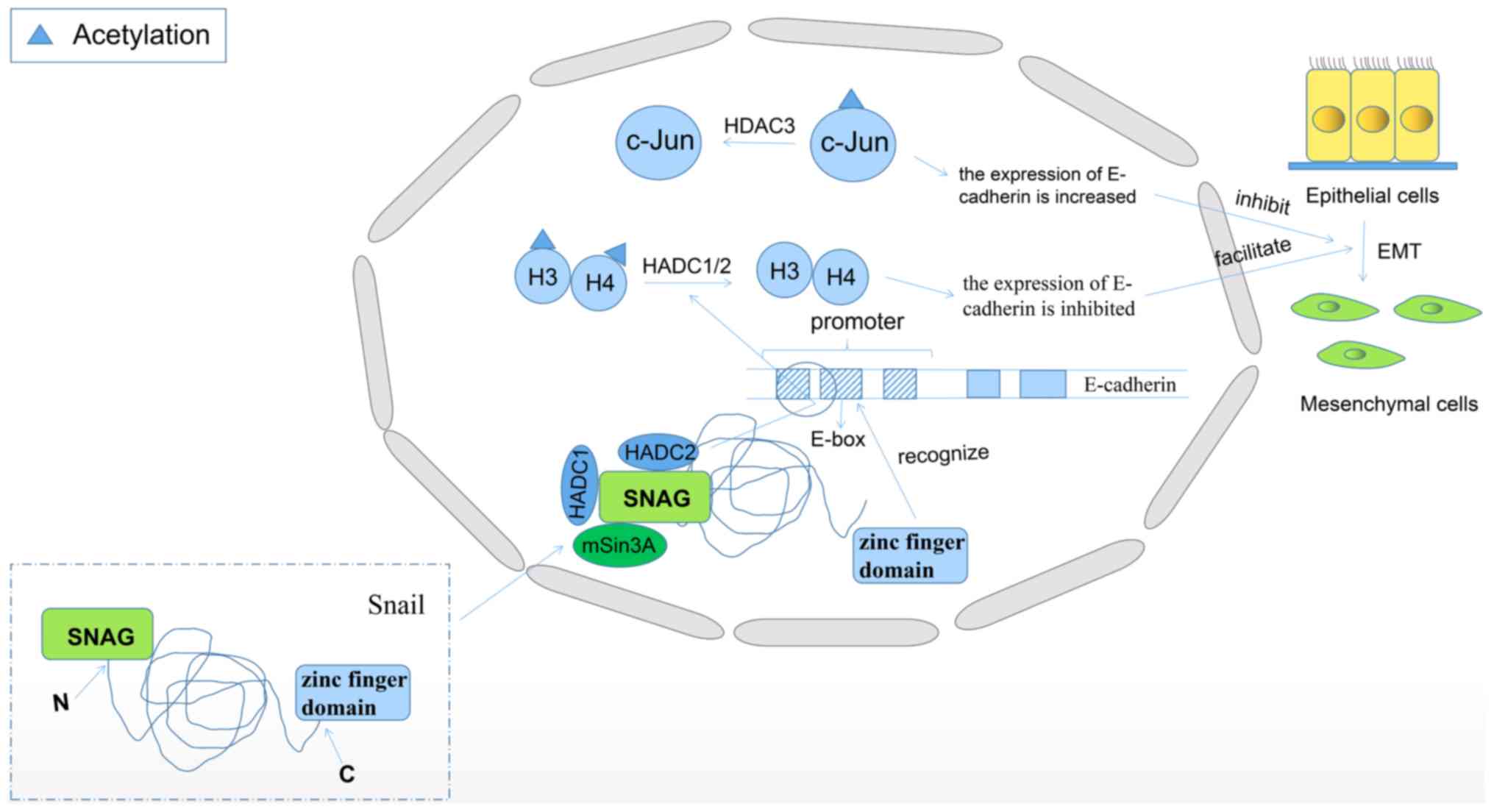
Zinc finger protein SNAI1 (Snail) is involved in the
induction of epithelial-mesenchymal transition (EMT) and plays a
crucial role in the metastasis of malignant tumors (116). The Snail protein is usually
comprised of a C-terminal zinc finger domain and an N-terminal SNAG
domain. The C-terminal zinc finger domain recognizes the E-box
sequence in the promoter region of E-cadherin. The SNAG domain
associates with HDAC1/2 and corepressor mSin3A, and then recruits
the repressor complex to E-cadherin promoter, here HDAC1/2
deacetylates histone H3 and H4, inhibiting the expression of
E-cadherin (Fig. 2) (117). In previous years, numerous studies
have reported that there are lysine acetylation sites on Snail and
Snail acetylation activates the expression of Snail gene (40,118).
It has been demonstrated that the interaction of CBP and Snail can
cause the acetylation of K146 and K187 on Snail in the way of
Nε-acetylation. Snail can be used as a transcriptional activator to
induce the expression of cytokines in the tumor microenvironment
during tumor metastasis, or as a transcriptional repressor to
inhibit the expression of E-cadherin during tumor metastasis
(118). Cancer cells containing
acetylated Snail have increased metastatic ability compared with
primary cancer cells (116,118). In addition, it has been found that
Snail binds to the E-box motif on the E-cadherin promoter and
recruits HDACs to suppress the expression of E-cadherin. In lung
cancer cells, if recombinant Snail and p300 are incubated with
acetyl-CoA, acetylation of Snail will be observed (40). In addition to CBP and p300, KDACIs
can also promote the expression of Snail and induce EMT in hepatoma
cells. One of the possible reasons is that KDACIs regulate the
stability of Snail by upregulating the expression of COP9
signalosome 2 (CSN2). CSN2 binds to Snail and exposes its
acetylation site, which then promotes the acetylation of Snail.
Therefore, its phosphorylation and ubiquitination are inhibited and
its degradation is inhibited (119). Human c-Jun is a transcriptional
regulator of JUN proto-oncogene, and c-Jun N-terminal kinase gene
is associated with the expression of E-cadherin and snail. It has
been reported that the downregulation of HDAC3 expression can
increase the acetylation of c-Jun and may lead to the degradation
of c-Jun, which ultimately increases the expression of E-cadherin
and decreases the expression of snail, thus inhibiting the process
of EMT (Fig. 2) (120). Due to the promotion of tumor
metastasis by acetylated Snail, Snail could become an effective
target for cancer therapy.
When an organism is exposed to high temperatures,
heat shock protein 90 (Hsp90) is synthesized by thermal excitation
to protect the organism, and it has molecular chaperone activity
(126). The deacetylation of Hsp90
is effectively regulated by HDAC6. The inactivation of HDAC6 leads
to a higher degree of acetylation of Hsp90, which causes Hsp90 to
separate from P23 and lose its chaperone activity (127). If K294 on Hsp90 is not normally
acetylated, the life cycle of Hsp90 will be shorter and the
function will be weakened (128).
In breast cancer cells, carbamazepine can inhibit the effect of
HDAC6 on Hsp90 and further promote the degradation of HER2 protein
(129). This finding is expected to
contribute to the development of breast cancer treatment
strategies.
Additionally, it has been demonstrated that
acetylation at lys1053 of the activation loop of the kinase domain
may positively regulate kinase activity of VEGFR-2(130). Also, KAT7, a member of MYST family,
colocalizes with vascular endothelial growth factor receptor 2
(VEGFR-2), directly regulating the chromatin structure of the
VEGFR-2 locus and affecting VEGFR-2 transcriptional activity. KAT7
depletion was demonstrated to reduce the expression of
VEGFR-2(131). Also, using KAT7
morpholino inhibitor in zebrafish embryos lead to abnormal vessel
formation (131). All these results
indicate that KAT7 plays a critical role in endothelial
function.
ESCO1 and ESCO2 are two KATs that are involved in
the aggregation of sister chromatids in the S phase of the cell
cycle. K105 and K106 on human structural maintenance of chromosomes
3 (SMC3) are two conserved amino acid residues that can be
acetylated by ESCO1 and ESCO2(46).
If these sites are mutated to non-acetylated, the sister chromatid
will lose cohesion and the human genome will be unstable (46,132).
Further study revealed that the acetylation of SMC3 altered the
function of the N-terminal ATPase of SMC3 and transformed the
chromosome-bound cohesin complex into a cohesive complex (133). These results indicate that
acetylation of SMC3 regulates the aggregation of sister chromatids
and keeps the cell cycle functioning normally. In addition, some of
the proteins and acetylation sites involved in this review are
listed in Table III (72,73,77,85,97,106,118,122,129,133-138).
Although protein acetylation has been well studied
in eukaryotes, new insights into protein acetylation in prokaryotes
has gained more attention in recent years. Shewanella
baltica is one of the specific spoilage organism of aquatic
products and numerous lysine acetylation sites have been detected
in its protein, such as key enzymes involved in fat metabolism and
putrescine biosynthesis that are related to the spoilage ability of
Shewanella baltica (139).
Previously, putrescine was demonstrated to have effects on
proliferation, migration and apoptosis of human skin fibroblasts
(140). Cyanobacteria are the only
prokaryotes capable of performing oxygenic photosynthesis.
Experimental data have demonstrated that lysine acetylation in
cyanobacteria plays an important role in the regulation of
photosynthesis (141). Also,
acetylation is abundant in Escherichia coli (142).
As mitochondria carry out a number of essential
functions in metabolism, the study of Mechanisms and Dynamics of
Protein Acetylation in Mitochondria becomes necessary. PCAF
functions as a lysine acetyltransferase inside mitochondria
(143). PCAF affects intermediary
metabolism by acetylating isocitrate dehydrogenase 2 (IDH2) at K180
in the mitochondrial matrix, which interferes with the catalytic
mechanisms of isocitrate binding and oxidation (143). A number of central enzymes in
mitochondria are deacetylated by SIRT3, which reverses the
suppressive effect of acetylation, leading to enhanced oxidative
metabolism (144). It was proposed
that most protein lysine acetylation in mitochondria is due to
non-enzymatic modification of protein lysine residues. The
environment of the mitochondrial matrix has an alkaline pH and
abundant acetyl-CoA, which increases the number of amino groups
acting as nucleophiles towards the inherently reactive acetyl-CoA,
resulting in an acetylated lysine (145). Pyruvate is a principal source of
acetyl-CoA. The data suggest excessive lysine acetylation in the
mitochondrial matrix can be prevented by decreasing the matrix
acetyl-CoA formation (146). In the
mitochondria, when acetyl-CoA levels exceed physiological
requirements, a signal is generated to slow flux through oxidative
energy production (144). A
substrate-level braking system is established via the induction of
acetyl-CoA-dependent protein acetylation. When energy demands
require increased oxidative metabolism, SIRT3 expression is
induced, which removes the brake and allows the cell to increase
energy production (144).
Acetylation in mitochondria is primarily the result of nonenzymatic
modification of lysine residues, some enzyme-mediated acetylation
also exists in mitochondria. PCAF acetylates isocitrate
dehydrogenase 2 (IDH2) at lysine 180, which may reduce IDH2
affinity for isocitrate. In this way, PACF influences myoblast
differentiation (143).
CCAAT/enhancer-binding protein a (C/EBPα), which can be acetylated
by p300, regulates the transcription of metabolic genes and further
enhances its transactivation activity (147). In addition, C/EBPα can be
deacetylated by SIRT1 and low acetylation levels of C/EBPα enhances
mitochondrial function. When energy is required, SIRT1 is activated
by high levels of nicotinamide adenine dinucleotide; mitochondrial
biogenesis and functions are regulated in this way (147).
AKI refers to a rapid decline in renal function in a
short period of time and leads to the accumulation of metabolic
waste (148). AKI primarily affects
renal tubules. Tubular cells are rich in mitochondria, and changes
in mitochondria of the tubules are an important indicator of the
occurrence and development of renal diseases (149). AKI can be caused by various
factors, such as bacterial infection, drugs and sepsis (150). At present, the clinical diagnosis
of AKI primarily depends on the detection of elevated serum
creatinine levels and decreased urine output (151). It has been demonstrated that the
KDACI participates in the process of renal regeneration and repair,
and plays different roles in AKI models (152). Differences in cell type and
etiology will determine activation of KDACs, thus leading to cell
survival or death (153). Using
SIRT1, which is currently studied more, as an example, SIRT1 can
participate in the regulation of a variety of signaling pathways,
and plays a role in anti-oxidative stress and anti-apoptotic
effects to protect kidney function (154). For example, in the mouse model of
AKI induced by sepsis, SIRT1 activity is significantly decreased.
However, if the SIRT1 activity was increased by resveratrol, the
damage of mouse mitochondria will be reduced and the survival time
will be significantly prolonged (155). Cisplatin is a commonly used
anti-tumor drug, but it is also associated with an increased risk
of causing serious side effects, such as AKI. Its pathogenesis is
related to a number of factors, such as mitochondrial damage,
oxidative stress and apoptosis (156). Inducing AKI in SIRT3-deficient mice
and wild-type mice using cisplatin revealed that the
SIRT3-deficient mice suffered more severe kidney damage and even
death (157). Further study
reported that the expression of SIRT3 was significantly decreased
in kidney cells of cisplatin-induced AKI. Due to the loss of SIRT3
regulation, mitochondria are damaged and unable to carry out normal
functions (158). If the activity
of SIRT3 is restored by treating with the adenosine
monophosphate-activated protein kinase activator AICAR or
Acetyl-L-Carnitine, the symptoms of AKI are relieved to some extent
(157). Ning et al (159) demonstrated that short-term caloric
restriction could protect AKI induced by cisplatin in aged rats
because it has anti-apoptotic effects and promotes the expression
of SIRT1. Although the current treatment of AKI is still limited to
intravenous rehydration, diuretic therapy and continuous renal
replacement therapy, the study of the relationship between protein
acetylation and AKI is useful (160). These results suggest that restoring
the activity of SIRT1/3 may be a novel therapeutic target for AKI.
Using resveratrol, the activity of SIRT1/3 can be restored
efficiently (161). In addition,
oxidative stress and mitochondrial function of renal tubular
epithelial cells tend to be ameliorated (155). Also, dexmedetomidine plays a role
in treating AKI because it induces the upregulation SIRT3(162).
Congenital heart disease (CHD) is the most common
type of congenital malformation (163). The main cause of CHD is the failure
of heart or blood vessel formation and dysplasia during embryonic
development (164). Additionally,
it is also the result of structural and functional abnormalities
caused by the channel failing to close automatically after birth
(164). Wu et al (165) induced the abnormal expression of
crucial genes in cardiac development by exposing mice to sodium
valproate to decrease the activity of KDACs. This experiment led to
malformation of the heart, and indicated that acetylation
modification may be related to CHD. There have also been
experiments that use ethanol and metabolites of ethanol to increase
the degree of acetylation of H3K9(166). It has been found that cardiac
precursor cells are abnormally differentiated (165). Study has also reported that
valproic acid may cause teratogenic effects by directly inhibiting
expression levels and activity of KDAC, thus leading to an
imbalance in the ratio of acetylation/deacetylation. As a result of
this, the expression levels of VANGL planar cell polarity protein
2, scribble planar cell polarity protein and Rac family small
GTPase 1, the key genes of the H9C2 cardiomyocyte planar cell
polarity pathway, are decreased (134), which can lead to CHD. Schlesinger
et al (135) screened
multiple acetylation sites and found that the acetylation levels of
H3K9 and H3K14 had significant effects on the expression of NK2
homeobox 5, methyltransferase like 2A, GATA binding protein 4 and
serum response factor, which are important factors involved in
cardiac development. If the expression of KAT2A in H3K9 is
downregulated, the development process of mesenchymal stem cells
into myocardium will be blocked, resulting in abnormal myocardial
development (167).
In addition to CHD, protein acetylation may also be
associated with heart disease caused by oxidative damage (168). During oxidative stress, the content
of SIRT3 in mitochondria and nucleus of cardiomyocytes increased
significantly (169). It is
speculated that SIRT3 is associated with heart disease (168). In a previous study, ku70 was used
as a target protein of SIRT3, thereby promoting the interaction
between ku70 and the proapoptotic protein Bax. When a stress
response is present, SIRT3 protects cardiomyocytes effectively by
blocking the translocation of Bax to the mitochondria and
preventing cytotoxic stress-mediated cell death (169). Meanwhile, it has been demonstrated
that the decrease in cardiac metabolic activity in patients with
diabetes may be related to the decreased pyruvate transport
activity induced by acetylation of mitochondrial pyruvate carriers
2(137). As a crucial regulator for
myocardial ischemia and reperfusion injury, HDAC4 overexpression
increases autophagy microtubule-associated protein light chain 3
and active caspase 3, decreases superoxide dimutase 1 in the
myocardium, and ultimately promotes myocardial ischemia/reperfusion
injuries (170). Myocardial
fibrosis is common in patients with CHD (171); it has reported that HDAC
overactivation causes atrial fibrosis and HDAC inhibitors have been
demonstrated to be useful in the treatment of heart diseases.
Therefore, regulating the activity of HDACs may be a possible
therapeutic target of CHD (172).
Curcumin, the main ingredient of turmeric, which inhibits p300
activity, prevents the development of cardiomyocyte hypertrophy
that leads to heart dysfunction. As a result of this, curcumin
could be a positive therapeutic method to aid in the treatment of
heart diseases (173).
The acetylation of some proteins may have an impact
on the occurrence of cancer. Colon cancer-associated transcription
factor 1 (CCAT1) is significantly higher in esophageal squamous
cell carcinoma (ESCC) cells compared with corresponding non-tumor
tissue cells (174). As commonly
known, high expression of CCAT1 promotes cell proliferation and
invasion, while downregulation of CCAT1 can inhibit cell
proliferation and invasion (175).
It has been demonstrated that the acetylation of H3K27 can
partially upregulate the expression of CCAT1, which has the
potential to induce cancer (176).
Apart from TFs, the proliferation of cancer cells requires
glycolysis to provide a large amount of energy. Phosphoglycerate
kinase 1 (PGK1) is an important reductase in the glycolysis
process, and the functional changes as a result of its acetylation
may also be closely related to the changes in cancer cell
proliferation. Using liver cancer cells, acetylation of K323 at
PGK1 upregulates its activity and enhances its proliferative
capacity (138).
The deacetylation of certain proteins is also very
important in the occurrence of cancer. Forkhead box protein O1
(FoxO1) is a tumor suppressor that mediates autophagy, specifically
autophagy that is produced by oxidative stress and serum starvation
in cancer cells (177). In the
cytosol, SIRT2 binds to FoxO1 to deacetylate it and inhibit
FoxO1-mediated autophagy. When a stress response occurs, SIRT2 is
separated from FoxO1 and results in the acetylation of FoxO1. This
process will promote autophagy and finally lead to cell death. This
mechanism links the autophagy signal pathway to cancer and is
regulated by the acetylation of FoxO1(178). SIRT7 was demonstrated to be
overexpressed in colorectal cancer cells compared with normal
cells. Additionally, SIRT7 is an important facilitator of
metastasis in human colorectal cancers, whose overexpression leads
to lung and skin metastases (179).
Abnormal SIRT7 overexpression accelerates cancer cell growth and
enhances invasiveness, and leads to the upregulation of mesenchymal
markers vimentin and fibronectin (179). It has reported that SIRT7 can cause
carcinogenic transformation of human cancer cells by deacetylating
H3K18(66). In pancreatic cancer,
upregulation or downregulation of HDAC6 expression has no
significant effect on cancer cell proliferation and cell cycle
progression, but overexpression of HDAC6 in combination with
cytoplasmic linker protein-170 can enhance cancer cell migration
activity significantly (180).
In recent years, KDACIs have been found to be
effective in treating diseases such as diabetes, heart disease,
chronic fibrosis and cancer (181-184).
If the balance of acetylation/deacetylation becomes dysregulated,
several physiological and pathological cellular processes will be
disrupted and some genes will be abnormally expressed and become
carcinogenic factors, eventually leading to diseases (184,185).
KDACs can deacetylate histones, which positively charges them
again. This will result in tight binding of histones to DNA and
make this section of the gene difficult to transcribe. However,
KDACIs can selectively inhibit the deacetylation of certain cancer
suppressor genes, including p53, TGF β type II receptor gene, and
restores their transcriptional activity (184). Therefore, indicating that KDACIs
may show anticancer effects.
A number of studies have shown that a large
proportion of histones have a low degree of acetylation, and this
discovery has led to the use of KDACIs in the treatment of cancer
(186-189).
KDACIs can slow down the proliferation rate of cancer cells, which
may inhibit the growth of cancer cells and eventually lead to
apoptosis (190). Comparing the
targets of two HDAC inhibitors, SAHA and MS-275, KDACIs have a high
substrate specificity and provide an important basis for the
treatment application of KDACIs (14). In addition, a number of studies have
demonstrated that KDACIs can inhibit cancer cells and reduce the
resistance of cancer cells to other drugs, which makes it possible
to use KDACIs in combination with other drugs to treat cancer
(191). There are studies showing
that KDACIs can promote the differentiation of CSCs to treat cancer
(192,193). It has reported that HDACIs can
induce the differentiation of cancer cells to assist cancer therapy
by affecting the developmental signaling pathway. These findings
could suggest that the underlying mechanisms of KDACIs in cancer
therapy may be diverse (193).
Resveratrol is a type of natural polyphenolic KDACI that has been
studied a large amount. The main target of resveratrol is SIRT1,
which has achieved initial success in the treatment of tumor
diseases, such as liver cancer and breast cancer (194). Li et al (195) cultured PC-3 and LNCaP prostate
cancer cell lines, which were induced by lipopolysaccharides to
produce EMT. After being treated with resveratrol, these cells
showed significant changes. The results revealed that EMT no longer
occurred, and the mesenchymal cells restored the epithelial cell
phenotype. This study further promoted the treatment application of
resveratrol in cancer (195).
However, due to the lack of development of detection techniques,
the use of KDACIs to treat cancer has only achieved some initial
results, and numerous mechanisms have not been studied in detail.
Moreover, KDACIs have limitations in the inhibition of cancer. So,
most of the drugs based on this have stayed in the clinical trial
stage and have not been officially put into use.
Stem cells are a type of pluripotent cell that can
self-replicate and differentiate to produce other types of cells or
divide to produce a large number of cells of the same type. They
can be used for tissue and organ regeneration, and they have very
broad clinical application prospects. It is currently known that
acetylation of histones can regulate physiological activities, such
as glycolysis, and thereby regulate the differentiation ability of
stem cells (196).
Embryonic stem cells (ESCs) are isolated from early
embryos or primitive gonads. Previous study has reported that the
level of acetylation of H3K9 is related to the reprogramming
ability of ESCs by analyzing multiple chromatin markers. The
reprogramming ability of ESCs, of which the level of H3K9
acetylation is low, is also relatively weak. If KDACI is added to
ESC colonies, its reprogramming ability is significantly improved
(197). Actin-like protein 6a is a
component of the ATP-dependent histone acetylation complexes. If it
is knocked out, the pluripotency of ESCs will decrease (198). In neural stem cells cultured in
vitro, the level of acetylation of H3K9 decreased first and
then increased with the differentiation process. If KDACI is added
to increase acetylation levels during the first 4 days of
differentiation, cell pluripotency will be promoted and neural
differentiation will be inhibited (199). The radiosensitivity of normal stem
cells is higher compared with their isogenic differentiated
progeny, and the DNA damage response of normal stem cells is
stronger compared with their isogenic differentiated progeny.
However, if H3K56 acetylation in stem cells is downregulated, their
radiosensitivity is significantly decreased and survival rate is
significantly increased (200).
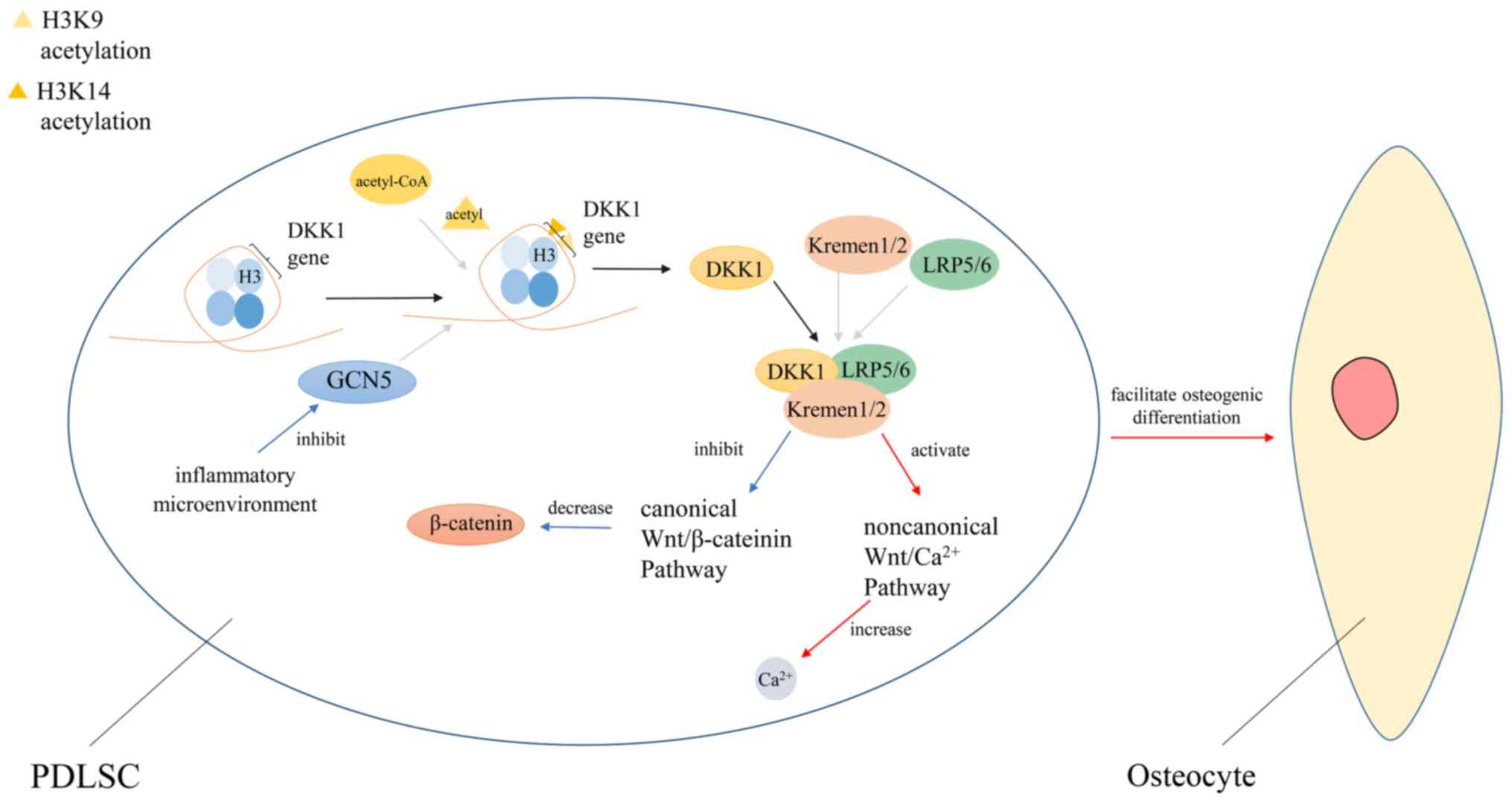
Here is a particular example of acetylation
regulating stem cell differentiation. In patients with
periodontitis, periodontal ligament stem cells (PDLSCs), which are
a novel population of mesenchymal stem cells, exhibit defects in
osteogenic differentiation (201).
Also, this may be due to the downregulation of GCN5 expression in a
micro-inflammatory environment. GCN5 can induce H3K9 and H3K14
acetylation in the dickkopf-related protein 1 (DKK1) promoter
region, thereby regulating the expression level of DKK1. DKK1 can
inhibit the Wnt/β-catenin signaling pathway by binding to
LDL-receptor-related protein 5/6 (LRP5/6) and Kremen1/2, and
enhance the osteogenic differentiation of PDLSCs. If GCN5 is
knocked out, the expression level of DKK1 decreases, resulting in
decreased osteogenic differentiation of PDLSCs (Fig. 3) (202-204).
Acetylation can also indirectly regulate the proliferation of stem
cells. For example, SIRT6 is involved in maintaining the stability
of the Wnt signaling pathway and SIRT6 deletion results in aberrant
activation of the Wnt signaling pathway and promote the
proliferation of hematopoietic stem cells (205).
At present, methods to affect stem cell activity by
acetylation have been applied to clinical treatments. Imatinib is a
tyrosine kinase inhibitor for the treatment of chronic myeloid
leukemia (CML), but has poor clearance for inactive leukemia stem
cells (211). SIRT1 protein was
labeled with an anti-SIRT1 antibody and SIRT1 expression was
detected by western blotting. It was found that the expression of
SIRT1 in CML CD34+ cells in chronic and blast phase was
much greater than that of normal cells. It is speculated that SIRT1
inhibitors can be used in combination with imatinib to treat CML to
enhance efficacy (212). Different
acetylation sites are regulated to varying degrees during
differentiation, which plays an important role in regulating the
differentiation direction of pluripotent stem cells and monitoring
stem cell differentiation (213).
In addition to the role mentioned above, acetylation may also play
vital role in regulating stem cell self-renewal and cell cycle
(214).
In the process of studying protein acetylation, it
was found that the acetylation site is first step to understand
acetylation mechanism (215).
Therefore, it is critical to predict acetylation sites with
relatively simple methods. Bioinformatics is a new subject derived
from the rapid development of biological sciences and computer
science. It uses computer programs as a tool to retrieve and
analyze biological data. The method obtains relevant information
via the databases and processes the data to achieve the goal. As
commonly known, bioinformatics methods have played an important
role in the field of genomics and proteomics (216). There are some computational models
that have been developed to predict acetylation sites.
One of these methods is called LAceP. The first
step in this method, researchers need to collect data on protein
and acetylation sites in the SysPTM2 and PhosphoSitePlus databases.
By this way, researchers can obtain a certain number of acetylation
sites in the protein after eliminating redundancy, and use this
data as positive data. Then, a peptide containing lysine is
selected from the acetylated protein, and negative data is obtained
after knocking out the fragment containing the lysine acetylation
site. After that, a sliding window strategy is used to determine
the optimal length of the acetylated peptide. The homology of the
peptides is carried out via the CD-hit software to avoid model over
fitting. If the similarity of the two peptides exceeds 70%, they
will be classified as one class. Only one of them will be retained
while the other peptides will be discarded. The model uses three
types of features, which are amino acid physicochemical property,
transition probability matrix and position-specific symbol
composition, to predict lysine acetylation sites. Because the
calculations are performed by different algorithms, the probability
of acetylation may be analyzed from three aspects. Then, the
classification of the peptides in the training datasets, their
class tags and features were used as input of the logistic
regression model. After model training, the optimized parameters
are generated as outputs. Researchers can analyze the probability
of amino acid acetylation base on these data (215).
Another method is called ASEB. Firstly, it is
necessary to collect acetylated human proteins from different
families using PubMed. In this step, researchers need to query the
detailed acetylation sites and KATs, and go to the UniProt database
to obtain the UniProt IDs corresponding to the acetylated proteins.
Then, using the idea of Gene Set Enrichment Analysis, the ASEB
method was developed to form an acetylated polypeptide, which
consists of the acetylation site, its first eight amino acids and
its last eight amino acids. Different KAT families form different
acetylated peptide groups. In order to determine whether a given
peptide can be acetylated by a certain KAT family, it is only
necessary to analyze how similar the given peptide is to the
acetylated peptide in that family. For analysis, a set of
predefined KAT specific polypeptides, including N-polypeptides, is
inputted. Then, the similarity between the given polypeptide
fragment and the peptide contained therein is searched in the set.
The background peptide set can be calculated by BLOSUM 62 matrix.
If the similarity with the polypeptide in a certain KAT family is
extremely high, it will be possible that the polypeptide is a new
substrate of the KAT family. Then, the enrichment score is
calculated and estimated to obtain the relatively significant
chance that the given peptides were acetylated by the KAT family
(217).
GPS-PAIL, one of these methods, contains 702 known
HAT-specific acetylation sites in 205 proteins for seven HATs,
including CREBBP, p300, HAT1, KAT2A, KAT2B, KAT5 and KAT8,
developed from the scientific literature and public data resources.
The method predicts acetylation sites based on the principle that
different HATs have distinct sequence specificities for the
substrate modifications. GPS-PAIL develop a computational model for
each HAT by training a previously established algorithm of
Group-Based Prediction System. Online service and stand-alone
packages of GPS-PAIL are also provided. The two tools mentioned
above have their own distinct advantages on the inputting of the
online service, which contained three parts: i) HAT types; ii) the
protein sequences; and iii) four thresholds, including ‘High’,
‘Medium’, ‘Low’ and ‘All’ (218).
Moreover, other methods, such as N-Ace and PLMLA,
are applied to predict acetylation sites. N-Ace predicts the
protein acetylation sites based on the support of vector machine.
The training of N-Ace depends on the amino acid sequence and other
structural characteristics; it has higher predictive accuracy
compared with models trained using only amino acid sequences
(8). PLMLA combines protein
sequences, secondary structures and amino acid properties to
predict the methylation and acetylation of lysine residues in
protein sequences (219).
In conclusion, LAceP, N-Ace and PLMLA are unable to
predict HAT-specific acetylation sites. Compared with ASEB, LAceP
can carry out acetylation peptide length assays and takes more
consideration to peptide redundancy and residue property, which may
have an important impact on acetylation. In addition, LAceP has the
potential for improved performance in the future when considering
the Matthews correlation coefficient measurement. Conversely, ASEB
have the ability to determine KATs are responsible for the
acetylation of given proteins. The present version only predicts
acetylation catalyzed by two KAT families, including CBP/p300 and
GCN5/PCAF. Both GPS-PAIL and ASEB can predict HAT-specific
acetylation sites. However, in general, GPS-PAIL generated an
improved performance compared with ASEB. All of them have a high
level of accuracy (8,215,217-219).
The use of bioinformatics to predict acetylation
sites greatly facilitates the study of acetylation, and saves a lot
of invaluable research time. The integration of the properties of
different acetylated peptides is also conducive to the study of the
functions and characteristics of protein acetylation.
Histone acetylation and non-histone protein
acetylation are important for human biological functions, and are
closely related to the mechanisms of various diseases. At present,
the study process of PTM is still steadily advancing. The process
of acetylation together with other modifications, such as
methylation and glycosylation, regulates biological activities and
its role in the whole metabolic network still needs further
research. Moreover, the constant innovation of theory is destined
to provide more space and possibility for clinical application.
In-depth study of protein acetylation could lead to novel prospects
in the clinical treatment of a number of diseases. The regulation
of TFEB acetylation and deacetylation may be a useful biological
process in the development of Alzheimer's disease and renal injury.
The acetylation of E2F1 is associated with the treatment of lung
cancer. Additionally, Snail has also been demonstrated as an
effective target for cancer therapy. The effect of HDAC6 on Hsp90
is expected to promote the development strategies of breast cancer
treatment. Activation of KDACs participates in the process of renal
regeneration and repair, and plays different roles in AKI. By
mediating the expression of KAT2A, HDAC and p300, the development
process of CHD can be inhibited. Moreover, CCAT1 acetylation is
associated with ESCC, and the PGK1 acetylation and deacetylation
may be a promising target for cancer treatment. Also, FoxO1
acetylation and the expression of SIRT7 are associated with cancer.
At present, KDACIs have been used in the treatment of diseases,
such as diabetes, heart disease, chronic fibrosis and cancer.
Protein acetylation is increasingly related to clinical treatment,
which provides novel ideas and methods for the treatment of
numerous intractable diseases.
Not applicable.
This work was supported by the Distinguished
Professorship Program of National Research Program on prevention
and control of major birth defects in reproductive health (grant
no. 2017YFC1000900) and Special-funded Programme on National Key
Scientific Instruments and Equipment Development (grant no.
2016YFF0103800).
Not applicable.
CX and YT conceived and drafted the manuscript. ML
and TC contributed to the literature retrieval and manuscript
modification. JQ supervised and designed the present study and
contributed to the approval of the final version of the manuscript.
All authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Marsh JA and Forman-Kay JD: Sequence
determinants of compaction in intrinsically disordered proteins.
Biophysical J. 98:2383–2390. 2010.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bah A and Forman-Kay JD: Modulation of
intrinsically disordered protein function by post-translational
modifications. J Biol Chem. 291:6696–6705. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Adaniya SM, O-Uchi J, Cypress MW, Kusakari
Y and Jhun BS: Posttranslational modifications of mitochondrial
fission and fusion proteins in cardiac physiology and
pathophysiology. Am J Physiol Cell Physiol. 316:C583–C604.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ametzazurra A, Larrea E, Civeira MP,
Prieto J and Aldabe R: Implication of human
N-alpha-acetyltransferase 5 in cellular proliferation and
carcinogenesis. Oncogene. 27:7296–7306. 2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Christensen DG, Baumgartner JT, Xie X, Jew
KM, Basisty N, Schilling B, Kuhn ML and Wolfe AJ: Mechanisms,
detection, and relevance of protein acetylation in prokaryotes.
mBio. 10:e02708–18. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Drazic A, Myklebust LM, Ree R and Arnesen
T: The world of protein acetylation. Biochim Biophys Acta.
1864:1372–1401. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Verdin E and Ott M: 50 years of protein
acetylation: From gene regulation to epigenetics, metabolism and
beyond. Nat Rev Mol Cell Biol. 16:258–264. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lee TY, Hsu JB, Lin FM, Chang WC, Hsu PC
and Huang HD: N-Ace: Using solvent accessibility and
physicochemical properties to identify protein N-acetylation sites.
J Comput Chem. 31:2759–2771. 2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hollebeke J, Van Damme P and Gevaert K:
N-terminal acetylation and other functions of
Nalpha-acetyltransferases. Biol Chem. 393:291–298. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Thao S, Chen CS, Zhu H and
Escalante-Semerena JC: Nepsilon-lysine acetylation of a bacterial
transcription factor inhibits Its DNA-binding activity. PLoS One.
5(e15123)2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yang XJ and Gregoire S: Metabolism,
cytoskeleton and cellular signalling in the grip of protein
Nepsilon- and O-acetylation. EMBO Rep. 8:556–562. 2007.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Allfrey VG, Faulkner R and Mirsky AE:
Acetylation and methylation of histones and their possible role in
the regulation of RNA synthesis. Proc Natl Acad Sci USA.
51:786–794. 1964.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Verdone L, Caserta M and Di Mauro E: Role
of histone acetylation in the control of gene expression. Biochem
Cell Biol. 83:344–353. 2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Choudhary C, Kumar C, Gnad F, Nielsen ML,
Rehman M, Walther TC, Olsen JV and Mann M: Lysine acetylation
targets protein complexes and co-regulates major cellular
functions. Science. 325:834–840. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Allis CD, Berger SL, Cote J, Dent S,
Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar
R, et al: New nomenclature for chromatin-modifying enzymes. Cell.
131:633–636. 2007.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li P, Ge J and Li H: Lysine
acetyltransferases and lysine deacetylases as targets for
cardiovascular disease. Nat Rev Cardiol. 17:96–115. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Song L, Wang G, Malhotra A, Deutscher MP
and Liang W: Reversible acetylation on Lys501 regulates the
activity of RNase II. Nucleic Acids Res. 44:1979–1988.
2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sterner DE and Berger SL: Acetylation of
histones and transcription-related factors. Microbiol Mol Biol Rev.
64:435–459. 2000.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ruhlmann F, Windhof-Jaidhauser IM, Menze
C, Beißbarth T, Bohnenberger H, Ghadimi M and Dango S: The
prognostic capacities of CBP and p300 in locally advanced rectal
cancer. World J Surg Oncol. 17(224)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Narita T, Weinert BT and Choudhary C:
Functions and mechanisms of non-histone protein acetylation. Nat
Rev Mol Cell Biol. 20:156–174. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hwang CS, Shemorry A and Varshavsky A:
N-terminal acetylation of cellular proteins creates specific
degradation signals. Science. 327:973–977. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Vetting MW, S de Carvalho LP, Yu M, Hegde
SS, Magnet S, Roderick SL and Blanchard JS: Structure and functions
of the GNAT superfamily of acetyltransferases. Arch Biochem
Biophys. 433:212–226. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ruiz-Garcia AB, Sendra R, Galiana M,
Pamblanco M, Perez-Ortin JE and Tordera V: HAT1 and HAT2 proteins
are components of a yeast nuclear histone acetyltransferase enzyme
specific for free histone H4. J Biol Chem. 273:12599–12605.
1998.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Miskiewicz K, Jose LE, Bento-Abreu A,
Fislage M, Taes I, Kasprowicz J, Swerts J, Sigrist S, Versées W,
Robberecht W and Verstreken P: ELP3 controls active zone morphology
by acetylating the ELKS family member Bruchpilot. Neuron.
72:776–788. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sampath V, Liu B, Tafrov S, Srinivasan M,
Rieger R, Chen EI and Sternglanz R: Biochemical characterization of
Hpa2 and Hpa3, two small closely related acetyltransferases from
Saccharomyces cerevisiae. J Biol Chem. 288:21506–21513.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sapountzi V and Cote J: MYST-family
histone acetyltransferases: Beyond chromatin. Cell Mol Life Sci.
68:1147–1156. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Reiter C, Heise F, Chung HR and
Ehrenhofer-Murray AE: A link between Sas2-mediated H4 K16
acetylation, chromatin assembly in S-phase by CAF-I and Asf1, and
nucleosome assembly by Spt6 during transcription. FEMS Yeast Res.
15(fov073)2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Church M, Smith KC, Alhussain MM, Pennings
S and Fleming AB: Sas3 and Ada2(Gcn5)-dependent histone H3
acetylation is required for transcription elongation at the
de-repressed FLO1 gene. Nucleic Acids Res. 45:4413–4430.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yan Y, Barlev NA, Haley RH, Berger SL and
Marmorstein R: Crystal structure of yeast Esa1 suggests a unified
mechanism for catalysis and substrate binding by histone
acetyltransferases. Mol Cell. 6:1195–1205. 2000.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang F, Marshall CB and Ikura M:
Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis:
Structural and functional versatility in target recognition. Cell
Mol Life Sci. 70:3989–4008. 2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hu LI, Lima BP and Wolfe AJ: Bacterial
protein acetylation: The dawning of a new age. Mol Microbiol.
77:15–21. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Pelletier N, Champagne N, Stifani S and
Yang XJ: MOZ and MORF histone acetyltransferases interact with the
Runt-domain transcription factor Runx2. Oncogene. 21:2729–2740.
2002.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Rokudai S, Laptenko O, Arnal SM, Taya Y,
Kitabayashi I and Prives C: MOZ increases p53 acetylation and
premature senescence through its complex formation with PML. Proc
Natl Acad Sci USA. 110:3895–3900. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fournier M, Orpinell M, Grauffel C, Scheer
E, Garnier JM, Ye T, Chavant V, Joint M, Esashi F, Dejaegere A, et
al: KAT2A/KAT2B-targeted acetylome reveals a role for PLK4
acetylation in preventing centrosome amplification. Nat Commun.
7(13227)2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bao X, Liu H, Liu X, Ruan K, Zhang Y,
Zhang Z, Hu Q, Liu Y, Akram S, Zhang J, et al: Mitosis-specific
acetylation tunes Ran effector binding for chromosome segregation.
J Mol Cell Biol. 10:18–32. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ghosh TK, Aparicio-Sanchez JJ, Buxton S,
Ketley A, Mohamed T, Rutland CS, Loughna S and Brook JD:
Acetylation of TBX5 by KAT2B and KAT2A regulates heart and limb
development. J Mol Cell Cardiol. 114:185–198. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cheng X, Ma X, Zhu Q, Song D, Ding X, Li
L, Jiang X, Wang X, Tian R, Su H, et al: Pacer is a mediator of
mTORC1 and GSK3-TIP60 signaling in regulation of autophagosome
maturation and lipid metabolism. Mol Cell. 73:788–802.e7.
2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Miotto B and Struhl K: HBO1 histone
acetylase is a coactivator of the replication licensing factor
Cdt1. Genes Dev. 22:2633–2638. 2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yuan H, Rossetto D, Mellert H, Dang W,
Srinivasan M, Johnson J, Hodawadekar S, Ding EC, Speicher K,
Abshiru N, et al: MYST protein acetyltransferase activity requires
active site lysine autoacetylation. EMBO J. 31:58–70.
2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chang R, Zhang Y, Zhang P and Zhou Q:
Snail acetylation by histone acetyltransferase p300 in lung cancer.
Thoracic Cancer. 8:131–137. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Yang Y, Cui J, Xue F, Zhang C, Mei Z, Wang
Y, Bi M, Shan D, Meredith A, Li H and Xu ZQ: Pokemon (FBI-1)
interacts with Smad4 to repress TGF-β-induced transcriptional
responses. Biochim Biophys Acta. 1849:270–281. 2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cazzalini O, Sommatis S, Tillhon M, Dutto
I, Bachi A, Rapp A, Nardo T, Scovassi AI, Necchi D, Cardoso MC, et
al: CBP and p300 acetylate PCNA to link its degradation with
nucleotide excision repair synthesis. Nucleic Acids Res.
42:8433–8448. 2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Senf SM, Sandesara PB, Reed SA and Judge
AR: p300 Acetyltransferase activity differentially regulates the
localization and activity of the FOXO homologues in skeletal
muscle. Am J Physiol Cell Physiol. 300:C1490–C1501. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lee K and Seo PJ: The HAF2 protein shapes
histone acetylation levels of PRR5 and LUX loci in Arabidopsis.
Planta. 248:513–518. 2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Nakakura T, Nemoto T, Suzuki T,
Asano-Hoshino A, Tanaka H, Arisawa K, Nishijima Y, Kiuchi Y and
Hagiwara H: Adrenalectomy facilitates ATAT1 expression and
alpha-tubulin acetylation in ACTH-producing corticotrophs. Cell
Tissue Res. 366:363–370. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang
Z, Yang T, Fu X, Jung SY, Wang Y, et al: Acetylation of Smc3 by
Eco1 is required for S phase sister chromatid cohesion in both
human and yeast. Mol Cell. 31:143–151. 2008.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wu H, Moshkina N, Min J, Zeng H, Joshua J,
Zhou MM and Plotnikov AN: Structural basis for substrate
specificity and catalysis of human histone acetyltransferase 1.
Proc Natl Acad Sci USA. 109:8925–8930. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
de Ruijter AJ, van Gennip AH, Caron HN,
Kemp S and van Kuilenburg AB: Histone deacetylases (HDACs):
Characterization of the classical HDAC family. Biochem J.
370:737–749. 2003.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Banik D, Moufarrij S and Villagra A:
Immunoepigenetics combination therapies: An overview of the role of
HDACs in cancer immunotherapy. Int J Mol Sci.
20(2241)2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Parra M: Class IIa HDACs-new insights into
their functions in physiology and pathology. FEBS J. 282:1736–1744.
2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Muller BM, Jana L, Kasajima A, Lehmann A,
Prinzler J, Budczies J, Winzer KJ, Dietel M, Weichert W and Denkert
C: Differential expression of histone deacetylases HDAC1, 2 and 3
in human breast cancer-overexpression of HDAC2 and HDAC3 is
associated with clinicopathological indicators of disease
progression. BMC Cancer. 13(215)2013.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Miller KM, Tjeertes JV, Coates J, Legube
G, Polo SE, Britton S and Jackson SP: Human HDAC1 and HDAC2
function in the DNA-damage response to promote DNA nonhomologous
end-joining. Nat Struct Mol Biol. 17:1144–1151. 2010.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Saito S, Zhuang Y, Suzuki T, Ota Y,
Bateman ME, Alkhatib AL, Morris GF and Lasky JA: HDAC8 inhibition
ameliorates pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol.
316:L175–L186. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Winbanks CE, Wang B, Beyer C, Koh P, White
L, Kantharidis P and Gregorevic P: TGF-beta regulates miR-206 and
miR-29 to control myogenic differentiation through regulation of
HDAC4. J Biol Chem. 286:13805–13814. 2011.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Cho Y, Sloutsky R, Naegle KM and Cavalli
V: Injury-induced HDAC5 nuclear export is essential for axon
regeneration. Cell. 155:894–908. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Bradley EW, Carpio LR, Olson EN and
Westendorf JJ: Histone deacetylase 7 (Hdac7) suppresses chondrocyte
proliferation and β-catenin activity during endochondral
ossification. J Biol Chem. 290:118–126. 2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Hu Y, Sun L, Tao S, Dai M, Wang Y, Li Y
and Wu J: Clinical significance of HDAC9 in hepatocellular
carcinoma. Cell Mol Biol (Noisy-le-Grand). 65:23–28.
2019.PubMed/NCBI
|
|
58
|
Bitler BG, Wu S, Park PH, Hai Y, Aird KM,
Wang Y, Zhai Y, Kossenkov AV, Vara-Ailor A, Rauscher FJ III, et al:
ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell
Biol. 19:962–973. 2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Radhakrishnan R, Li Y, Xiang S, Yuan F,
Yuan Z, Telles E, Fang J, Coppola D, Shibata D, Lane WS, et al:
Histone deacetylase 10 regulates DNA mismatch repair and may
involve the deacetylation of MutS homolog 2. J Biol Chem.
290:22795–22804. 2015.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zerr P, Palumbo-Zerr K, Huang J, Tomcik M,
Sumova B, Distler O, Schett G and Distler JH: Sirt1 regulates
canonical TGF-β signalling to control fibroblast activation and
tissue fibrosis. Ann Rheum Dis. 75:226–233. 2016.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Yuan Q, Zhan L, Zhou QY, Zhang LL, Chen
XM, Hu XM and Yuan XC: SIRT2 regulates microtubule stabilization in
diabetic cardiomyopathy. Eur J Pharmacol. 764:554–561.
2015.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Ahn BH, Kim HS, Song S, Lee IH, Liu J,
Vassilopoulos A, Deng CX and Finkel T: A role for the mitochondrial
deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad
Sci USA. 105:14447–14452. 2008.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Jeong SM, Xiao C, Finley LW, Lahusen T,
Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al:
SIRT4 has tumor-suppressive activity and regulates the cellular
metabolic response to DNA damage by inhibiting mitochondrial
glutamine metabolism. Cancer Cell. 23:450–463. 2013.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Rardin MJ, He W, Nishida Y, Newman JC,
Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, et al: SIRT5
regulates the mitochondrial lysine succinylome and metabolic
networks. Cell Metab. 18:920–933. 2013.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Kaluski S, Portillo M, Besnard A, Stein D,
Einav M, Zhong L, Ueberham U, Arendt T, Mostoslavsky R, Sahay A and
Toiber D: Neuroprotective functions for the histone Deacetylase
SIRT6. Cell Rep. 18:3052–3062. 2017.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Barber MF, Michishita-Kioi E, Xi Y,
Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL,
Chen K, et al: SIRT7 links H3K18 deacetylation to maintenance of
oncogenic transformation. Nature. 487:114–118. 2012.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Yuan L, Chen X, Cheng L, Rao M, Chen K,
Zhang N, Meng J, Li M, Yang LT, Yang PC, et al: HDAC11 regulates
interleukin-13 expression in CD4+T cells in the heart. J Mol Cell
Cardiol. 122:1–10. 2018.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Sahakian E, Chen J, Powers JJ, Chen X,
Maharaj K, Deng SL, Achille AN, Lienlaf M, Wang HW, Cheng F, et al:
Essential role for histone deacetylase 11 (HDAC11) in neutrophil
biology. J Leukoc Biol. 102:475–486. 2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Chatterjee SS, Saj A, Gocha T, Murphy M,
Gonsalves FC, Zhang X, Hayward P, Akgöl Oksuz B, Shen SS, Madar A,
et al: Inhibition of β-catenin-TCF1 interaction delays
differentiation of mouse embryonic stem cells. J Cell Biol.
211:39–51. 2015.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Abu-Elmagd M, Robson L, Sweetman D, Hadley
J, Francis-West P and Munsterberg A: Wnt/Lef1 signaling acts via
Pitx2 to regulate somite myogenesis. Dev Biol. 337:211–219.
2010.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Wapenaar H and Dekker FJ: Histone
acetyltransferases: Challenges in targeting bi-substrate enzymes.
Clin Epigenetics. 8(59)2016.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Chen GD, Yu WD and Chen XP: SirT1
activator represses the transcription of TNFα in THP1 cells of a
sepsis model via deacetylation of H4K16. Mol Med Rep. 14:5544–5550.
2016.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Michishita E, McCord RA, Berber E, Kioi M,
Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL,
Barrett JC, et al: SIRT6 is a histone H3 lysine 9 deacetylase that
modulates telomeric chromatin. Nature. 452:492–496. 2008.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Kim TK and Shiekhattar R: Architectural
and functional commonalities between enhancers and promoters. Cell.
162:948–959. 2015.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Rada-Iglesias A, Bajpai R, Swigut T,
Brugmann SA, Flynn RA and Wysocka J: A unique chromatin signature
uncovers early developmental enhancers in humans. Nature.
470:279–283. 2011.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Pradeepa MM, Grimes GR, Kumar Y, Olley G,
Taylor GC, Schneider R and Bickmore WA: Histone H3 globular domain
acetylation identifies a new class of enhancers. Nat Genet.
48:681–686. 2016.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Pradeepa MM: Causal role of histone
acetylations in enhancer function. Transcription. 8:40–47.
2017.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Dhar S, Gursoy-Yuzugullu O, Parasuram R
and Price BD: The tale of a tail: Histone H4 acetylation and the
repair of DNA breaks. Philos Trans R Soc Lond B Biol Sci.
372(20160284)2017.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Noguchi C, Singh T, Ziegler MA, Peake JD,
Khair L, Aza A, Nakamura TM and Noguchi E: The NuA4
acetyltransferase and histone H4 acetylation promote replication
recovery after topoisomerase I-poisoning. Epigenetics Chromatin.
12(24)2019.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Vadla R, Chatterjee N and Haldar D:
Cellular environment controls the dynamics of histone H3 lysine 56
acetylation in response to DNA damage in mammalian cells. J Biosci.
45(19)2020.PubMed/NCBI
|
|
81
|
Koprinarova M, Schnekenburger M and
Diederich M: Role of histone acetylation in cell cycle regulation.
Curr Top Med Chem. 16:732–744. 2016.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Gao FH, Hu XH, Li W, Liu H, Zhang YJ, Guo
ZY, Xu MH, Wang ST, Jiang B, Liu F, et al: Oridonin induces
apoptosis and senescence in colorectal cancer cells by increasing
histone hyperacetylation and regulation of p16, p21, p27 and c-myc.
BMC Cancer. 10(610)2010.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Kim E, Bisson WH, Löhr CV, Williams DE, Ho
E, Dashwood RH and Rajendran P: Histone and non-histone targets of
dietary Deacetylase inhibitors. Curr Top Med Chem. 16:714–731.
2016.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Glozak MA, Sengupta N, Zhang X and Seto E:
Acetylation and deacetylation of non-histone proteins. Gene.
363:15–23. 2005.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Suzuki M, Ikeda A and Bartlett JD: Sirt1
overexpression suppresses fluoride-induced p53 acetylation to
alleviate fluoride toxicity in ameloblasts responsible for enamel
formation. Arch Toxicol. 92:1283–1293. 2018.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Ito A, Lai CH, Zhao X, Saito S, Hamilton
MH, Appella E and Yao TP: p300/CBP-mediated p53 acetylation is
commonly induced by p53-activating agents and inhibited by MDM2.
EMBO J. 20:1331–1340. 2001.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Iyer NG, Xian J, Chin SF, Bannister AJ,
Daigo Y, Aparicio S, Kouzarides T and Caldas C: p300 is required
for orderly G1/S transition in human cancer cells. Oncogene.
26:21–29. 2007.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Shi D, Dai C, Qin J and Gu W: Negative
regulation of the p300-p53 interplay by DDX24. Oncogene.
35:528–536. 2016.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Morton DJ, Patel D, Joshi J, Hunt A,
Knowell AE and Chaudhary J: ID4 regulates transcriptional activity
of wild type and mutant p53 via K373 acetylation. Oncotarget.
8:2536–2549. 2017.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Ou HL and Schumacher B: DNA damage
responses and p53 in the aging process. Blood. 131:488–495.
2018.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Miyajima C, Kawarada Y, Inoue Y, Suzuki C,
Mitamura K, Morishita D, Ohoka N, Imamura T and Hayashi H:
Transcriptional Coactivator TAZ negatively regulates tumor
suppressor p53 activity and cellular senescence. Cells.
9(171)2020.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Reed SM and Quelle DE: p53 Acetylation:
Regulation and consequences. Cancers. 7:30–69. 2014.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Shan W, Jiang Y, Yu H, Huang Q, Liu L, Guo
X, Li L, Mi Q, Zhang K and Yang Z: HDAC2 overexpression correlates
with aggressive clinicopathological features and DNA-damage
response pathway of breast cancer. Am J Cancer Res. 7:1213–1226.
2017.PubMed/NCBI
|
|
94
|
Zhang L, Kang W, Lu X, Ma S, Dong L and
Zou B: Weighted gene co-expression network analysis and
connectivity map identifies lovastatin as a treatment option of
gastric cancer by inhibiting HDAC2. Gene. 681:15–25.
2019.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Brandl A, Wagner T, Uhlig KM, Knauer SK,
Stauber RH, Melchior F, Schneider G, Heinzel T and Krämer OH:
Dynamically regulated sumoylation of HDAC2 controls p53
deacetylation and restricts apoptosis following genotoxic stress. J
Mol Cell Biol. 4:284–293. 2012.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Li D, Marchenko ND and Moll UM: SAHA shows
preferential cytotoxicity in mutant p53 cancer cells by
destabilizing mutant p53 through inhibition of the HDAC6-Hsp90
chaperone axis. Cell Death Differ. 18:1904–1913. 2011.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Marrogi AJ, Khan MA, van Gijssel HE, Welsh
JA, Rahim H, Demetris AJ, Kowdley KV, Hussain SP, Nair J, Bartsch
H, et al: Oxidative stress and p53 mutations in the carcinogenesis
of iron overload-associated hepatocellular carcinoma. J Natl Cancer
Inst. 93:1652–1655. 2001.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Zhou Y, Que KT, Zhang Z, Yi ZJ, Zhao PX,
You Y, Gong JP and Liu ZJ: Iron overloaded polarizes macrophage to
proinflammation phenotype through ROS/acetyl-p53 pathway. Cancer
Med. 7:4012–4022. 2018.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Tu W, Zhang Q, Liu Y, Han L, Wang Q, Chen
P, Zhang S, Wang A and Zhou X: Fluoride induces apoptosis via
inhibiting SIRT1 activity to activate mitochondrial p53 pathway in
human neuroblastoma SH-SY5Y cells. Toxicol Appl Pharmacol.
347:60–69. 2018.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Wingelhofer B, Neubauer HA, Valent P, Han
X, Constantinescu SN, Gunning PT, Müller M and Moriggl R:
Implications of STAT3 and STAT5 signaling on gene regulation and
chromatin remodeling in hematopoietic cancer. Leukemia.
32:1713–1726. 2018.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Chen Y, Wu R, Chen HZ, Xiao Q, Wang WJ, He
JP, Li XX, Yu XW, Li L, Wang P, et al: Enhancement of hypothalamic
STAT3 acetylation by nuclear receptor Nur77 dictates leptin
sensitivity. Diabetes. 64:2069–2081. 2015.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Zheng J, van de Veerdonk FL, Crossland KL,
Smeekens SP, Chan CM, Al Shehri T, Abinun M, Gennery AR, Mann J,
Lendrem DW, et al: Gain-of-function STAT1 mutations impair STAT3
activity in patients with chronic mucocutaneous candidiasis (CMC).
Eur J Immunol. 45:2834–2846. 2015.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Ma L, Huang C, Wang XJ, Xin DE, Wang LS,
Zou QC, Zhang YS, Tan MD, Wang YM, Zhao TC, et al: Lysyl oxidase 3
is a dual-specificity enzyme involved in STAT3 Deacetylation and
Deacetylimination modulation. Mol Cell. 65:296–309. 2017.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Hansen DV, Hanson JE and Sheng M:
Microglia in Alzheimer's disease. J Cell Biol. 217:459–472.
2018.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Dani M, Wood M, Mizoguchi R, Fan Z, Walker
Z, Morgan R, Hinz R, Biju M, Kuruvilla T, Brooks DJ and Edison P:
Microglial activation correlates in vivo with both tau and amyloid
in Alzheimer's disease. Brain. 141:2740–2754. 2018.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Eufemi M, Cocchiola R, Romaniello D,
Correani V, Di Francesco L, Fabrizi C, Maras B and Schininà ME:
Acetylation and phosphorylation of STAT3 are involved in the
responsiveness of microglia to beta amyloid. Neurochem Int.
81:48–56. 2015.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Xu YS, Liang JJ, Wang Y, Zhao XJ, Xu L, Xu
YY, Zou QC, Zhang JM, Tu CE, Cui YG, et al: STAT3 undergoes
acetylation-dependent mitochondrial translocation to regulate
pyruvate metabolism. Sci Rep. 6(39517)2016.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Gough DJ, Corlett A, Schlessinger K,
Wegrzyn J, Larner AC and Levy DE: Mitochondrial STAT3 supports
Ras-dependent oncogenic transformation. Science. 324:1713–1716.
2009.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Wang Y, Huang Y, Liu J, Zhang J, Xu M, You
Z, Peng C, Gong Z and Liu W: Acetyltransferase GCN5 regulates
autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep.
21(e48335)2020.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Bao J, Zheng L, Zhang Q, Li X, Zhang X, Li
Z, Bai X, Zhang Z, Huo W, Zhao X, et al: Deacetylation of TFEB
promotes fibrillar Aβ degradation by upregulating lysosomal
biogenesis in microglia. Protein Cell. 7:417–433. 2016.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Zhang J, Wang J, Zhou Z, Park JE, Wang L,
Wu S, Sun X, Lu L, Wang T, Lin Q, et al: Importance of TFEB
acetylation in control of its transcriptional activity and
lysosomal function in response to histone deacetylase inhibitors.
Autophagy. 14:1043–1059. 2018.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Brijmohan AS, Batchu SN, Majumder S,
Alghamdi TA, Thieme K, McGaugh S, Liu Y, Advani SL, Bowskill BB,
Kabir MG, et al: HDAC6 inhibition promotes transcription factor EB
activation and is protective in experimental kidney disease. Front
Pharmacol. 9(34)2018.PubMed/NCBI View Article : Google Scholar
|
|
113
|
She A, Kurtser I, Reis SA, Hennig K, Lai
J, Lang A, Zhao WN, Mazitschek R, Dickerson BC, Herz J and Haggarty
SJ: Selectivity and kinetic requirements of HDAC inhibitors as
progranulin enhancers for treating frontotemporal dementia. Cell
Chem Biol. 24:892–906.e5. 2017.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Manickavinayaham S, Velez-Cruz R, Biswas
AK, Bedford E, Klein BJ, Kutateladze TG, Liu B, Bedford MT and
Johnson DG: E2F1 acetylation directs p300/CBP-mediated histone
acetylation at DNA double-strand breaks to facilitate repair. Nat
Commun. 10(4951)2019.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Chen L, Wei T, Si X, Wang Q, Li Y, Leng Y,
Deng A, Chen J, Wang G, Zhu S and Kang J: Lysine acetyltransferase
GCN5 potentiates the growth of non-small cell lung cancer via
promotion of E2F1, cyclin D1, and cyclin E1 expression. J Biol
Chem. 288:14510–14521. 2013.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Lin Y, Dong C and Zhou BP: Epigenetic
regulation of EMT: The snail story. Curr Pharm Des. 20:1698–1705.
2014.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Hsu DS, Wang HJ, Tai SK, Chou CH, Hsieh
CH, Chiu PH, Chen NJ and Yang MH: Acetylation of snail modulates
the cytokinome of cancer cells to enhance the recruitment of
macrophages. Cancer Cell. 26:534–548. 2014.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Xu W, Liu H, Liu ZG, Wang HS, Zhang F,
Wang H, Zhang J, Chen JJ, Huang HJ, Tan Y, et al: Histone
deacetylase inhibitors upregulate Snail via Smad2/3 phosphorylation
and stabilization of Snail to promote metastasis of hepatoma cells.
Cancer Lett. 420:1–13. 2018.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Zhang L, Shan X, Chen Q, Xu D, Fan X, Yu
M, Yan Q and Liu J: Downregulation of HDAC3 by ginsenoside Rg3
inhibits epithelial-mesenchymal transition of cutaneous squamous
cell carcinoma through c-Jun acetylation. J Cell Physiol.
234:22207–22219. 2019.PubMed/NCBI View Article : Google Scholar
|
|
121
|
McMahon SB: MYC and the control of
apoptosis. Cold Spring Harb Perspect Med. 4(a014407)2014.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Kenneth NS, Ramsbottom BA, Gomez-Roman N,
Marshall L, Cole PA and White RJ: TRRAP and GCN5 are used by c-Myc
to activate RNA polymerase III transcription. Proc Natl Acad Sci
USA. 104:14917–14922. 2007.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Mao B, Zhao G, Lv X, Chen HZ, Xue Z, Yang
B, Liu DP and Liang CC: Sirt1 deacetylates c-Myc and promotes
c-Myc/Max association. Int J Biochem Cell Biol. 43:1573–1581.
2011.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Bhadury J, Nilsson LM, Muralidharan SV,
Green LC, Li Z, Gesner EM, Hansen HC, Keller UB, McLure KG and
Nilsson JA: BET and HDAC inhibitors induce similar genes and
biological effects and synergize to kill in Myc-induced murine
lymphoma. Proc Natl Acad Sci USA. 111:E2721–E2730. 2014.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Nebbioso A, Carafa V, Conte M, Tambaro FP,
Abbondanza C, Martens J, Nees M, Benedetti R, Pallavicini I,
Minucci S, et al: c-Myc modulation and acetylation is a key HDAC
inhibitor target in cancer. Clin Cancer Res. 23:2542–2555.
2017.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Wang F, Wang Z, Li D and Chen Q:
Identification and characterization of a Bursaphelenchus xylophilus
(Aphelenchida: Aphelenchoididae) Thermotolerance-related gene:
Bx-HSP90. Int J Mol Sci. 13:8819–8833. 2012.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Kovacs JJ, Murphy PJ, Gaillard S, Zhao X,
Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB and Yao TP: HDAC6
regulates Hsp90 acetylation and chaperone-dependent activation of
glucocorticoid receptor. Mol Cell. 18:601–607. 2005.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Scroggins BT, Robzyk K, Wang D, Marcu MG,
Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, et al:
An acetylation site in the middle domain of Hsp90 regulates
chaperone function. Mol Cell. 25:151–159. 2007.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Meng Q, Chen X, Sun L, Zhao C, Sui G and
Cai L: Carbamazepine promotes Her-2 protein degradation in breast
cancer cells by modulating HDAC6 activity and acetylation of Hsp90.
Mol Cell Biochem. 348:165–171. 2011.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Rahimi N and Costello CE: Emerging roles
of post-translational modifications in signal transduction and
angiogenesis. Proteomics. 15:300–309. 2015.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Yan MS, Turgeon PJ, Man HJ, Dubinsky MK,
Ho JJD, El-Rass S, Wang YD, Wen XY and Marsden PA:
Acetyltransferase 7 (KAT7)-dependent intragenic histone acetylation
regulates endothelial Histone cell gene regulation. J Biol Chem.
293:4381–4402. 2018.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Alomer RM, da Silva EML, Chen J, Piekarz
KM, McDonald K, Sansam CG, Sansam CL and Rankin S: Esco1 and Esco2
regulate distinct cohesin functions during cell cycle progression.
Proc Natl Acad Sci USA. 114:9906–9911. 2017.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Rowland BD, Roig MB, Nishino T, Kurze A,
Uluocak P, Mishra A, Beckouët F, Underwood P, Metson J, Imre R, et
al: Building sister chromatid cohesion: Smc3 acetylation
counteracts an antiestablishment activity. Mol Cell. 33:763–774.
2009.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Gallinari P, Di Marco S, Jones P, Pallaoro
M and Steinkuhler C: HDACs, histone deacetylation and gene
transcription: From molecular biology to cancer therapeutics. Cell
Res. 17:195–211. 2007.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Schlesinger J, Schueler M, Grunert M,
Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I and
Sperling SR: The cardiac transcription network modulated by Gata4,
Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS
Genet. 7(e1001313)2011.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Leszczynska KB, Foskolou IP, Abraham AG,
Anbalagan S, Tellier C, Haider S, Span PN, O'Neill EE, Buffa FM and
Hammond EM: Hypoxia-induced p53 modulates both apoptosis and
radiosensitivity via AKT. J Clin Invest. 125:2385–2398.
2015.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Vadvalkar SS, Matsuzaki S, Eyster CA,
Giorgione JR, Bockus LB, Kinter CS, Kinter M and Humphries KM:
Decreased mitochondrial pyruvate transport activity in the diabetic
heart: Role of mitochondrial pyruvate carrier 2 (MPC2) Acetylation.
J Biol Chem. 292:4423–4433. 2017.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Hu H, Zhu W, Qin J, Chen M, Gong L, Li L,
Liu X, Tao Y, Yin H, Zhou H, et al: Acetylation of PGK1 promotes
liver cancer cell proliferation and tumorigenesis. Hepatology.
65:515–528. 2017.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Wang Y, Wang F, Bao X and Fu L: Systematic
analysis of lysine acetylome reveals potential functions of lysine
acetylation in Shewanella baltica, the specific spoilage organism
of aquatic products. J Proteomics. 205(103419)2019.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Chen J, Rong X, Fan G, Li S and Li Q:
Effects of different concentrations of putrescine on proliferation,
migration and apoptosis of human skin fibroblasts. Nan Fang Yi Ke
Da Xue Xue Bao. 35:758–762. 2015.PubMed/NCBI(In Chinese).
|
|
141
|
Mo R, Yang M, Chen Z, Cheng Z, Yi X, Li C,
He C, Xiong Q, Chen H, Wang Q and Ge F: Acetylome analysis reveals
the involvement of lysine acetylation in photosynthesis and carbon
metabolism in the model cyanobacterium Synechocystis sp. PCC 6803.
J Proteome Res. 14:1275–1286. 2015.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Yu BJ, Kim JA, Moon JH, Ryu SE and Pan JG:
The diversity of lysine-acetylated proteins in Escherichia coli. J
Microbiol Biotechnol. 18:1529–1536. 2008.PubMed/NCBI
|
|
143
|
Savoia M, Cencioni C, Mori M, Atlante S,
Zaccagnini G, Devanna P, Di Marcotullio L, Botta B, Martelli F,
Zeiher AM, et al: P300/CBP-associated factor regulates
transcription and function of isocitrate dehydrogenase 2 during
muscle differentiation. FASEB J. 33:4107–4123. 2019.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Baeza J, Smallegan MJ and Denu JM:
Mechanisms and dynamics of protein acetylation in mitochondria.
Trends Biochem Sci. 41:231–244. 2016.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Wagner GR and Hirschey MD: Nonenzymatic
protein acylation as a carbon stress regulated by sirtuin
deacylases. Mol Cell. 54:5–16. 2014.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Ronowska A, Szutowicz A, Bielarczyk H,
Gul-Hinc S, Klimaszewska-Łata J, Dyś A, Zyśk M and Jankowska-Kulawy
A: The regulatory effects of Acetyl-CoA distribution in the healthy
and diseased brain. Front Cell Neurosci. 12(169)2018.PubMed/NCBI View Article : Google Scholar
|
|
147
|
Zaini MA, Muller C, de Jong TV, Ackermann
T, Hartleben G, Kortman G, Gührs KH, Fusetti F, Krämer OH, Guryev V
and Calkhoven CF: A p300 and SIRT1 regulated acetylation switch of
C/EBPα controls mitochondrial function. Cell Rep. 22:497–511.
2018.PubMed/NCBI View Article : Google Scholar
|
|
148
|
Priante G, Gianesello L, Ceol M, Del Prete
D and Anglani F: Cell death in the kidney. Int J Mol Sci.
20(3598)2019.PubMed/NCBI View Article : Google Scholar
|
|
149
|
De Rasmo D, Signorile A, De Leo E,
Polishchuk EV, Ferretta A, Raso R, Russo S, Polishchuk R, Emma F
and Bellomo F: Mitochondrial dynamics of proximal tubular
epithelial cells in Nephropathic Cystinosis. Int J Mol Sci.
21(192)2019.PubMed/NCBI View Article : Google Scholar
|
|
150
|
Silva Junior G, Liborio AB, Mota RM, Abreu
KLS, Silva AEB, Silva SMHA and Daher EF: Acute kidney injury in
AIDS: Frequency, RIFLE classification and outcome. Brazilian J Med
Biol Res. 43:1102–1108. 2010.PubMed/NCBI View Article : Google Scholar
|
|
151
|
Kellum JA, Sileanu FE, Murugan R, Lucko N,
Shaw AD and Clermont G: Classifying AKI by urine output versus
serum creatinine level. J Am Soc Nephrol. 26:2231–2238.
2015.PubMed/NCBI View Article : Google Scholar
|
|
152
|
Cianciolo Cosentino C, Skrypnyk NI, Brilli
LL, Chiba T, Novitskaya T, Woods C, West J, Korotchenko VN,
McDermott L, Day BW, et al: Histone deacetylase inhibitor enhances
recovery after AKI. J Am Soc Nephrol. 24:943–953. 2013.PubMed/NCBI View Article : Google Scholar
|
|
153
|
Ranganathan P, Hamad R, Mohamed R,
Jayakumar C, Muthusamy T and Ramesh G: Histone deacetylase-mediated
silencing of AMWAP expression contributes to cisplatin
nephrotoxicity. Kidney Int. 89:317–326. 2016.PubMed/NCBI View Article : Google Scholar
|
|
154
|
Fan H, Yang HC, You L, Wang YY, He WJ and
Hao CM: The histone deacetylase, SIRT1, contributes to the
resistance of young mice to ischemia/reperfusion-induced acute
kidney injury. Kidney Int. 83:404–413. 2013.PubMed/NCBI View Article : Google Scholar
|
|
155
|
Xu S, Gao Y, Zhang Q, Wei S, Chen Z, Dai
X, Zeng Z and Zhao KS: SIRT1/3 activation by resveratrol attenuates
acute kidney injury in a septic rat model. Oxid Med Cell Longev.
2016(7296092)2016.PubMed/NCBI View Article : Google Scholar
|
|
156
|
Ozkok A, Ravichandran K, Wang Q,
Ljubanovic D and Edelstein CL: NF-κB transcriptional inhibition
ameliorates cisplatin-induced acute kidney injury (AKI). Toxicol
Lett. 240:105–113. 2016.PubMed/NCBI View Article : Google Scholar
|
|
157
|
Morigi M, Perico L, Rota C, Longaretti L,
Conti S, Rottoli D, Novelli R, Remuzzi G and Benigni A: Sirtuin
3-dependent mitochondrial dynamic improvements protect against
acute kidney injury. J Clin Invest. 125:715–726. 2015.PubMed/NCBI View Article : Google Scholar
|
|
158
|
Zhang Q, Liu X, Li N, Zhang J, Yang J and
Bu P: Sirtuin 3 deficiency aggravates contrast-induced acute kidney
injury. J Transl Med. 16(313)2018.PubMed/NCBI View Article : Google Scholar
|
|
159
|
Ning YC, Cai GY, Zhuo L, Gao JJ, Dong D,
Cui SY, Shi SZ, Feng Z, Zhang L, Sun XF and Chen XM: Beneficial
effects of short-term calorie restriction against cisplatin-induced
acute renal injury in aged rats. Nephron Exp Nephrol. 124:19–27.
2013.PubMed/NCBI View Article : Google Scholar
|
|
160
|
Scharman EJ and Troutman WG: Prevention of
kidney injury following rhabdomyolysis: A systematic review. Ann
Pharmacother. 47:90–105. 2013.PubMed/NCBI View Article : Google Scholar
|
|
161
|
Jian B, Yang S, Chaudry IH and Raju R:
Resveratrol restores sirtuin 1 (SIRT1) activity and pyruvate
dehydrogenase kinase 1 (PDK1) expression after hemorrhagic injury
in a rat model. Mol Med. 20:10–16. 2014.PubMed/NCBI View Article : Google Scholar
|
|
162
|
Si Y, Bao H, Han L, Chen L, Zeng L, Jing
L, Xing Y and Geng Y: Dexmedetomidine attenuation of renal
ischaemia-reperfusion injury requires sirtuin 3 activation. Br J
Anaesth. 121:1260–1271. 2018.PubMed/NCBI View Article : Google Scholar
|
|
163
|
Chaix MA, Andelfinger G and Khairy P:
Genetic testing in congenital heart disease: A clinical approach.
World J Cardiol. 8:180–191. 2016.PubMed/NCBI View Article : Google Scholar
|
|
164
|
Yuan S, Zaidi S and Brueckner M:
Congenital heart disease: Emerging themes linking genetics and
development. Curr Opin Genet Dev. 23:352–359. 2013.PubMed/NCBI View Article : Google Scholar
|
|
165
|
Wu G, Nan C, Rollo JC, Huang X and Tian J:
Sodium valproate-induced congenital cardiac abnormalities in mice
are associated with the inhibition of histone deacetylase. J Biomed
Sci. 17(16)2010.PubMed/NCBI View Article : Google Scholar
|
|
166
|
Zhong L, Zhu J, Lv T, Chen G, Sun H, Yang
X, Huang X and Tian J: Ethanol and its metabolites induce histone
lysine 9 acetylation and an alteration of the expression of heart
development-related genes in cardiac progenitor cells. Cardiovasc
Toxicol. 10:268–274. 2010.PubMed/NCBI View Article : Google Scholar
|
|
167
|
Li L, Zhu J, Tian J, Liu X and Feng C: A
role for Gcn5 in cardiomyocyte differentiation of rat mesenchymal
stem cells. Mol Cell Biochem. 345:309–316. 2010.PubMed/NCBI View Article : Google Scholar
|
|
168
|
Pillai VB, Sundaresan NR, Jeevanandam V
and Gupta MP: Mitochondrial SIRT3 and heart disease. Cardiovasc
Res. 88:250–256. 2010.PubMed/NCBI View Article : Google Scholar
|
|
169
|
Sundaresan NR, Samant SA, Pillai VB,
Rajamohan SB and Gupta MP: SIRT3 is a stress-responsive deacetylase
in cardiomyocytes that protects cells from stress-mediated cell
death by deacetylation of Ku70. Mol Cell Biol. 28:6384–6401.
2008.PubMed/NCBI View Article : Google Scholar
|
|
170
|
Zhang L, Wang H, Zhao Y, Wang J,
Dubielecka PM, Zhuang S, Qin G, Chin YE, Kao RL and Zhao TC:
Myocyte-specific overexpressing HDAC4 promotes myocardial
ischemia/reperfusion injury. Mol Med. 24(37)2018.PubMed/NCBI View Article : Google Scholar
|
|
171
|
Möller T and de Lange C: Myocardial
fibrosis in congenital heart disease. Tidsskr Nor Laegeforen: 138:
2018 (In Norwegian) doi: 10.4045/tidsskr.18.0864.
|
|
172
|
Pang M and Zhuang S: Histone deacetylase:
A potential therapeutic target for fibrotic disorders. J Pharmacol
Exp Ther. 335:266–272. 2010.PubMed/NCBI View Article : Google Scholar
|
|
173
|
Morimoto T, Sunagawa Y, Fujita M and
Hasegawa K: Novel heart failure therapy targeting transcriptional
pathway in cardiomyocytes by a natural compound, curcumin. Circ J.
74:1059–1066. 2010.PubMed/NCBI View Article : Google Scholar
|
|
174
|
Hu M, Zhang Q, Tian XH, Wang JL, Niu YX
and Li G: lncRNA CCAT1 is a biomarker for the proliferation and
drug resistance of esophageal cancer via the miR-143/PLK1/BUBR1
axis. Mol Carcinog. 58:2207–2217. 2019.PubMed/NCBI View Article : Google Scholar
|
|
175
|
Li J and Qi Y: Ginsenoside Rg3 inhibits
cell growth, migration and invasion in Caco-2 cells by
downregulation of lncRNA CCAT1. Exp Mol Pathol. 106:131–138.
2019.PubMed/NCBI View Article : Google Scholar
|
|
176
|
Zhang E, Han L, Yin D, He X, Hong L, Si X,
Qiu M, Xu T, De W, Xu L, et al: H3K27 acetylation activated-long
non-coding RNA CCAT1 affects cell proliferation and migration by
regulating SPRY4 and HOXB13 expression in esophageal squamous cell
carcinoma. Nucleic Acids Res. 45:3086–3101. 2017.PubMed/NCBI View Article : Google Scholar
|
|
177
|
Zhang H, Xie C, Yue J, Jiang Z, Zhou R,
Xie R, Wang Y and Wu S: Cancer-associated fibroblasts mediated
chemoresistance by a FOXO1/TGFβ1 signaling loop in esophageal
squamous cell carcinoma. Mol Carcinog. 56:1150–1163.
2017.PubMed/NCBI View Article : Google Scholar
|
|
178
|
Zhao Y, Yang J, Liao W, Liu X, Zhang H,
Wang S, Wang D, Feng J, Yu L and Zhu WG: Cytosolic FoxO1 is
essential for the induction of autophagy and tumour suppressor
activity. Nat Cell Biol. 12:665–675. 2010.PubMed/NCBI View Article : Google Scholar
|
|
179
|
Yu H, Ye W, Wu J, Meng X, Liu RY, Ying X,
Zhou Y, Wang H, Pan C and Huang W: Overexpression of sirt7 exhibits
oncogenic property and serves as a prognostic factor in colorectal
cancer. Clin Cancer Res. 20:3434–3445. 2014.PubMed/NCBI View Article : Google Scholar
|
|
180
|
Li D, Sun X, Zhang L, Yan B, Xie S, Liu R,
Liu M and Zhou J: Histone deacetylase 6 and cytoplasmic linker
protein 170 function together to regulate the motility of
pancreatic cancer cells. Protein Cell. 5:214–223. 2014.PubMed/NCBI View Article : Google Scholar
|
|
181
|
Sharma S and Taliyan R: Histone
deacetylase inhibitors: Future therapeutics for insulin resistance
and type 2 diabetes. Pharmacol Res. 113:320–326. 2016.PubMed/NCBI View Article : Google Scholar
|
|
182
|
Berry JM, Cao DJ, Rothermel BA and Hill
JA: Histone deacetylase inhibition in the treatment of heart
disease. Expert Opin Drug Saf. 7:53–67. 2008.PubMed/NCBI View Article : Google Scholar
|
|
183
|
Yoon S, Kang G and Eom GH: HDAC
Inhibitors: Therapeutic potential in fibrosis-associated human
diseases. Int J Mol Sci. 20(1329)2019.PubMed/NCBI View Article : Google Scholar
|
|
184
|
Clawson GA: Histone deacetylase inhibitors
as cancer therapeutics. Ann Transl Med. 4(287)2016.PubMed/NCBI View Article : Google Scholar
|
|
185
|
Peserico A and Simone C: Physical and
functional HAT/HDAC interplay regulates protein acetylation
balance. J Biomed Biotechnol. 2011(371832)2011.PubMed/NCBI View Article : Google Scholar
|
|
186
|
Qiu X, Xiao X, Li N and Li Y: Histone
deacetylases inhibitors (HDACis) as novel therapeutic application
in various clinical diseases. Prog Neuropsychopharmacol Biol
Psychiatry. 72:60–72. 2017.PubMed/NCBI View Article : Google Scholar
|
|
187
|
Xu W, Ngo L, Perez G, Dokmanovic M and
Marks PA: Intrinsic apoptotic and thioredoxin pathways in human
prostate cancer cell response to histone deacetylase inhibitor.
Proc Natl Acad Sci USA. 103:15540–15545. 2006.PubMed/NCBI View Article : Google Scholar
|
|
188
|
Rosato RR and Grant S: Histone deacetylase
inhibitors: Insights into mechanisms of lethality. Expert Opin Ther
Targets. 9:809–824. 2005.PubMed/NCBI View Article : Google Scholar
|
|
189
|
Minucci S and Pelicci PG: Histone
deacetylase inhibitors and the promise of epigenetic (and more)
treatments for cancer. Nat Rev Cancer. 6:38–51. 2006.PubMed/NCBI View Article : Google Scholar
|
|
190
|
Lane AA and Chabner BA: Histone
deacetylase inhibitors in cancer therapy. J Clin Oncol.
27:5459–5468. 2009.PubMed/NCBI View Article : Google Scholar
|
|
191
|
Eckschlager T, Plch J, Stiborova M and
Hrabeta J: Histone deacetylase inhibitors as anticancer drugs. Int
J Mol Sci. 18(1414)2017.PubMed/NCBI View Article : Google Scholar
|
|
192
|
Di Pompo G, Salerno M, Rotili D, Valente
S, Zwergel C, Avnet S, Lattanzi G, Baldini N and Mai A: Novel
histone deacetylase inhibitors induce growth arrest, apoptosis, and
differentiation in sarcoma cancer stem cells. J Med Chem.
58:4073–4079. 2015.PubMed/NCBI View Article : Google Scholar
|
|
193
|
Debeb BG, Lacerda L, Larson R, Wolfe AR,
Krishnamurthy S, Reuben JM, Ueno NT, Gilcrease M and Woodward WA:
Histone deacetylase inhibitor-induced cancer stem cells exhibit
high pentose phosphate pathway metabolism. Oncotarget.
7:28329–28339. 2016.PubMed/NCBI View Article : Google Scholar
|
|
194
|
Cao D, Wang M, Qiu X, Liu D, Jiang H, Yang
N and Xu RM: Structural basis for allosteric, substrate-dependent
stimulation of SIRT1 activity by resveratrol. Genes Dev.
29:1316–1325. 2015.PubMed/NCBI View Article : Google Scholar
|
|
195
|
Li J, Chong T, Wang Z, Chen H and Li H,
Cao J, Zhang P and Li H: A novel anticancer effect of resveratrol:
Reversal of epithelial-mesenchymal transition in prostate cancer
cells. Mol Med Rep. 10:1717–1724. 2014.PubMed/NCBI View Article : Google Scholar
|
|
196
|
Moussaieff A, Rouleau M, Kitsberg D, Cohen
M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M,
et al: Glycolysis-mediated changes in acetyl-CoA and histone
acetylation control the early differentiation of embryonic stem
cells. Cell Metab. 21:392–402. 2015.PubMed/NCBI View Article : Google Scholar
|
|
197
|
Hezroni H, Tzchori I, Davidi A, Mattout A,
Biran A, Nissim-Rafinia M, Westphal H and Meshorer E: H3K9 histone
acetylation predicts pluripotency and reprogramming capacity of ES
cells. Nucleus. 2:300–309. 2011.PubMed/NCBI View Article : Google Scholar
|
|
198
|
Zhang Y, Cui P, Li Y, Feng G, Tong M, Guo
L, Li T, Liu L, Li W and Zhou Q: Mitochondrially produced ATP
affects stem cell pluripotency via Actl6a-mediated histone
acetylation. FASEB J. 32:1891–1902. 2018.PubMed/NCBI View Article : Google Scholar
|
|
199
|
Qiao Y, Wang R, Yang X, Tang K and Jing N:
Dual roles of histone H3 lysine 9 acetylation in human embryonic
stem cell pluripotency and neural differentiation. J Biol Chem.
290:2508–2520. 2015.PubMed/NCBI View Article : Google Scholar
|
|
200
|
Jacobs KM, Misri S, Meyer B, Raj S, Zobel
CL, Sleckman BP, Hallahan DE and Sharma GG: Unique epigenetic
influence of H2AX phosphorylation and H3K56 acetylation on normal
stem cell radioresponses. Mol Biol Cell. 27:1332–1345.
2016.PubMed/NCBI View Article : Google Scholar
|
|
201
|
Wen Y, Yang H, Wu J, Wang A, Chen X, Hu S,
Zhang Y, Bai D and Jin Z: COL4A2 in the tissue-specific
extracellular matrix plays important role on osteogenic
differentiation of periodontal ligament stem cells. Theranostics.
9:4265–4286. 2019.PubMed/NCBI View Article : Google Scholar
|
|
202
|
Li B, Sun J, Dong Z, Xue P, He X, Liao L,
Yuan L and Jin Y: GCN5 modulates osteogenic differentiation of
periodontal ligament stem cells through DKK1 acetylation in
inflammatory microenvironment. Sci Rep. 6(26542)2016.PubMed/NCBI View Article : Google Scholar
|
|
203
|
Liu N, Shi S, Deng M, Tang L, Zhang G, Liu
N, Ding B, Liu W, Liu Y, Shi H, et al: High levels of β-catenin
signaling reduce osteogenic differentiation of stem cells in
inflammatory microenvironments through inhibition of the
noncanonical Wnt pathway. J Bone Miner Res. 26:2082–2095.
2011.PubMed/NCBI View Article : Google Scholar
|
|
204
|
Ishitani T, Kishida S, Hyodo-Miura J, Ueno
N, Yasuda J, Waterman M, Shibuya H, Moon RT, Ninomiya-Tsuji J and
Matsumoto K: The TAK1-NLK mitogen-activated protein kinase cascade
functions in the Wnt-5a/Ca(2+) pathway to antagonize
Wnt/beta-catenin signaling. Mol Cell Biol. 23:131–139.
2003.PubMed/NCBI View Article : Google Scholar
|
|
205
|
Wang H, Diao D, Shi Z, Zhu X, Gao Y, Gao
S, Liu X, Wu Y, Rudolph KL, Liu G, et al: SIRT6 controls
hematopoietic stem cell homeostasis through epigenetic regulation
of Wnt signaling. Cell Stem Cell. 18:495–507. 2016.PubMed/NCBI View Article : Google Scholar
|
|
206
|
Wagner VP, Martins MD and Castilho RM:
Histones acetylation and cancer stem cells (CSCs). Methods Mol
Biol. 1692:179–193. 2018.PubMed/NCBI View Article : Google Scholar
|
|
207
|
Liu C, Liu L, Shan J, Shen J, Xu Y, Zhang
Q, Yang Z, Wu L, Xia F, Bie P, et al: Histone deacetylase 3
participates in self-renewal of liver cancer stem cells through
histone modification. Cancer Lett. 339:60–69. 2013.PubMed/NCBI View Article : Google Scholar
|
|
208
|
Liu D, Zhou P, Zhang L, Gong W, Huang G,
Zheng Y and He F: HDAC1/DNMT3A-containing complex is associated
with suppression of Oct4 in cervical cancer cells. Biochemistry
(Mosc). 77:934–940. 2012.PubMed/NCBI View Article : Google Scholar
|
|
209
|
Choi HJ, Park JH, Park M, Won HY, Joo HS,
Lee CH, Lee JY and Kong G: UTX inhibits EMT-induced breast CSC
properties by epigenetic repression of EMT genes in cooperation
with LSD1 and HDAC1. EMBO Rep. 16:1288–1298. 2015.PubMed/NCBI View Article : Google Scholar
|
|
210
|
Salvador MA, Wicinski J, Cabaud O, Toiron
Y, Finetti P, Josselin E, Lelièvre H, Kraus-Berthier L, Depil S,
Bertucci F, et al: The histone deacetylase inhibitor abexinostat
induces cancer stem cells differentiation in breast cancer with low
Xist expression. Clin Cancer Res. 19:6520–6531. 2013.PubMed/NCBI View Article : Google Scholar
|
|
211
|
Corbin AS, Agarwal A, Loriaux M, Cortes J,
Deininger MW and Druker BJ: Human chronic myeloid leukemia stem
cells are insensitive to imatinib despite inhibition of BCR-ABL
activity. J Clin Invest. 121:396–409. 2011.PubMed/NCBI View Article : Google Scholar
|
|
212
|
Li L, Wang L, Li L, Wang Z, Ho Y, McDonald
T, Holyoake TL, Chen W and Bhatia R: Activation of p53 by SIRT1
inhibition enhances elimination of CML leukemia stem cells in
combination with imatinib. Cancer Cell. 21:266–281. 2012.PubMed/NCBI View Article : Google Scholar
|
|
213
|
Hsu YC, Wu YT, Tsai CL and Wei YH: Current
understanding and future perspectives of the roles of sirtuins in
the reprogramming and differentiation of pluripotent stem cells.
Exp Biol Med (Maywood). 243:563–575. 2018.PubMed/NCBI View Article : Google Scholar
|
|
214
|
Sun J, Wei HM, Xu J, Chang JF, Yang Z, Ren
X, Lv WW, Liu LP, Pan LX, Wang X, et al: Histone H1-mediated
epigenetic regulation controls germline stem cell self-renewal by
modulating H4K16 acetylation. Nat Commun. 6(8856)2015.PubMed/NCBI View Article : Google Scholar
|
|
215
|
Hou T, Zheng G, Zhang P, Jia J, Li J, Xie
L, Wei C and Li Y: LAceP: Lysine acetylation site prediction using
logistic regression classifiers. PLoS One. 9(e89575)2014.PubMed/NCBI View Article : Google Scholar
|
|
216
|
Chen C, Huang H and Wu CH: Protein
bioinformatics databases and resources. Methods Mol Biol.
1558:3–39. 2017.PubMed/NCBI View Article : Google Scholar
|
|
217
|
Wang L, Du Y, Lu M and Li T: ASEB: A web
server for KAT-specific acetylation site prediction. Nucleic Acids
Res. 40:W376–379. 2012.PubMed/NCBI View Article : Google Scholar
|
|
218
|
Deng W, Wang C, Zhang Y, Xu Y, Zhang S,
Liu Z and Xue Y: GPS-PAIL: Prediction of lysine
acetyltransferase-specific modification sites from protein
sequences. Sci Rep. 6(39787)2016.PubMed/NCBI View Article : Google Scholar
|
|
219
|
Shi SP, Qiu JD, Sun XY, Suo SB, Huang SY
and Liang RP: PLMLA: Prediction of lysine methylation and lysine
acetylation by combining multiple features. Mol Biosyst.
8:1520–1527. 2012.PubMed/NCBI View Article : Google Scholar
|