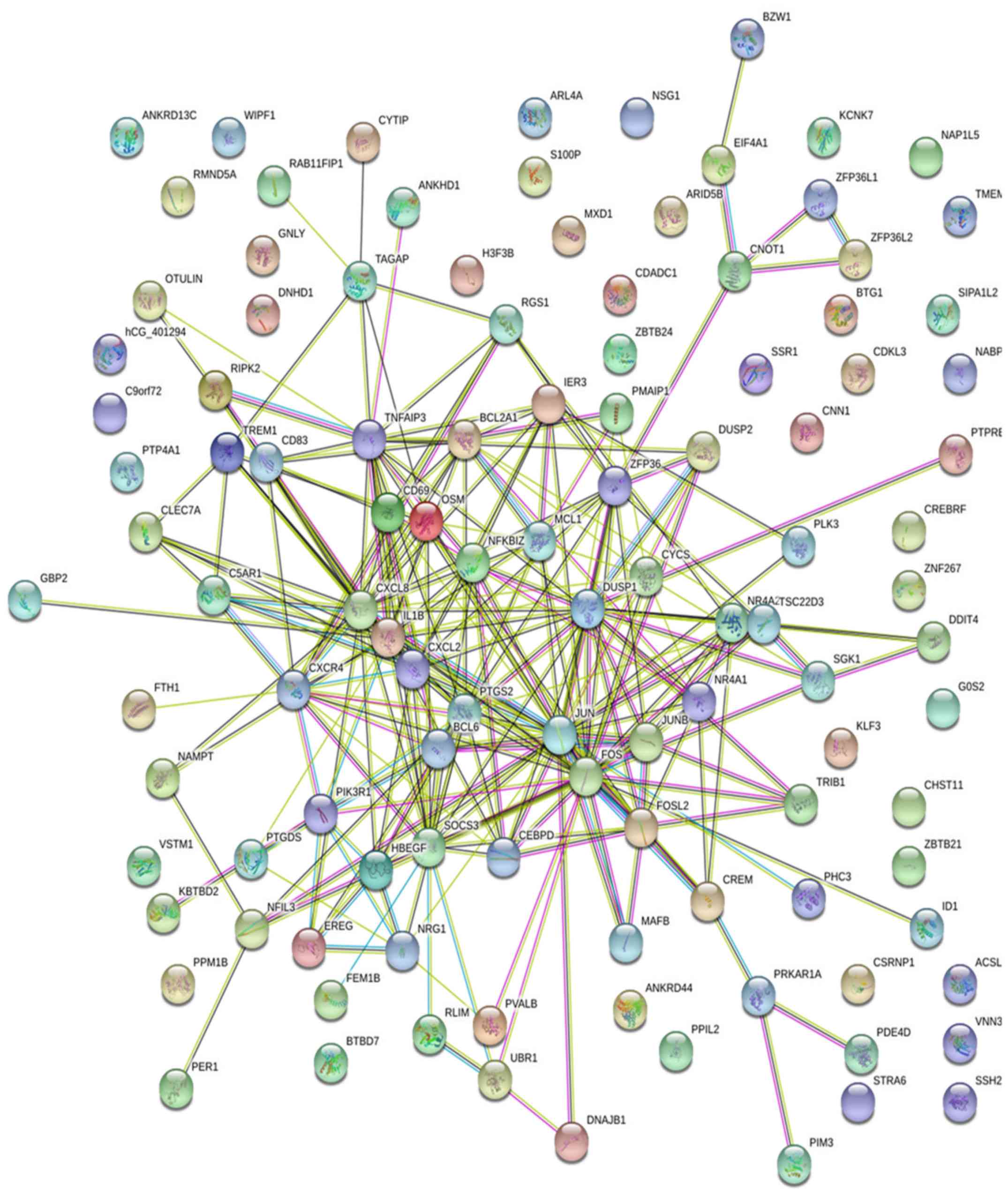
| 11725632 | 3.963 | NR4A2 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | Rna polymerase II
regulatory region sequence-specific DNA binding |
| 11716771 | 2.98 | LOC102724428 | Negative regulation
of transcription from RNA polymerase II promoter | Intracellular | Nucleotide
binding |
| 11719898 | 3.593 | HBEGF | MAPK CasCHDe receptor
binding | Extracellular
region | Epidermal growth
factor |
| 11718841 | 6.035 | CXCL8 | Angiogenesis | Extracellular
region | Cytokine
activity |
| 11761272 | 2.91 | BCL2A1 | Apoptotic
process | Cytoplasm | Protein binding |
| 11743972 | 2.917 | DDIT4 | Response to
hypoxia | Intracellular | 14-3-3 Protein
binding |
| 11732719 | 4.2 | EREG | MAPK CasCHDe
receptor binding | Extracellular
region | Epidermal growth
factor |
| 11744219 | 6.925 | G0S2 | Apoptotic
process | Mitochondrion | Protein
binding |
| 11721695 | 3.385 | DUSP2 | Inactivation of
MAPK activity | Nucleus | Phosphoprotein
phosphatase activity |
| 11742765 | 4.1 | RGS1 | Immune
response | Cytoplasm | GTPase activator
activity |
| 11724037 | 5.573 | PTGS2 | Prostaglandin
biosynthetic process | Nucleus | Peroxidase
activity |
| 11746721 | 2.647 | TREM1 | Positive regulation
of defense response to virus by host | Extracellular
region | Receptor
activity |
| 11759749 | 3.44 | KLF3 | Transcription,
DNA-templated | Nucleus | Nucleic acid
binding |
| 11744850 | 1.885 | SSH2 | Protein
dephosphorylation | Extracellular
space | DNA binding |
| 11743000 | 2.982 | CD83 | Regulation of
cytokine production | Plasma
membrane | Protein
binding |
| 11715931 | 3.478 | SGK1 | Regulation of cell
growth | Nucleus | Nucleotide
binding |
| 11724447 | 2.157 | PDE4D | Regulation of heart
rate | Cytoplasm | Cyclic-nucleotide
phosphodiesterase activity |
| 11737750 | 3.272 | SGK1 | Regulation of cell
growth | Nucleus | Nucleotide
binding |
| 11728190 | 2.058 | CXCR4 | Activation of MAPK
activity | Cytoplasm | Virus receptor
activity |
| 11743110 | 2.268 | NAMPT | Vitamin metabolic
process (carboxylating) activity | Extracellular
region |
Nicotinate-nucleotide diphosphorylase |
| 11763170 | 2.103 | FOSL2 | Keratinocyte
development sequence-specific DNA binding | Nucleus | RNA polymerase II
regulatory region |
| 11733698 | 3.143 | SGK1 | Regulation of cell
growth | Nucleus | Nucleotide
binding |
| 11744932 | 2.245 | CREBRF | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | Transcription
factor activity, Sequence-specific DNA binding |
| 11757721 | 2.265 | CSRNP1 | Transcription,
DNA-templated RNA polymerase II transcription regulatory region
sequence-specific binding | Nucleus | Transcriptional
activator activity, |
| 11715766 | 2.552 | DUSP1 | Inactivation of
MAPK activity | Nucleus | Phosphoprotein
phosphatase activity |
| 11728191 | 2.465 | CXCR4 | Activation of MAPK
activity | Cytoplasm | Virus receptor
activity |
| 11719218 | 2.567 | SOCS3 | Response to
hypoxia | Intracellular | Protein kinase
inhibitor activity |
| 11739540 | 2.195 | PIK3R1 | Cellular glucose
homeostasis | Nucleus | Transmembrane
receptor protein tyrosine kinase adaptor activity |
| 11728189 | 2.54 | CXCR4 | Activation of MAPK
activity | Cytoplasm | Virus receptor
activity |
| 11743596 | 1.75 | PTPRE | Protein
dephosphorylation | Nucleus | Phosphoprotein
phosphatase activity |
| 11717830 | 1.69 | TSC22D3 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | Transcription
factor activity, sequence-specific DNA binding |
| 11752993 | 3.005 | DUSP1 | Inactivation of
MAPK activity | Nucleus | Phosphoprotein
phosphatase activity |
| 11717897 | 1.79 | PTP4A1 | Protein
dephosphorylation | Nucleus | Phosphoprotein
phosphatase activity |
| 11752039 | 1.468 | PHC3 | Multicellular
organismal development | Nucleus | DNA Binding |
| 11756587 | 2.1 | PTGDS | Prostaglandin
biosynthetic process | Extracellular
region | Prostaglandin-D
synthase activity |
| 11723679 | 2.607 | CD69 | Signal
transduction | Integral component
of plasma membrane | Transmembrane
signaling Receptor activity |
| 11718939 | 2.723 | TNFAIP3 | B-1 B cell
homeostasis | Nucleus | Protease
binding |
| 11736467 | 2.2 | TAGAP | Signal
transduction | Cytosol | Guanyl-nucleotide
exchange factor activity |
| 11739094 | 2.712 | CXCR4 | Activation of MAPK
activity | Cytoplasm | Virus receptor
activity |
| 11763367 | 1.535 | NABP1 | Double-strand break
repair via homologous recombination | Nucleus | DNA binding |
| 11743111 | 2.857 | NAMPT | Vitamin metabolic
process | Extracellular
region |
Nicotinate-nucleotide diphosphorylase
(Carboxylating) activity |
| 11715673 | 2.002 | JUNB | Negative regulation
of transcription from RNA polymerase II pomoter | Chromatin | RNA polymerase II
regulatory region Sequence-specific DNA binding |
| 11717994 | 2.812 | NR4A1 | Positive regulation
of endothelial cell proliferation | Nucleus | Transcriptional
activator activity, RNA polymerase II core promoter proximal region
sequence-specific binding |
| 11715691 | 2.008 | ZFP36 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | DNA binding |
| 11733022 | 2.29 | BTG1 | Regulation of
transcription, DNA-templated | Nucleus | Transcription
cofactor activity |
| 11724446 | 1.738 | PDE4D | Regulation of heart
rate | Cytoplasm | Cyclic-nucleotide
phosphodiesterase activity |
| 11737176 | 2.052 | C9ORF72 | Endocytosis | Extracellular
region | Protein
binding |
| 11715487 | 1.405 | MCL1 | Cell fate
determination | Intracellular | Protein
binding |
| 11722615 | 2.275 | HCAR2 ///
HCAR3 | Neutrophil
apoptotic process response to virus by host | Plasma
membrane | Signal transducer
activity |
| 11719862 | 2.093 | TREM1 | Positive regulation
of defense response to virus by host | Extracellular
region | Receptor
activity |
| 11734799 | 1.505 | RLIM | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | Transcription
corepressor activity |
| 11763169 | 1.667 | FOSL2 | Keratinocyte
development | Nucleus | RNA polymerase II
regulatory region sequence-specific DNA binding |
| 11718394 | 3.535 | JUN | Angiogenesis | Nuclear
chromosome | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11749652 | 1.69 | ZBTB21 | Transcription,
DNA-templated | Nucleus | RNA polymerase II
regulatory region sequence-specific DNA binding |
| 11724038 | 4.715 | PTGS2 | Prostaglandin
biosynthetic process | Nucleus | Peroxidase
activity |
| 11753803 | 1.37 | CYCS | Response to
reactive oxygen species | Protein phosphatase
type 2A complex | Protein serine |
| 11725631 | 3.285 | NR4A2 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | RNA polymerase II
regulatory region sequence-specific DNA binding |
| 11751242 | 1.28 | FCGR2A ///
FCGR2C | Immune system
process | Cytoplasm | Transmembrane
signaling receptor activity |
| 11721629 | 2.755 | MAFB | Transcription,
DNA-templated | Nucleus | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11733355 | 1.655 | C5AR1 | Activation of MAPK
activity | Cytosol | Complement
component C5a binding |
| 11759628 | 1.838 | WIPF1 | Actin cortical
patch assembly | Ruffle | Actin binding |
| 11726889 | 1.505 | ZFP36L1 | Nuclear-transcribed
mRNA catabolic process, deadenylation-dependent decay | Nucleus | DNA binding |
| 11750700 | 1.375 | ACSL1 | Long-chain fatty
acid metabolic process | Mitochondrion | Nucleotide
binding |
| 11716602 | 1.895 | KBTBD2 | Protein
ubiquitination | Cul3-RING ubiquitin
ligase complex | Ubiquitin-protein
transferase activity |
| 11751415 | 1.688 | TSC22D3 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | Transcription
factor activity, sequence-specific DNA binding |
| 11744775 | 1.497 | BZW1 | Transcription,
DNA-templated | Cytoplasm | Binding |
| 11747736 | 1.2 | CNN1 | Regulation of
smooth muscle contraction | Cytoskeleton | Actin binding |
| 11727757 | 3.808 | OSM | Positive regulation
of acute inflammatory response | Extracellular
region | Cytokine
activity |
| 11758730 | 1.39 | DUSP1 | Inactivation of
MAPK activity | Nucleus | Phosphoprotein
phosphatase activity |
| 11744810 | 1.312 | ZBTB24 | Hematopoietic
progenitor cell differentiation | Nucleus | Nucleic acid
binding |
| 11737147 | 1.357 | CLEC7A | Response to
yeast | Nucleoplasm | Opsonin
Binding |
| 11719713 | 1.662 | PPM1B | Protein
dephosphorylation | Cytoplasm | Magnesium ion
binding |
| 11715445 | 1.248 | DNAJB1 | Protein
folding | Nucleus | Atpase activator
activity |
| 11720062 | 2.98 | IER3 | Response to
protozoan | Nucleus | Protein
binding |
| 11757513 | 2.262 | NFKBIZ | Transcription,
DNA-templated | Nucleus | Transcription
cofactor activity |
| 11758522 | 1.92 | CREM | Glucose metabolic
process | Nucleus | Core promoter
sequence-specific DNA binding |
| 11724835 | 1.355 | HCAR2 ///
HCAR3 | Neutrophil
apoptotic process | Plasma
membrane | Signal transducer
activity |
| 11762406 | 1.695 | GBP2 | Immune
response | Golgi membrane | Nucleotide
binding |
| 11752577 | 1.438 | FTH1 | Iron ion
transport | Cell | Ferroxidase
activity |
| 11744128 | 5.325 | CXCL2 | Response to
molecule of bacterial origin | Extracellular
region | Cytokine
activity |
| 11760678 | 1.337 | PPIL2 | Protein
polyubiquitination | Nucleus | Peptidyl-prolyl
cis-trans isomerase activity |
| 11727569 | 1.328 | OTULIN | Angiogenesis | Cytoplasm | Ubiquitin-specific
protease activity |
| 11719916 | 4.9 | IL1B | Negative regulation
of transcription from RNA polymerase II promoter | Extracellular
region | Receptor
binding |
| 11715817 | 1.18 | ZFP36L2 | Nuclear-transcribed
mRNA catabolic process, deadenylation-dependent decay | Nucleus | DNA binding |
| 11743434 | 1.4 | CHST11 | Chondrocyte
development | Golgi membrane |
N-acetylgalactosamine 4-O-sulfotransferase
activity |
| 11724236 | 1.272 | RIPK2 | Activation of MAPK
activity | Cytoplasm | Nucleotide
binding |
| 11720745 | 1.815 | BCL6 | Protein import into
nucleus, translocation | Nucleus | RNA polymerase II
regulatory region sequence-specific DNA binding |
| 11749291 | 1.75 | FOS | Conditioned taste
aversion | Nucleus | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11764029 | 1.58 | CEBPD | Transcription from
RNA polymerase II promoter | Nucleus | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11718347 | 1.585 | S100P | Response to organic
substance | Nucleus | Magnesium ion
binding |
| 11724509 | 2.365 | PMAIP1 | Release of
cytochrome c from mitochondria | Nucleus | Protein
binding |
| 11734690 | 1.252 | CYTIP | Regulation of cell
adhesion | Nucleoplasm | Protein
binding |
| 11736782 | 1.395 | RAB11FIP1 | Transport | Cytoplasm | Protein
binding |
| 11764030 | 1.27 | CEBPD | Transcription from
RNA polymerase II promoter | Nucleus | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11743344 | 1.67 | RMND5A | | | Protein
binding |
| 11716048 | 2.357 | TRIB1 | Protein
phosphorylation | Nucleus | Protein kinase
activity |
| 11756387 | 1.558 | ARL4A | Intracellular
protein transport | Intracellular | Nucleotide
binding |
| 11724510 | 2.298 | PMAIP1 | Release of
cytochrome c from mitochondria | Nucleus | Protein
binding |
| 11732665 | 1.782 | VSTM1 | Immune system
process | Extracellular
region | Cytokine
activity |
| 11759780 | 1.348 | ANKRD13C | Protein retention
in ER lumen | Endoplasmic
reticulum | Receptor
binding |
| 11737148 | 1.333 | CLEC7A | Response to
yeast | Nucleoplasm | Opsonin
binding |
| 11758593 | 1.325 | H3F3B | Chromatin silencing
at rdna | Nuclear
chromosome | RNA polymerase II
core promoter sequence-specific DNA binding |
| 11763972 | 1.2 | SSR1 | Translation | Endoplasmic
reticulum | Protein
binding |
| 11727523 | 1.36 | ZNF267 | Transcription,
DNA-templated | Intracellular | Nucleic acid
binding |
| 11718395 | 4.04 | JUN | Angiogenesis | Nuclear
chromosome | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11727032 | -2.47 | NSG1 | Posittive
regulation of transcription from | Nucleus | Transcription
regulatory region RNA polymerase II promoter sequence-specific DNA
binding |
| 11718061 | 1.88 | PVALB | Cytosolic calcium
ion homeostasis | Nucleus | Calcium ion
binding |
| 11721630 | 2.138 | MAFB | Transcription,
DNA-templated | Nucleus | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11763755 | 1.068 | GNLY | Cellular defense
response | Extracellular
region | |
| 11757924 | 1.542 | SIPA1L2 | Positive regulation
of gtpase activity | | Gtpase activator
activity |
| 11732859 | 1.03 | DNHD1 | Microtubule-based
movement | Dynein complex | Microtubule motor
activity |
| 11750016 | 1.498 | MXD1 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11744127 | 4.107 | CXCL2 | Response to
molecule of bacterial origin | Extracellular
region | Cytokine
activity |
| 11763556 | 1.777 | EIF4A1 | Nuclear-transcribed
mrna catabolic process, deadenylation-dependent decay | Nucleus | Nucleotide
binding |
| 11745466 | 1.252 | CDADC1 | Metabolic
process | | Catalytic
activity |
| 11756358 | 1.87 | PLK3 | G1 | Chromatin | Nucleotide
binding |
| 11718397 | 3.16 | JUN | Angiogenesis | Nuclear
chromosome | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11763954 | 1.04 | SCARNA9L | | | |
| 11730655 | 1.12 | CNOT1 | Negative regulation
of transcription from RNA polymerase II promoter | Cytoplasmic mrna
processing body | Poly(A)-specific
ribonuclease activity |
| 11754074 | 4.195 | G0S2 | Apoptotic
process | Mitochondrion | Protein
binding |
| 11739230 | 1.132 | ARL4A | Intracellular
protein transport | Intracellular | Nucleotide
binding |
| 11743010 | 2.372 | NFIL3 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | RNA polymerase II
regulatory region sequence-specific DNA binding |
| 11724036 | 4.27 | PTGS2 | Prostaglandin
biosynthetic process | Nucleus | Peroxidase
activity |
| 11747474 | 1.64 | NR4A2 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | RNA polymerase II
regulatory region sequence-specific DNA binding |
| 11740892 | 1 | KCNK7 | Transport | Plasma
membrane | Voltage-gated ion
channel activity |
| 11733140 | 1.617 | ARL4A | Intracellular
protein transport | Intracellular | Nucleotide
binding |
| 11724769 | 0.978 | FCGR2A ///
FCGR2C | Immune system
process | Cytoplasm | Transmembrane
signaling receptor activity |
| 11760192 | 0.987 | TMEM68 | Metabolic
process | Membrane | Transferase
activity, transferring acyl groups |
| 11734988 | 1.285 | FEM1B | Epithelial cell
maturation | Nucleus | Ubiquitin-protein
transferase activity |
| 11757798 | 2.205 | MAFB | Transcription,
DNA-templated | Nucleus | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11740347 | 1.2 | NRG1 | MAPK casCHDe | Extracellular
region | Transcription
cofactor activity |
| 11753791 | 2.535 | PRKAR1A | Mesoderm
formation | Immunological
synapse | Nucleotide
binding |
| 11725114 | 1.45 | ANKHD1 | Regulation of
translation | Nucleoplasm | Nucleic acid
binding |
| 11734659 | 1.54 | FOS | Conditioned taste
aversion | Nucleus | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11716071 | 1.3 | PIM3 | Protein
phosphorylation | Cytoplasm | Nucleotide
binding |
| 11720612 | 1.04 | NAP1L5 | Nucleosome
assembly | Nucleus | |
| 11755987 | 0.98 | ANKRD44 | | | Protein
binding |
| 11717895 | 1.285 | PTP4A1 | Protein
dephosphorylation | Nucleus | Phosphoprotein
phosphatase activity |
| 11725199 | 1.525 | BTBD7 | Multicellular
organismal development | Nucleus | Protein
binding |
| 11746681 | 1.88 | VNN3 | Nitrogen compound
metabolic process | Extracellular
space | Hydrolase
activity |
| 11720726 | 1.413 | UBR1 | Ubiquitin-dependent
protein catabolic process | Ubiquitin ligase
complex | Ubiquitin-protein
transferase activity |
| 11760818 | 1.05 | CDKL3 | Cellular protein
modification process | Cytoplasm | Nucleotide
binding |
| 11733686 | 1.052 | STRA6 | Retinoid metabolic
process | Plasma
membrane | Receptor
activity |
| 11724768 | 0.958 | FCGR2A ///
FCGR2C | Immune system
process | Cytoplasm | Transmembrane
signaling receptor activity |
| 11748516 | 0.932 | NAP1L5 | Nucleosome
assembly | Nucleus | |
| 11718927 | 0.975 | ARID5B | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | RNA polymerase II
regulαtory region sequence-specific DNA binding |
| 11716195 | 2.692 | ID1 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | Transcription
factor activity, sequence-specific DNA binding |
| B, Down
regulation |
| 11720657 | -3.328 | HLA-DRB5 | Immune system
process | Golgi membrane | Protein
binding |
| 11724843 | -2.185 | CISH | Regulation of cell
growth | Cytoplasm | protein kinase
inhibitor activity |
| 11762593 | -2.075 | NUMA1 | G2 | | Structural molecule
activity |
| 11742832 | -1.783 | ASPM | Neuron
migration | Spindle pole | Binding |
| 11758261 | -2.223 | CEP55 | Mitotic
cytokinesis | Cytoplasm | Protein
binding |
| 11758089 | -1.885 | HMMR | Carbohydrate
metabolic process | Cytoplasm | Protein
binding |
| 11721145 | -1.542 | MKI67 | DNA metabolic
process | Chromosome,
centromeric region | Nucleotide
binding |
| 11743423 | -2.188 | NSG1 | Positive regulation
of receptor recycling | Golgi membrane | Receptor
binding |
| 11736674 | -1.588 | KLHL35 | | | Protein
binding |
| 11759710 | -1.417 | TXNDC9 | Cell redox
homeostasis | Cell | Protein
binding |
| 11735385 | -1.752 | DACT1 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | Protein kinase C
binding |
| 11748198 | -1.55 | NSG1 | Positive regulation
of receptor recycling | Golgi membrane | Receptor
binding |
| 11732363 | -1.775 | ZNF2 | Transcription,
DNA-templated | Intracellular | Nucleic acid
binding |
| 11741074 | -1.407 | METTL18 | Methylation | | Methyltransferase
activity |
| 11722367 | -1.55 | DLGAP5 | Protein
dephosphorylation | Nucleus | Phosphoprotein
phosphatase activity |
| 11751805 | -1.83 | TYMS | G1 | Nucleus | Nucleotide
binding |
| 11764270 | -1.498 | PLGLB1 ///
PLGLB2 | | Extracellular
region | |
| 11747230 | -1.518 | BUB1 | Mitotic cell
cycle | Chromosome,
centromeric region | Nucleotide
binding |
| 11723209 | -1.732 | KBTBD6 | Protein
ubiquitination | Cul3-RING ubiquitin
ligase complex | Ubiquitin-protein
transferase activity |
| 11732390 | -1.465 | CCR9 | Chemotaxis | Cytosol | Signal transducer
activity |
| 11716666 | -1.623 | ID3 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | Transcription
factor activity, sequence-specific DNA binding |
| 11763252 | -1.705 | PSPH | Protein
dephosphorylation | Cytoplasm | Magnesium ion
binding |
| 11720240 | -1.307 | TMSB15A | Actin filament
organization | Cytoplasm | Actin binding |
| 11716793 | -1.518 | CCNB2 | G2 | Nucleus | Protein
binding |
| 11717163 | -1.375 | CDC20 | Mitotic cell
cycle | Spindle pole | Protein
binding |
| 11716427 | -1.71 | POMC | Generation of
precursor metabolites and energy | Extracellular
region | G-protein coupled
receptor binding |
| 11755958 | -1.4 | ZNF691 | Regulation of
transcription, DNA-templated | Nucleus | RNA polymerase II
regulatory region sequence-specific DNA binding |
| 11724022 | -2.403 | TRIM13 | Signal
transduction | Intracellular | Ubiquitin-protein
transferase activity |
| 11730821 | -1.295 | CDKN3 | Regulation of
cyclin-dependent protein serine | Nucleus | Phosphoprotein
phosphatase activity |
| 11726302 | -1.245 | DTL | Protein
polyubiquitination | Nucleus | Ubiquitin-protein
transferase activity |
| 11744793 | -1.44 | DLGAP5 | Protein
dephosphorylation | Nucleus | Phosphoprotein
phosphatase activity |
| 11718599 | -1.502 | TM2D2 | | Membrane | |
| 11760734 | -1.272 | GULP1 | Transport | Cytoplasm | Signal transducer
activity |
| 11718058 | -1.782 | TYMS | G1 | Nucleus | Nucleotide
binding |
| 11734748 | -1.245 | LOC100507547 ///
PRRT1 | Response to biotic
stimulus | Plasma
membrane | |
| 11730796 | -1.165 | PSPH | Protein
dephosphorylation | Cytoplasm | Magnesium ion
binding |
| 11727968 | -1.148 | ESCO2 | Mitotic cell
cycle | Chromatin | Lysine
N-acetyltransferase activity, acting on acetyl phosphate as
donor |
| 11758219 | -1.48 | RRM2 | G1 | Nucleus |
Ribonucleoside-diphosphate reductase
activity thioredoxin disulfide as acceptor |
| 11755381 | -1.73 | PLGLA /// PLGLB1
/// PLGLB2 | | Extracellular
region | |
| 11762018 | -1.635 | DCLRE1C | Nucleotide-excision
repair, telomeric region | Nuclear
chromosome, | Single-stranded
DNA |
| | | | | DNA damage
recognition |
endodeoxyribonuclease activity |
| 11734263 | -1.97 | ZNF780A | Transcription,
DNA-templated | Intracellular | Nucleic acid
binding |
| 11733149 | -1.62 | DDX58 | Positive regulation
of defense response to virus by host | Cytoplasm | Nucleotide
binding |
| 11754360 | -1.762 | RRM2 | G1 | Nucleus |
Ribonucleoside-diphosphate reductase
activity, thioredoxin disulfide as acceptor |
| 11758872 | -2.118 | CDC37L1 | Protein
folding | Cytoplasm | Protein
binding |
| 11728830 | -1.47 | RAB3IP | Protein targeting
to membrane | Nucleus | Guanyl-nucleotide
exchange factor activity |
| 11759287 | -1.37 | DNAJB4 | Protein
folding | Nucleoplasm | Protein
binding |
| 1172130009 | -1.157 | ZNF174 | Negative regulation
of transcription from RNA polymerase II promoter | Nucleus | RNA polymerase II
transcription factor activity, sequence-specific DNA binding |
| 11756778 | -1.195 | NEFL | MAPK casCHDe | Cytoplasm | Structural molecule
activity |
| 11740504 | -1.218 | ZNF680 | Transcription,
DNA-templated | Intracellular | RNA polymerase II
core promoter proximal region sequence-specific DNA binding |
| 11758615 | -1.177 | FRMD4B | Establishment of
epithelial cell polarity | Ruffle | Protein
binding |
| 11738883 | -1.055 | TNFSF14 | Apoptotic
process | Extracellular
region | Receptor
binding |
| 11726443 | -1.183 | KCTD6 | Protein
homooligomerization | | Protein
binding |
| 11727112 | -1.397 | SIT1 | Adaptive immune
response | Plasma
membrane | Protein
binding |
| 11717301 | -1.218 | TACSTD2 | Cell cycle | Extracellular
space | Receptor
activity |
| 11734218 | -1.38 | ZNF681 | Positive regulation
of defense response to virus by host | Intracellular | Nucleic acid
binding |
| 11728339 | -1.667 | CENPBD1 | | Nucleus | DNA binding |
| 11764234 | -1.865 | INTS7 | DNA damage
checkpoint | Nucleus | Binding |
| 11721932 | -1.09 | KIF23 | Mitotic spindle
elongation | Nucleus | Nucleotide
binding |
| 11753763 | -1.18 | CDKN3 | Regulation of
cyclin-dependent protein serine | Nucleus | Phosphoprotein
phosphatase activity |
| 11721190 | -1.312 | TTC9C | Protein
peptidyl-prolyl isomerization | Cytoplasm | Peptidyl-prolyl
cis-trans isomerase activity |
| 11721418 | -1.2 | SH3RF3 | | | Protein
binding |
| 11733069 | -1.825 | WDR5B | | | Protein
binding |
| 11730969 | -1.765 | THAP2 | | Nucleolus | Nucleic acid
binding |
| 11719780 | -1.728 | TNFAIP8L2 | Immune system
process | Cytoplasm | Protein
binding |
| 11758125 | -1.32 | DEPDC1B | Signal
transduction | Intracellular | Gtpase activator
activity |
| 11733695 | -1.055 | UBE2C | Mitotic cell
cycle | Nucleus | Nucleotide
binding |
| 11756100 | -1.227 | TMEM60 | | Membrane | |
| 11732295 | -1.455 | ZNF566 | Transcription,
DNA-templated | Intracellular | Nucleic acid
binding |
| 11724984 | -1.255 | EXPH5 | Keratinocyte
development | Intracellular | Protein
binding |
| 11726757 | -1.165 | CDC25A | Regulation of
cyclin-dependent protein serine | Intracellular | Phosphoprotein
phosphatase activity |
| 11727604 | -1.278 | EPB41L4A | | Cytoplasm | Cytoskeletal
protein binding |
| 11716226 | -1.147 | LIMA1 | Negative regulation
of actin filament depolymerization | Stress fiber | Actin binding |
| 11760155 | -1.105 | FUBP1 | Transcription,
DNA-templated | Nucleus | Transcriptional
activator activity, RNA polymerase II distal enhancer
sequence-specific binding |
| 11725833 | -1.087 | ALKBH1 | In utero embryonic
development | Nucleus | Catalytic
activity |
| 11723939 | -1.115 | CCNB1 | G1 | Spindle pole | Patched
binding |
| 11722160 | -1.218 | GRB10 | Signal
transduction | Cytoplasm | Sh3 |
| 11759488 | -1.133 | EYA3 | DNA repair | Nucleus | Chromatin
binding |
| 11739828 | -1.188 | CYS1 | Kidney
development | Cytoplasm | Protein
binding |
| 11740637 | -1.258 | GPR19 | Signal
transduction | Plasma
membrane | Signal transducer
activity |
| 11717629 | -1.82 | KIF1BP | Mitochondrial
transport | Cytoplasm | Protein
binding |
| 11753965 | -1.15 | MSL3P1 | Transcription,
DNA-templated | Nucleus | |
| 11758149 | -0.975 | RACGAP1 | Mitotic
cytokinesis | Acrosomal
vesicle | Gtpase activator
activity |
| 11732175 | -1.44 | FANCF | Ovarian follicle
development | Nucleus | Ubiquitin-protein
transferase activity |
| 11750856 | -1.062 | CCR2 | Blood vessel
remodeling | Cytoplasm | Signal transducer
activity |
| 11757036 | -1.278 | SAC3D1 | Cell cycle | Nucleus | Protein
binding |
| 11758529 | -1.192 | CENPA | Establishment of
mitotic spindle orientation | Chromosome,
centromeric region | DNA binding |
| 11732544 | -1.292 | GPR18 | Signal
transduction | Plasma
membrane | Signal transducer
activity |
| 11718213 | -1.118 | SLC27A2 | Very long-chain
fatty acid metabolic process | Mitochondrion | Nucleotide
binding |
| 11725709 | -0.968 | WDHD1 | RNA processing | Chromosome,
centromeric region | DNA binding |
| 11739531 | -1.598 | PLGLB2 | | Extracellular
region | |
| 11745077 | -1.282 | CRNKL1 | Spliceosomal
complex assembly | Prp19 complex | RNA binding |
| 11730590 | -1.198 | KCTD21 | Protein
homooligomerization | | Protein
binding |
| 11727196 | -1.37 | ZNF202 | Negative regulation
of transcription from RNA polymerase II promoter | Intracellular | RNA polymerase II
transcription factor activity, sequence-specific DNA binding |
| 11731676 | -1.567 | CCR2 | Blood vessel
remodeling | Cytoplasm | Signal transducer
activity |
| 11721473 | -1.388 | HCCS | Metabolic
process | Mitochondrion | Holocytochrome-c
synthase activity |
| 11737395 | -1.292 | SOWAHD | | | Protein
binding |
| 11737108 | -1.05 | ACKR4 | Receptor-mediated
endocytosis | Endosome | Signal transducer
activity |
| 11728404 | -1.113 | SHCBP1 | Fibroblast growth
factor receptor signaling pathway | | Protein
binding |
| 11760278 | -0.968 | | HCG8 ///
ZNRD1-AS1 | | |
| 11728360 | -1.655 | BCDIN3D | RNA
methylation | Nucleus | Protein
binding |
| 11743686 | -1.005 | ZNF436 | Transcription,
DNA-templated | Intracellular | Nucleic acid
binding |
| 11762571 | -1.425 | GNPDA2 | Carbohydrate
metabolic process | Nucleus |
Glucosamine-6-phosphate deaminase
activity |