Introduction
Gastric cancer is the second most common malignancy
and cause of cancer-related mortality in China, ranking with the
highest incidence and mortality among all malignancies in Northwest
China (1,2). Although surgery is the primary
therapeutic strategy for gastric cancer, studies have suggested
that an enhanced understanding of the molecular mechanisms
underlying gastric tumorigenesis may enable the identification of
novel diagnostic and prognostic markers, as well as the development
of novel molecular drugs (3,4). The
molecular pathogenesis underlying gastric cancer is not completely
understood and requires further investigation, which is
particularly important for patients with gastric cancer in high
prevalence areas.
The human ubiquitin protein ligase E3 component
N-recognin 5 (UBR5) gene, which is localized to chromosome 8q22,
encodes an ~10 kb mRNA and a >300 kDa protein, which is
abundantly expressed in various types of cells (5-7).
The UBR5 protein belongs to the E6-AP carboxyl terminus family,
which serve as E3 ubiquitin-protein ligases and target specific
proteins for ubiquitin-induced proteolysis (5-7).
UBR5 is an important nuclear phosphate protein, which is involved
in DNA damage response, cell proliferation, cell cycle arrest,
metabolism and apoptosis (8-13).
Previous studies have implicated UBR5 in several types of cancer,
including ovarian cancer, gallbladder cancer and lymphoma (5-7,14,15).
Originally, UBR5 was identified in a screen for progestin-regulated
genes in breast cancer cells as a novel isolate progestin-regulated
gene (16). UBR5 was also reported
to serve an important role in the pathogenesis, development and
therapeutic resistance of other progestin-related types of cancer,
such as ovarian cancer (17-19).
The functions of UBR5 have been reported in several types of
cancer, indicating the role of UBR5 in the pathogenesis of cancer
(5). However, although disrupted
UBR5 expression has been reported in gastric cancer (20), the role of UBR5 in gastric cancer
cell proliferation, migration and invasion is not completely
understood.
In the present study, UBR5 mRNA and protein
expression levels were investigated in patients with gastric
cancer, and the association between survival time and UBR5
expression was examined. The aim of the present study was to
improve the current understanding of the role of UBR5 in the
biological functions of gastric cancer cells, and UBR5 knockdown
was employed to explore the effects of UBR5 in vitro.
Materials and methods
Patients and cancer samples
The present study was approved by the Institutional
Review Board of Lanzhou University First Hospital (approval no.
LDYYLL2020-235). All procedures were conducted in accordance with
the principles outlined in the Declaration of Helsinki, and
exception to the requirement of informed consent was approved by
the Institutional Review Board of Lanzhou University First
Hospital. Gastric cancer and para-carcinoma tissue (5 cm from the
tumor margin) samples were collected at the Lanzhou University
First Hospital between January 2012 and December 2012. A section of
the samples were sent to the Department of Pathology for
clinicopathological examination and immunohistochemical staining.
The remainder of the samples were transferred to the lab and stored
in liquid nitrogen until further analysis. The patients included in
the present study were initially diagnosed with gastric cancer, and
none had received chemotherapy or radiotherapy prior to surgery.
The clinicopathological information of the patients was also
collected for analysis and is presented in Table I.
 | Table IClinicopathological characteristics
and UBR5 expression in patients with gastric cancer. |
Table I
Clinicopathological characteristics
and UBR5 expression in patients with gastric cancer.
| | UBR5 expression
(n) | |
|---|
| Variable | Total (n) | Low | High | P-value |
|---|
| Gender | | | | 0.7009 |
|
Male | 75 | 25 | 50 | |
|
Female | 37 | 11 | 26 | |
| Age (years) | | | | 0.5826 |
|
≥56 | 54 | 16 | 38 | |
|
<56 | 58 | 20 | 38 | |
| Tumor size (cm) | | | | <0.05 |
|
≥3 | 97 | 25 | 72 | |
|
<3 | 15 | 11 | 4 | |
| TNM stage | | | | <0.05 |
|
I | 19 | 10 | 9 | |
|
II, III | 93 | 26 | 77 | |
| Lymph node
metastasis | | | | <0.05 |
|
Positive | 79 | 19 | 60 | |
|
Negative | 33 | 17 | 16 | |
| UBR5
expression | | | | |
|
≥0.9945 | 76 | | | |
|
<0.9945 | 36 | | | |
Cell culture and small interfering
(si)RNA knockdown of UBR5
The gastric cell line HGC-27 (21,22)
and the gastric mucosal epithelial cell line GES-1 (23,24)
were purchased from China Infrastructure of Cell Line Resources,
Institute of Basic Medical Sciences, Chinese Academy of Medical
Sciences. Cells were cultured in RPMI-1640 (HyClone; Cytiva)
supplemented with 10% FBS (HyClone; Cytiva) and 1%
penicillin-streptomycin (Beijing Solarbio Science & Technology
Co., Ltd.). UBR5 siRNA and negative control (NC) siRNA were
purchased from Wuhan GeneCreate Biological Engineering Co., Ltd.
The sequences of the UBR5 siRNAs transfected were as follows: 1,
forward 5'-CGGGAAAGGGAGAGAGAAATT-3' and reverse,
5'-UUUCUCUCUCCCUUUCCCGTT-3'; 2 forward, 5'-GGUCAAUAGUAGAGAAGAUTT-3'
and reverse, 5'-AUCUUCUCUACUAUUGACCTT-3'; 3 forward,
5'-GAAAUAUCCUCAAGUGAAATT-3' and reverse,
5'-UUUCACUUGAGGAUAUUUCTT-3'. The sequences of the NC siRNA wereas
follows: Forwad, 5'-UUCUCCGAACGUGUCACGUTT-3' and reverse,
5'-ACGUGACACGUUCGGAGAATT-3'. Cells were seeded
(1x106/ml) into 6-well (for the migration assay) or
96-well plates (for the cell proliferation assay). Following
culture for 24 h, when cell confluence reached 70-80%, cells were
transfected with 50 nm UBR5 siRNA or 50 nm NC siRNA using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). At 48-72 h post-transfection, cells were used
for subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR analysis (2-∆∆Cq method) was
performed to detect the mRNA expression level of UBR5(25). Total RNA was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The concentration and purity of the extracted RNA was
determined using a NanoDrop 2000 Spectrophotometer (Thermo Fisher
Scientific, Inc.). Subsequently, total RNA was reverse transcribed
into cDNA using the high-capacity cDNA Reverse Transcriptase kit
(Invitrogen; Thermo Fisher Scientific, Inc.) with the following
temperature protocol: 25˚C for 10 min, 37˚C for 120 min and 85˚C
for 5 min. Subsequently, qPCR was performed in a final volume of 20
µl consisting of cDNA, qPCR mix (Beijing Transgen Biotech Co.,
Ltd.), primers and water, with the Real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used for qPCR: 94˚C for 30 sec; 40
cycles of 94˚C for 5 sec and 60˚C for 30 sec. The following primers
were used for qPCR: UBR5 forward, 5'-TGATCCTGAGCCTTTGCCAG-3' and
reverse, 5'-GCGCTTTCGGTTTTCCTGTA-3'; and GAPDH forward,
5'-AATGGGCAGCCGTTAGGAAA-3' and reverse, 5'-GCGCCCAATACGACCAAATC-3'.
The receiver operating characteristics curve analysis was applied
to identify the optimal cut off for grouping patients by UBR6
expression. The area under the curve was 0.7022 and the cut off was
0.9945. Therefore, expression <0.9945 was defined as low level,
whereas >0.9945 was defined as high level.
Western blotting
Total protein was extracted from clinical samples
and cells using RIPA buffer (Sigma-Aldrich; Merck KGaA) with
protease inhibitor cocktail (Roche Diagnostics). Protein
concentration was determined by using a BicinChoninic Acid kit
(Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of proteins
(human tissue, 10 µg/lane; cell line, 30 µg/lane) were separated
via 10% SDS-PAGE and transferred to a nitrocellulose membrane
(Amersham; Cytiva), followed by blocking with 5% not-fat milk
powder in PBS for 1 h at room temperature. The membranes were
incubated overnight at 4˚C with primary antibodies targeted
against: UBR5 (cat. No. ab70311; 1:1,000; Abcam) and β-actin (cat.
No. sc-47778; 1:1,000; Santa Cruz Biotechnology, Inc.). Following
washing three times with PBST (0.1% Tween-20) for 5 min, the
membranes were incubated with horseradish peroxidase-labeled goat
anti-mouse and goat anti-rabbit secondary antibodies (cat. Nos.
sc-2005 and sc-2004; Santa Cruz Biotechnology, Inc.) for 2 h at
room temperature. Protein bands were visualized using a
chemiluminescence kit (Merck KGaA). Protein analysis was performed
by using Image J (version 2.1.4; National Institutes of
Health).
Immunohistochemistry
Prior to embedding with paraffin, gastric cancer and
para-carcinoma tissues were fixed using 10% paraformaldehyde for
48-72 h at room temperature. Then they were cut into 4-µm sections.
The sections were deparaffinized in xylene and rehydrated in
descending alcohol series. After rehydration, sections were washed
with PBS at room temperature. Then, following antigen retrieval at
92˚C for 2 h using Citrate Antigen Retrieval Solution (Beijing
Leagene Biotechnology Co., Ltd.) and blocking with 5% BSA (Beijing
Leagene Biotechnology Co., Ltd.) at 4˚C for 2 h, the sections were
incubated overnight at 4˚C with an anti-UBR5 primary antibody (cat.
No. ab70311; 1:500; Abcam). Subsequently, the sections were rinsed
three times with PBS and incubated with a goat anti rabbit
secondary antibody for 1 h at room temperature (cat. no. SP 2001;
1:200; OriGene Technologies, Inc.). Chromogen detection was
performed using a DAB kit (OriGene Technologies, Inc.). Images were
obtained using DP72 digital light microscope (Olympus Corporation;
magnification, x200).
Cell proliferation assay
Cells (1x106/ml) were cultured in 96-well
plates with RPMI-1640/10% FBS medium. After cell confluence reached
70-80%, transfection was performed. Following transfection for 48
h, the MTS assay (Promega Corporation) was performed according to
the manufacturer's protocol. Briefly, following incubation with MTS
at 37˚C for 4 h, the absorbance of each well was measured at a
wavelength of 490 nm using a microplate reader (BioTek Instruments,
Inc.).
Cell invasion and migration
assays
Cell invasion was assessed using 6-well Transwell
chambers (Corning, Inc.). Cells were seeded into the upper chambers
(5x104 cells/ml) with serum-free RPMI-1640 medium. The
lower chambers were pre-coated with Matrigel (50 mg/l) for 37˚C for
1 h. RPMI-1640 medium containing 10% FBS was subsequently added to
the lower chamber and then incubated at 37˚C for 24 h. Invading
cells were stained with hematoxylin for 5 min at room temperature.
Invading cells were visualized using a light microscope (Olympus
Corporation; magnification, x200) in five randomly selected fields
of view. Cell migration was assessed using a wound healing assay.
At 48 h post-transfection, the cell layer was scratched with a
sterile 200-µl pipette tip. After washing with PBS, cells were
cultured with serum-free RPMI-1640 for 24 h at 37˚C. Images of the
wound were captured at 0 and 24 h using a DP72 digital light
micropscope (Olympus Corporation; magnification, x200). Cell
migration was quantified by calculating the percentage of migration
area covered with Image J (v2.1.4; National Institutes of
Health).
Statistical analysis
Data are presented as the mean ± standard deviation
and at least three independent experiments were performed per
experiment. Comparisons between two groups were analyzed using an
unpaired Student's t-test. Comparisons among multiple groups were
analyzed using one-way ANOVA followed by Tukey's post hoc test. The
predicted significance of UBR5 was assessed using Kaplan-Meier
analysis and the log-rank test. The association between UBR5
expression and clinicopathological variables was examined using the
χ2 test. Statistical analyses were conducted using
GraphPad Prism software (v5; GraphPad Software, Inc.).
Results
Characteristics of patients with
gastric cancer
As shown in Table I,
112 patients (75 male patients and 37 female patients; median age,
56 years; age range, 24-80 years) with gastric cancer were included
in the present study. The size of the tumor was ≥3 cm in 97
patients and <3 cm in 15 patients. A total of 19 patients had
TNM (26) stage I, whereas 93
patients had advanced TNM stage (II and III). Lymph node metastasis
was detected in 79 patients, whereas 33 patients were negative for
lymph node metastasis.
Expression of UBR5 in human gastric
cancer
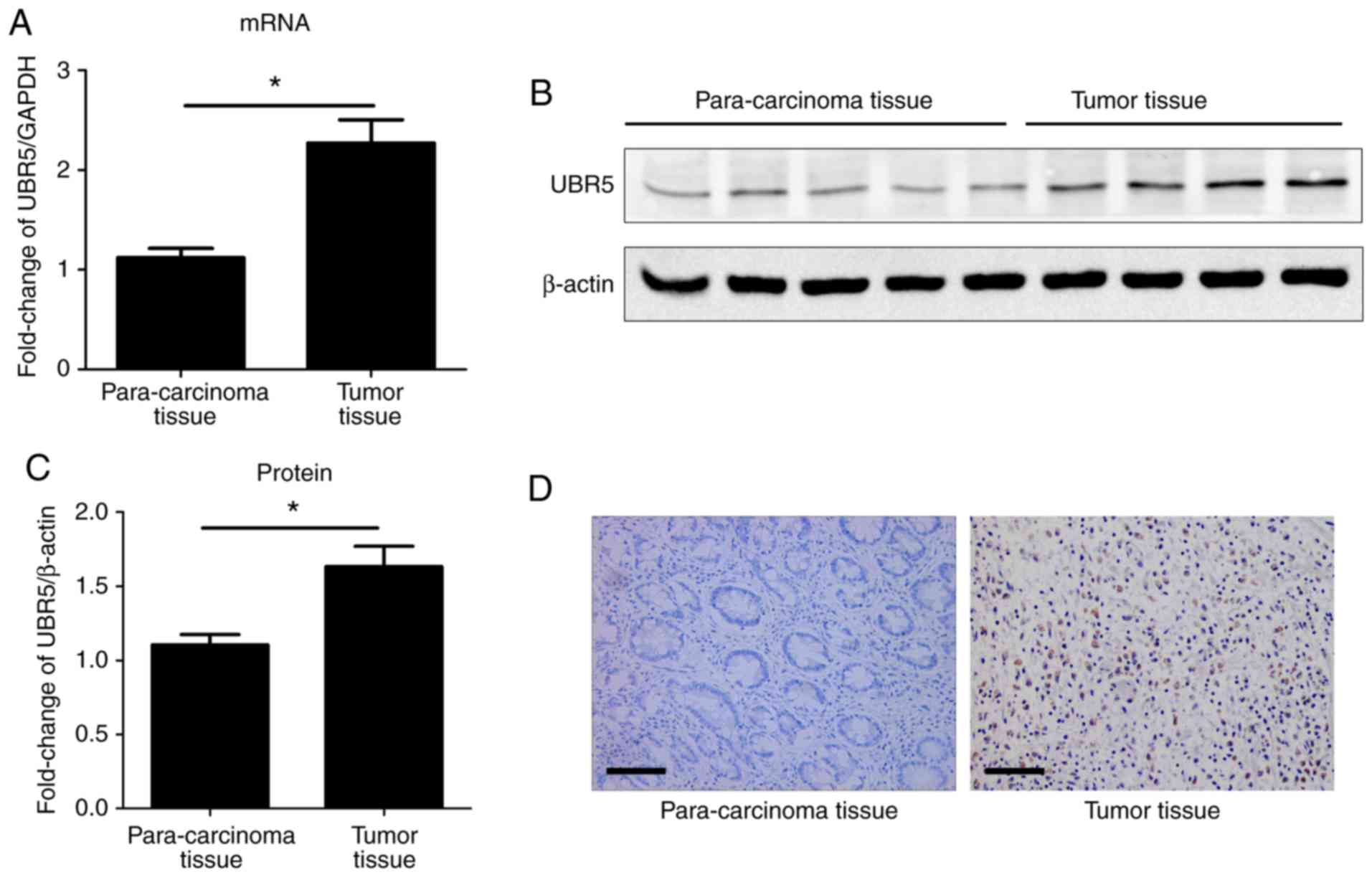
The mRNA and protein expression levels of UBR5 were
examined in gastric cancer and para-carcinoma tissue samples. The
mRNA expression levels of UBR5 were significantly increased in
gastric cancer tissue samples compared with para-carcinoma tissue
samples (Fig. 1A). Regarding the
mRNA expression of UBR5, 76 cases were in the high expression group
(>0.9945 fold) and 36 cases were in the low expression group
(<0.9945 fold). A total of 43 gastric cancer samples and 20
para-carcinoma samples were examined via western blotting. The
protein expression level of UBR5 was significantly increased in
gastric cancer tissues compared with para-carcinoma tissue samples
(Fig. 1B). The protein expression
of UBR5 was also detected via immunohistochemistry. The results
indicated that the expression of UBR5 was increased in gastric
cancer samples compared with para-carcinoma samples (Fig. 1D).
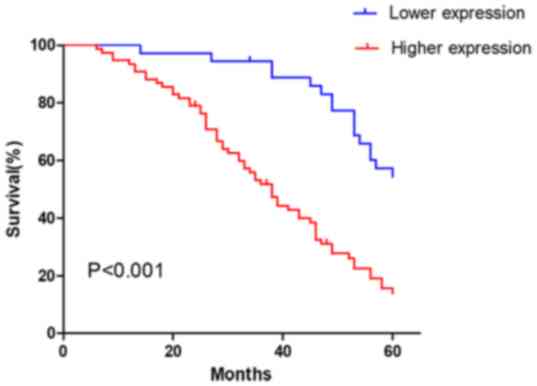
Association between UBR5 expression,
clinicopathological characteristics and prognosis of patients with
gastric cancer
Increased expression of UBR5 in gastric cancer
tissues suggested that UBR5 may serve an important role in the
development of gastric cancer. High UBR5 expression was
significantly associated with larger tumor size, advanced TNM stage
and lymph node metastasis (P<0.05; Table I). To further explore the role of
UBR5 in the prediction of patient survival, the association between
the mRNA expression level of UBR5 and survival was analyzed. The
results indicated that higher UBR5 expression levels were
associated with poorer prognosis in patients with gastric cancer
compared with lower UBR5 expression levels (P<0.001; Fig. 2).
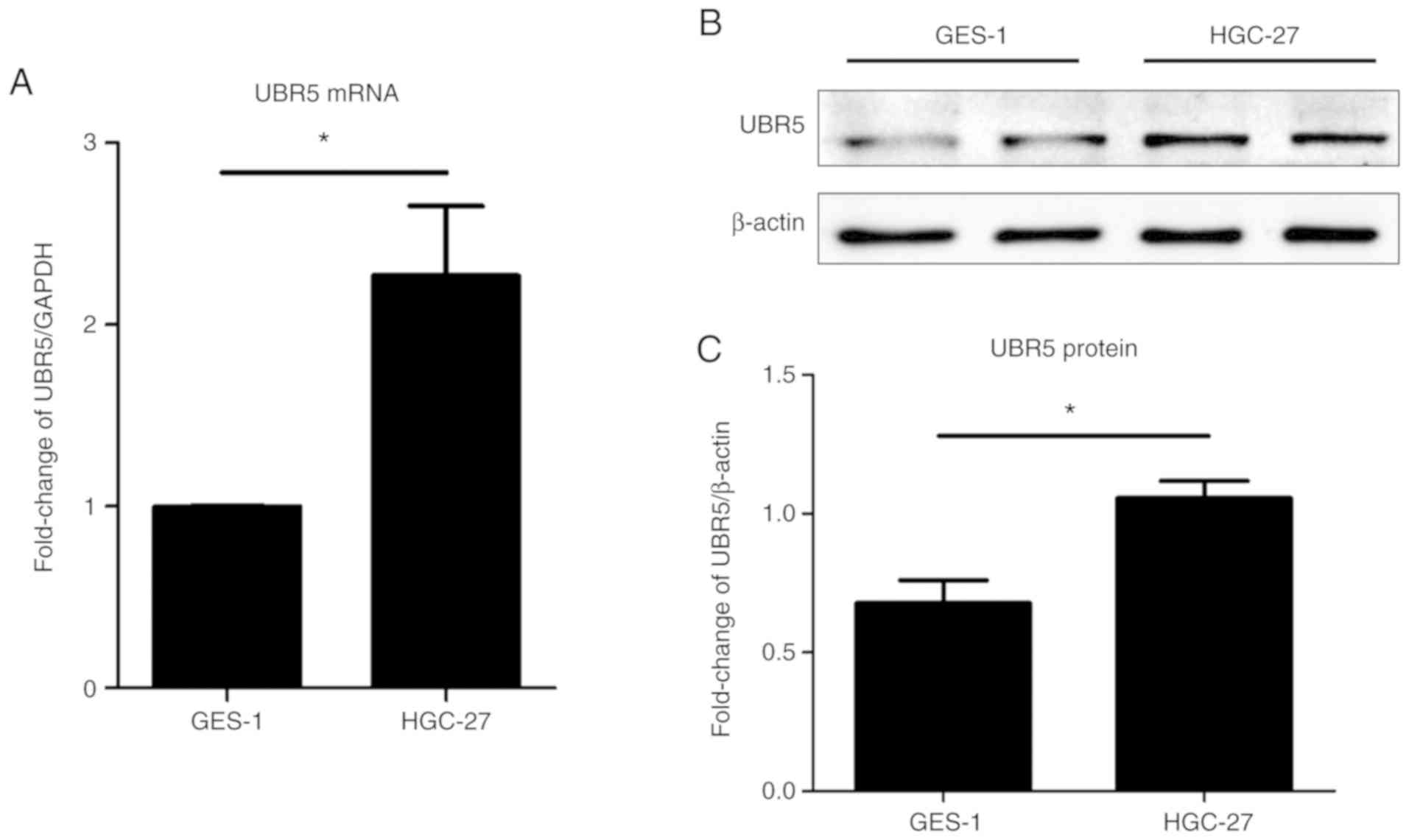
mRNA and protein expression levels of
UBR5 in cancer and control cell lines
To understand the role of UBR5 in the pathogenesis
of gastric cancer, the HGC-27 gastric cell line and the GES-1
gastric mucosal epithelial cell line were used in present study.
UBR5 mRNA and protein expression levels were examined in
vitro. The mRNA and protein expression levels of UBR5 were
significantly increased in gastric cancer cells compared with
control gastric mucosal epithelial cells (Fig. 3).
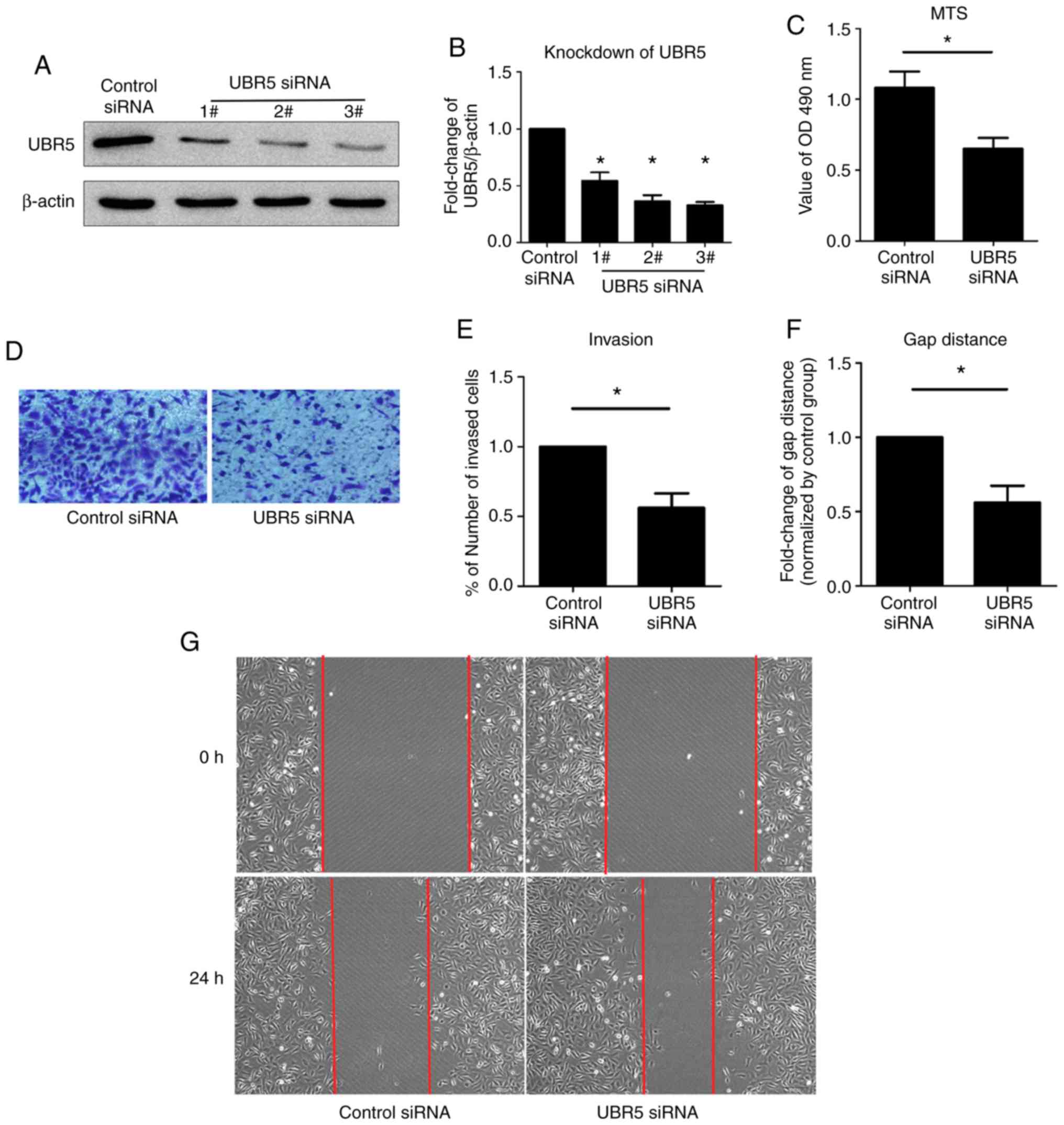
Effect of UBR5 knockdown on gastric
cancer cell proliferation, invasion and migration
According to the aforementioned results, increased
expression of UBR5 indicated that inhibition of UBR5 might be
beneficial for the treatment of gastric cancer. Therefore, UBR5
knockdown was performed using three UBR5-targeting siRNAs in the
present study. All three siRNAs significantly decreased UBR5
expression levels compared with the control siRNA group; however,
the lowest level of UBR5 expression was observed in the 3# siRNA
group. Thus, UBR5-targeting 3# siRNA was used in subsequent
experiments. Strong proliferation, invasion and migration abilities
are the classical biological properties of cancer cells (27). To examine whether UBR5 knockdown
effectively inhibited malignant biological properties, MTS,
Transwell and wound healing assays were performed. A higher optical
density value at 490 nm was observed in the UBR5 siRNA group
compared with the control siRNA group, which indicated that UBR5
knockdown decreased cell proliferation. Additionally, a reduced
number of invading cells and fewer changes in the gap distance were
observed in the UBR6 siRNA group compared with the control siRNA
group, suggesting that UBR5 knockdown decreased cancer cell
invasion and migration (Fig.
4D-G).
Discussion
UBR5 serves as an important regulator in several
types of cancer, including ovarian cancer, gallbladder cancer and
lymphoma (5-7,14,15).
The present study investigated the role of UBR5 in the pathogenesis
and development of gastric cancer. The results indicated that UBR5
mRNA and protein expression levels were significantly increased in
gastric cancer samples compared with para-carcinoma samples.
Similarly, UBR5 expression levels were upregulated in the HGC-27
gastric cancer cell line compared with control mucosal epithelial
cells. The dysregulated expression indicated that UBR5 may be
involved in the pathogenesis of gastric cancer.
To further elucidate the role of UBR5 in gastric
cancer, the association between the expression of UBR5 and
clinicopathological characteristics in gastric cancer samples was
investigated in the present study. Higher UBR5 expression was
associated with larger tumor size, advanced TNM stages and lymph
node metastasis, suggesting that increased expression of UBR5
indicated poorer prognosis. Moreover, the association between the
expression of UBR5 and survival was investigated. The higher the
expression of UBR5 in cancer tissues, the shorter the survival time
of patients with gastric cancer. Previous studies indicated a
similar role of UBR5 in other types of cancer, such as ovarian
cancer and gallbladder cancer (5-7,14).
The results indicated that increased UBR5 expression levels may
serve an important role in the development and progression of
gastric cancer. Therefore, UBR5 may serve as an oncogenic factor
and may be considered as a biomarker or prognosis predictor in
gastric cancer.
Based on the finding that increased UBR5 expression
favored tumor development, UBR5 inhibition may serve as a potential
therapeutic strategy for gastric cancer. In addition, the HGC-27
gastric cancer cell line was used in the present study. After
selecting the most effective siRNA, basic tumor properties were
examined in vitro. Compared with the control siRNA group,
UBR5 knockdown inhibited cancer cell proliferation, invasion and
migration. However, a minor limitation of the migration assay was
that it could not be guaranteed that the images obtained at 0 and
24 h were from the same field of view. Despite the limitation, the
results further supported the use of UBR5 inhibition as a
therapeutic strategy for gastric cancer.
UBR5 serves an important role in DNA damage
response, metabolism, transcription and apoptosis (8-13).
In the present study, the in vitro results demonstrated that
UBR5 may also serve a role in cell proliferation, invasion and
migration. UBR5 is a member of the E3 ubiquitin ligases, which are
key regulators in the ubiquitin-proteasome system (6,7). In
addition, UBR5 interacts with several proteins and signaling
pathways involved in a wide variety of cellular processes,
including the cell cycle, transcriptional and translational
machinery (9,14,28,29).
The PI3K/Akt signaling pathway is associated with cell survival and
proliferation in various types of cancer (14,30),
indicating that the role of UBR5 in gastric cancer proliferation
may be mediated via the PI3K/Akt signaling pathway. In addition,
UBR5, acting as an ubiquitin ligase, can directly degrade modulator
of apoptosis protein 1 (MOAP-1) by ubiquitylation, thereby
inhibiting MOAP-1 stability, and MOAP-1 exerts an effect by
enhancing the expression of the proapoptotic protein-Bax (9,19).
Therefore, whether inhibition of UBR5 can decrease gastric cancer
cell proliferation via MOAP-1 requires further investigation.
Future studies should focus on the molecular mechanism underlying
the role of UBR5 in the development and progression of gastric
cancer.
In summary, the present study demonstrated that
increased expression levels of UBR5 were associated with poor
prognosis in patients with gastric cancer, whereas UBR5 knockdown
decreased gastric cancer cell proliferation, invasion and
migration.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Health
Industry Research Project of Gansu Province (grant no.
GSWSKY2017-26), the Gansu Province Science Foundation for
Distinguished Young Scholars (grant no. 1606RJDA317), the Key
Laboratory of Biotherapy and Regenerative Medicine of Gansu
Province (grant no. zdsyskfkt- 201704) and the Foundation of The
First Hospital of Lanzhou University (grant no. ldyyyn2015-16).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FD, WZ and XL designed the study. FD, XZ, XS and LR
collected the samples, analyzed the data and wrote the manuscript.
FD, PY and CC conducted the literature search and collected the
data. FD, PY and XS performed the experiments and collected the
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Lanzhou University First Hospital (approval no.
LDYYLL2020-235). All procedures were conducted in accordance with
the principles outlined in the Declaration of Helsinki, and
exception to the requirement of informed consent was approved by
the Institutional Review Board of Lanzhou University First
Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Global Burden of Disease Cancer
Collaboration. Fitzmaurice C, Abate D, Abbasi N, Abbastabar H,
Abd-Allah F, Abdel-Rahman O, Abdelalim A, Abdoli A, Abdollahpour I,
et al: Global, regional, and national cancer incidence, mortality,
years of life lost, years lived with disability, and
disability-adjusted life-years for 29 cancer groups, 1990 to 2017:
A systematic analysis for the global burden of disease study. JAMA
Oncol. 5:1749–1768. 2019.PubMed/NCBI View Article : Google Scholar : (Online ahead of
print).
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Song Z, Wu Y, Yang J, Yang D and Fang X:
Progress in the treatment of advanced gastric cancer. Tumour Biol.
39(1010428317714626)2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Digklia A and Wagner AD: Advanced gastric
cancer: Current treatment landscape and future perspectives. World
J Gastroenterol. 22:2403–2414. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Shearer RF, Iconomou M, Watts CK and
Saunders DN: Functional roles of the E3 ubiquitin ligase UBR5 in
cancer. Mol Cancer Res. 13:1523–1532. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li W, Bengtson MH, Ulbrich A, Matsuda A,
Reddy VA, Orth A, Chanda SK, Batalov S and Joazeiro CA: Genome-wide
and functional annotation of human E3 ubiquitin ligases identifies
MULAN, a mitochondrial E3 that regulates the organelle's dynamics
and signaling. PLoS One. 3(e1487)2008.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Callaghan MJ, Russell AJ, Woollatt E,
Sutherland GR, Sutherland RL and Watts CK: Identification of a
human HECT family protein with homology to the Drosophila tumor
suppressor gene hyperplastic discs. Oncogene. 17:3479–3491.
1998.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Henderson MJ, Russell AJ, Hird S, Muñoz M,
Clancy JL, Lehrbach GM, Calanni ST, Jans DA, Sutherland RL and
Watts CK: EDD, the human hyperplastic discs protein, has a role in
progesterone receptor coactivation and potential involvement in DNA
damage response. J Biol Chem. 277:26468–26478. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Matsuura K, Huang NJ, Cocce K, Zhang L and
Kornbluth S: Downregulation of the proapoptotic protein MOAP-1 by
the UBR5 ubiquitin ligase and its role in ovarian cancer resistance
to cisplatin. Oncogene. 36:1698–1706. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Scialpi F, Mellis D and Ditzel M: EDD, a
ubiquitin-protein ligase of the N-end rule pathway, associates with
spindle assembly checkpoint components and regulates the mitotic
response to nocodazole. J Biol Chem. 290:12585–12594.
2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cojocaru M, Bouchard A, Cloutier P, Cooper
JJ, Varzavand K, Price DH and Coulombe B: Transcription factor IIS
cooperates with the E3 ligase UBR5 to ubiquitinate the CDK9 subunit
of the positive transcription elongation factor B. J Biol Chem.
286:5012–5022. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Munoz MA, Saunders DN, Henderson MJ,
Clancy JL, Russell AJ, Lehrbach G, Musgrove EA, Watts CK and
Sutherland RL: The E3 ubiquitin ligase EDD regulates S-phase and
G(2)/M DNA damage checkpoints. Cell Cycle. 6:3070–3077.
2007.PubMed/NCBI View Article : Google Scholar
|
|
13
|
McDonald WJ, Sangster SM, Moffat LD,
Henderson MJ and Too CK: alpha4 phosphoprotein interacts with EDD
E3 ubiquitin ligase and poly(A)-binding protein. J Cell Biochem.
110:1123–1129. 2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang Z, Zheng X, Li J, Duan J, Cui L,
Yang L, Zhang L, Zhang Q and Wang X: Overexpression of UBR5
promotes tumor growth in gallbladder cancer via PTEN/PI3K/Akt
signal pathway. J Cell Biochem, Feb 18, 2019 (Online ahead of
print).
|
|
15
|
Meissner B, Kridel R, Lim RS, Rogic S, Tse
K, Scott DW, Moore R, Mungall AJ, Marra AM, Connors JM, et al: The
E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell
lymphoma. Blood. 121:3161–3164. 2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Clancy JL, Henderson MJ, Russell AJ,
Anderson DW, Bova RJ, Campbell IG, Choong DY, Macdonald GA, Mann
GJ, Nolan T, et al: EDD, the human orthologue of the hyperplastic
discs tumour suppressor gene, is amplified and overexpressed in
cancer. Oncogene. 22:5070–5081. 2003.PubMed/NCBI View Article : Google Scholar
|
|
17
|
O'Brien PM, Davies MJ, Scurry JP, Smith
AN, Barton CA, Henderson MJ, Saunders DN, Gloss BS, Patterson KI,
Clancy JL, et al: The E3 ubiquitin ligase EDD is an adverse
prognostic factor for serous epithelial ovarian cancer and
modulates cisplatin resistance in vitro. Br J Cancer. 98:1085–1093.
2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bradley A, Zheng H, Ziebarth A, Sakati W,
Branham-O'Connor M, Blumer JB, Liu Y, Kistner-Griffin E,
Rodriguez-Aguayo C, Lopez-Berestein G, et al: EDD enhances cell
survival and cisplatin resistance and is a therapeutic target for
epithelial ovarian cancer. Carcinogenesis. 35:1100–1109.
2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Eblen ST and Bradley A: MOAP-1, UBR5 and
cisplatin resistance in ovarian cancer. Transl Cancer Res. 6 (Suppl
1):S18–S21. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yang M, Jiang N, Cao QW, Ma MQ and Sun Q:
The E3 ligase UBR5 regulates gastric cancer cell growth by
destabilizing the tumor suppressor GKN1. Biochem Biophys Res
Commun. 478:1624–1629. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Akagi T, Yoshino T, Motoi M, Takata H,
Yano S, Miyoshi I, Oka T and Ohtsuki Y: Isolation of
virus-producing transformants from human gastric cancer cell line,
HGC-27, infected with human T-cell leukemia virus type I. Jpn J
Cancer Res. 79:836–842. 1988.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Akagi T and Kimoto T: Human cell line
(HGC-27) derived from the metastatic lymph node of gastric cancer.
Acta Med Okayama. 30:215–219. 1976.PubMed/NCBI
|
|
23
|
Ding X, Zhu J and Wang X, Zhou W, Wu K,
Zhou Z, Zhou K, Wu D, Jiao J, Xia Y and Wang X: Different
cytotoxicity of disinfection by-product haloacetamides on two
exposure pathway-related cell lines: Human gastric epithelial cell
line GES-1 and immortalized human keratinocyte cell line HaCaT. Sci
Total Environ. 692:1267–1275. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ke Y, Ning T and Wang B: Establishment and
characterization of a SV40 transformed human fetal gastric
epithelial cell line-GES-1. Zhonghua Zhong Liu Za Zhi. 16:7–10.
1994.PubMed/NCBI(In Chinese).
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Agnes A, Biondi A, Laurino A, Persiani R
and D'Ugo D: Global updates in the treatment of gastric cancer: A
systematic review. Part 1: Staging, classification and surgical
treatment. Updates Surg. 72:341–353. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Socovich AM and Naba A: The cancer
matrisome: From comprehensive characterization to biomarker
discovery. Semin Cell Dev Biol. 89:157–166. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Flack JE, Mieszczanek J, Novcic N and
Bienz M: Wnt-dependent inactivation of the groucho/TLE co-repressor
by the HECT E3 ubiquitin ligase Hyd/UBR5. Mol Cell. 67:181–193.e5.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li CG, Mahon C, Sweeney NM, Verschueren E,
Kantamani V, Li D, Hennigs JK, Marciano DP, Diebold I, Abu-Halawa
O, et al: PPARγ interaction with UBR5/ATMIN promotes DNA repair to
maintain endothelial homeostasis. Cell Rep. 26:1333–1343.e7.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lee YR, Chen M and Pandolfi PP: The
functions and regulation of the PTEN tumour suppressor: New modes
and prospects. Nat Rev Mol Cell Biol. 19:547–562. 2018.PubMed/NCBI View Article : Google Scholar
|