Introduction
The non-predatory storage mite Tyrophagus
putrescentiae is distributed worldwide (1) and can be found in a variety of stored
goods, particularly in high-fat and protein-rich stores (2). In addition to causing damage to food
stores, T. putrescentiae can cause human allergic diseases
(3,4). Allergic diseases caused by T.
putrescentiae are more common in Asia (5) and Europe (6).
At present, the diagnosis and allergen immunotherapy
(AIT) of mite allergic diseases depend on the use of natural
allergen extracts. However, natural allergen extracts are
associated with a number of issues, including unpredictable amounts
of allergens and non-allergenic components in natural extracts,
which can lead to serious immediate and late side effects (7,8). The
majority of these issues in allergy diagnosis and AIT can be
circumvented with the use of recombinant allergens and peptides
(9). In order to improve the safety
of AIT, researchers have turned their attention to recombinant
allergens with modified B-cell epitopes, which can prevent IgE
binding, but with intact T-cell epitopes to produce an
immunotherapeutic benefit (10).
With regards to allergy diagnosis, studies on food allergens have
revealed the potential role of B-cell epitopes (IgE-binding
epitopes) as biomarkers for characterizing various phenotypes
(11). Thus, peptide-ELISA or
epitope-ELISA has the potential to lead to advances in the clinical
diagnosis of allergic diseases.
A better understanding of T.
putrescentiae-produced allergens may aid the diagnosis and
treatment of allergies to this mite. A series of studies on the
T. putrescentiae allergens from China have thus been
conducted. Using molecular cloning techniques, the sequencing and
characterization of group 4, 28, 35 and 36 T. putrescentiae
allergens from China were reported in our previous studies
(12,13). The T. putrescentiae group 13
allergen (Tyr p 13) was another allergic component in our series of
studies. The identification of B-cell epitopes of Tyr p 13 may be
helpful in designing sequences for more accurate and safer
peptide-based allergen diagnosis and immunotherapeutic agents.
However, the tertiary structure and epitope of Tyr p 13 remain
unclear.
The aim of the present study was to clone and
express the full-length sequence encoding Tyr p 13 from China.
Combined with the prediction analysis of Tyr p 13 structure and
B-cell epitopes, these results were intended for use in the
diagnosis and treatment of anaphylaxis caused by T.
putrescentiae and may lay the foundation for the design of
recombinant Tyr p 13 (rTyr p 13) with modified B-cell epitopes.
Materials and methods
Patient serum
Peripheral blood samples were collected from
patients with allergic asthma at the Respiratory Department, Wuxi
People's Hospital (Wuxi, China). The peripheral blood samples were
placed at room temperature for 1 h, and then centrifuged at 1,000 x
g for 10 min at 4˚C to obtain the serum. Patients had not received
any treatment and asthma diagnosis was performed following the
World Health Organization criteria (14). The diagnosis of allergy to T.
putrescentiae allergens was established on the basis of a
suggestive clinical history and positive serum IgE (sIgE) to T.
putrescentiae extracts, as determined by the Allergy Screening
test panel for atopy (Mediwiss Analytic GmbH). The inclusion
criteria were as follows: i) Allergy to T. putrescentiae
extracts with >1 year of follow-up, and ii) levels of sIgE to
T. putrescentiae extract >3 kU/l at the time of inclusion
in the study. A total of 38 patients (male:female ratio, 18:20),
with a mean age of 35.263±8.374 years, were enrolled as
serum-positive for T. putrescentiae and their serum was used
to detect specific IgE-binding to recombinant allergens. In
addition, serum from five non-atopic healthy people (malefemale
ratio=2:3), with a mean age of 35.8±8.727 years, was used as the
negative control. All participants provided written informed
consent. The study protocol was approved by the Ethics Committee
for Clinical Investigation of Wuxi People's Hospital Affiliated to
Nanjing Medical University (Wuxi, China).
T. putrescentiae culture and isolation
of adult mites
To isolate T. putrescentiae, flour and dust
were obtained from the floors of a flour storage warehouse in
Yancheng, China. The collected sample was placed in a 9-cm glass
petri dish and the sample was observed under a stereomicroscope.
The suspected pregnant T. putrescentiae were picked under a
stereomicroscope and placed separately in different culture flasks
containing culture medium for cultivation. Under the
stereomicroscope, pregnant mites moved slowly, crept on the bottom
of the petri dish, formed egg cells in the body and their body
shape was larger. After ~2 months, the whole culture in the flask
was examined under the stereomicroscope. According to the movement
speed, body type and body surface of the T. putrescentiae,
the species of mites in the culture medium could be simply judged
(15). Subsequently, mites
considered to be T. putrescentiae were cultivated in small
culture flasks for pure culture. After a further 2 months, the
mites previously isolated and cultured were identified again. The
identification of T. putrescentiae was performed in strict
accordance with the morphological characteristics of T.
putrescentiae (15). If the
mite was confirmed to be T. putrescentiae, all mites in that
flask were considered to be T. putrescentiae. During the
cultivation process, these T. putrescentiae were raised in
an atmosphere of 25˚C and 85% relative humidity. In the culture
process of T. putrescentiae, the culture medium consisted of
rice bran medium, flour and yeast powder (11). The isolation method of pure T.
putrescentiae mites from the culture medium was the same as
that of pure Dermatophagoides farinae mites (16).
Preparation of Tyr p 13 cDNA
Total RNA was obtained from T. putrescentiae
using the Takara MiniBEST Universal RNA Extraction Kit (cat. no.
9767; Takara Biotechnology Co., Ltd.). Based on the published
sequence of Tyr p 13 (GenBank accession no. DQ983316; https://www.ncbi.nlm.nih.gov/nuccore/DQ983316.1/),
a pair of primers were designed: Forward,
5'-GGAATTCCATATGTCGGTCGAAGAAC-3' and reverse,
5'-CCGCTCGAGTTAATCACCAGTCATCATCTCC-3', with an NdeI and an
XhoI site at their 5' ends (underlined), respectively.
Reverse transcription (RT)-PCR was performed using total mite RNA
and the High-Fidelity PrimeScript RT Reagent Kit with gDNA Eraser
(cat. no. DRR047A; Takara Biotechnology Co., Ltd.) on a PCR Thermal
Cycler Dice (cat. no. TP600; Takara Biotechnology Co., Ltd.). gDNA
was removed from the total RNA samples; briefly, the reaction
mixture (10 µl) contained total RNA (1 µl), 5X gDNA Eraser Buffer
(2 µl), gDNA Eraser (1 µl) and RNase-free H2O (6 µl).
The mixture was incubated at 42˚C for 2 min, and RT was performed
according to the manufacturer's instructions. PrimeScript RT Enzyme
Mix I (1 µl), RT Primer Mix (1 µl), 5X PrimeScript Buffer 2 (4 µl)
and RNase-free dH2O (4 µl) were added to the
aforementioned reaction mixture. The reaction mixture (20 µl) was
incubated at 37˚C for 15 min and 85˚C for 5 sec. The RT product was
used as the template for PCR using PrimeSTAR® HS DNA
Polymerase (cat. no. R010A; Takara Biotechnology Co., Ltd.). The
PCR mix comprised RT products (1 µl), 5X PrimeSTAR PCR buffer (10
µl), 2.5 mmol/l dNTP mixture (4 µl), 20 µmol/l forward primer (1
µl), 20 µmol/l reverse primer (1 µl), 2.5 U/µl PrimeSTAR HS DNA
polymerase (0.5 µl) and dH2O (32.5 µl). For PCR, samples
were incubated for 2 min at 94˚C, followed by 30 cycles at 98˚C for
10 sec, 65˚C for 30 sec and 72˚C for 40 sec. Finally, samples were
incubated for 10 min at 72˚C and 5 min at 10˚C, and amplicons (5
µl) were analyzed by agarose gel electrophoresis (1.0%) containing
ethidium bromide (cat. no. E7637; Sigma-Aldrich, Merck KGaA) and
visualized with ImageMaster® VDS (Pharmacia
Biotech).
Cloning and DNA sequencing
The PCR-amplified DNA was recovered from the gel
using the Agarose Gel DNA Purification kit v2.0 (cat. no. DV805;
Takara Biotechnology Co., Ltd.) and a poly-A tail was added using
the DNA A-Tailing Kit (cat. no. 6109; Takara Biotechnology Co.,
Ltd.). The poly-A tailed product was cloned into the simple vector
pMD20-T (cat. no. D107A; Takara Biotechnology Co., Ltd. according
to the manufacturer's instructions. Competent Escherichia coli
(E. coli) JM109 cells (cat. no. 9052; Takara Biotechnology Co.,
Ltd.) were transformed with the recombinant plasmid pMD20T-Tyr p 13
using the heat-shock method. Positive clones were selected by
blue/white screening on Luria-Bertani (LB) plates containing 100
µg/ml ampicillin and samples were sequenced using Bca BEST
sequencing primer RV-M/M13-47 (Takara Biotechnology Co., Ltd.) on
an ABI PRISM 377XL DNA sequencer (Applied Biosystems; Thermo Fisher
Scientific, Inc.).
DNA sequence retrieval and
analysis
The sequences were edited to remove the vector
sequence and extra restriction sites using SeqMan on DNASTAR
Lasergene software suite v7.1 (DNASTAR, Inc.). The open reading
frame (ORF) was identified using the ORF finder (http://www.bioinformatics.org/sms2/orf_find.html)
from the National Center for Biotechnology Information.
Physicochemical analysis and analysis
of patterns
The sequence of Tyr p 13 was obtained by sequencing
after pMD20T-Tyr p 13 was transformed into E. coli JM109
cells. The amino acid sequence of Tyr p 13 was predicted using
Translate Tools in ExPaSy (https://web.expasy.org/protparam/). The family
classification of rTyr p 13 was analyzed using InterPro v56.0
(www.ebi.ac.uk/interpro/) (17). The signal peptide sequence of rTyr p
13 was predicted by SignalP 4.1 software (http://www.cbs.dtu.dk/services/SignalP/), as
previously described (17).
Physiochemical analyses, including theoretical pI, aliphatic index,
grand average of hydropathicity (GRAVY) and instability index of
rTyr p 13, were predicted using ProtParam (web.expasy.org/protparam/) (17) and ProtScale tools (https://web.expasy.org/protscale/) (18). The phosphorylation sites of rTyr p
13 were determined using the NetPhos3.1 server (http://www.cbs.dtu.dk/services/NetPhos/), as
previously described (17). The
secondary structure was predicted using GOR4.0 (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html),
as previously described (19). The
TMHMM server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used to
predict transmembrane protein helices (20).
Homology modeling
Homology modeling was used to construct a tertiary
structure of Tyr p 13. A BLASTP (blast.ncbi.nlm.nih.gov/Blast.cgi) search with default
parameters was performed against the Protein Data Bank (PDB;
www.rcsb.org/pdb/) to identify a suitable
template for Tyr p 13. Based on this template, the tertiary
structure of Tyr p 13 was predicted using SWISS-MODEL (https://www.swissmodel.expasy.org/). The
generated model was evaluated using QMEAN, Global Model Quality
Estimation (GMQE), ERRAT, VERIFY_3D, PROCHECK and ProSA-Web. QMEAN
was used to to provide the global (for the entire structure) and
local (per residue) error estimates on the basis of a single model,
while GMQE was used to reflect the expected accuracy of a model
built with that alignment and template and the coverage of the
target, ERRAT (services.mbi.ucla.edu/SAVES) was used to analyze the
statistics of non-bond interactions between different atom types,
VERIFY_3D (services.mbi.ucla.edu/SAVES) was used to determine the
compatibility of the atomic model (3D) with its amino acid sequence
(1D) and compare the results to an advantageous structure, PROCHECK
(services.mbi.ucla.edu/SAVES) was used
to verify the stereochemical quality of the Tyr p 13 structure, and
ProSA-Web (https://prosa.services.came.sbg.ac.at/prosa.php) was
used to analyze whether the interaction between each residue of the
model was within a reasonable range (21). Superimposition of the query and
template structure and visualization of the generated models was
performed using UCSF Chimera 1.10.2 (www.cgl.ucsf.edu/chimera/).
Structure-based prediction of Bcell
epitopes
Although the reliable prediction of epitopes is
challenging, the prediction of allergen epitopes is beneficial to
the immunological diagnosis of allergic diseases. Linear and
discontinuous B-cell epitopes were predicted using ElliPro:
Antibody Epitope Prediction; IEDB analysis resource (http://tools.immuneepitope.org/tools/ElliPro/tutorial.jsp)
(22). The rTyr p 13 protein
sequence was analyzed using different analytical parameters, such
as secondary structure, hydropathy, antigenicity, amphilicity,
surface probability and flexibility in the Protean software of the
DNASTAR Lasergene software suite v7.1(23).
Construction of expression plasmids
pET28a(+)-Tyr p 13
The recombinant pMD20-T-Tyr p 13 plasmid was
digested with NdeI and XhoI to release the Tyr p 13
cDNA. The cDNA was separated by agarose gel electrophoresis and
purified from the gel using the Agarose Gel DNA Purification kit
v2.0. The DNA Ligation kit (cat. no. D6023; Takara Biotechnology
Co., Ltd.) was used to sub-clone the cDNA into the expression
vector pET28a(+) (cat. no. N72770; Novagen; Merck KGaA) to create
pET28a(+)-Tyr p 13. E. coli DH5α cells (cat. no. D9057;
Takara Biotechnology Co., Ltd.) were transformed with pET28a(+)-Tyr
p 13 plasmids. Positive clones were selected by blue/white
screening on LB plates containing 50 µg/ml kanamycin and verified
by restriction enzyme analysis with NdeI and
XhoI.
Production and purification of
recombinant allergen
The pET-28a (+)-Tyr p 13 plasmid (0.5 µl) was
purified using the MiniBEST Plasmid Purification Kit Ver. 4.0 (cat.
no. 9760; Takara Biotechnology Co., Ltd.) and used to transform 100
µl E. coli BL21 (DE3) cells (cat. no. 200131; Agilent
Technologies, Inc.). The pET28a(+)-Tyr p13-transformed E.
coli BL21 cells were cultured in 3 ml LB liquid medium
containing 50 µg/ml kanamycin for 8 h at 37˚C. After 8 h of cell
culture, 2 ml cell culture was diluted into 100 ml LB + kanamycin
in a glass tube and cultured at 37˚C until an approximate
absorbance of 0.5 at 600 nm was reached.
Isopropyl-β-D-thiogalactopyranoside was added to a final
concentration of 0.1 mmol/l. The sample was incubated for 2 h at
30˚C. E. coli cells were then harvested by centrifugation
(8,000 x g) at 4˚C for 10 min, resuspended in 15 ml Tris-HCl (pH
7.5) buffer and sonicated (20 kHz; ultrasound duration 3 sec,
interval 10 sec) in an ice bath until the suspension became
transparent. The cell debris and supernatant were separated by
centrifugation (5,000 x g) at 4˚C for 40 min. The presence of the
recombinant protein was verified by 10% SDS-PAGE and Coomassie
Brilliant Blue R-250 (CBB-R250) staining.
The supernatant was applied to a Ni-NTA affinity
chromatography column (cat. no. 70971; Novagen; Merck KGaA)
pre-equilibrated with buffer [50 mmol/l Tris-HCl (pH 7.5), 0.3
mol/l NaCl]. The column was washed with 10 ml washing buffer 1 [50
mmol/l Tris-HCl (pH 7.5), 10 mmol/l imidazole and 0.3 mol/l NaCl]
to remove unbound proteins. Recombinant protein was eluted by a
gradient of increasing concentrations of imidazole elution buffer.
The elution flow rate was 1 ml/min. Eluents at different imidazole
concentrations were collected and verified by 10% SDS-PAGE. The
eluents containing high purity recombinant protein samples were
mixed and desalinated by 12-h dialysis with several rounds of
buffer replacement. The purified product was separated by 10%
SDS-PAGE and observed by Coomassie Brilliant Blue R-250 (CBB-R250)
staining.
Specific IgE-binding to recombinant
allergen
The ability of rTyr p 13 to bind to IgE was assessed
by ELISA. The rTyr p 13 used for ELISA was purified by Ni NTA
affinity chromatography. For antigen coating, 100 µl rTyr p 13 (1
µg/ml) diluted in coating buffer (0.1 mol/l PBS)was added to a
96-well microtiter plate and incubated at 8˚C overnight. Wells were
washed with 300 µl PBS-0.05%Tween-20 (PBS-T) buffer and blocked
with 3% bovine serum albumin (cat. no. A1933, Sigma-Aldrich; Merck
KGaA) in PBS-T for 1 h at 37˚C. Subsequently, 100 µl patient serum
and 100 µl healthy donor serum (diluted at 1:4 with PBS-T) were
added, followed by overnight incubation at 4˚C. Secondary antibody
(50 µl; HRP-mouse anti-human IgE; 1:5,000; Sigma-Aldrich; Merck
KGaA) was added and samples were incubated at 37˚C for 1 h. To
detect binding, 100 µl TMB substrate solution (cat. no. P0209;
Beyotime Institute of Biotechnology) was added and samples were
incubated for 15 min at room temperature. A total of 2 mol/l
H2SO4 (50 µl) was added to stop the
reactions. Optical density (OD) was measured for triplicate samples
at a wavelength of 450 nm using an iMark Microplate Absorbance
Reader (Bio-Rad Laboratories, Inc.). The reactivity was considered
positive if the OD values of the detected serum were higher than
the cutoff ELISA value (mean ELISA value of healthy donors + 3
SD).
Results
Cloning Tyr p 13
Total RNA was isolated from adult mites, and Tyr p
13 cDNA was amplified by RT-PCR. A product of the expected size
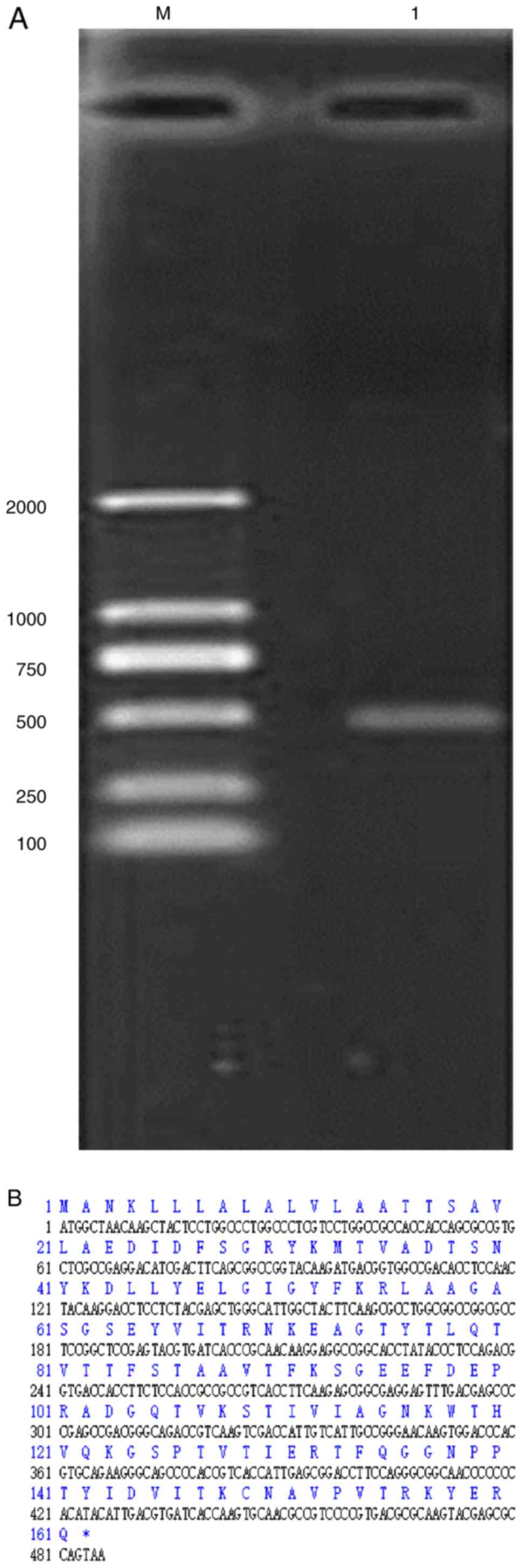
(486 bp) was produced (Fig. 1A).
The product was cloned into the pMD20T vector and sequenced. Using
the ORF Finder, a complete ORF was identified in the Tyr p 13 cDNA.
The length from the start codon ATG to the stop codon TAA was 486
bp (Fig. 1B).
Inferred amino acid sequence, and its
structural and functional prediction
Family classification revealed that Tyr p 13 belongs
to the intracellular lipid-binding protein family. An amino acid
sequence of 161 residues was predicted for Tyr p 13 using the
Translate Tools on the ExPaSy web server. A signal peptide sequence
from amino acids 1 to 22 was predicted using SignalP 4.1 software.
The removal of the signal peptide sequence predicted by the SignalP
4.0 software (Fig. S1) yielded a
predicted mature Tyr p 13 protein of 139 amino acid residues with a
theoretical pI of 8.07. The instability index was 26.7, indicating
a stable amino acid sequence. The grand average of GRAVY was -0.49,
indicating that Tyr p 13 was hydrophilic. The NetPhos3.1 server was
used to search the Tyr p 13 amino acid sequence for phosphorylation
sites. Nineteen sites were predicted (Fig. S2), including seven serine sites
positioned at residues 6, 17, 39, 63, 71, 87 and 103, nine
threonine sites at residues 12, 53, 58, 61, 68, 84, 97, 107 and
133, and three tyrosine sites at residues 19, 43 and 120. The
secondary structure of Tyr p 13 was predicted to comprise an
α-helix (15 peptides, 10.79%), an extension chain (47 peptides,
33.81%) and a random coil (77 peptides, 55.40%; Fig. 2). The Tyr p 13 protein sequences
were entered into the TMHMM Server 2.0, which predicted no
transmembrane helices, and inferred that all protein sequences were
located outside the membrane.
Homology modeling and model
evaluation
The fatty acid-binding protein in brain tissue of
Drosophila melanogaster (PDB accession no. 5GKB) belongs to
the intracellular lipid-binding protein family (InterPro no.
IPR031259), has a high sequence identity with Tyr p 13 (36.43%),
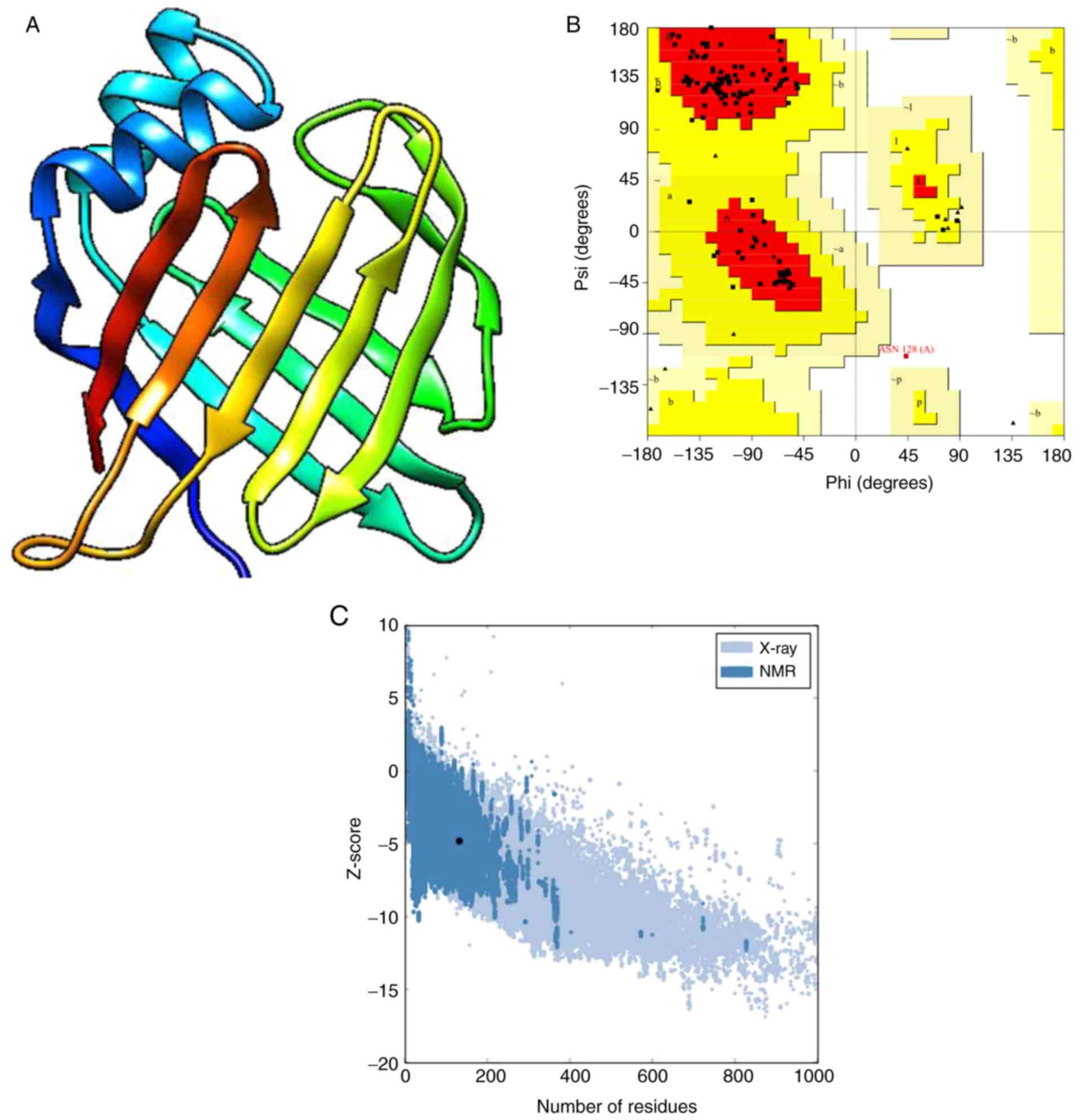
and was used as the template for homology modeling (Fig. 3A). In the Ramachandran plot, 96.6%
of the residues were in favored regions, 2.6% were in additional
allowed parts, 0.8% were in generously allowed regions and 0%
percent were in outlier regions (Fig.
3B). Based on the ERRAT results, the overall quality factor was
96.64%, indicating that the tertiary structure of Tyr p 13 had a
high resolution. The Verify3D test requires an average 3D-1D score
of ≥80% of the amino acids to be ≥0.2. The VERIFY 3D results showed
that 84.44% of residues had an average 3D1D score of ≥0.2, which
suggested that the structures were favorable. ProSA analysis
revealed that the predicted model was comparable to other
acceptable proteins with a z score of -4.78 (Fig. 3C). The QMEAN server results revealed
that the QMEAN Zscore was 1.61, thus suggesting that the protein
model variation rate was low, overall folding and local structures
had high accuracy rates, and stereochemistry was reasonable.
Furthermore, the Q value was 0.7, which indicated that the
predicted model of Tyr p 13 was reliable. These findings revealed
that the tertiary structure model of Tyr p 13 was reliable and was
suitable for use in the current study.
Prediction of B-cell epitopes by an
integrated strategy
α-Helices and β-sheets have been reported to have
high chemical bond energies and to be unlikely to form epitope
sequences (24). By contrast,
β-turns and random coils are located in surface-exposed regions of
a protein, which often contain epitope sequences (21). The secondary structure,
hydrophilicity, antigenicity, amphilicity, surface probability and
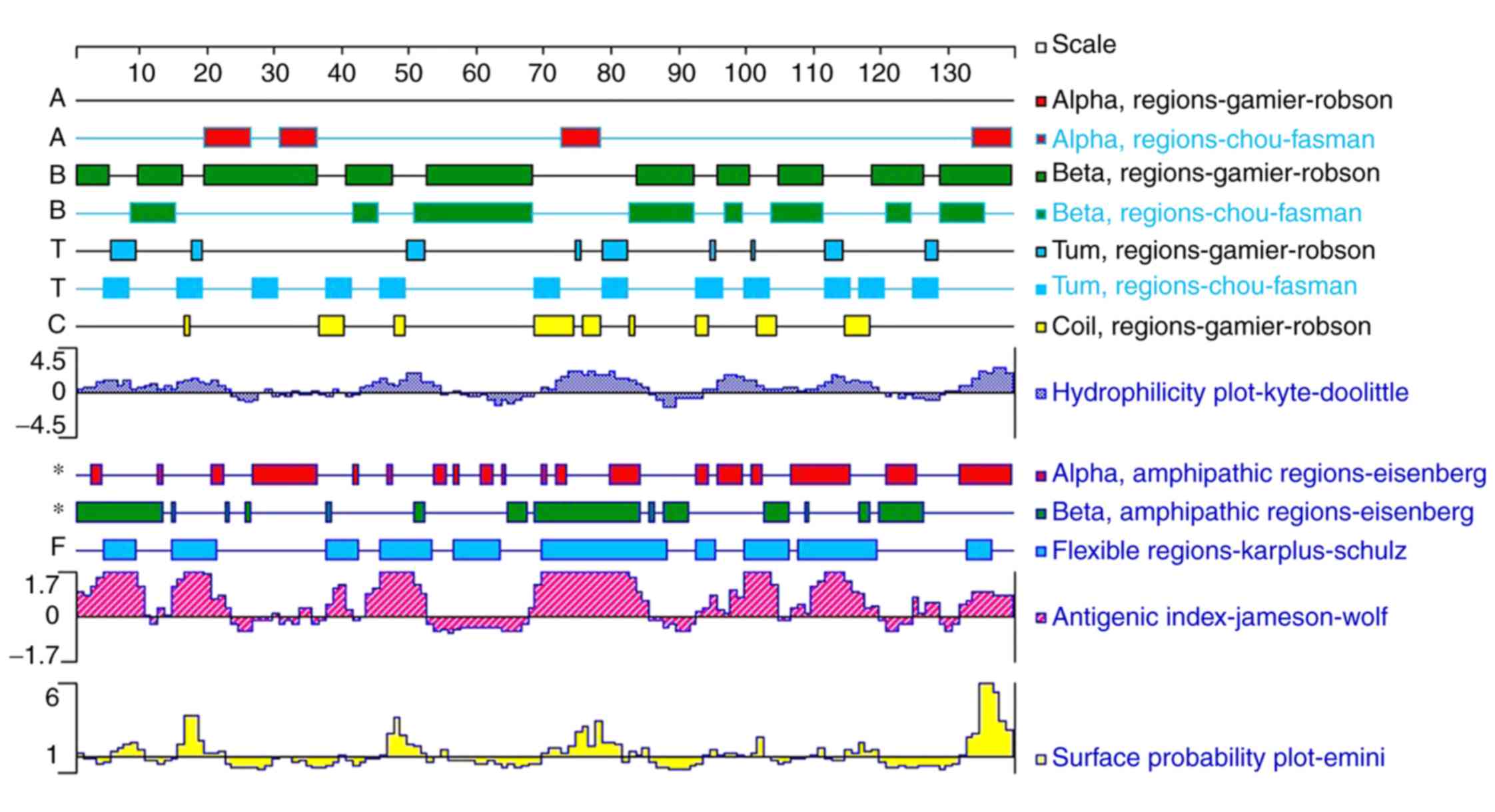
flexibility of Tyr p 13 were analyzed using the Protean Software
(for Protein Structure Analysis and Prediction) of the DNASTAR
Lasergene software suite Ver. 7.1 (Fig.
4). Based on these results, linear and discontinuous B-cell
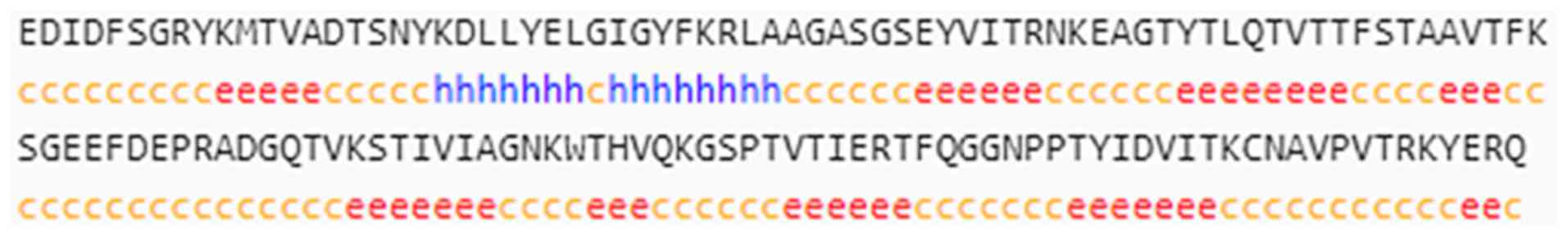
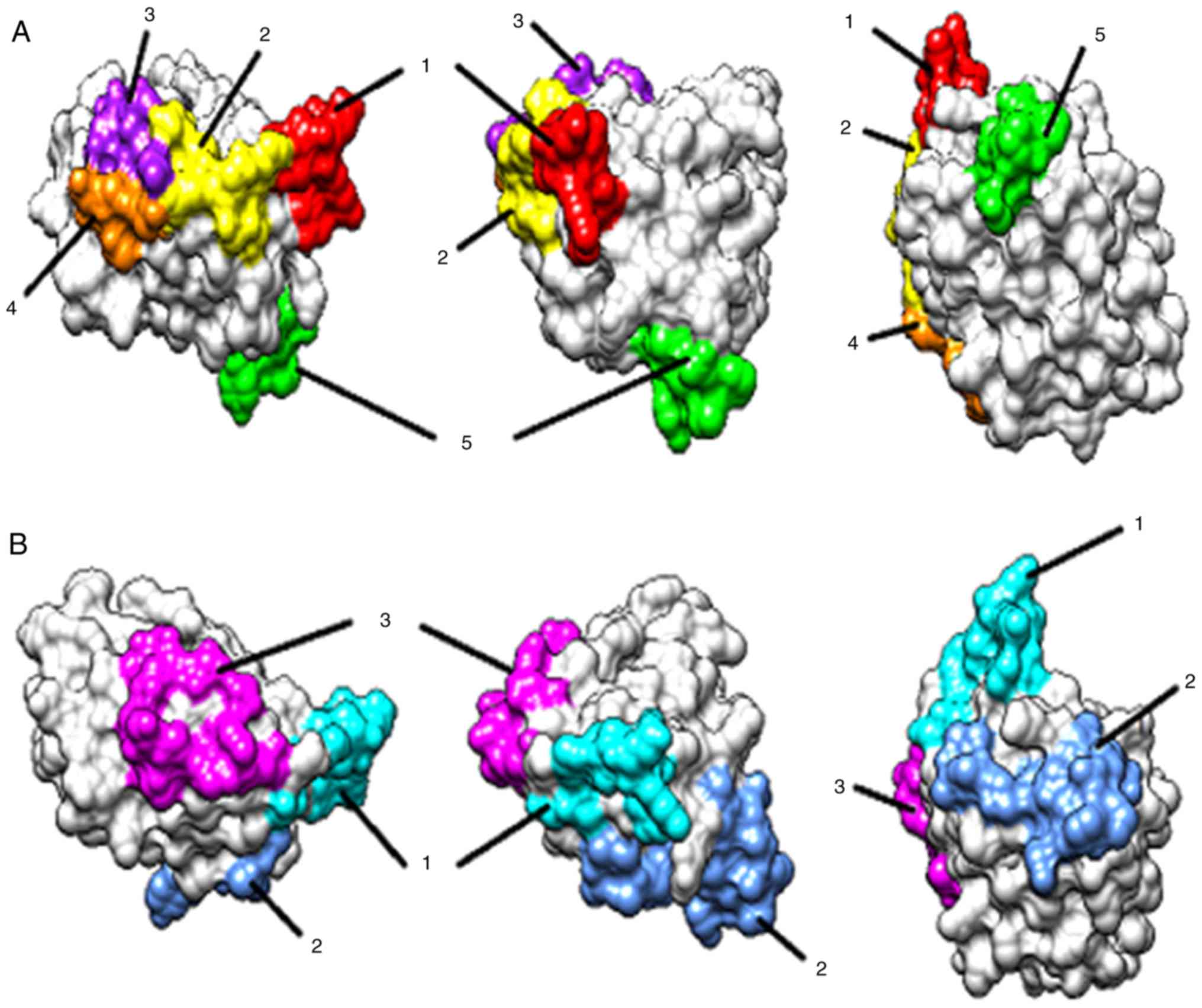
epitopes were predicted by ElliPro software (Table I). Five amino acid peptide sequences
were identified as promising linear epitopes (Fig. 5A) and three clusters were predicted
to form discontinuous epitopes (Fig.
5B).
 | Table IPredicted Tyrophagus
putrescentiae group 13 allergen Bcell epitopes. |
Table I
Predicted Tyrophagus
putrescentiae group 13 allergen Bcell epitopes.
| Epitopes | Peptide and
position | Number of
residues |
|---|
| Predicted linear
epitopes | | |
|
1 | RNKEAGT
(47-53) | 7 |
|
2 | KSGEEFD
(70-76) | 8 |
|
3 | DGQTVK (81-86) | 7 |
|
4 | KGSPT
(101-105) | 5 |
|
5 | FQGGNPPTY
(112-120) | 9 |
| Predicted
discontinuous epitopes | | |
|
1 | R 47, N 48, K 49, E
50, A 51, G 52, T 53, K 70, S 71, G 72, E 73 | 11 |
|
2 | I 91, A 92, G 93, N
94, F 112, Q 113, G 114, G 115, N 116, P 117, P 118, T 119, Y 120,
I 121, R 138 | 15 |
|
3 | E 74, D 76, R 79, D
81, G 82, Q 83, T 84, K 86, K 101, G 102, S 103, P 104, T 105 | 13 |
Expression and purification of
recombinant allergen (rTyr p 13)
Tyr p 13 cDNA was sub-cloned into the pET-28a (+)
vector to produce a recombinant His-tagged protein. The recombinant
His-tagged protein (rTyr p 13) was successfully purified by Ni-NTA
affinity chromatography (Fig.
6).
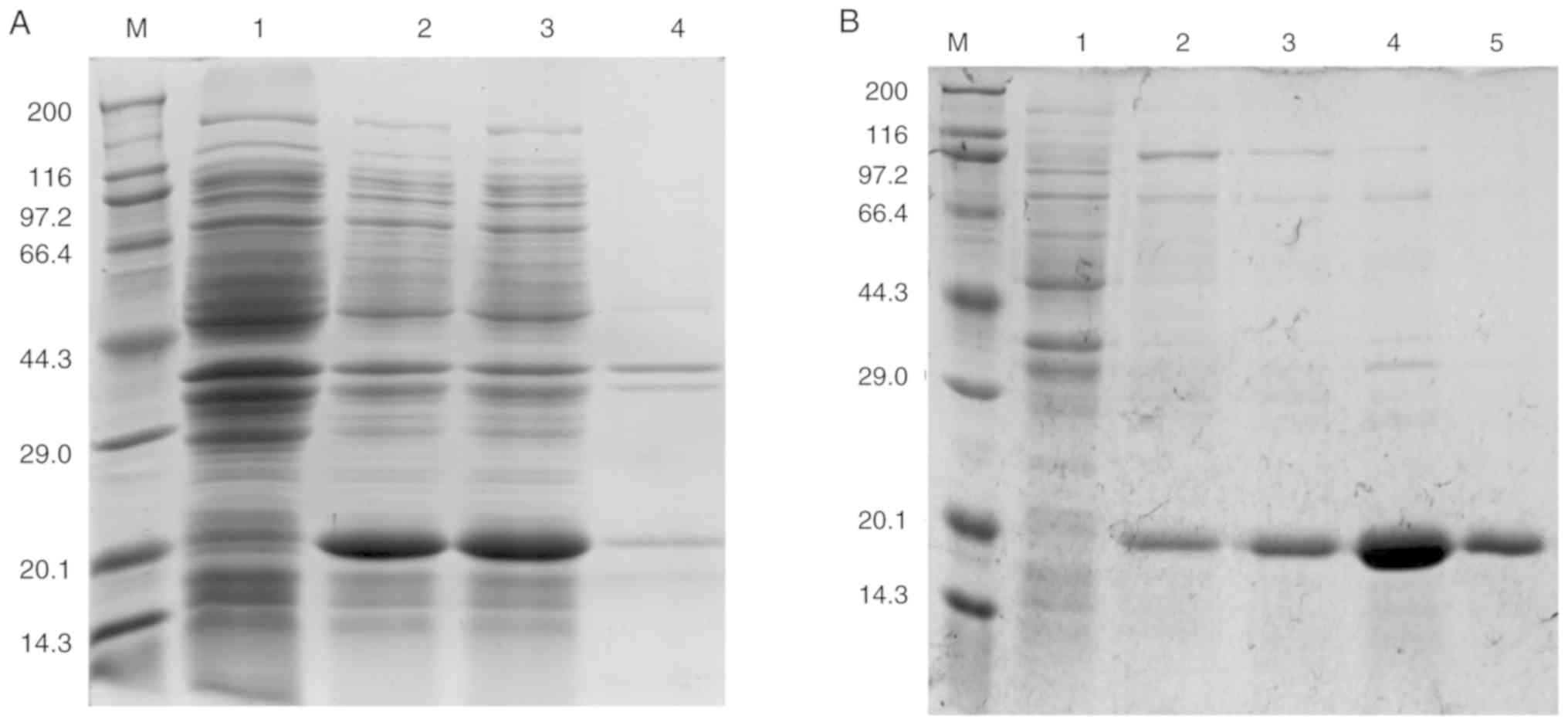 | Figure 6Expression of rTyr p 13 in E.
coli BL21 cells. (A) SDS-PAGE of rTyr p 13 expressed in E.
coli BL21 cells. Lane M, Takara Protein Marker (Broad); Lane 1,
whole cell lysate of E. coli BL21 cells containing pET-28a
as a negative control; Lane 2, whole-cell lysate of E.coli
BL21 cells containing pET-28a (+)-Tyr p 13; Lane 3, supernatant of
cells containing pET-28a (+)-Tyr p 13; Lane 4, pellet of cells
containing pET-28a (+)-Tyr p 13. (B) SDS-PAGE of purified rTyr p
13. Lane M, Takara Protein Marker (Broad); Lane 1, protein
flow-through from the column; Lanes 2, 3, 4 and 5, eluted fractions
with 0, 10, 50 and 250 mmol/l imidazole elution buffer,
respectively. rTyr p 13, recombinant Tyrophagus
putrescentiae group 13 allergen. |
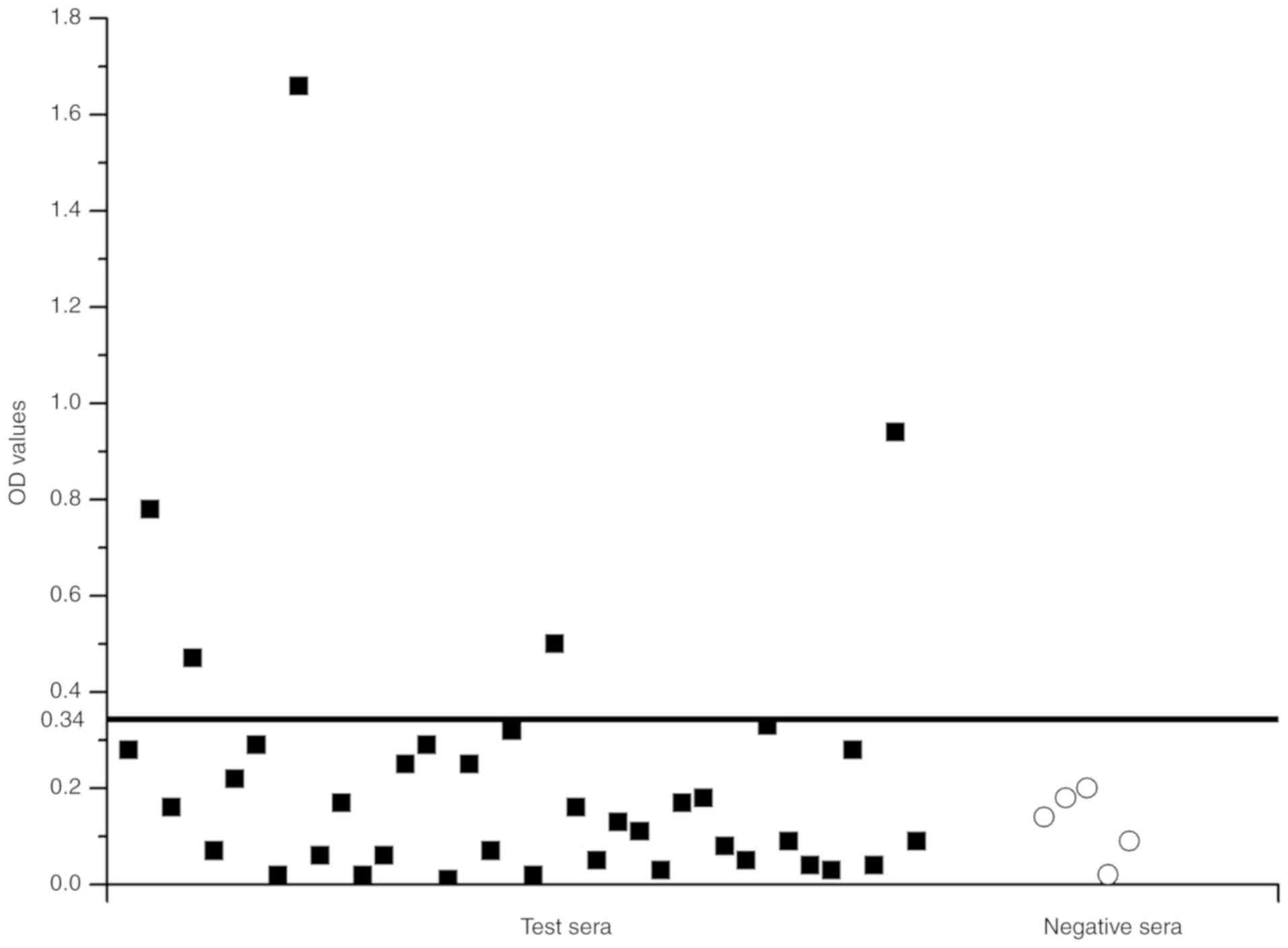
IgE reactivity to rTyr p 13
ELISA was used to determine whether E.
coli-produced rTyr p 13 had an IgE-binding ability with
purified rTyr p 13 as a coat antigen. The mean OD value of the five
healthy donors was 0.12 and the standard deviation (SD) was 0.0726.
The postulated cutoff value was 0.34 (the mean OD value of healthy
donors + three SD) (Fig. 7). Those
with OD values >0.34 were judged to be IgE-positive. Positive
IgE reactions to rTyr p 13 were detected in the serum of 13.2%
(5/38) of the T. putrescentiae-allergic patients. The
results indicated that rTyr p 13 had the ability to bind to
IgE.
Discussion
Allergic diseases caused by T. putrescentiae,
a type of storage mite, are common in several countries worldwide,
including China. The positive skin prick test prevalence for T.
putrescentiae in 2012 was 63% in Guangzhou, China (25). A better understanding of T.
putrescentiae allergens may aid the diagnosis and treatment of
T. putrescentiae allergies. For example, polymorphisms have
been described for several storage mite allergens from different
regions (25,26). Polymorphisms can have an important
effect on the epitopes recognized by T lymphocytes, monoclonal
antibodies and IgE of allergic patients (27). Therefore, in the present study, cDNA
encoding Tyr p 13 was amplified from mites collected from a flour
storage warehouse in China; subsequently, rTyr p 13 was expressed
in an E. coli expression system and was purified by affinity
chromatography.
Bioinformatics tools are crucial to allergy
research, particularly the characterization of allergens by
identification of structural motifs and epitopes and are
complementary to experimental studies of allergens. Bioinformatics
analysis of the sequence of Tyr p 13 revealed it to be a
hydrophilic and stable protein, with no transmembrane helices and
all protein sequences located outside the membrane. In addition,
homology modeling based on a template was used to predict the
tertiary structure of rTyr p 13. The key to homology modeling is to
identify the right template; notably, quality of the homology model
is dependent on high quality sequence alignment and template
structure. The fatty acid-binding protein in brain tissue of D.
melanogaster has a high sequence identity with Tyr p 13
(36.43%), and both proteins belong to the intracellular
lipid-binding protein family (IPR031259). Therefore, the fatty
acid-binding protein in brain tissue of D. melanogaster can
be used as a modeling template. Following homology modeling using
SWISS-MODEL, various additional parameters/programs were
incorporated to establish a reliable model of Tyr p 13. Future work
on these features can improve our understanding of Tyr p 13.
The specific interaction of allergens with IgE
antibodies is a key event in allergic diseases. The present study
assessed the allergenicity of rTyr p 13 and revealed that rTyr p 13
bound with serum from 13.2% (5/38) of patients allergic to T.
putrescentiae, according to the results of ELISA. Furthermore,
studies have determined a correlation between the severity or
persistence of allergic diseases and the diversity of B-cell
epitopes (IgE-binding epitopes) (27). The identification of B-cell epitopes
of allergens is valuable for accurate and safe peptide-based
allergen diagnosis, such as peptide-ELISA or epitope-ELISA, and
immunotherapy. The in-silico prediction of B-cell epitopes
is considered a useful tool for selecting B-cell epitopes from
immunologically relevant proteins and has been reported to be well
correlated with the experimental approach (23). In most cases, B-cell epitopes are
located on the surface of antigen molecules. Secondary and tertiary
protein structures also contain important information regarding
B-cell epitope prediction. For example, β-turns and random coils
are detected in surface-exposed protein regions of, which often
contain epitope sequences (21).
Numerous algorithms have been generated to predict B-cell epitopes
on protein sequences; these algorithms are based on the propensity
values of the amino-acid properties of hydrophilicity,
antigenicity, flexibility and accessibility. By integrating DNAStar
Protean and ElliPro analysis results, and combining information
from secondary and tertiary structures, the present study
identified potential B-cell linear and discontinuous epitopes.
However, further experimental verification is required for these
predicted epitopes. Therefore, the findings of the present study
may be useful not only for further work on Tyr p 13 but may also
lay the foundation for the study of peptide-based allergen
diagnosis and immunotherapy.
In conclusion, the present study demonstrated that
the cloning, expression, characterization and B cell epitopes of
recombinant Tyr p 13 protein provided initial evaluation of its
potency as an allergen in mite-allergic individuals. These findings
provided a foundation for which to explore the structural biology
and biochemistry of Tyr p 13 protein, thereby enabling future work
with ASIT.
Supplementary Material
Signal peptide prediction (eukaryotic
networks) of Tyrophagus putrescentiae group 13 allergen by
SignalP 4.0 software.
Phosphorylation site analysis of
Tyrophagus putrescentiae group 13 allergen using NetPhos 3.1
Server.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Sciences
Foundation of China (grant nos. NSFC30060166, NSFC81001330 and
NSFC31272369).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
NW and YC conceived and designed the study. NW and
YZ performed the experiments and wrote the manuscript. YC revised
the manuscript critically. MW and HZ collected the patients'
samples and analyzed the data. All authors contributed to the
preparation of the final manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All participants provided written informed consent.
The present study was approved by the Ethics Committee for Clinical
Investigation of Wuxi People's Hospital Affiliated to Nanjing
Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jeong KY, Lee H, Lee JS, Lee J, Lee IY,
Ree HI, Hong CS and Yong TS: Molecular cloning and the allergenic
characterization of tropomyosin from Tyrophagus
putrescentiae. Protein Pept. Lett. 14:431–436. 2007.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Aygun O, Yaman M and Durmaz H: A survey on
occurrence of Tyrophagus putrescentiae (Acari: Acaridae) in
Surk, a traditional Turkish dairy product. J Food Eng. 78:878–881.
2007.
|
|
3
|
Uzunoğlu Karagöz E, Akdemir C, Direkel Ş
and Cebeci Guler N: The investigation of the presence of mites in
some served dry foodstuffs. Turkiye Parazitol Derg. 41:92–95.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Liao EC, Hsu EL, Tsai JJ and Ho CM:
Immunologic characterization and allergenicity of recombinant Tyr p
3 allergen from the storage mite Tyrophagus putrescentiae. Int Arch
Allergy Immunol. 150:15–24. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mondal P, Dey D, Sarkar T, Laha A, Moitra
S, Bhattacharyya S, Saha NC, Saha GK and Podder S: Evaluation of
sensitivity toward storage mites and house dust mites among
nasobronchial allergic patients of kolkata, india. J Med Entomol.
56:347–352. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Szilman E, Szilman P, Solarz K,
Brewczyński P and Sieroń AL: Sensitization to the storage mite
Tyrophagus putrescentiae in urban population of Upper
Silesia (Poland). Wiad Parazytol. 50:471–476. 2004.PubMed/NCBI
|
|
7
|
Tabesh S, Fanuel S, Fazlollahi MR,
Yekaninejad MS, Kardar GA and Razavi SA: Design and evaluation of a
hypoallergenic peptide-based vaccine for Salsola kali allergy. Int
Immunopharmacol. 66:62–68. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Niederberger V and Valenta R: Molecular
approaches for new vaccines against allergy. Expert Rev Vaccines.
5:103–110. 2006.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ramos JD, Valmonte GR and de Guia RM:
Recombinant proteins and peptides as diagnostic and therapeutic
reagents for arthropod allergies. Protein Pept Lett. 14:992–1002.
2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yang L and Kulis M: Hypoallergenic
proteins for the treatment of food allergy. Curr Allergy Asthma
Rep. 19(15)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cui Y: Immunoglobulin E-binding epitopes
of mite allergens: from characterization to immunotherapy. Clin Rev
Allergy Immunol. 47:344–353. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Teng FX, Huang HF, Ge DZ, Yu LL, Xu C and
Cui YB: Tyrophagus putrescentiae group 4 allergen
allergenicity and epitope prediction. Allergol Immunopathol (Madr):
2020.
|
|
13
|
Cui Y, Yu L, Teng F, Zhang C, Wang N, Yang
L and Zhou Y: Transcriptomic/proteomic identification of allergens
in the mite Tyrophagus putrescentiae. Allergy. 71:1635–1639.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Nicklas RA: National and international
guidelines for the diagnosis and treatment of asthma. Curr Opin
Pulm Med. 3:51–55. 1997.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li Y, Guifang M, Qisong L, Yubao C and
Jinfang S: Use of a multimedia diagnostic microscope to
morphologically identify adult Tyrophagus putrescentiae. J
Pathogen Biology. 13:164–167. 2018.
|
|
16
|
Cui YB, Zhou P, Peng JL, Peng M, Zhou Y,
Lin YZ and Liu L: Cloning, sequence analysis, and expression of
cDNA coding for the major house dust mite allergen, Der f 1, in
Escherichia coli. Braz J Med Biol Res. 41:380–388.
2008.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Mitchell A, Chang HY, Daugherty L, Fraser
M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S,
et al: The InterPro protein families database: The classification
resource after 15 years. Nucleic Acids Res. 43:D213–D221.
2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
de Castro E, Sigrist CJ, Gattiker A,
Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A and Hulo
N: ScanProsite: Detection of PROSITE signature matches and
ProRule-associated functional and structural residues in proteins.
Nucleic Acids Res. 34:W362–W365. 2006.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Cui YB, Yu LL, Teng FX, Wang N, Zhou Y,
Yang L and Zhang CB: Dust mite allergen Der f 4: Expression,
characterization, and IgE binding in pediatric asthma. Pediatr
Allergy Immunol. 27:391–397. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Krogh A, Larsson B, von Heijne G and
Sonnhammer EL: Predicting transmembrane protein topology with a
hidden Markov model: Application to complete genomes. J Mol Biol.
305:567–580. 2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Teng F, Yu L, Sun J, Wang N and Cui Y:
Homology modeling and prediction of Bcell and Tcell epitopes of the
house dust mite allergen Der f 20. Mol Med Rep. 17:1807–1812.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shahsavani N, Sheikhha MH, Yousefi H and
Sefid F: In silico homology modeling and epitope prediction of NadA
as a potential vaccine candidate in neisseria meningitidis. Int J
Mol Cell Med. 7:53–68. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tong X, Guo M, Jin M, Chen H, Li Y and Wei
JF: In silico epitope prediction, expression and functional
analysis of Per a 10 allergen from the American cockroach. Int J
Mol Med. 38:1806–1814. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sikic K, Tomic S and Carugo O: Systematic
comparison of crystal and NMR protein structures deposited in the
protein data bank. Open Biochem J. 4:83–95. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang C, Li J, Lai X, Zheng Y, Gjesing B,
Spangfort MD and Zhong N: House dust mite and storage mite IgE
reactivity in allergic patients from Guangzhou, China. Asian Pac J
Allergy Immunol. 30:294–300. 2012.PubMed/NCBI
|
|
26
|
Zakzuk J, Jiménez S, Cheong N, Puerta L,
Lee BW, Chua KY and Caraballo L: Immunological characterization of
a Blo t 12 isoallergen: identification of immunoglobulin E
epitopes. Clin Exp Allergy. 39:608–616. 2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cui Y, Zhou Y, Wang Y, Ma G and Yang L:
The group 10 allergen of Dermatophagoides farinae (Acari:
Pyroglyphidae): cDNA cloning, sequence analysis, and expression in
Escherichia coli BL21. J Med Entomol. 50:205–208.
2013.PubMed/NCBI View Article : Google Scholar
|