Introduction
Colorectal carcinoma is one of the major
malignancies that has a serious impact on human morbidity and
mortality (1). The Wnt signaling
pathway regulates a number of biological processes, ranging from
cell fate decision, cell differentiation, embryonic development and
carcinogenesis (2). Abnormal
activation of the Wnt signaling pathway has been identified in
numerous types of solid tumors, particularly in colorectal cancer
(CRC) (3), and dysregulation of the
Wnt signaling pathway has been found in 90% of CRC case (4,5).
Activation of the Wnt signaling pathway by both genetic and
epigenetic mechanisms is important for both the initiation and
progression of CRC. The Wnt/β-catenin signaling pathway results in
diverse downstream intracellular events, such as driving the
transcription of target genes, including c-myc, cyclin D and
surviving (2). The targeted
inhibition of this pathway at its early stages is a reasonable and
promising strategy for the development of CRC therapies (2).
Secreted frizzled-related proteins (sFRPs) are a
family of secreted proteins (sFRP1-5) and are inhibitors of the Wnt
signaling pathway. Several members, including sFRP1, sFRP2 and
sFPR4, possess a conserved frizzled (Fzd) receptors type
cysteine-rich domain that binds Wnt and typically antag onizes Wnt
signaling, presumably by preventing Wnt/Fzd interactions (6,7). Thus,
sFRPs, excluding sFRP3, function as antagonists of Wnt signaling by
competing with Wnt proteins via binding their receptor, Frizzled.
Moreover, sFRPs attenuate Wnt signaling even in the presence of
downstream gene mutations (8,9).
Therefore, silencing sFRP genes may be essential for the aberrant
activation of the Wnt pathway in colorectal tumorigenesis.
sFRP4, a protein with 346 amino acids, is the
largest member of the sFRP family and is expressed in various
tissue types, including endometrial stroma pancreas, stomach,
colon, lung, skeletal muscle, testis, ovary, kidney, heart, brain,
mammary gland, cervix, eye, bone, prostate and liver (10). Previous studies have suggested that
sFRP4 may serve a role as a tumor suppressor and function as a
modulator of cell proliferation in ovarian, prostate and breast
cancer (10,11). A previous study found that sFRP4 is
downregulated in 26.4% of colorectal carcinoma samples (P=0.023)
and in 9.1% of colorectal adenoma tissue (P=0.438) (12). The downregulation of sFRP4 is
more frequent in carcinomas than in adenomas, and sFRP4 was
hypermethylated in 36.1% (26/72) of colorectal carcinomas; 24.2%
(8/33) of colorectal adenomas and 2.6% (1/38) of adjacent normal
mucosas (10,12). The expression levels and the
underlying mechanisms of action behind the epigenetic regulation of
sFRP4 in CRC remain to be elucidated.
DNA methylation is one of the most well understood
epigenetic regulation mechanisms. Gene silencing by promoter DNA
hypermethylation is an important characteristic of colorectal
tumors (13). Aberrant
hypermethylation of the CpG islands in the gene promoter regions
has been found to be a primary mechanism of action behind the
inactivation of several tumor suppressor genes (14). DNA methylation is highly stable
across cell generations and promoter DNA hypermethylation can
silence gene expression without affecting the DNA sequences
(15). DNA methylation is catalyzed
by DNA methyltransferases, including DNMT1, DNMT3A and DNMT3B
(16). DNMT3A and DNMT3B are de
novo DNA methyltransferases and add methyl groups to the
unmethylated CpG sites, while DNMT1 is a DNA methyltransferase that
converts the hemi-methylated DNA into fully methylated DNA during
cell mitosis (15).
Polycomb group (PcG) proteins are a group of
proteins that were first discovered in fruit flies, that can
remodel chromatin, such that epigenetic silencing of genes takes
place. Direct interactions between PcG proteins and the DNA
methylation machinery have been reported, and co-occupations were
further supported by chromatin immunoprecipitation (ChIP)
experiments in cancer cells (17,18).
The aim of the present study was to analyze the epigenetic
regulation mechanisms of action for sFRP4, to determine whether a
specific PcG protein could help to establish the promoter DNA
methylation status of sFRP4, and finally to investigate approaches
to alter the expression levels of sFRP4.
Materials and methods
Cell culture
CRC cell lines SW1116 (Dukes A) (19), SW480 (Dukes B) and HCT116 (Dukes C)
were readily available in the present laboratory (Gastroenterology
Laboratory in Zhongnan Hospital of Wuhan University, purchased from
Wuhan University, Wuhan, China). The human embryo intestinal mucosa
cell line CCC-HIE-2 was purchased from the China Infrastructure of
Cell Line Resources. CRC cells were cultured in RPMI (HyClone; GE
Healthcare Life Sciences) supplemented with 10% FBS (HyClone; GE
Healthcare Life Sciences) and 1% penicillin/streptomycin (HyClone;
GE Healthcare Life Sciences). CCC-HIE-2 cells were cultured in DMEM
(HyClone; GE Healthcare Life Sciences) supplemented with 20% FBS,
0.01 mg/ml insulin (Shanghai No. 1 Biochemical Pharmaceutical Co.,
Ltd.), 10 ng/ml epidermal growth factor (EGF) and 1%
penicillin/streptomycin. All cells were incubated at 37˚C in the
presence of 5% CO2.
Methylation-specific PCR
The DNA methylation levels of the sFRP4 promoter
were assessed using methylation-specific PCR in several CRC cell
lines that represent different stages of CRC progression, according
to the Dukes classification: SW1116 (Dukes A), SW480 (Dukes B),
HCT116 (Dukes C) cells and the normal control cells CCC-HIE-2. DNA
was extracted from SW1116, SW480, HCT116 and CCC-HIE-2 cells using
the TIANamp Genomic DNA kit (Tiangen Biotech Co., Ltd.) according
to the manufacturer's instructions. Subsequently, the DNA was
treated with sodium bisulfite from the EZ Methylation-Gold™ kit
(Zymo Research Corp.) according to the manufacturer's instructions.
After bisulfite conversion, hot start DNA polymerase
(GoldStar® Taq DNA Polymerase; CWBio) was used to
amplify the converted DNA a the following thermocycling conditions:
30 cycles of 94˚C for 60 sec, 37˚C for 60 sec and 72˚C for 2 min. A
total of 10 µl PCR products were resolved and analyzed using
electrophoresis on a 3% agarose gel. The primers and their
annealing temperatures for the methylated and unmethylated
sequences are summarized in Table
I. sFRP1 is widely reported as frequently methylated and
silenced in CRC (20-22);
therefore, this was used as the methylation positive control in the
present study.
 | Table IPrimer sequences for methylation
specific PCR and RT-qPCR. |
Table I
Primer sequences for methylation
specific PCR and RT-qPCR.
| sFRP4 primers | Primer sequence
(5'→3') | Product length,
base pairs | Annealing
temperature, ˚C |
|---|
| Unmethylated | F:
GGGGGTGATGTTATTGTTTTTGTATTGAT | 115 | 54 |
| | R:
CACCTCCCCTAACATAAACTCAAAACA | | |
| Methylated | F:
GGGTGATGTTATCGTTTTTGTATCGAC | 111 | 60 |
| | R:
CCTCCCCTAACGTAAACTCGAAACG | | |
| RT-qPCR | F:
TGGCAACGTATCTCAGCAAA | 132 | 60 |
| | R:
GGATGGGTGATGAGGACTTG | | |
| GAPDH | F:
CAGCCTCAAGATCATCAGCA- | | |
| | R:
TGTGGTCATGAGTCCTTCCA | 106 | 60 |
| EZH2 | F:
AATCAGAGTACATGCGACTGAGA | 141 | 60 |
| | R:
GCTGTATCCTTCGCTGTTTCC | | |
Reverse transcription-quantitative PCR
(RT-qPCR)
RNA was extracted from SW1116, SW480, HCT116 and
CCC-HIE-2 cells using TRIzol® Reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The PrimeScript RT reagent kit (Takara Bio, Inc.) was
used for first-strand cDNA synthesis. For qPCR, the primers and
annealing temperature used for sFRP4 are listed in Table I. GAPDH was used as an internal
control. CFX Manager™ 3.0 software (Bio-Rad Laboratories, Inc.) was
used to analyze the results. Each 25 µl total reaction volume
contained 12.5 µl SYBR Green (PrimeScript RT reagent kit; Takara
Bio, Inc.), 5 µl cDNA, 0.8 µl primer and 6.7 µl water. The thermal
cycling conditions used were an initial step at 95˚C for 30 sec
followed by 40 cycles at 95˚C for 5 sec and 60˚C for 35 sec.
Melting curves were presented as single curves, ensuring that the
reaction products were single, specific products. Each cDNA sample
was analyzed in triplicate. Each reaction was carried out in
duplicate, and the 2-ΔΔCq method
was employed to calculate the relative expression levels (23).
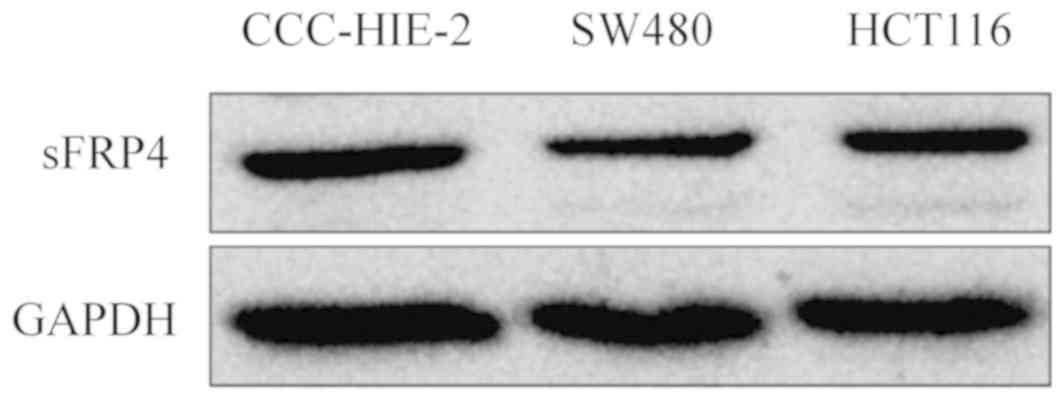
Western blot analysis
When we performing this experiment, we chose HCT116
and SW480 as represent for CRC cell lines. The colorectal cell
lines SW480 and HCT116 were lysed in lysis buffer [20 mM Tris-HCl
(pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 20 mM NaF, 1 mM
Na3VO4 and 0.1% protease inhibitor cocktail
(cat. no. P8340; Sigma-Aldrich; Merck KGaA)]. Protein
concentrations of cell lysates were measured using the DC protein
assay kit (Bio-Rad Laboratories, Inc.), with 10 µl (50 µg) of the
protein samples for each experiment separated using 12% SDS-PAGE
and transferred on to an Immobilon P PVDF membrane (EMD Millipore).
The membranes were blocked with 5% milk for 2 h at room
temperature. Membranes were then incubated with anti-SFRP4
(1:1,000; cat. no. A4189; ABclonal Biotech Co., Ltd.) and
anti-GAPDH (1:10,000; cat. no. AC036; ABclonal Biotech Co., Ltd.)
primary antibodies were added, and gently agitated for 1 h at room
temperature. Subsequently, membranes were incubated with
horseradish peroxidase-labeled anti-rabbit secondary antibody
(1:3,000; cat. no. AS014; ABclonal Biotech Co., Ltd.) and gently
agitated for 30 min at room temperature. ECL were used to visualize
protein signals. Where indicated, the signals were semi-quantified
using ImageJ (v1.8.0.112; National Institutes for Health).
Gene array
Illumina® Whole-Genome Gene Expression
Bead Chip (Illumina, Inc.), consisting of 47,322 probes, was used
to evaluate genes that were deferentially expressed. The whole
hybridization procedure was performed following the
Illumina® protocol. RNA was extracted from SW1116,
SW480, HCT116 and CCC-HIE-2 cells, using TRIzol® Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. An Illumina® TotalPrepRNA
amplification kit was used to synthesize cDNA following the
manufacturer's instructions, and reverse transcribed to synthesize
first strand cDNA at 42˚C for 2 h and stored at 4˚C. Second strand
cDNA was synthesized at 16˚C for 2 h and stored at 4˚C. cDNA
products were then purified using the Illumina TotalPrep RNA
Amplification kit (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. A total of 15 µg of
each biotinylated cDNA preparation was fragmented and placed in a
hybridization mixture. Samples were then hybridized onto a Sentrix
BeadChip (Human WG-6 gene expressionl Illumina, Inc.) at 58˚C for
16 h, according to the manufacturer's protocols. Microarray scanned
images were obtained using an Illumina BeadChip Reader, using the
default settings. Images were analyzed using Illumina Bead Station
500 system (Illumina, Inc.). Comparisons were made between
CCC-HIE-2 samples and the SW1116, SW480 and HCT116 samples, using
CCC-HIE-2 as the baseline. The fold-change values, indicating the
relative change in the expression levels between CCC-HIE-2 samples
and the SW1116, SW480 and HCT116 samples, were used to identify
genes that were differentially expressed. To generate a heat map
for the related PcG protein and Wnt signaling genes, the means of
the normalized values for each protein were subjected to
conditional formatting. The highest value was assigned a red color,
middle values a yellow color and the lowest values a green
color.
ChIP
ChIP was performed using the Magna
ChIP™A/G One-Day Chromatin immunoprecipitation kit (cat.
no. 17-10085; EMD Millipore) according to the manufacturer's
instructions, using 1% formaldehyde at 37˚C for 10 min and
neutralizing with glycine for 5 min at room temperature to achieve
crosslinking of the chromatin. All colorectal cell lines
(CCC-HIE-2, SW1116, SW480 and HCT116) were washed with cold 1 ml
PBS + protease inhibitors (1 mM PMSF, 1 mg aprotinin and 1 mg
pepstatin A). Cells were centrifuged at 4˚C at 716 x g for 5 min.
SDS Lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris-Hcl pH 8.0) was
then used to disrupt the cells, which were subsequently sonicated
at 150 Hz, sheared with 4 sets of 10 sec pulses on wet ice using a
high intensity ultrasonic processor (Cole-Parmer). An equal amount
of chromatin was immunoprecipitated at 4˚C overnight. ChIP was
performed using antibodies targeting chromobox 7 (CBX7; cat. no.
ab21873; Abcam), enhancer of zeste homolog 2 (EZH2; cat. no.
ab191250; Abcam) and jumonji and AT-rich interaction domain
containing 2 (JARID2; cat. no. ab192252; Abcam). Total chromatin
was used as the input. Immunoprecipitated products were collected
after incubation with magnetic beads coupled with anti-mouse IgG
(cat. no. ab18413; Abcam). The beads were washedusing a magnetic
separation rack and the bound chromatin was eluted in ChIP Elution
Buffer with Proteinase K mixer, according to the manufacturer's
instructions. The DNA fragments immunoprecipitated by CBX7, EZH2
and JARID2 were amplified using qPCR with the primers targeting the
sFRP4 promoter, which are listed in Table II.
 | Table IIPrimer sequences for chromatin
immunoprecipitation-quantitative PCR. |
Table II
Primer sequences for chromatin
immunoprecipitation-quantitative PCR.
| Gene | Primer sequence
(5'→3') | Annealing
temperature, ˚C | Product length,
base pairs |
|---|
| sFRP4-1 | F:
ACATTGTCCCAACTGTCCTCA3 | 60 | 82 |
| | R:
TTCTGCTGCCCTCTAATTCTG | | |
| sFRP4-2 | F:
GCAGAGGGAGCAAAGTTCAGT | 60 | 91 |
| | R:
TTTTCGACACCGGATACAAGA | | |
Small interfering (si)RNA
transfections
The siRNA sequences targeting EZH2 were designed and
synthesized by Guangzhou Ribobio Co., Ltd. The sequences used were
as follows: siRNA (si)-EZH2-1 forward, 5'-GACUCUGAAUGCAGUUGCUTT-3'
and reverse, 5'-AGCAACUGCAUUCAGAGUCTT-3'; si-EZH2-2 forward,
5'-GCAGCUUUCUGUUCAACUUTT-3' and reverse,
5'-AAGUUGAACAGAAAGCUGCTT-3'; si-EZH2-3 forward,
5'-CCUGACCUCUGUCUUACUUTT-3' and reverse,
5'-AAGUAAGACAGAGGUCAGGTT-3'; and negative control (24) forward, 5'-UUCUCCGAACGUGUCACGUTT-3'
and reverse, 5'-ACGUGACACGUUCGGAGAATT-3'. Cells were seeded in
6-well plates (50,000 cells/ml). Transfections were performed when
the cell confluence reached 50-60%. Cells were transfected with
si-EZH2-1, si-EZH2-2, si-EZH2-3 or si-NC (50 nM) using GenMuteTM
siRNA Transfection reagent (SignaGen Laboratories, LLC), according
to the manufacturer's instructions. At ~24 h after transfection,
cells were harvested for subsequent analyses.
MTT assays
After transfection for 24 h, SW480 cells were seeded
in 96-well plates at a density of 3,000 cells/well. Briefly, 10 µl
of MTT was added into cells to incubate for 4 h and replaced with
200 µl of DMSO. The absorbance values at 490 nm of each well was
recorded. Each set was repeated at least three times. The viability
of SW480 cells was detected at different time points (0, 24, 48 and
72 h).
Statistical analysis
SPSS 19.0 (IBM Corp.) was used to analyze the
RT-qPCR data. One-way ANOVAs with post-hoc Dunnett's tests were
used to analyze the expression levels of sFRP4 mRNA and the data
from the MTT assays. The statistical method, Normalization and
Differential Analysis, developed by Illumina, Inc., was used to
analyze the gene array data. P<0.05, diffscore <-20 or
diffscore >20 was considered to indicate a statistically
significant difference. The 2-ΔΔCq
method was used to analyze ChIP-qPCR data, which were normalized to
the input DNA fraction Cq value. The % Input for each ChIP fraction
was calculated as: % Input=2 x (Cqinput -
CqChIP) x Fd x100%. In this instance, Fd is the input
dilution factor. For example, if 100 µl of sonicated sample was
used for ChIP, and 10 µl of sonicated sample is used as the input,
Fd=1/10. Fold Enrichment=[% (ChIP/input))]/[% (negative
control/input)]. The normalized ChIP fraction Cq value was adjusted
for the normalized background (mock immunoprecipitation (25) fraction Cq value. ΔΔCq (ChIP/mock
IP)=ΔCq (normalized ChIP)-ΔCq (normalized mock IP).
Results
Promoter DNA methylation status of
sFRP4 during CRC progression from Dukes A to Dukes C
As indicated in Fig.
1, methylation of the sFRP4 promoter region was detected in
SW1116, SW480 and HCT116 cells, but not in the CCC-HIE-2 cells.
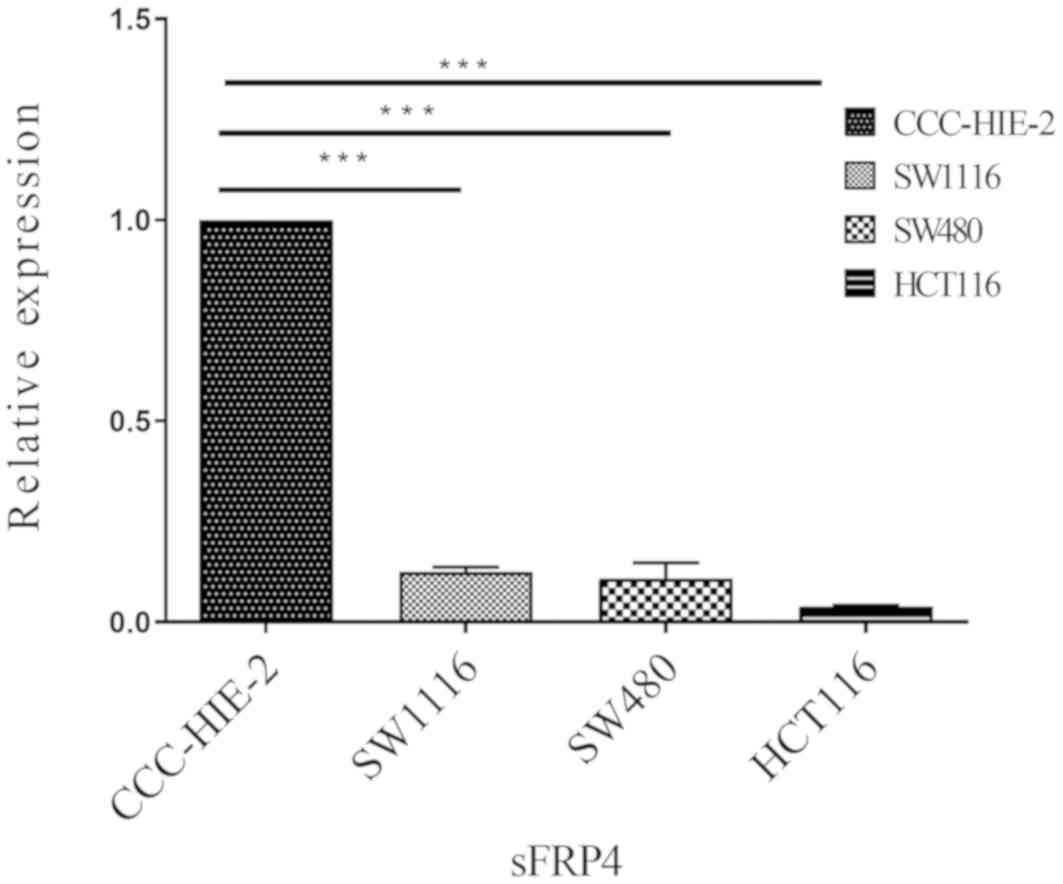
sFRP4 is downregulated in CRC
cells
To determine whether DNA methylation of the sFRP4
promoter coincided with decreased expression levels, the mRNA and
protein expression levels of sFRP4 were measured in all four cell
lines using RT-qPCR and western blotting analysis (Figs. 2 and 3, respectively). Compared with the
CCC-HIE-2 cells, the mRNA expression levels of sFRP4 were
significantly lower in the SW1116, SW480 and HCT116 cells (Fig. 2;
P<0.001), while in western blot analysis, sFRP4 appeared to be
downregulated (Fig. 3). These
results appear to be negatively associated with the methylation
status of the sFRP4 (Fig. 1). The
expression levels of sFRP4 were downregulated by more than
nine-fold in the CRC cell lines compared with the control cell
line.
Expression of EZH2, CBX7 and JARID2 is
upregulated and may be positively associated with the activities of
Wnt/β-catenin signaling in the CRC cell lines
sFRPs may block Wnt signaling either by interacting
with Wnt proteins to prevent them from binding Fzd proteins or by
forming non-functional complexes with Fzd (21). The transcriptional expression levels
of components of the Wnt signaling pathway were assessed using gene
array analysis of whole genome expression in SW1116, SW480, HCT116
and CCC-HIE-2 cells. The Wnt signaling pathway may be abnormally
activated in all three CRC cell lines, exhibiting upregulated
expression levels of the downstream genes, FZD9, Low-density
lipoprotein related protein-5 (LRP5), β-catenin, T-cell factor 3
(TCF3), T-cell factor 4 (TCF4), c-myc and cycD2 (Table III, Fig. 4), and downregulated expression of
glycogen synthase kinase 3β (GSK3β). GSK3β is considered to serve a
negative role in this pathway by promoting the degradation of
β-catenin (2).
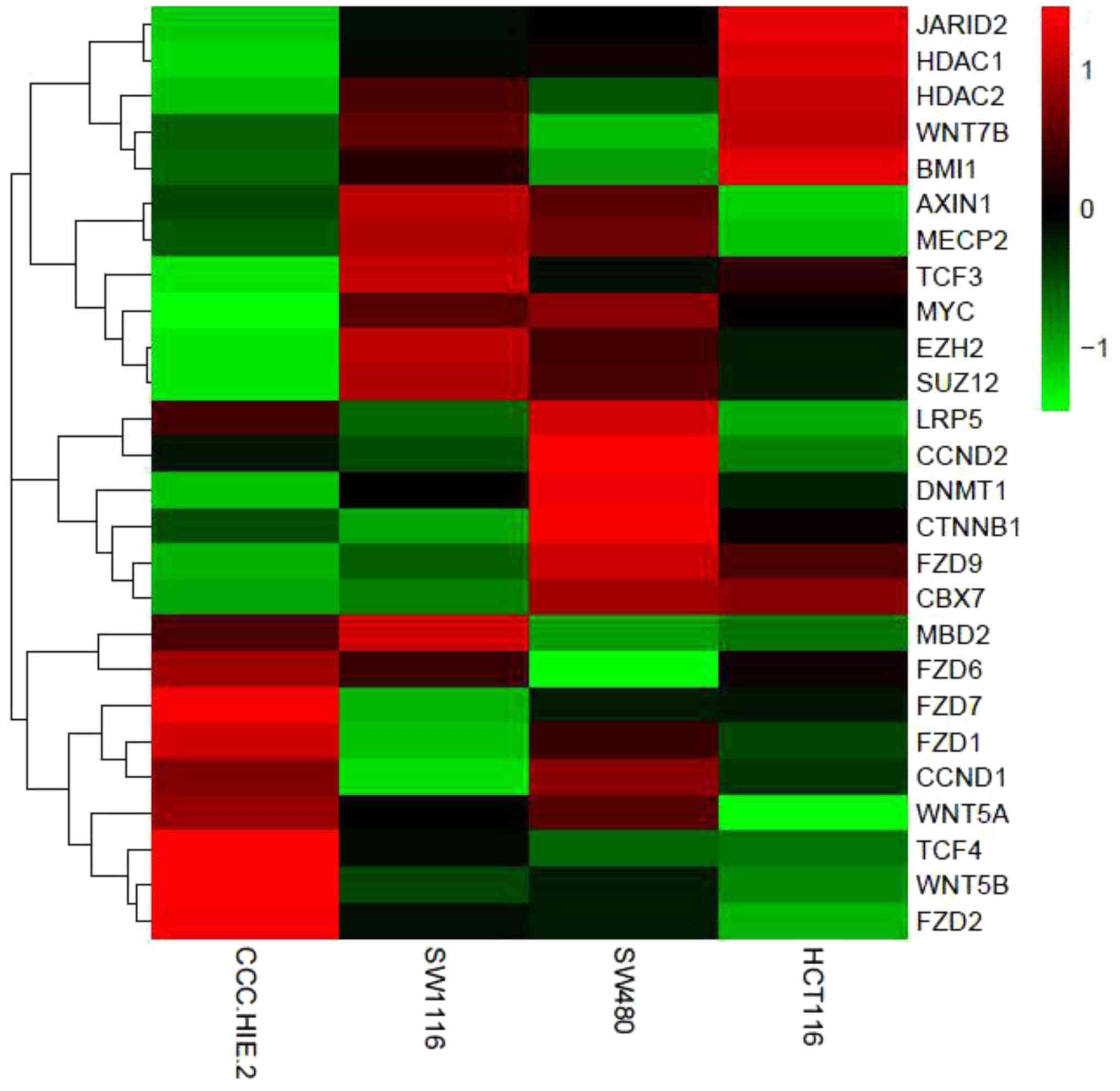 | Figure 4Heat map of differentially expressed
genes in the four colorectal cell lines. PcG proteins (BMI1, EZH2,
Suz12, CBX7 and JARID2) and DNMT1 were all upregulated in the
cancer cell lines. Meanwhile, the mRNA expression of genes in the
Wnt signaling pathway (FZD9, LRP5, β-catenin, TCF3, TCF4, c-myc and
cycD2) were upregulated in CRC cell lines compared with normal
colorectal cells. Red indicates high expression. Green indicates
low expression. PcG, polycomb group; BMI1, B-lymphoma Mo-MLV
insertion region 1; EZH2, enhancer of zeste homolog 2; Suz12,
suppressor of zeste 12 homolog; CBX7, chromobox protein homolog 7;
JARID2, jumonji and AT-rich interaction domain containing 2; DNMT1,
DNA methyltransferase 1; cyD2, cytochrome bd-I ubiquinol oxidase
subunit II; FZD9, frizzled receptors 9; LRP5, low-density
lipoprotein related protein-5; TCF3, T-cell factor 3; TCF4, T-cell
factor 4. |
 | Table IIIGenes differentially expressed
between colorectal cancer cell lines and the CCC-HIE-2 cell
line. |
Table III
Genes differentially expressed
between colorectal cancer cell lines and the CCC-HIE-2 cell
line.
| | Fold-change in
expression | | |
|---|
| Gene symbol | SW1116 | SW480 | HCT116 | mRNA Accession | Description |
|---|
| FZD9 | 1.57 | 8.66 | 4.56 | NM_003508.2 | Homo sapiens
frizzled homolog 9 |
| LRP5 | 0.52 | 1.51 | 0.45 | NM_002335.1 | Homo sapiens
low-density lipoprotein rece ptor-related protein 5 |
| β-catenin | 0.72 | 3.65 | 1.60 | XM_945654.1 | Homo sapiens
catenin (cadherin-associated protein) β1 |
| TCF3 | 2.76 | 1.66 | 1.99 | NM_003200.1 | Homo sapiens
transcription factor 3 |
| TCF4 | 4.32 | 1.30 | 5.34 | NM_003199.1 | Homo sapiens
transcription factor 4 |
| c-myc | 9.24 | 15.37 | 4.97 | NM_002467.3 | Homo sapiens v-myc
myelocytomatosis viral oncogene homolog |
| cycD2 | 0.33 | 18.18 | 0.02 | NM_001759.2 | Homo sapiens cyclin
D2 |
| JARID2 | 2.51 | 2.72 | 8.07 | NM_004973.2 | Homo sapiens
jumonji and AT-rich interaction domain containing 2 |
| CBX7 | 1.21 | 5.86 | 5.28 | NM_175709.2 | Homo sapiens
chromobox 7 |
| EZH2 | 59.84 | 30.27 | 13.69 | NM_004456.3 | Homo sapiens
enhancer of zeste homolog 2 |
| DNMT1 | 1.55 | 2.56 | 1.41 | NM_001379.1 | Homo sapiens DNA
(cytosine-5-)-methyltransferase 1 |
| HDAC1 | 1.39 | 1.49 | 2.05 | NM_004964.2 | Homo sapiens
histone deacetylase 1 |
| HDAC2 | 2.09 | 1.29 | 2.74 | NM_001527.2 | Homo sapiens
histone deacetylase 2 |
To screen for important regulators that modulate the
promoter DNA methylation of sFRP4, the expression levels of
components in the PcG protein family in the three CRC cell lines
were analyzed. Table III lists
certain PcG protein-related genes that were significantly
upregulated or downregulated >1.5-fold (P<0.05; differ score
>20 or differ score <20). For example, EZH2, CBX7 and JARID2
were all upregulated in cancer cells. Among these genes, EZH2 was
dramatically upregulated in the SW1116 cells (59.84-fold), SW480
cells (30.27-fold) and HCT116 cells (13.69-fold). In addition, the
expression levels of histone deacetylase (HDAC) 1 were higher in
the SW480 cells (1.49-fold) and HCT116 cells (2.05-fold) compared
with the CCC-HIE-2 control, while HDAC2 was upregulated in the
SW1116 cells (2.09-fold) and HCT116 cells (2.74-fold). DNMT1 was
upregulated in all CRC cell lines (1.55-fold in SW1116, 2.56-fold
in SW480 and 1.41-fold in HCT116). These results suggested that
histone modification and DNA methylation may serve important roles
in the advanced stages of CRC and that the expression of the PcG
proteins EZH2, CBX7 and JARID2 may be associated with the
expression of sFRP4. RT-qPCR was showed similar results to the gene
array results (Fig. S1).
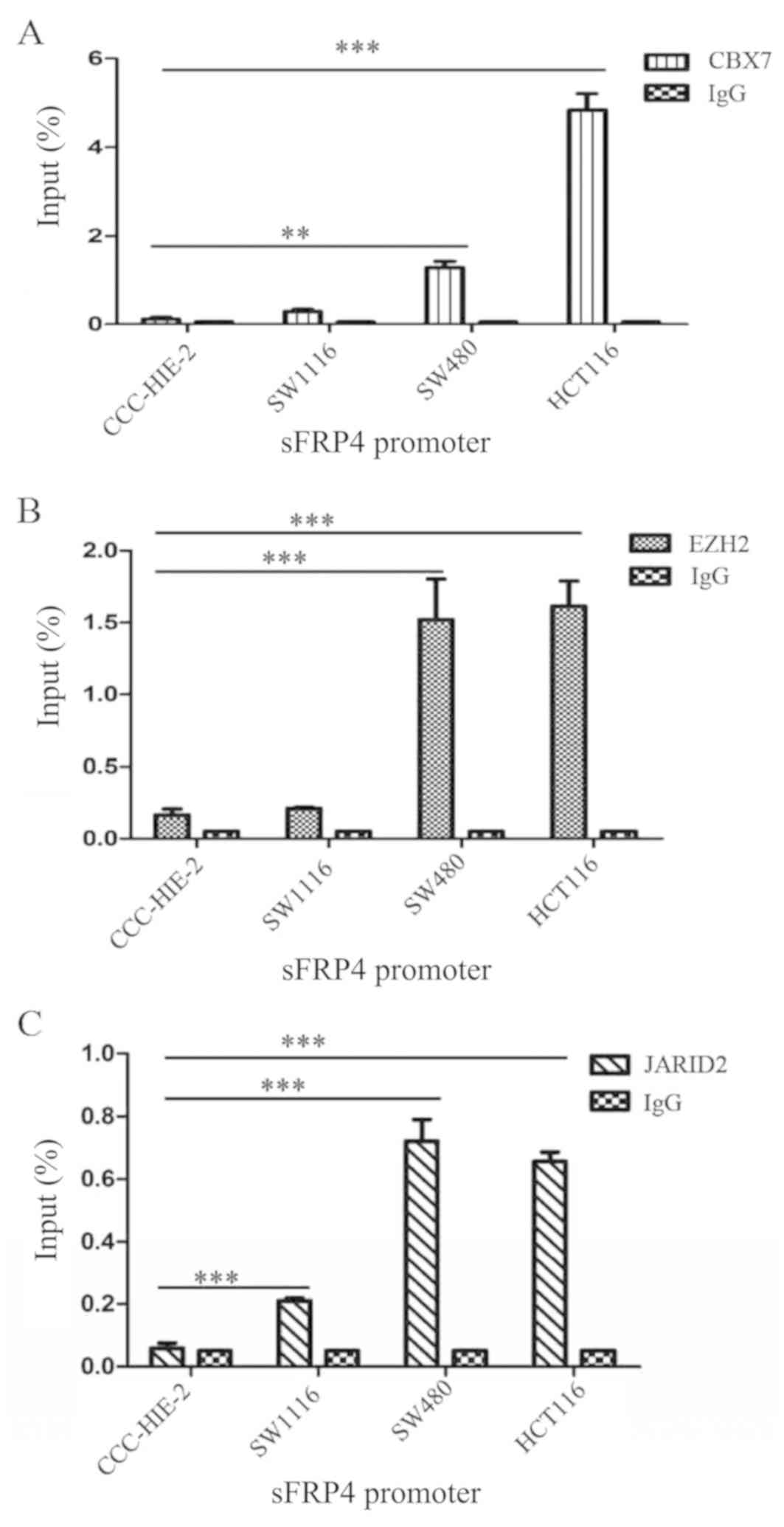
CBX7, EZH2 and JARID2 are enriched in
the sFRP4 promoter region in CRC cells
Since the PcG proteins have been demonstrated to be
associated with the promoter DNA methylation status of targeted
genes, whether the selected PcG proteins (CBX7, EZH2 and JARID2)
are enriched in or bind the promoter region of sFRP4 in CRC was
investigated. ChIP-qPCR using anti-CBX7, anti-EZH2 and anti-JARID2
antibodies confirmed the enrichment of these proteins at the sFRP4
promoter region (Table II).
Fig. 5 demonstrated that CBX7, EZH2
and JARID2 were enriched in the sFRP4 promoter region in all four
of the colorectal cell lines, while in the CRC cell lines, CBX7,
EZH2 and JARID2 were enriched.
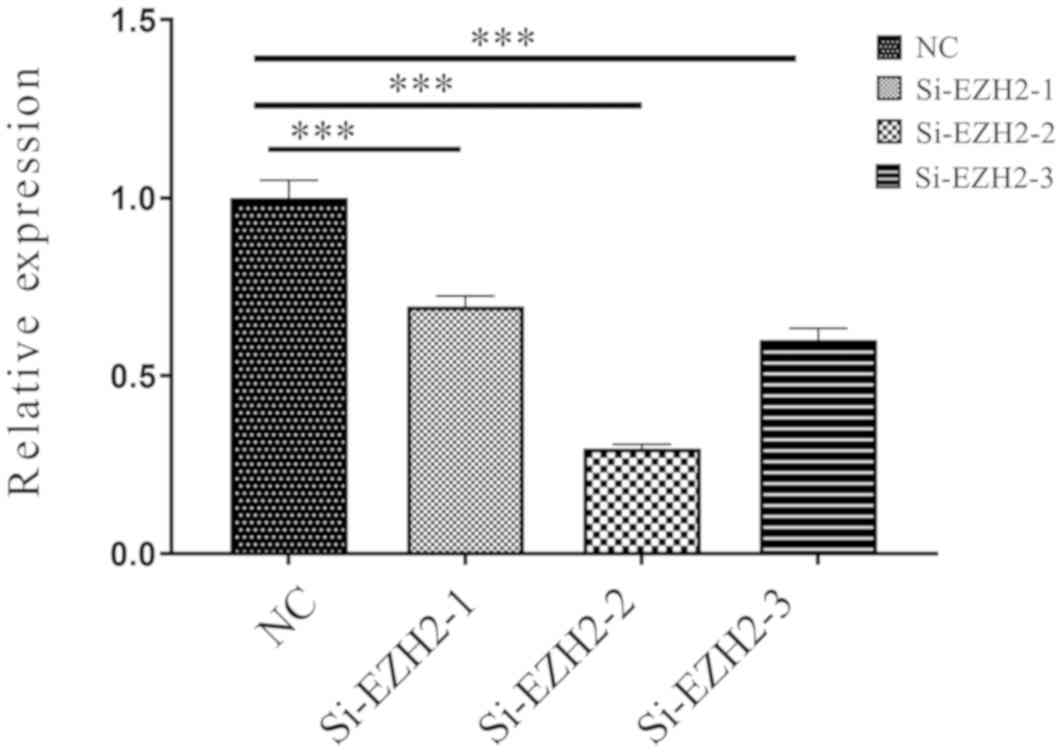
Silencing of EZH2 rescues sFRP4
expression levels in CRC
Inhibition of EZH2 was performed to determine
whether EZH2 affected the promoter DNA methylation of sFRP4.
Various si-EZH2 were tested, si-EZH2-1, si-EZH2-2 and si-EZH2-3.
si-EZH2-2 was selected because the EZH2 expression levels were the
lowest following transfection (Fig.
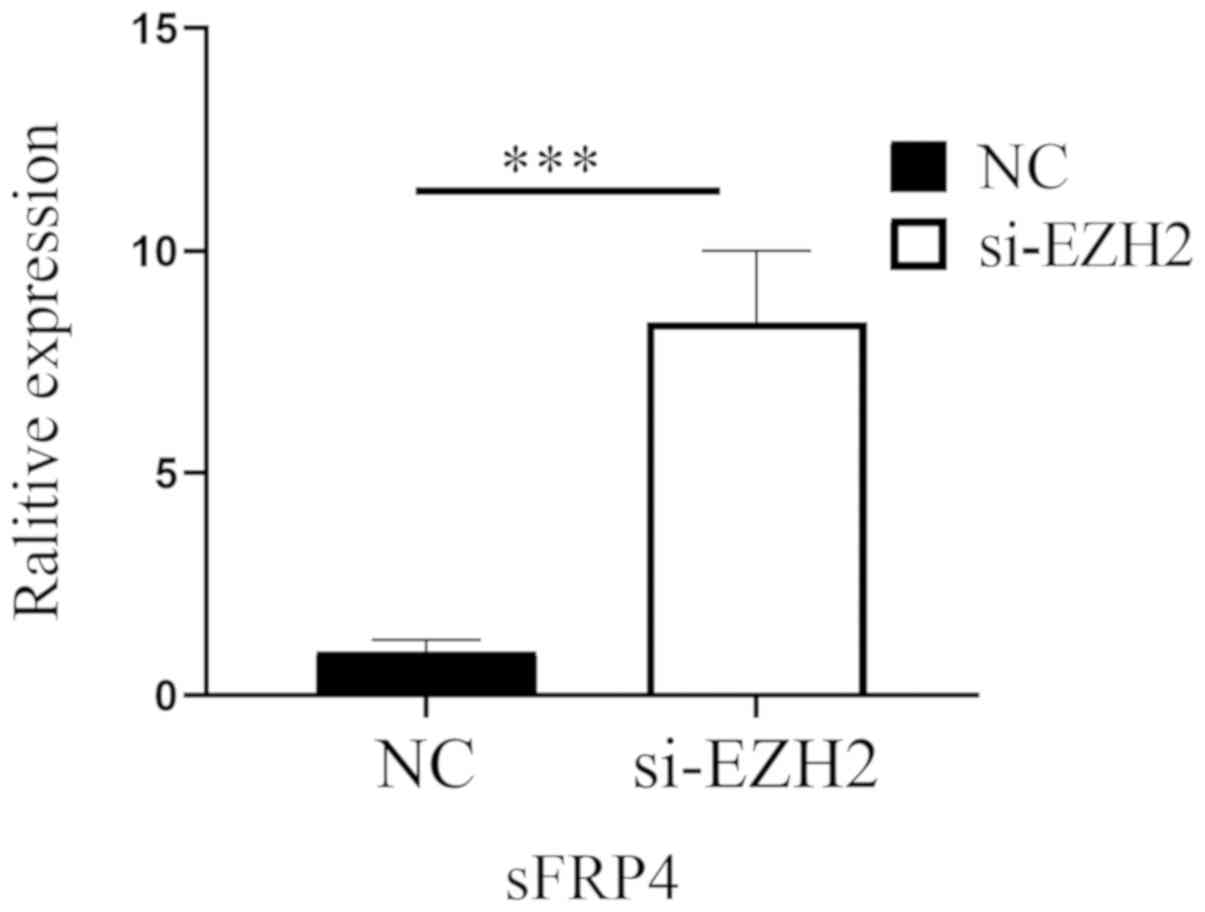
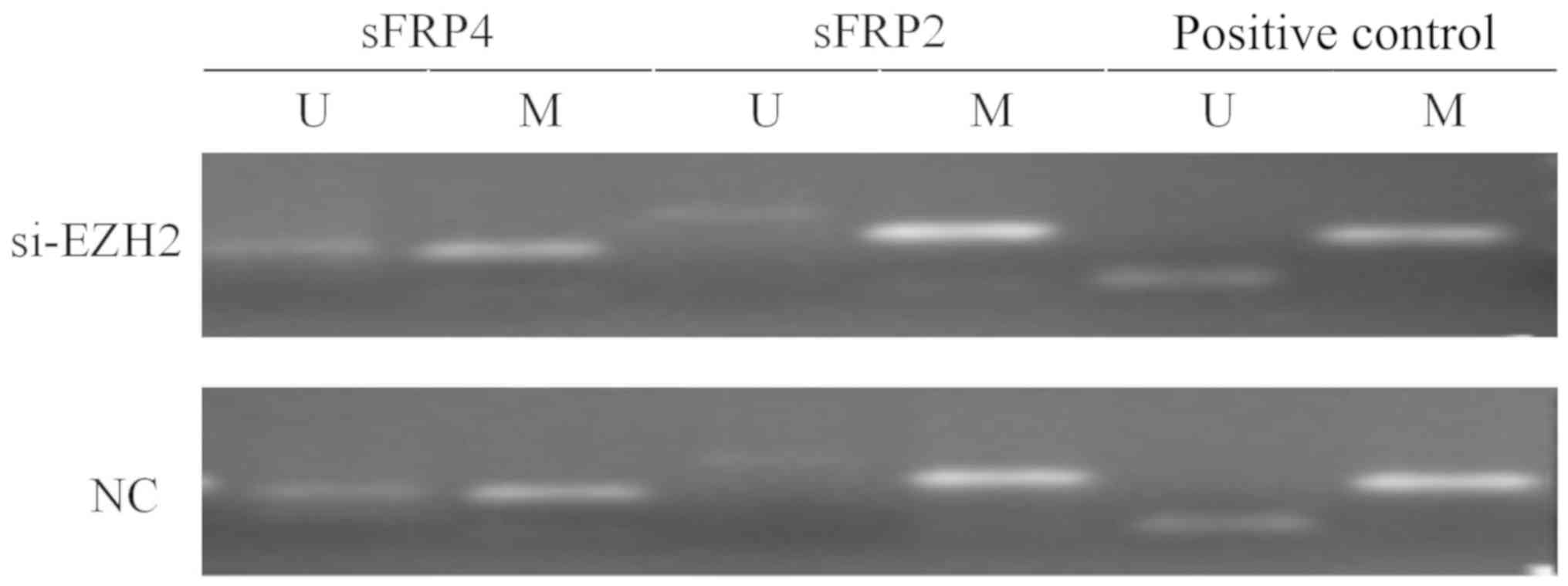
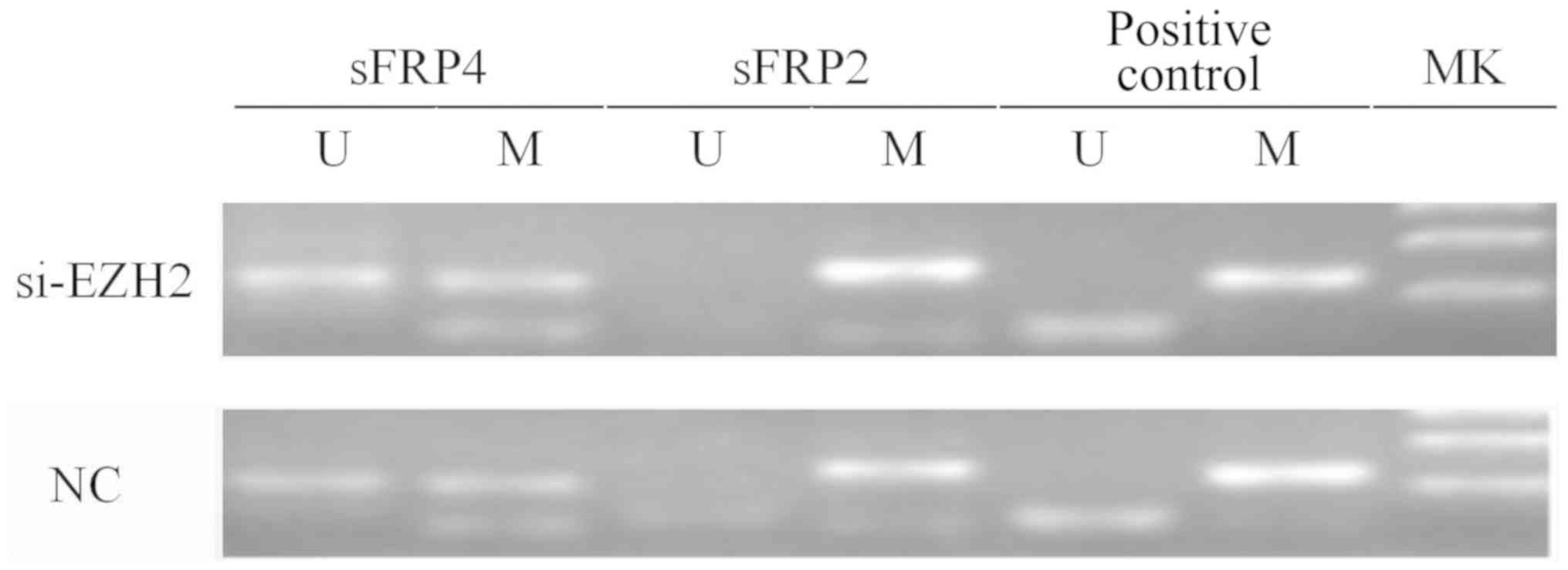
6). Fig. 7 indicates that after
EZH2 was inhibited in SW480 cells, the mRNA expression levels of
sFRP4 were upregulated (P<0.01), but the methylation status of
sFRP4 was still hypermethylated (Figs.
8 and 9).
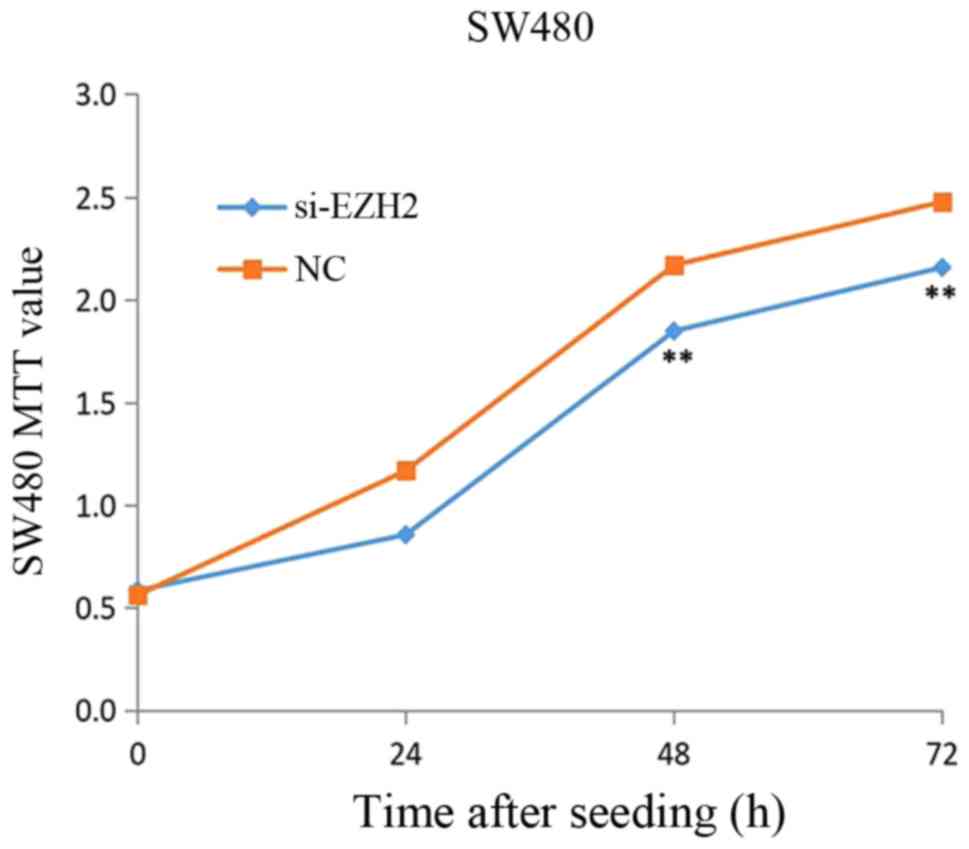
Knockdown of EZH2 influences CRC cell
proliferation
To investigate the effects of EZH2 on the
proliferation of CRC, Knockdown of EZH2 was performed in SW480
cells for 24, 48 and 72 h and, subsequently, the cell viability was
measured using the MTT assays. Fig.
10 revealed that a significant decrease in CRC cell viability
was observed when EZH2 was knocked down (P<0.05), with a
reduction of 31.0% in the CRC cell line treated with si-EZH2 for 24
h, 32.0% at 48 h and 32.0% at 72 h.
Discussion
CRC is a leading cause of cancer-associated
mortality worldwide. One of the fundamental mechanisms driving the
initiation and progression of CRC is the accumulation of a variety
of genetic and epigenetic changes (20). Over the past several decades,
advances have been made in the understanding of cancer epigenetics,
particularly for aberrant DNA methylation, microRNA and non-coding
RNA dysregulation, and alterations in histone modification states
(14).
sFRPs have been recognized for their potential to
sequester Wnt ligands away from their receptor complexes and
ultimately antagonize Wnt signaling (4,11,21).
Analysis of sFRP hypermethylation may have diagnostic and
prognostic value for the detection and management of CRC (24,26).
To the best of our knowledge, no mutation in any sFRP gene has been
reported to be associated with tumors. Previous studies have
documented the loss of sFRP expression due to promoter methylation;
thus, silencing sFRP expression through epigenetics may be the
mechanism behind colorectal tumorigenesis (11,12,16,27).
In the present study, the methylation-specified PCR
results indicated that sFRP4 was hypermethylated at the advanced
stage in CRC cells. In RT-qPCR and western blot analysis, sFRP4 was
downregulated in CRC cell lines compared with CCC-HIE-2 cells. In a
previous study, high-dose DNMT inhibitor DAC treatment increased
the expression levels of sFRP4 in CRC cells (12). These results indicated that promoter
DNA hypermethylation represents a possible mechanism of sFRP4 gene
silencing in CRC. By analyzing the data from the gene array, the
expression of DNMT1 in CRC was found to be upregulated and it was
also revealed that Wnt signaling may be aberrantly activated in
CRC. DNA methylation variations have been reported to be associated
with carcinogenesis (16,27), and may occur before or at the
beginning of carcinogenesis. DNA methylation may silence sFRP4
expression and induce CRC initiation and progression.
DNA methylation and histone lysine methylation are
dynamic chemical modifications that serve a crucial role in the
establishment of gene expression patterns during development, which
are considered to be tightly coordinated (28). Methylation of lysine residues on
histones can initiate, target or maintain DNA methylation. Histone
methylation is more commonly observed on lysine residues of histone
tails H3 and H4. Common histone methylation sites associated with
gene activation include H3K4, H3K48 and H3K79, while common sites
for gene inactivation include H3K9 and H3K27(28).
PcG proteins are transcriptional repressors that
regulate several crucial developmental and physiological processes
in the cell (29). More recently,
these proteins have been revealed to serve important roles in human
carcinogenesis and cancer development and progression (29,30).
In particular, the PcG proteins and other epigenetic regulators,
participate in regulation of gene transcription (29,31).
PcG proteins form two major protein complexes, polycomb repressive
complex 1 and 2. EZH2, the core component of PRC2, is an epigenetic
regulatory protein associated with tumor aggressiveness and poor
survival outcomes in several types of humancancer (32). EZH2 catalyzes the trimethylation on
Lys27 of histone H3 (H3K27me3), which recruits transcriptionally
repressive complexes involved in chromatin compaction and results
in stable gene silencing (33).
EZH2 has also been reported to interact with both DNMT1 and DNMT3
which, in turn, methylate the target DNA where EZH2 was enriched
(34). Although the mechanistic
contributions of EZH2 to cancer progression are not yet determined,
functional links between EZH2-mediated histone methylation and DNA
methylation (35) suggest a
partnership with the gene silencing machinery implicated in tumor
suppressor loss.
In the present study, EZH2 was enriched in the sFRP4
gene promoter region, and knockdown of EZH2 did not influence the
promoter DNA methylation levels of sFRP4. These results suggested
that EZH2 regulated sFRP4 expression without affecting the promoter
DNA methylation levels in CRC cells. Moreover, in SW480 CRC cells,
si-EZH2 treatment restored sFRP4 expression levels and decreased
CRC cell viability. These results provided evidence that EZH2
serves an important role in enhancing the proliferation of human
CRC cells. Given that EZH2 did not affect the DNA methylation level
of sFRP4, it was speculated that EZH2 may regulate sFRP4 expression
via histone methylation.
Additionally, the Wnt signaling pathway has several
key components, such as GSK3β and β-catenin (2). GSK3β is considered to serve negative
roles in this pathway by promoting the degradation of β-catenin
(2). The mRNA expression levels of
GSK3β were confirmed to be downregulated in CRC cell lines via a
gene array. EZH2 was revealed to have a significant influence on
the downstream activity in the Wnt signaling pathway. Chen et
al (36), reported that the
expression levels of β-catenin, vimentin and c-Myc were detected
via western blotting after EZH2 knockdown in SW480 cells. These
data reveal that knockdown of EZH2 decreased the expression of
c-Myc and vimentin, which are known downstream target genes of the
Wnt/β-catenin signaling pathway. In hepatic cancer, EZH2 reduces
the expression levels of nuclear β-catenin, TCF transcriptional
activity and downstream proliferate targets cyclin D1 and epidermal
growth factor receptor (37). In
cervical carcinoma, EZH2-mediates the repression of GSK3β and tumor
suppressor p53 (TP53), which promotes Wnt/β-catenin
signaling-dependent cell expansion (38). Inhibition of GSK 3β activity is
associated with excessive EZH2 expression levels and enhanced tumor
invasion in nasopharyngeal carcinoma (39).
In the present study, the enrichment of CBX7 and
JARID2 at the sFRP4 promoter was also increased in all three
colorectal cell lines in which the sFRP4 promoter was
hypermethylated. CBX7 is a canonical component in PRC1, which can
physically interact with H3K27me3 via its N-terminal chromodomain
(40). CBX7 is also known to be
involved in repressive chromatin modifications and to be able to
form complexes with DNA methyltransferase enzymes, but knockdown of
CBX7 is unable to relieve the suppression of silenced genes in
cancer cells (37,38). JARID2, a member of the Jumonji
family, is a DNA binding protein that binds to the promoter region
of specific myogenic genes in rhabdomyosarcoma cells in conjunction
with a PRC2 protein and regulates H3K27me3(25). CBX7, JARID2 and EZH2 all show close
connections with histone methylation (25,29).
The functions of CBX7 and JARID2 still need to be analyzed.
In conclusion, based on the current results, the
expression levels of sFRP4 appear to be associated with its DNA
methylation. CBX7, EZH2 and JARID2 were enriched in the sFRP4
promoter region when sFRP4 was hypermethylated, and there was a
greater level of enrichment with more malignant CRC cell lines. The
PcG protein may have participated in sFRP4 promoter DNA
hypermethylation and induced sFRP4 silencing. In particular, EZH2
regulated sFRP4 expression without affecting the hypermethylation
of sFRP4 in CRC cells, while there may be a potential regulation of
EZH2 and sFRP4. This mechanism of action may be of interest for
developing a new therapy for CRC.
Supplementary Material
Figure S1. Validation of the gene
array analysis in colorectal cancer cell lines. The expression
levels of CBX7, Jarid2 and EZH2 coincide with the gene array
analysis. **P<0.01, ***P<0.001. CBX7,
chromobox 7; EZH2, enhancer of zeste homolog 2; Jarid2, jumonji and
AT.rich interaction domain containing 2.
Acknowledgements
The abstract was presented at the Digest Disease
Week May 31-June 3, 2018 in Washington, DC, USA, and published as
an abstract Gastroenterology Volume 154, Issue 6, Supplement 1, May
2018, page s-332. The authors would like to thank Dr Wenzhong Shen
(College of Life Science, Wuhan University) for his useful
suggestions. The authors would also like to thank Dr Bo Li from The
Seventh Affiliated Hospital of Sun Yat-sen University (Guangmin,
China) for his help.
Funding
The present project was supported by Wuhan Applied
Basic Research Projects (grant no. 2015061701011642).
Availability of data and material
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JQ collected the data and designed the study. YL
performed the experiments, analyzed the data and wrote the
manuscript. JY, YX, ML and FW aided in experiments, analyzed the
data and revised it critically for important intellectual content.
JZ aided in experiments, analyzed the data and revised it
critically for important intellectual content.JQ supervised the
project. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480.
2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017.
|
|
3
|
Krishnamurthy N and Kurzrock R: Targeting
the Wnt/beta-catenin pathway in cancer: Update on effectors and
inhibitors. Cancer Treat Rev. 62:50–60. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cruciat CM and Niehrs C: Secreted and
transmembrane wnt inhibitors and activators. Cold Spring Harb
Perspect Biol. 5(a015081)2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Farooqi AA, de la Roche M, Djamgoz MBA and
Siddik ZH: Overview of the oncogenic signaling pathways in
colorectal cancer: Mechanistic insights. Semin Cancer Biol.
58:65–79. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Liu TH, Raval A, Chen SS, Matkovic JJ,
Byrd JC and Plass C: CpG island methylation and expression of the
secreted frizzled-related protein gene family in chronic
lymphocytic leukemia. Cancer Res. 66:653–658. 2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rubin JS and Bottaro DP: Loss of secreted
frizzled-related protein-1 expression in renal cell carcinoma
reveals a critical tumor suppressor function. Clin Cancer Res.
13:4660–4663. 2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chong JM, Uren A, Rubin JS and Speicher
DW: Disulfide bond assignments of secreted frizzled-related
protein-1 provide insights about frizzled homology and netrin
modules. J Biol Chem. 277:5134–5144. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Deshmukh A, Arfuso F, Newsholme P and
Dharmarajan A: Epigenetic demethylation of sFRPs, with emphasis on
sFRP4 activation, leading to Wnt signalling suppression and histone
modifications in breast, prostate, and ovary cancer stem cells. Int
J Biochem Cell Biol. 109:23–32. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pawar NM and Rao P: Secreted frizzled
related protein 4 (sFRP4) update: A brief review. Cell Signal.
45:63–70. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pohl S, Scott R, Arfuso F, Perumal V and
Dharmarajan A: Secreted frizzled-related protein 4 and its
implications in cancer and apoptosis. Tumour Biol. 36:143–152.
2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Qi J, Zhu YQ, Luo J and Tao WH:
Hypermethylation and expression regulation of secreted
frizzled-related protein genes in colorectal tumor. World J
Gastroenterol. 12:7113–7117. 2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Michalak EM, Burr ML, Bannister AJ and
Dawson MA: The roles of DNA, RNA and histone methylation in ageing
and cancer. Nat Rev Mol Cell Biol. 20:573–589. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Dor Y and Cedar H: Principles of DNA
methylation and their implications for biology and medicine.
Lancet. 392:777–786. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Schübeler D: Function and information
content of DNA methylation. Nature. 517:321–326. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Samaei NM, Yazdani Y, Alizadeh-Navaei R,
Azadeh H and Farazmandfar T: Promoter methylation analysis of
WNT/β-catenin pathway regulators and its association with
expression of DNMT1 enzyme in colorectal cancer. J Biomed Sci.
21(73)2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Takeshima H, Wakabayashi M, Hattori N,
Yamashita S and Ushijima T: Identification of coexistence of DNA
methylation and H3K27me3 specifically in cancer cells as a
promising target for epigenetic therapy. Carcinogenesis.
36:192–201. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ning X, Shi Z, Liu X, Zhang A, Han L,
Jiang K, Kang C and Zhang Q: DNMT1 and EZH2 mediated methylation
silences the microRNA-200b/a/429 gene and promotes tumor
progression. Cancer Lett. 359:198–205. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sarma DP: The Dukes classification of
colorectal cancer. JAMA. 256(1447)1986.PubMed/NCBI
|
|
20
|
Coppedè F: Epigenetic biomarkers of
colorectal cancer: Focus on DNA methylation. Cancer Lett.
342:238–247. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Baharudin R, Tieng FYF, Lee LH and Ab
Mutalib NS: Epigenetics of SFRP1: The dual roles in human cancers.
Cancers (Basel). 12(445)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu R, Su X, Long Y, Zhou D, Zhang X, Ye
Z, Ma J, Tang T, Wang F and He C: A systematic review and
quantitative assessment of methylation biomarkers in fecal DNA and
colorectal cancer and its precursor, colorectal adenoma. Mutat Res.
779:45–57. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Vincent KM and Postovit LM: Matricellular
proteins in cancer: A focus on secreted frizzled-related proteins.
J Cell Commun Signal. 12:103–112. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Walters ZS, Villarejo-Balcells B, Olmos D,
Buist TW, Missiaglia E, Allen R, Al-Lazikani B, Garrett MD, Blagg J
and Shipley J: JARID2 is a direct target of the PAX3-FOXO1 fusion
protein and inhibits myogenic differentiation of rhabdomyosarcoma
cells. Oncogene. 33:1148–1157. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yang Q, Huang T, Ye G, Wang B and Zhang X:
Methylation of SFRP2 gene as a promising noninvasive biomarker
using feces in colorectal cancer diagnosis: A systematic
meta-analysis. Sci Rep. 6(33339)2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Suzuki H, Watkins DN, Jair KW, Schuebel
KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van
Engeland M, et al: Epigenetic inactivation of SFRP genes allows
constitutive WNT signaling in colorectal cancer. Nat Genet.
36:417–422. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Stoll S, Wang C and Qiu H: DNA methylation
and histone modification in hypertension. Int J Mol Sci.
19(1174)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang W, Qin JJ, Voruganti S, Nag S, Zhou J
and Zhang R: Polycomb group (PcG) proteins and human cancers:
Multifaceted functions and therapeutic implications. Med Res Rev.
35:1220–1267. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chan HL and Morey L: Emerging roles for
polycomb-group proteins in stem cells and cancer. Trends Biochem
Sci. 44:688–700. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Piunti A and Shilatifard A: Epigenetic
balance of gene expression by polycomb and COMPASS families.
Science. 352(aad9780)2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Colón-Caraballo M, Monteiro JB and Flores
I: H3K27me3 is an epigenetic mark of relevance in endometriosis.
Reprod Sci. 22:1134–1142. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu M, Zhou J, Chen Z and Cheng ASL:
Understanding the epigenetic regulation of tumours and their
microenvironments: Opportunities and problems for epigenetic
therapy. J Pathol. 241:10–24. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Bogdanovic O, Long SW, van Heeringen SJ,
Brinkman AB, Gómez-Skarmeta JL, Stunnenberg HG, Jones PL and
Veenstra GJ: Temporal uncoupling of the DNA methylome and
transcriptional repression during embryogenesis. Genome Res.
21:1313–1327. 2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Emran AA, Chatterjee A, Rodger EJ, Tiffen
JC, Gallagher SJ, Eccles MR and Hersey P: Targeting DNA methylation
and EZH2 activity to overcome melanoma resistance to immunotherapy.
Trends Immunol. 40:328–344. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Chen JF, Luo X, Xiang LS, Li HT, Zha L, Li
N, He JM, Xie GF, Xie X and Liang HJ: EZH2 promotes colorectal
cancer stem-like cell expansion by activating p21cip1-Wnt/β-catenin
signaling. Oncotarget. 7:41540–41558. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cheng AS, Lau SS, Chen Y, Kondo Y, Li MS,
Feng H, Ching AK, Cheung KF, Wong HK, Tong JH, et al: EZH2-mediated
concordant repression of Wnt antagonists promotes
β-catenin-dependent hepatocarcinogenesis. Cancer Res. 71:4028–4039.
2011.
|
|
38
|
Chen Q, Zheng PS and Yang WT:
EZH2-mediated repression of GSK-3β and TP53 promotes Wnt/β-catenin
signaling-dependent cell expansion in cervical carcinoma.
Oncotarget. 7:36115–36129. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ma R, Wei Y, Huang X, Fu R, Luo X, Zhu X,
Lei W, Fang J, Li H and Wen W: Inhibition of GSK 3β activity is
associated with excessive EZH2 expression and enhanced tumour
invasion in nasopharyngeal carcinoma. PLoS One.
8(e68614)2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Clermont PL, Lin D, Crea F, Wu R, Xue H,
Wang Y, Thu KL, Lam WL, Collins CC, Wang Y and Helgason CD:
Polycomb-mediated silencing in neuroendocrine prostate cancer. Clin
Epigenetics. 7(40)2015.PubMed/NCBI View Article : Google Scholar
|