Introduction
Osteosarcoma (OS) is a type of malignant tumor that
often occurs in children and adolescents (1). As previously reported, the prevalence
rate of OS increases by 0.3% per year (2). Similar to several other human
malignancies, a high mortality rate, recurrence and metastasis are
key factors leading to the poor prognosis of patients with OS
(3). Surgical resection combined
with adjuvant therapy, including chemotherapy and radiotherapy, has
been reported to improve the prognosis of patients with OS to a
certain degree (4). However, over
the past two decades, no evident improvements have been observed in
the survival rates of these patients (5).
Oxidative stress refers to an increased cell
production of free radicals, including reactive oxygen species,
ultimately resulting in damage to cells and tissues (6). A previous study demonstrated that
tumor cells can be destroyed by increased oxidative stress
(7). Inducing tumor cell apoptosis
via increased oxidative stress may therefore be consider as a
promising therapeutic strategy for malignancies (7). Hydrogen peroxide
(H2O2) is a main intermediate of oxidative
stress and is considered as a potent oxidant that induces cell
senescence (8). In OS, it has been
reported that treatment with 100 µM H2O2
decreases the viability and enhances the apoptosis of MG63 cells
(9). However, the underlying
mechanisms of H2O2 interference with OS cell
phenotype remains unclear.
Circular RNA (circRNA) is a type of non-coding RNA
that was discovered decades ago (10,11).
Increasing evidence indicates that circRNAs play an important role
in the development of human tumors (12). Salmena et al (13) proposed the competitive endogenous
RNA mechanism (ceRNA mechanism) theory that gene transcripts, such
as long-chain non-coding RNA, can influence mRNA expression by
binding microRNAs (miRs) through competitive sponge attachment. In
recent years, circRNAs have also been found to regulate mRNA
expression through miRNA adsorption (14). A previous study has demonstrated
that circ_0002052 inhibits OS progression by sponging
miR-1205(15). Huang et al
(16) reported that circNASP
upregulates Forkhead box F1 expression by sponging miR-1253 in OS
cells, thereby regulating the development of OS. These findings
suggest that circRNAs could play a crucial role in the development
of OS.
circKMT2D (also known as circRNA9920) is a recently
discovered circRNA (17); however,
its role in human tumors, including OS, remains unknown. In
particular, we previously determined that circKMT2D expression is
abnormally upregulated in H2O2-treated OS
cells in vitro and that circKMT2D knockdown in
H2O2-treated OS cells markedly decreased OS
cell viability. Subsequently, it was hypothesized that circKMT2D
could serve a regulatory role in the progression of OS. The effect
of circKMT2D on H2O2-treated OS cell
phenotype was therefore investigated in the present study.
Furthermore, the role of miRNAs in regulating key pathways related
to tumorigenesis may provide a powerful novel tool for diagnosis
and targeted therapy. miR-210 has been reported to be involved in
the occurrence and development of various types of cancer,
including bladder cancer, glioblastoma and hepatocellular carcinoma
(18-20).
The clinical treatment of human tumors is in urgent need for the
development of novel strategies to provide novel and reliable
biomarkers and therapeutic targets (21). Therefore, the mechanisms through
which circKMT2D may affect H2O2-attenuated OS
cell progression were investigated by exploring the
miR-210/autophagy pathway in the present study.
To the best of our knowledge, the present study was
the first to investigate the role of circKMT2D in the progression
of H2O2-attenuated OS cells. The findings
from the present study may provide new insights into OS progression
and may therefore provide a novel theoretical basis and a potential
effective target for OS therapy.
Materials and methods
Cell lines
MG63 and U2OS cell lines were provided by The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
Cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin (Thermo Fisher Scientific, Inc.) and 100 µg/ml
streptomycin (Thermo Fisher Scientific, Inc.) and placed at 37˚C in
a humidified incubator containing 5% CO2.
Cell treatment with
H2O2
MG63 and U2OS cells in the logarithmic phase were
collected and prepared as dispersed cell suspensions in DMEM
complete medium containing 100 µmol/l H2O2
(Beijing Solarbio Science & Technology Co., Ltd.) for 24 h at
37˚C (22). The density of each
cell suspension was 2x106 cells/ml and each cell
suspension was seeded into 6-well plates at a volume of 1 ml. All
plates were maintained in an incubator at 37˚C and 5%
CO2.
Transfection
MG63 and U2OS cells in the logarithmic phase were
collected and seeded in 6-well plates at a density of
5x105 cells/well. A total of 1 ml serum-free DMEM medium
was then added into each well. When the cell confluence rate
reached ~70%, MG63 and U2OS cells were transfected with 10 nM
circKMT2D short hairpin (sh)RNA (shcircKMT2D group;
5'-UGAUGAAUGCCGGCUCUAG-3') and negative control (shNC group;
5'-UGACGGAUGCACGCUCUAG-3') using Lipofectamine™ 2000 transfection
reagent (Thermo Fisher Scientific, Inc.). In addition, MG63 cells
were subjected to transfection with 10 nM miR-210 mimic (miR-210
mimic group; 5'-UCAGUCCAUGGUAGAACUUCG-3'), miR-210 inhibitor
(miR-210 inhibitor group; 5'-UCGCUUGUGUCAGGUCCGCAA-3') and miR-210
NC (NC group; 5'-GGACCGUAGCCACUGUGAGUU-3'), respectively.
Furthermore, pcDNA3.1-circKMT2D vector (10 nM) was used to
transfect the MG63 cells (pcDNA3.1-circKMT2D group).
Co-transfection was also performed on the MG63 cells using 10 nM
circKMT2D shRNA and 10 nM miR-210 inhibitor simultaneously
(shcircKMT2D + miR-210 inhibitor group). All plates were maintained
in an incubator at 37˚C and 5% CO2 for 6 h of
transfection. Transfection was conducted using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
transfection efficiency was measured by reverse
transcription-quantitative (RT-q)PCR. Subsequently, transfected
cells were cultured in DMEM containing 10% FBS at a volume of 1 ml
and were placed at 37˚C and 5% CO2 for 48 h. circKMT2D
shRNA, shRNA NC and pcDNA3.1-circKMT2D vector were purchased from
Genechem, Inc. miR-210 mimic, miR-210 inhibitor and miR-210 NC were
provided by Shangai GenePharma Co., Ltd..
Autophagy inhibitor treatment
MG63 cells in the miR-210 mimic group were incubated
with DMEM complete medium containing 12.5 µg/ml 3-methyladenine
(3-MA; Sigma-Aldrich; Merck KGaA) for 12 h at 37˚C and 5%
CO2. These cells constituted the miR-210 mimic + 3-MA
group.
Cell counting kit-8 (CCK-8) assay
Cells cultured at 48 h post-transfection were
harvested and seeded in 96-well plates at a density of
2x103 cells/well. Cells were incubated with 100 µl DMEM
complete medium containing or not 100 µmol/l
H2O2 for 48 h at 37˚C. Subsequently, 10 µl
CCK-8 solution (Dojindo Molecular Technologies, Inc.) was added to
each well at 37˚C for 2 h before reading the absorbance at 450 nm
using a microplate reader. Each sample was assessed in triplicate
and the experiments were repeated three times.
Transwell assay
Transwell assay was performed using an 8-µm
polycarbonate filter culture chamber (96-well plates; EMD
Millipore) pre-plated with Matrigel for 1 h at room temperature. At
48 h post-transfection, cells were harvested and were seeded in the
upper chamber of the chamber at the density of 2x105
cells per well. Serum-free DMEM (100 µl, with or without 100 µM
H2O2) was also added to the upper chamber.
The lower chamber was filled with 500 µl DMEM complete medium with
three replicate wells per group. Cells were cultured for 36 h at
37˚C and 5% CO2. After the non-invasive cells were
removed, the invading cells were washed twice with PBS, fixed for
10 min with 4% paraformaldehyde at room temperature for 20 min and
stained with 0.1% crystal violet for 5 min. The invading cells were
placed under a light microscope (Nikon Corporation; magnification,
x200) and photographed.
Apoptosis detection
Cells cultured for 48 h were washed with PBS,
harvested and centrifuged at room temperature for 5 min at 1,000 x
g. A total of 500 µl binding buffer was used to resuspend the
cells, and 5 µl AnnexinV-fluorescein isothiocyanate (FITC; cat. no.
40302ES20; Shanghai Yeasen Biotechnology Co., Ltd.) and 10 µl
propidium iodide (PI) were then added to the cells. Cells were
evenly mixed with the reagent and were stored in the dark for 15
min at room temperature. The apoptosis rate of each cell sample was
measured by flow cytometry (BD Biosciences) and analyzed using
FlowJo software (version 7.6.1; FlowJo LLC).
RT-qPCR
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse
transcribed into a cDNA template busing the reverse transcription
kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according
to the manufacturers' instructions. The PCR amplification reaction
was carried out using cDNA as template with the following
conditions: 95˚C for 10 min, 95˚C for 15 sec, 62˚C for 30 sec, and
72˚C for 30 sec. This process was cycled 40 times, followed by an
extension at 72˚C for 10 min. The relative expression levels of
circKMT2D, miR-210, Beclin1 and p62 were calculated using the
2-ΔΔCq method (23). U6 was used as
the internal control for circKMT2D and miR-210, while GAPDH served
as the internal control for Beclin1 and p62. The primer (Shanghai
GeneChem) sequences used in the present study were as follows:
circKMT2D forward, 5'-GATTTAATATTAACTGACG-3', reverse,
5'-GTTATGGATCCCGGCATGGC-3'; miR-210 forward,
5'-ATGCCTGTGCGTGTGA-3', reverse, 5'-GTGCGTGTCGTGGAGTC-3'; U6
forward, 5'-CGCTTCGGCAGCACATATACTA-3', reverse,
5'-CGCTTCACGAATTTGCGTGTCA-3'; Beclin1 forward,
5'-GATGGAAGGGTCTAAGACGTCCAA-3', reverse,
5'-TTTCGCCTGGGCTGTGGTAAG-3'; p62 forward,
5'-CCAGCACCAAGAGCACGGACAGCG-3', reverse,
5'-TGGGGAGAAGAAGGGGACCACGAA-3'; and GAPDH forward,
5'-AAGGTCATCCCTGAGCTGAAC-3' and reverse,
5'-ACGCCTGCTTCACCACCTTCT-3'.
Bioinformatic prediction and dual
luciferase reporter assay
StarBase 2.0 (http://starbase.sysu.edu.cn) was used to predict
whether the miRNAs could bind to circKMT2D. MG63 cells
(1.5x104 cells/well) in the NC group, miR-210 mimic
group and miR-210 inhibitor group were seeded into 6-well plates at
logarithmic growth phase. Following 24 h culture,
pmirGLO-circKMT2D-Mutant-type (Promega Corporation) and
pmirGLO-circKMT2D-Wild-type (Promega Corporation) luciferase
reporter vectors were used to transfect these cells. Transfection
was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Three replicate wells were set in each group.
Following 24 h of incubation at 37˚C and 5% CO2. The
relative firefly and Renilla luciferase activities were
detected using a Dual-Luciferase Reporter assay system (Promega
Corporation). Firefly luciferase activity was normalized to that of
Renilla luciferase.
Western blotting
Cells were harvested and lysed using RIPA buffer
(Thermo Fisher Scientific, Inc.). Protein concentration was
detected using a BCA kit (Beyotime Institute of Biotechnology).
Proteins (20 µg) were separated by 10% SDS-PAGE and transferred to
PVDF membranes. Membranes were washed three times with TBST (0.1%)
and blocked with 5% non-fat milk for 2 h at room temperature.
Membranes were incubated with rabbit anti-mouse Beclin1 (1:1,000;
Cell Signaling Technology, Inc.) and p62 (1:1,000; Cell Signaling
Technology, Inc.) primary antibodies for 12 h at 4˚C. Membranes
were washed with TBST and were incubated with goat anti-rabbit
secondary antibody (1:2,000, Beijing Solarbio) for 2 h at room
temperature. Enhanced chemiluminescence chromogenic solution
(Beijing Solarbio Science & Technology Co., Ltd.) was used to
detect the signal on the membrane. The data were analyzed via
densitometry and normalized to expression of the internal control
GAPDH. Protein expression was evaluated using Image-Pro®
Plus software (version 6.0; Media Cybernetics, Inc.).
Statistical analysis
The data were expressed as the means ± standard
deviation. SPSS 19.0 software (IBM Corp.) was used for statistical
analysis. Comparison between two groups was carried out using
Student's t-test. Comparison between three groups was performed
using ANOVA followed by Tukey's test. P<0.05 was considered to
indicate a statistically significant difference.
Results
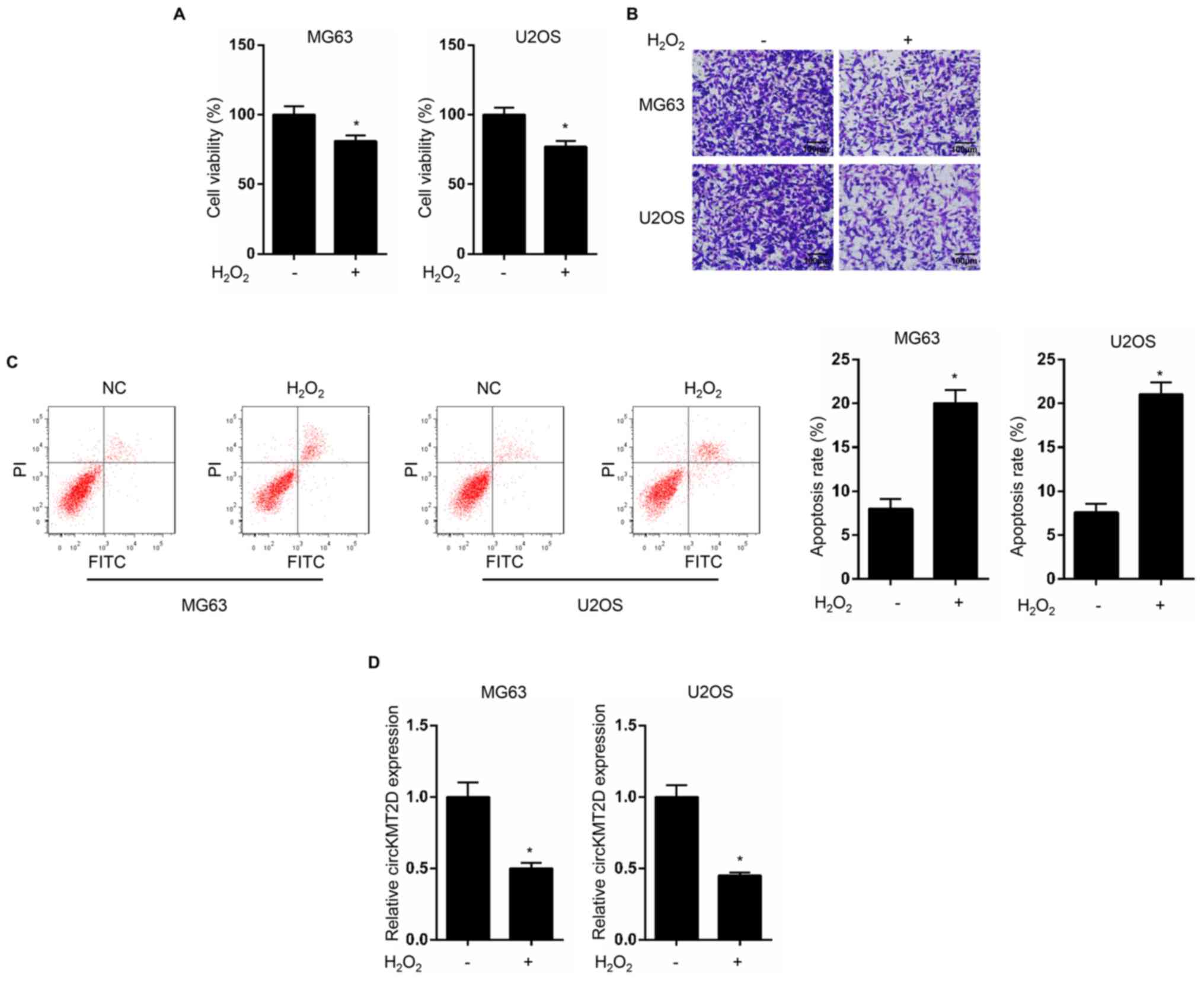
H2O2 treatment
inhibits OS progression and decreases circKMT2D expression in OS
cells
MG63 and U2OS cells were treated with
H2O2, and cell viability, invasive ability
and apoptosis were detected. The results demonstrated that MG63 and
U2OS cells treated with H2O2 exhibited a
significantly lower cell viability and invasive ability, and a
significantly increased apoptotic rate compared with untreated
cells (Fig. 1A-C). In addition,
circKMT2D expression level was significantly decreased in
H2O2-treated MG63 and U2OS cells compared
with control cells (Fig. 1D).
circKMT2D knockdown accentuates
H2O2-treated OS cell viability and invasion
inhibition
Compared with the shNC group, MG63 and U2OS cells in
the shcircKMT2D group exhibited a significantly lower circKMT2D
expression level (Fig. 2A),
indicating that circKMT2D was successfully knocked down in MG63 and
U2OS cells. The results of CCK-8 assay, Transwell assay and
apoptosis detection indicated that MG63 and U2OS cells in the
shcircKMT2D group exhibited a significantly lower cell viability
and invasion ability, and a significantly higher apoptotic rate
compared with cells in the shNC group (Fig. 2B-D).
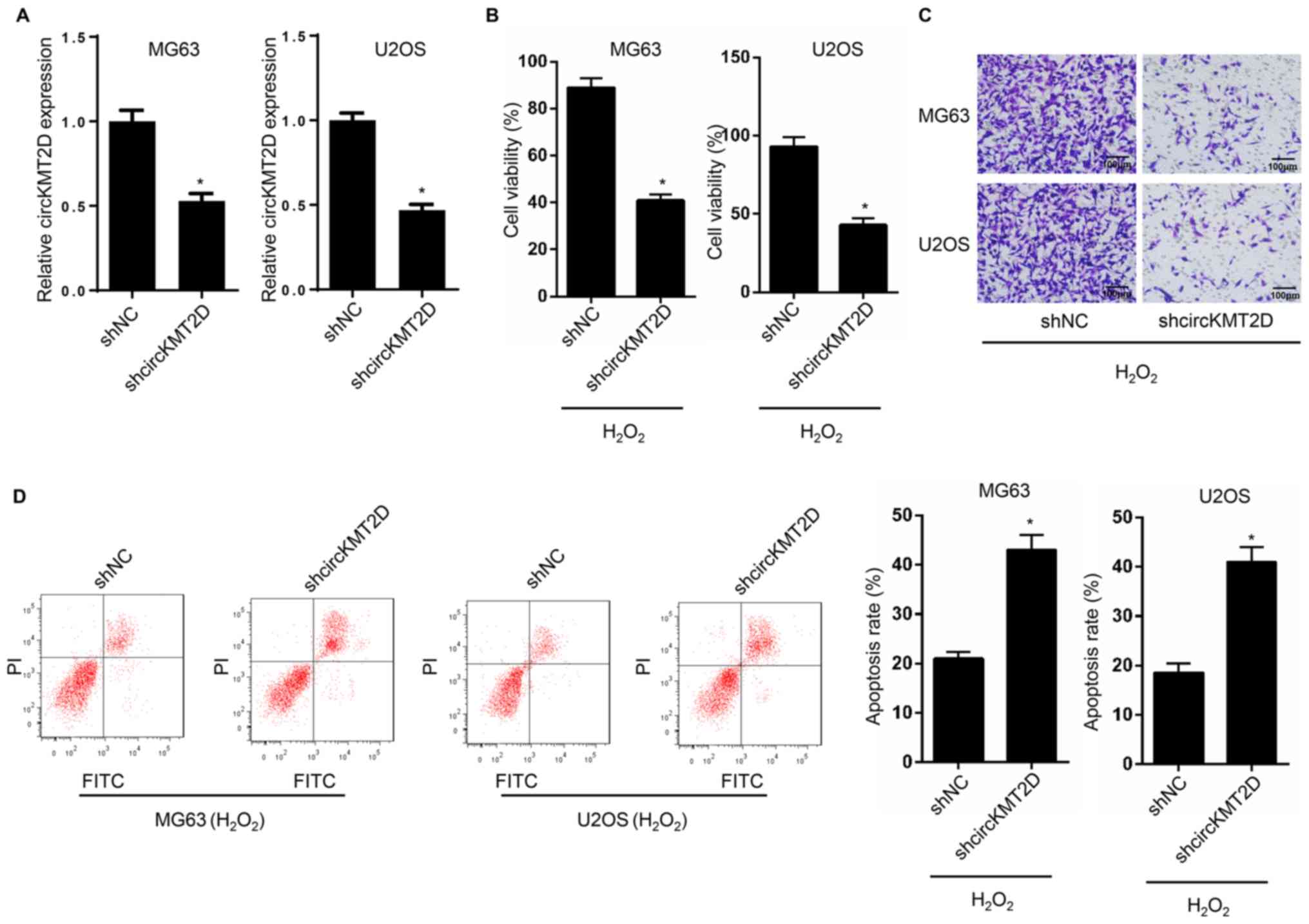 | Figure 2Knockdown of circKMT2D contributed to
H2O2-induced OS cells viability and invasion
inhibition. (A) MG63 and U2OS cells were transfected and circKMT2D
expression was detected by reverse transcription quantitative PCR.
(B) After transfection, MG63 and U2OS cell viability was evaluated
using the Cell Counting Kit-8 assay. (C) After transfection, MG63
and U2OS cell invasion ability was detected by Transwell experiment
(Magnification, x100; scale bar, 100 µm). (D) Following
transfection, MG63 and U2OS cell apoptosis was detected by flow
cytometry. *P<0.05. NC, negative control; FITC,
fluorescein isothiocyanate; PI, propidium iodide;
H2O2, hydrogen peroxide; sh, short
hairpin. |
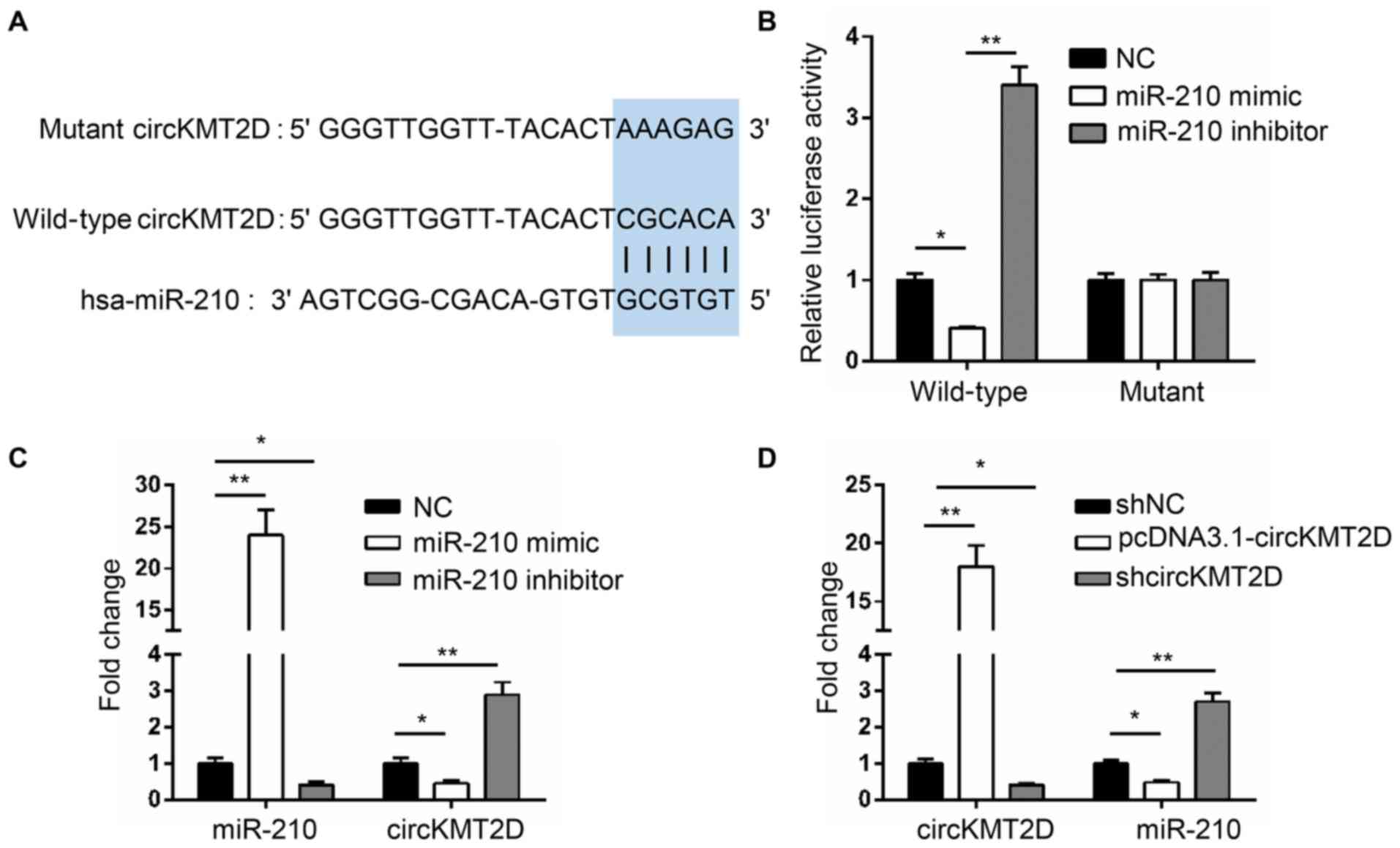
circKMT2D possesses binding sites for
miR-210 and directly inhibits miR-210 expression
The binding site between circKMT2D and miR-210 was
predicted by Starbase V2.0 and is illustrated in Fig. 3A. The results from dual luciferase
reporter assay reported that, compared with the luciferase activity
of wild-type circKMT2D in MG63 cells of the NC group, a
significantly decreased luciferase activity was observed in the
miR-210 mimic group, and a significantly increased luciferase
activity was demonstrated in the miR-210 inhibitor group. However,
compared with the luciferase activity of mutant circKMT2D in the NC
group, no changes were observed in the miR-210 mimic and miR-210
inhibitor groups (Fig. 3B). Thus,
miR-210 is a target gene of circKMT2D, and its expression is
directly inhibited by circKMT2D.
To further verify the regulatory association between
the two genes, MG63 cells were transfected with NC, miR-210 mimic
and miR-210 inhibitor and the expression of circKMT2D and miR-210
was detected. The results demonstrated that, compared with the NC
group, a significantly higher miR-210 expression and a
significantly lower circKMT2D expression were found in the miR-210
mimic group. However, when compared with the NC group, MG63 cells
transfected with miR-210 inhibitor exhibited a significantly lower
miR-210 expression and a higher circKMT2D expression (Fig. 3C). In addition, MG63 cells in the
pcDNA3.1-circKMT2D group exhibited a significantly higher circKMT2D
expression and a significantly lower miR-210 expression compared
with those in the shNC group; however, cells in the shcircKMT2D
group exhibited a lower circKMT2D expression and a significantly
increased miR-210 expression compared with those in the shNC group
(Fig. 3D). These findings further
confirmed that miR-210 expression may directly be inhibited by
circKMT2D.
miR-210 silencing partially reverses
the OS cell phenotype induced by circKMT2D knockdown
miR-210 expression in shNC, shcircKMT2D and
shcircKMT2D + miR-210 inhibitor groups was assessed. A
significanlty higher miR-210 expression was observed in the
shcircKMT2D group compared with the shNC group. However, compared
with the shcircKMT2D group, the expression of miR-210 was
significantly decreased in the shcircKMT2D + miR-210 inhibitor
group (Fig. 4A). Furthermore,
compared with the shNC group, MG63 cells in the shcircKMT2D group
exhibited a significantly lower cell viability and invasive
ability, and a significantly higher apoptotic rate. However, MG63
cells in the shcircKMT2D + miR-210 inhibitor group exhibited a
significantly higher cell viability and invasive ability, and a
significantly lower apoptotic rate compared with those in the
shcircKMT2D group (Fig. 4C and
D).
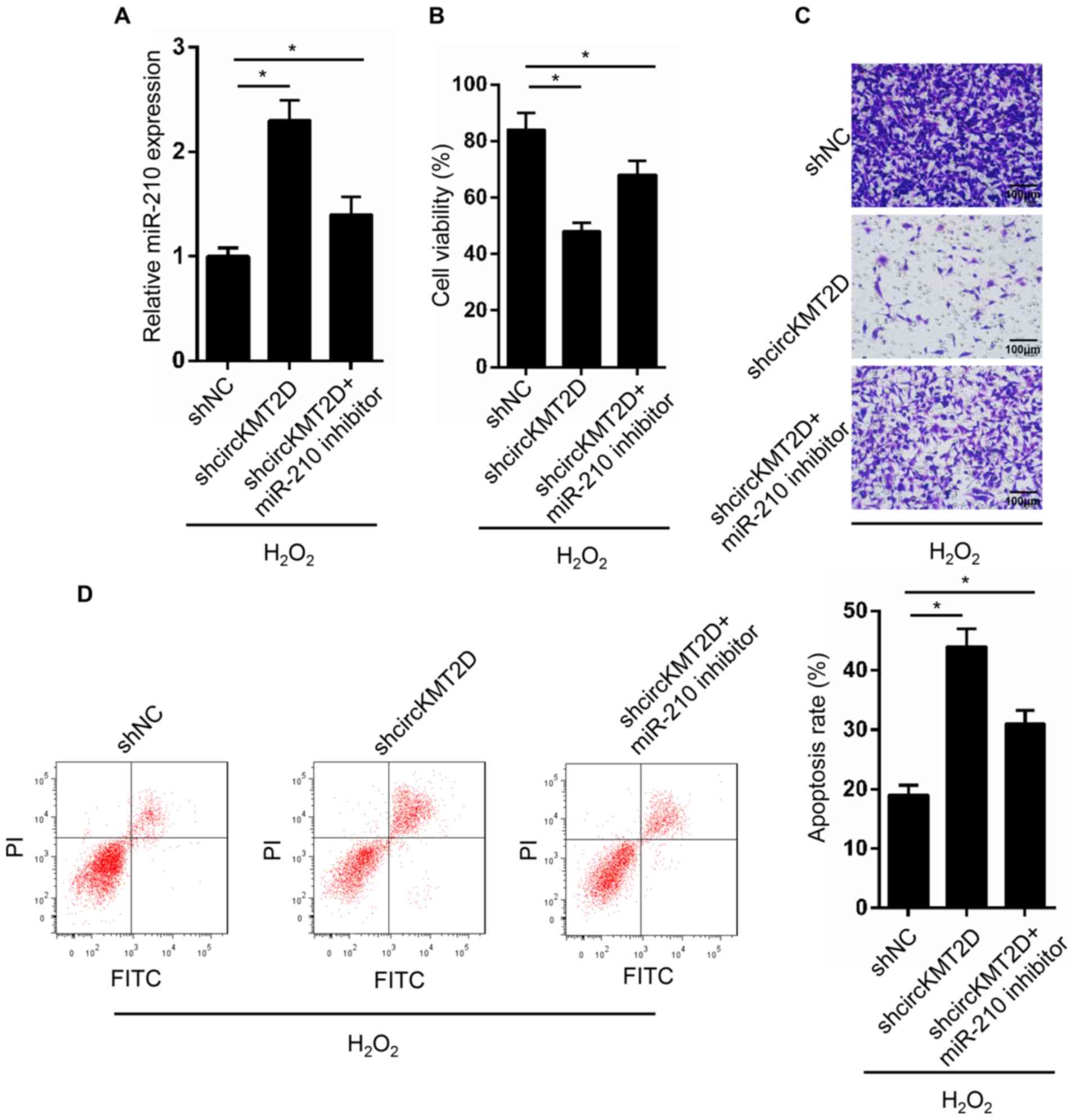 | Figure 4Silencing of miR-210 partially
reversed OS cells phenotype induced by circKMT2D knockdown. (A)
MG63 cells were transfected and miR-210 expression in MG63 cells of
each group was detected by reverse transcription quantitative PCR.
(B) After transfection, MG63 cell viability was evaluated using the
Cell Counting Kit-8 assay. (C) After transfection, MG63 cell
invasive ability was detected by Transwell experiment
(Magnification, x100; scale bar, 100 µm). (D) After transfection,
MG63 cell apoptosis was detected by flow cytometry.
*P<0.05. NC, negative control; FITC, fluorescein
isothiocyanate; PI, propidium iodide; H2O2,
hydrogen peroxide; sh, short hairpin; miR, microRNA. |
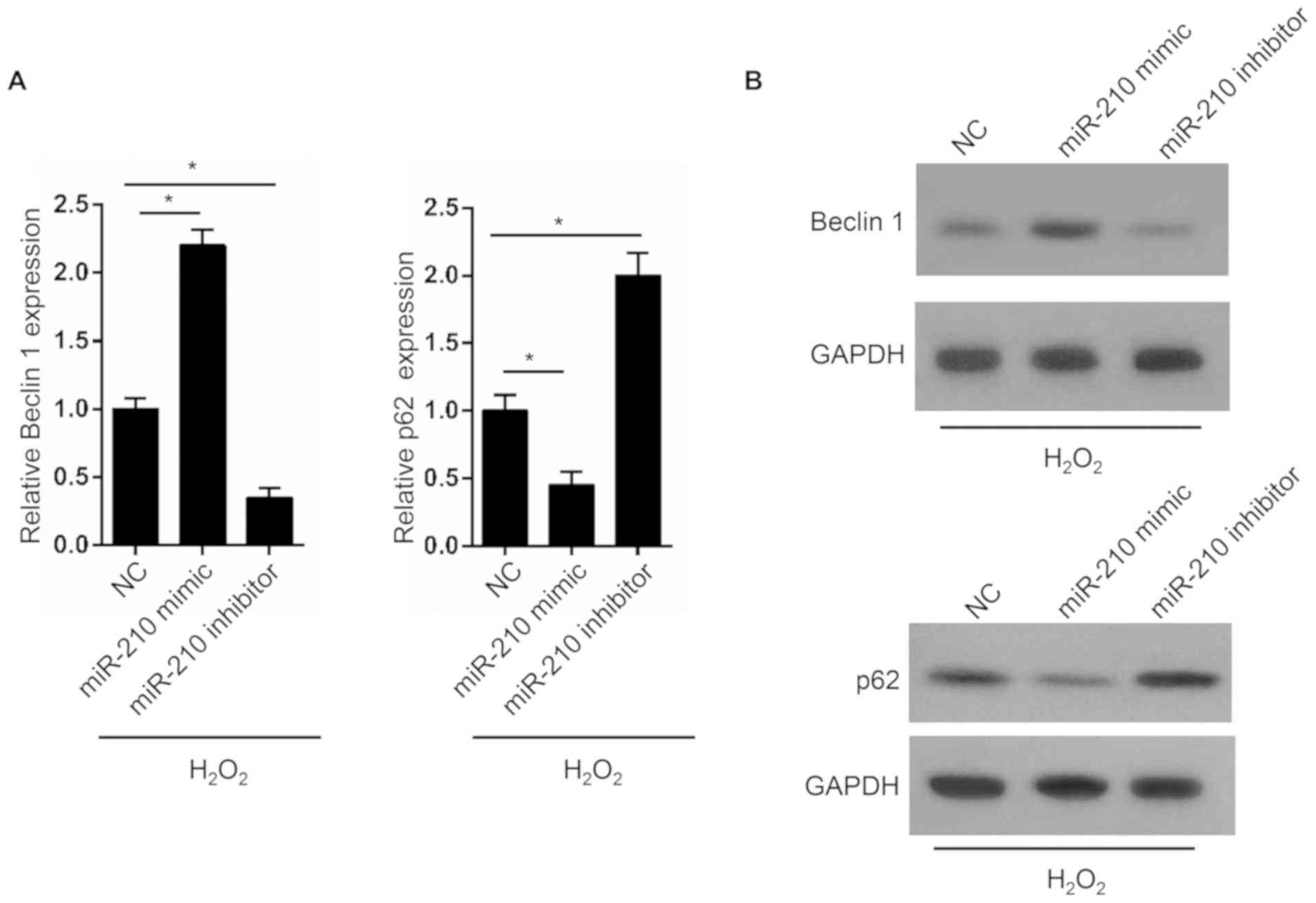
miR-210 promotes OS cell
autophagy
Beclin1 and p62 are genes associated with autophagy.
In the present study, the expression of Beclin1 and p62 in MG63
cells was determined. The results from RT-qPCR demonstrated that
MG63 cells in the miR-210 mimic group exhibited a significantly
higher Beclin1 expression level and a significantly decreased p62
expression level compared with those in the NC group. However,
compared with the NC group, a significantly lower Beclin1
expression level and a significantly decreased p62 expression level
were observed in the miR-210 inhibitor group (Fig. 5A). These results were similar to
those observed following western blotting analysis (Fig. 5B).
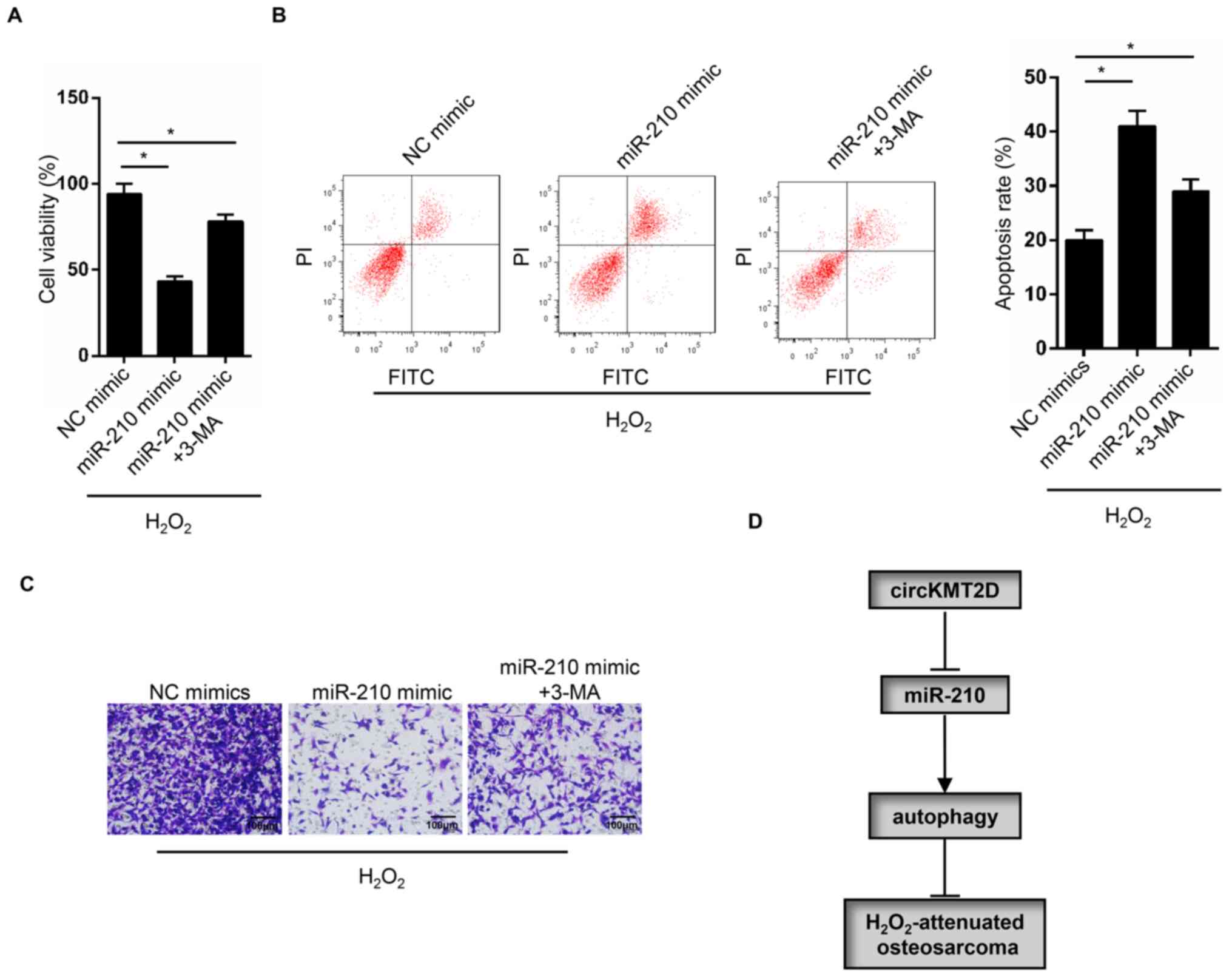
Autophagy inhibitor reverses the OS
cell phenotype induced by miR-210 overexpression
The autophagy inhibitor 3-MA was used to treat MG63
cells transfected with miR-210 mimic. MG63 cells in the miR-210
mimic group exhibited a significantly decreased cell viability and
invasive ability, and a significantly higher apoptotic rate
compared with those in the NC group. However, compared with the
miR-210 mimic group, a significantly elevated viability and
invasive ability, and a significantly decreased apoptotic rate were
observed in the miR-210 mimic + 3-MA group (Fig. 6A-C). These findings suggested that
circKMT2D knockdown may contribute to
H2O2-attenuated OS progression inhibition via
the miR-210/autophagy pathway (Fig.
6D).
Discussion
OS is the most common primary malignant bone tumor.
It mostly occurs in individuals under 20 years of age and is
characterized by a rapid speed of deterioration (24,25).
The prognosis of patients with OS is poor due to invasion and
metastasis occurring in the early stages of the disease (26). Currently, OS treatment with
chemotherapy has limited therapeutic efficacy as well as
unsatisfactory side effects, in particular for relapsed and
advanced OS (27).
H2O2 is one main contributor of oxidative
damage (28). Enhanced metabolic
activity is one of the key features of tumor cells, and
overproduction of H2O2 induces increased
oxidative stress (29).
Continuously elevated H2O2 levels can promote
the survival of tumor cells and may therefore promote tumor cell
ability to adapt to the tumor microenvironment. However, high
levels of H2O2 can also induce cell cycle
arrest and cell death by apoptosis, thereby inhibiting the
progression of tumors (30). Park
(31) reported that 100 µM
H2O2 suppressed the proliferation of lung
cancer cells by arresting the cell cycle and inducing cells
necrosis or apoptosis. In 2003, Ogawa et al (32) reported that potent oxidative stress
caused by exogenous H2O2 induces the
apoptosis of human OS cells. A recent study also demonstrated that
treatment with H2O2 at a concentration of 100
µM resulted in a significant decrease in OS cell viability and an
increase in apoptosis (9). The
present study also confirmed that 100 µM H2O2
inhibited OS cell viability and invasive ability and promoted OS
cell apoptosis. The underlying mechanism may involve the inhibition
of circKMT2D expression.
To the best of our knowledge, the effects of
circKMT2D on human tumors have not yet been reported. The present
study demonstrated for the first time that circKMT2D may function
as a cancer-promoting gene in OS. The regulatory effects of
circRNAs on OS progression have been previously reported. Zhang
et al (33) indicated that
circUBAP2 enhanced the progression of OS by acting as a sponge of
miR-143. Liu et al (34)
reported that circNT5C2 promoted the metastasis of OS by targeting
miR-448. However, circ0002052 can inhibit OS development by
inhibiting OS cell proliferation and invasion, and by promoting
apoptosis via targeting miR-1205(15). In the present study, the
cancer-promoting effects of circKMT2D in OS were discovered for the
first time, and circKMT2D knockdown contributed to the inhibition
of H2O2-attenuated OS progression by
targeting miR-210.
Accumulating evidence indicates that miRNAs, as new
types of diagnostic molecules and markers, have great prospects in
targeted therapy of tumors, including OS (35,36).
Fan et al (37) screened
some key genes involved in the development of OS, including miRNAs,
which may be related to the metastasis of OS. In several human
malignancies, such as breast cancer, malignant peripheral nerve
sheath tumors and renal cell carcinoma, miR-210 has been found to
promote tumor development due to its upregulation and the
enhancement of tumor cell proliferation or invasive ability
(38-40).
In OS, it has been reported that miR-210 functions as an oncogene.
It promotes the migration and invasion of OS cells and is closely
related to the aggressive development of OS (41,42).
miR-210 has also been demonstrated to promote the dedifferentiation
of OS cells (43). However, in the
present study, rescue experiments revealed that the silencing of
miR-210 partially reversed the H2O2-induced
OS cell phenotypes induced by circKMT2D knockdown. miR-210 may
therefore function as a tumor suppressor in OS and may serve as a
potential target for targeted therapy and diagnosis of OS. Further
experiments indicated that miR-210 may regulate the development of
OS by interfering with autophagy in OS cells.
Autophagy is present in eukaryotes and plays an
important role in cellular homeostasis, which is usually activated
during adverse microenvironmental stress (44,45).
During the autophagy process, organelles with damaged or aged
structures and unwanted biomacromolecules are relocated to
lysosomes for digestion and degradation, and autophagic products
can be utilized by cell reconstruction to provide material basis
and energy for cell survival (46,47).
Numerous studies have demonstrated that the loss or inhibition of
autophagy may lead to the development of a variety of tumors,
including OS (48-50).
A previous study has reported that the damage of lysosomal
degradation function can lead to the accumulation of
autophagosomes, whereas excess autophagosome accumulation and
autophagy degradation blocking play important roles in OS cell
death (51). Beclin-1 is a key
regulatory protein in autophagy and its downregulation has been
confirmed to promote the progression of several human cancers,
including OS (52,53). Under normal conditions, the p62
level is very low in cells, since it is degraded in selective
autophagy. However, p62 will accumulate in cells in the state of
autophagy inhibition and autophagy defects (54). The accumulation of p62 can lead to
the activation of NF-κB and stabilize Nrf2, whereas activated NF-κB
can promote tumorigenesis and Nrf2 can enhance the tolerance of
tumor cells to hypoxia-induced stress (55,56).
In the present study, miR-210 overexpression elevated Beclin1
expression and decreased p62 expression in OS cells. miR-210 may
therefore function by promoting autophagy to hinder the development
of OS.
The PI3K/AKT/mTOR pathway has been demonstrated to
be transitionally activated in OS, and the silencing of Wilms tumor
gene 1 can inhibit the proliferation of OS cells by inactivating
the PI3K/AKT pathway (57,58). Researchers have discovered that the
enhancement of the PI3K/AKT/mTOR pathway inhibits
doxorubicin-induced autophagy in OS cells and the diallyl
disulfide-induced autophagic death of OS cells by inhibiting the
PI3K/Akt/mTOR signaling pathway (59-61).
It is therefore hypothesized that circKMT2D and miR-210 may affect
autophagy through the PI3K/AKT/mTOR signaling pathway, thereby
regulating the development of OS. This hypothesis will be further
investigated.
In conclusion, the present study reported the
potential molecular mechanisms of circKMT2D in
H2O2-attenuated OS development. To the best
of our knowledge, the present study was the first to demonstrate
that circKMT2D knockdown contributed to the inhibition of
H2O2-attenuated OS progression via the
miR-210/autophagy pathway. These findings provided a theoretical
basis for the treatment of OS with H2O2. In
addition, circKMT2D may be considered as a novel target for the
targeted therapy of OS, which will be further investigated in
future studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JZ, ZL and XC designed the present study. MZ, CZ and
DC performed the experiments. YH and JZ analysed the data and
prepared the figures. JZ and XC drafted the initial manuscript. ZL
reviewed and revised the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by Ethics Committee
of The Third Affiliated Hospital of Soochow University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang L, Ge D, Chen X, Qiu J, Yin Z, Zheng
S and Jiang C: FOXP4-AS1 participates in the development and
progression of osteosarcoma by downregulating LATS1 via binding to
LSD1 and EZH2. Biochem Biophys Res Commun. 502:493–500.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lin YH, Jewell BE, Gingold J, Lu L, Zhao
R, Wang LL and Lee DF: Osteosarcoma: Molecular Pathogenesis and
iPSC Modeling. Trends Mol Med. 23:737–755. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bashur L and Zhou G: Cancer stem cells in
osteosarcoma. Case Orthop J. 10:38–42. 2013.PubMed/NCBI
|
|
4
|
Wang Z, Li B, Ren Y and Ye Z: T-cell-based
immunotherapy for osteosarcoma: Challenges and opportunities. Front
Immunol. 7(353)2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
He C, Sun J, Liu C, Jiang Y and Hao Y:
Elevated H3K27me3 levels sensitize osteosarcoma to cisplatin. Clin
Epigenetics. 11(8)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Fridovich I: Oxygen free radicals and
tissue damage: Chairman's introduction. Ciba Found Symp. 1–4.
1978.PubMed/NCBI
|
|
7
|
Dong K, Yang C, Yan Y, Wang P, Sun Y, Wang
K, Lu T, Chen Q, Zhang Y, Xing J and Dong Y: Investigation of the
intracellular oxidative stress amplification, safety and anti-tumor
effect of a kind of novel redox-responsive micelle. J Mater Chem B.
6:1105–1117. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hahn HJ, Kim KB, An IS, Ahn KJ and Han HJ:
Protective effects of rosmarinic acid against hydrogen
peroxideinduced cellular senescence and the inflammatory response
in normal human dermal fibroblasts. Mol Med Rep. 16:9763–9769.
2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang Y, Wang W and Qiu E: Protection of
oxidative stress induced apoptosis in osteosarcoma cells by
dihydromyricetin through down-regulation of caspase activation and
up-regulation of BcL-2. Saudi J Biol Sci. 24:837–842.
2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pei W, Tao L, Zhang LW, Zhang S, Cao J,
Jiao Y, Tong J and Nie J: Circular RNA profiles in mouse lung
tissue induced by radon. Environ Health Prev Med.
22(36)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang HD, Jiang LH, Sun DW, Hou JC and Ji
ZL: CircRNA: A novel type of biomarker for cancer. Breast Cancer.
25:1–7. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Barrett SP and Salzman J: Circular RNAs:
Analysis, expression and potential functions. Development.
143:1838–1847. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: circRNA_100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wu Z, Shi W and Jiang C: Overexpressing
circular RNA hsa_circ_0002052 impairs osteosarcoma progression via
inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis.
Biochem Biophys Res Commun. 502:465–471. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Huang L, Chen M, Pan J and Yu W: Circular
RNA circNASP modulates the malignant behaviors in osteosarcoma via
miR-1253/FOXF1 pathway. Biochem Biophys Res Commun. 500:511–517.
2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li Z, Xuan W, Huang L, Chen N, Hou Z, Lu
B, Wen C and Huang S: Claudin 10 acts as a novel biomarker for the
prognosis of patients with ovarian cancer. Oncol Lett. 20:373–381.
2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yang X, Shi L, Yi C, Yang Y, Chang L and
Song D: MiR-210-3p inhibits the tumor growth and metastasis of
bladder cancer via targeting fibroblast growth factor receptor-like
1. Am J Cancer Res. 7:1738–1753. 2017.PubMed/NCBI
|
|
19
|
Liu S, Jiang T, Zhong Y and Yu Y: miR-210
inhibits cell migration and invasion by targeting the brain-derived
neurotrophic factor in glioblastoma. J Cell Biochem: Feb 11, 2019
(Epub ahead of print).
|
|
20
|
Tan W, Lim SG and Tan TM: Up-regulation of
microRNA-210 inhibits proliferation of hepatocellular carcinoma
cells by targeting YES1. World J Gastroenterol. 21:13030–13041.
2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gulino R, Forte S, Parenti R, Memeo L and
Gulisano M: MicroRNA and pediatric tumors: Future perspectives.
Acta Histochem. 117:339–354. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhao XH, Xu ZR, Zhang Q and Yang YM:
Simvastatin protects human osteosarcoma cells from oxidative
stress-induced apoptosis through mitochondrial-mediated signaling.
Mol Med Rep. 5:483–488. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang Y, Wang F, Li M, Yu Z, Qi R, Ding J,
Zhang Z and Chen X: Self-stabilized hyaluronate nanogel for
intracellular codelivery of doxorubicin and cisplatin to
osteosarcoma. Adv Sci (Weinh). 5(1700821)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li S, Zhang T, Xu W, Ding J, Yin F, Xu J,
Sun W, Wang H, Sun M, Cai Z and Hua Y: Sarcoma-targeting
peptide-decorated polypeptide nanogel intracellularly delivers
shikonin for upregulated osteosarcoma necroptosis and diminished
pulmonary metastasis. Theranostics. 8:1361–1375. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Durfee RA, Mohammed M and Luu HH: Review
of osteosarcoma and current management. Rheumatol Ther. 3:221–243.
2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang Y, Cai L, Li D, Lao YH, Liu D, Li M,
Ding J and Chen X: Tumor microenvironment-responsive
hyaluronate-calcium carbonate hybrid nanoparticle enables effective
chemotherapy for primary and advanced osteosarcomas. Nano Res.
11:4806–4822. 2018.
|
|
28
|
Wu PF, Long LH, Zeng JH, Guan XL, Zhou J,
Jin Y, Ni L, Wang F, Chen JG and Xie N: Protection of L-methionine
against H2O2-induced oxidative damage in
mitochondria. Food Chem Toxicol. 50:2729–2735. 2012.
|
|
29
|
Glasauer A and Chandel NS: Targeting
antioxidants for cancer therapy. Biochem Pharmacol. 92:90–101.
2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lennicke C, Rahn J, Lichtenfels R,
Wessjohann LA and Seliger B: Hydrogen peroxide-production, fate and
role in redox signaling of tumor cells. Cell Commun Signal.
13(39)2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Park WH: Hydrogen peroxide inhibits the
growth of lung cancer cells via the induction of cell death and
G1phase arrest. Oncol Rep. 40:1787–1794. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ogawa Y, Takahashi T, Kobayashi T, Kariya
S, Nishioka A, Mizobuchi H, Noguchi M, Hamasato S, Tani T, Seguchi
H, et al: Mechanism of hydrogen peroxide-induced apoptosis of the
human osteosarcoma cell line HS-Os-1. Int J Mol Med. 12:459–463.
2003.PubMed/NCBI
|
|
33
|
Zhang H, Wang G, Ding C, Liu P, Wang R,
Ding W, Tong D, Wu D, Li C, Wei Q, et al: Increased circular RNA
UBAP2 acts as a sponge of miR-143 to promote osteosarcoma
progression. Oncotarget. 8:61687–61697. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu X, Zhong Y, Li J and Shan A: Circular
RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation
and metastasis through targeting miR-448. Oncotarget.
8:114829–114838. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Liu H, Li P, Chen L, Jian C, Li Z and Yu
A: MicroRNAs as a novel class of diagnostic biomarkers for the
detection of osteosarcoma: A meta-analysis. OncoTargets Ther.
10:5229–5236. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Fujiwara T, Uotani K, Yoshida A, Morita T,
Nezu Y, Kobayashi E, Yoshida A, Uehara T, Omori T, Sugiu K, et al:
Clinical significance of circulating miR-25-3p as a novel
diagnostic and prognostic biomarker in osteosarcoma. Oncotarget.
8:33375–33392. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fan H, Lu S, Wang S and Zhang S:
Identification of critical genes associated with human osteosarcoma
metastasis based on integrated gene expression profiling. Mol Med
Rep. 20:915–930. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang Y, Yan J, Wang L, Dai H, Li N, Hu W
and Cai H: HIF-1α promotes breast cancer cell MCF-7 proliferation
and invasion through regulating miR-210. Cancer Biother Radiopharm.
32:297–301. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wang Z, Yin B, Wang B, Ma Z, Liu W and Lv
G: MicroRNA-210 promotes proliferation and invasion of peripheral
nerve sheath tumor cells targeting EFNA3. Oncol Res. 21:145–154.
2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Redova M, Poprach A, Besse A, Iliev R,
Nekvindova J, Lakomy R, Radova L, Svoboda M, Dolezel J, Vyzula R
and Slaby O: MiR-210 expression in tumor tissue and in vitro
effects of its silencing in renal cell carcinoma. Tumour Biol.
34:481–491. 2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Cai H, Lin L, Cai H, Tang M and Wang Z:
Prognostic evaluation of microRNA-210 expression in pediatric
osteosarcoma. Med Oncol. 30(499)2013.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu X, Zhang C, Wang C, Sun J, Wang D,
Zhao Y and Xu X: miR-210 promotes human osteosarcoma cell migration
and invasion by targeting FGFRL1. Oncol Lett. 16:2229–2236.
2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang H, Mai Q and Chen J: MicroRNA-210 is
increased and it is required for dedifferentiation of osteosarcoma
cell line. Cell Biol Int. 41:267–275. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ebner P, Poetsch I, Deszcz L, Hoffmann T,
Zuber J and Ikeda F: The IAP family member BRUCE regulates
autophagosome-lysosome fusion. Nat Commun. 9(599)2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ding WX, Ni HM, Gao W, Hou YF, Melan MA,
Chen X, Stolz DB, Shao ZM and Yin XM: Differential effects of
endoplasmic reticulum stress-induced autophagy on cell survival. J
Biol Chem. 282:4702–4710. 2007.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kim EH, Sohn S, Kwon HJ, Kim SU, Kim MJ,
Lee SJ and Choi KS: Sodium selenite induces superoxide-mediated
mitochondrial damage and subsequent autophagic cell death in
malignant glioma cells. Cancer Res. 67:6314–6324. 2007.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Terman A, Gustafsson B and Brunk UT:
Autophagy, organelles and ageing. J Pathol. 211:134–143.
2007.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Li L, Li Y, Zhao J, Fan S, Wang L and Li
X: CX-5461 induces autophagy and inhibits tumor growth via
mammalian target of rapamycin-related signaling pathways in
osteosarcoma. Onco Targets Ther. 9:5985–5997. 2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ranjan A and Srivastava SK: Penfluridol
suppresses pancreatic tumor growth by autophagy-mediated apoptosis.
Sci Rep. 6(26165)2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li C, Xu H, Chen X, Chen J, Li X, Qiao G,
Tian Y, Yuan R, Su S, Liu X and Lin X: Aqueous extract of clove
inhibits tumor growth by inducing autophagy through AMPK/ULK
pathway. Phytother Res. 33:1794–1804. 2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Xu J, Wang H, Hu Y, Zhang YS, Wen L, Yin
F, Wang Z, Zhang Y, Li S, Miao Y, et al: Inhibition of CaMKIIα
activity enhances antitumor effect of fullerene C60 nanocrystals by
suppression of autophagic degradation. Adv Sci (Weinh).
6(1801233)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Zhou W, Yue C, Deng J, Hu R, Xu J, Feng L,
Lan Q, Zhang W, Ji D, Wu J, et al: Autophagic protein Beclin 1
serves as an independent positive prognostic biomarker for
non-small cell lung cancer. PLoS One. 8(e80338)2013.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Xu R, Liu S, Chen H and Lao L:
MicroRNA-30a downregulation contributes to chemoresistance of
osteosarcoma cells through activating Beclin-1-mediated autophagy.
Oncol Rep. 35:1757–1763. 2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Mathew R, Karp CM, Beaudoin B, Vuong N,
Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al:
Autophagy suppresses tumorigenesis through elimination of p62.
Cell. 137:1062–1075. 2009.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ,
Tsai TF, Lin YJ, Wu CT and Liu HS: Autophagy suppresses
tumorigenesis of hepatitis B virus-associated hepatocellular
carcinoma through degradation of microRNA-224. Hepatology.
59:505–517. 2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Cheong H, Wu J, Gonzales LK, Guttentag SH,
Thompson CB and Lindsten T: Analysis of a lung defect in
autophagy-deficient mouse strains. Autophagy. 10:45–56.
2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Graziano AC, Cardile V, Avola R, Vicario
N, Parenti C, Salvatorelli L, Magro G and Parenti R: Wilms' tumor
gene 1 silencing inhibits proliferation of human osteosarcoma MG-63
cell line by cell cycle arrest and apoptosis activation.
Oncotarget. 8:13917–13931. 2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Zhang J, Yu XH, Yan YG, Wang C and Wang
WJ: PI3K/Akt signaling in osteosarcoma. Clin Chim Acta.
444:182–192. 2015.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Perry JA, Kiezun A, Tonzi P, Van Allen EM,
Carter SL, Baca SC, Cowley GS, Bhatt AS, Rheinbay E, Pedamallu CS,
et al: Complementary genomic approaches highlight the PI3K/mTOR
pathway as a common vulnerability in osteosarcoma. Proc Natl Acad
Sci USA. 111:E5564–E5573. 2014.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Wang L, Tang B, Han H, Mao D, Chen J, Zeng
Y and Xiong M: miR-155 affects osteosarcoma MG-63 cell autophagy
induced by adriamycin through regulating PTEN-PI3K/AKT/mTOR
signaling pathway. Cancer Biother Radiopharm. 33:32–38.
2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Yue Z, Guan X, Chao R, Huang C, Li D, Yang
P, Liu S, Hasegawa T, Guo J and Li M: Diallyl disulfide induces
apoptosis and autophagy in human osteosarcoma MG-63 cells through
the PI3K/Akt/mTOR pathway. Molecules. 24(2665)2019.PubMed/NCBI View Article : Google Scholar
|