Introduction
Breast cancer is the leading cause of death due to cancer in females worldwide (1). Although breast cancer-associated mortality declined in Europe between the years 2014 and 2019, 5,290,000 patients are estimated to have died of breast cancer in the past three decades (2). In 2018 alone, >60,000 cases of breast carcinoma in situ were diagnosed in the US (3). In China, the incidence of breast cancer in 2014 was 28.77 per 10,000 females and the mortality rate was 6.35 per 10,000 females. Furthermore, the economic and social burden of breast cancer has markedly increased over the past few decades (4). Although breast cancer-associated mortality has decreased due to advances in early screening and treatment, breast cancer remains a threat to females in China (4). Heterogeneity within the tumor and between tumor cells makes effective breast cancer treatment challenging.
Recent studies have indicated that molecular biomarkers based on high-throughput sequencing for breast cancer typing are valuable for predicting the prognosis for patients (5). Abnormal expression of certain molecular biomarkers is associated with breast cancer progression and drug resistance (6,7). Among tumor-associated transcriptional regulators, E74-like factor 1 (ELF1) is a member of the E26 transformation-specific (ETS) transcription family, which has >20 members that bind to genes bearing ETS DNA-binding sites to initiate gene transcription. ELF1 is expressed in a variety of tumor cells and has a tumor-promoting function (8). In breast cancer, ELF1 is a major factor driving the expression of the tumor-promoting gene Pygopus 2(9). Cold-shock domain-containing E1 (CSDE1), also known as upstream of N-ras, is a translational regulator with RNA- or DNA-binding ability (10,11). A previous study has indicated that CSDE1 promotes tumor cell migration and invasion (12).
Although studies have reported the functions of ELF1 and CSDE1, their regulatory networks in breast cancer have remained elusive. Chromatin immunoprecipitation sequencing (ChIP-seq) is widely used to search for downstream target genes of transcription factors and may help identify novel key genes regulated by transcription factors in breast cancer (13). In the present study, a series of experiments on the functions of ELF1 and CSDE1 in breast cancer cell lines were performed.
Materials and methods
Data collection
A total of two ChIP-seq datasets of the human breast cancer cell line MCF-7 were downloaded from the Gene Expression Omnibus (GEO) database (14). These two datasets (GEO accession numbers GSE105503 and GSE105291) contained data of genes bound to ELF1 (GSM2827208 and GSM2827209) and CSDE1 (GSM2825436 and GSM2825437), respectively, that had been obtained by high-throughput sequencing after ChIP experiments using ELF1 and CSDE1 antibodies. The high-throughput sequencing platform used was Illumina HiSeq 4000.
Differential target gene analysis
R package software (version 3.5.3; www.r-project.org) was used to normalize the downloaded data and identify the differential target genes between the GSE105503 and GSE105291 datasets. Target genes meeting the following criterion were identified as differential target genes: P<0.05. In addition, the R target ggplot2 package was used to visualize the differential target genes in order to generate heatmaps and volcano plots.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
GO and KEGG analyses were performed using the ClusterProfiler (version 3.1) in R to predict the potential function of ELF1-and CSDE1-associated target genes.
Protein-protein interaction (PPI) network construction and analysis of novel target genes
The R software package was used to predict the PPI network and the ELF1 and CSDE1 regulatory networks were visualized using Cytoscape software (https://cytoscape.org/).
Cell culture
A total of four breast cancer cell lines, namely MCF-7, BT-20, HS578T and MDA-MB-468, and one normal breast cell line, MCF-10A, were obtained from the American Type Culture Collection. All cell lines were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) containing 1% penicillin-streptomycin and 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at a temperature of 37˚C in a humidified atmosphere containing 5% CO2. Reverse transcription-quantitative PCR (RT-qPCR) was used to determine the ELF-1 and CSDE1 mRNA expression levels in different cell lines.
Transfection
The full ELF1 (NCBI Gene ID: 1997) and CSDE1 (NCBI Gene ID: 7812) gene sequences were cloned into the lentivirus (LV)003 plasmids which were purchased from General Biosystems, Inc. to construct ELF1 overexpression plasmid (LV003-ELF1) and CSDE1 overexpression plasmid (LV003-CSDE1), respectively. Empty LV003 plasmid vector was used for the negative control. The interfering fragment for small interfering RNA (siRNA) targeting ELF1 (si-ELF1) with the sequence 5'-GCGCTTGGTGTATCAGTTT-3' and siRNA (negative control) with the sequence 5'-GTTTTTGATGCGACCGTTG-3'; and si-CSDE1 with the sequence 5'-GGATCTACTTCTCCTCAAATA-3' and siRNA (negative control) with the sequence 5'-CATTTATCACTTTCACAACGG-3' were purchased from Shanghai GenePharma Co., Ltd. MCF-7 cells were seeded at 2x105 cells/well in 6-well plates. MCF-7 cells were transfected using the jetPRIME® transfection reagent from Polyplus Transfection with 2 µg plasmid DNA according to the manufacturer's instructions. Different siRNA fragments were transfected into cells using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) based on the manufacturer's protocol. The overexpression and silencing effect were determined by RT-qPCR after transfection for 48 h. The transfected cells were then used for a series of experiments.
RT-qPCR
Total RNA of MCF-7, BT-20, HS578T, MDA-MB-468 and MCF-10A cells; and total RNA of MCF-7 cells transfected with overexpression plasmids and different siRNA fragments were extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) based on the manufacturer's protocol. mRNA expression levels were detected by RT-qPCR. Reactions were performed in 50-µl volumes using SYBR Green PCR master mix (Beijing Solarbio Science & Technology Co., Ltd.) in an ABI-7900HT thermocycler (Thermo Fisher Scientific, Inc.). The RT thermal conditions were as follows: 42˚C for 15 min followed by 95˚C for 3 min using a PrimeScript RT reagent kit (Takara Bio, Inc.). The qPCR thermocycling conditions were as follows: 95˚C for 15 min followed by 40 cycles of 95˚C for 30 sec and 60˚C for 30 sec, with a final extension of 72˚C for 2 min. The 2-ΔΔcq method was used to analyze the mRNA expression levels (15). GAPDH was selected as an internal reference. All primers are listed in Table I.
 |
Table I
Information on primers used for PCR.
|
Table I
Information on primers used for PCR.
| Primer ID |
Sequence (5' to 3') |
Product length (bp) |
| H-ELF1-forward |
AACGAATAAATGGGATTCCTGAAG |
137 |
| H-ELF1-reverse |
GGAATCTGGTCGTGGTGGTT |
|
| H-CSDE1-forward |
CAGGCAAGGAACAAACCTCG |
183 |
| H-CSDE1-reverse |
TGTACCGGGCAATCTCACAG |
|
| H-CXCL10-forward |
CCTGCAAGCCAATTTTGTCCA |
170 |
| H-CXCL10-reverse |
TGCATCGATTTTGCTCCCCT |
|
| H-CXCL8-forward |
CACTGCGCCAACACAGAAAT |
181 |
| H-CXCL8-reverse |
GCTTGAAGTTTCACTGGCATC |
|
| H-CXCL6-forward |
TGCGTTGCACTTGTTTACGC |
72 |
| H-CXCL8-reverse |
CGGGGAACACCTGCAGTTTA |
|
| H-MRPL4-forward |
CGACCTGCACATCATGGACT |
181 |
| H-MRPL4-reverse |
AGCCGGGATCAAGTTGAAGG |
|
| H-NDUFB7-forward |
GGTAGGAGCTAGGTGACCCT |
140 |
| H-NDUFB7-reverse |
GTAGTCTGGCGGGAAGGTTG |
|
| H-SNRPE-forward |
AATTCCACCATGGCGTACCG |
104 |
| H-SNRPE-reverse |
CACCACAGTGGCTCCTAGTC |
|
| H-RPS26-forward |
GCCCATGTAAGGAGCTGAGTT |
152 |
| H-RPS26-reverse |
TGTGTCACTTCACACTTGGTCT |
|
| H-RPS11-forward |
TACCAAAAGCAGCCGACCAT |
122 |
| H-RPS11-reverse |
GCCTCCTTGGGTGTCTTGAA |
|
| H-RPS6-forward |
TGTTACTCCACGTGTCCTGC |
166 |
| H-RPS6-reverse |
AAGTCTGCGTCTCTTCGCAA |
|
| H-GAPDH-forward |
GAGTCAACGGATTTGGTCGT |
185 |
| H-GAPDH-reverse |
GACAAGCTTCCCGTTCTCAG |
|
Statistical analysis
Each group of experiments was performed in parallel three times. SPSS v.16.0 (SPSS, Inc.) was used for statistical analyses. Tukey's test was used to analyze the differences between groups. P<0.05 was considered to indicate statistical significance.
Results
Distribution of differential target genes between ELF1 and CSDE1
Genes that were present in the ELF1 ChIP-seq dataset were considered to be ELF1-associated target genes and genes present in the CSDE1 ChIP-seq dataset were considered to be CSDE1-associated target genes. The heatmap revealed that certain ELF1- and CSDE1-associated target genes had similar expression levels, while others were differentially expressed (Fig. 1A and Table SI). Volcano plots indicated a total of 921 differential target genes, including 95 significantly upregulated ELF1-associated target genes, where the fold change was >2 in comparison with the level in the CSDE1 dataset and 826 significantly CSDE1-associated target genes, which were expressed at <0.5 the level in the ELF1 dataset compared with the CSDE1 data set (Fig. 1B and Table SI). Genomic distribution analysis of these differential target genes suggested that the ELF1-associated target genes were mainly distributed in chromosome (chr)1, chr11 and chr12, while the CSDE1-associated target genes were mainly distributed in chr1, chr16 and chr19 (Fig. 2).
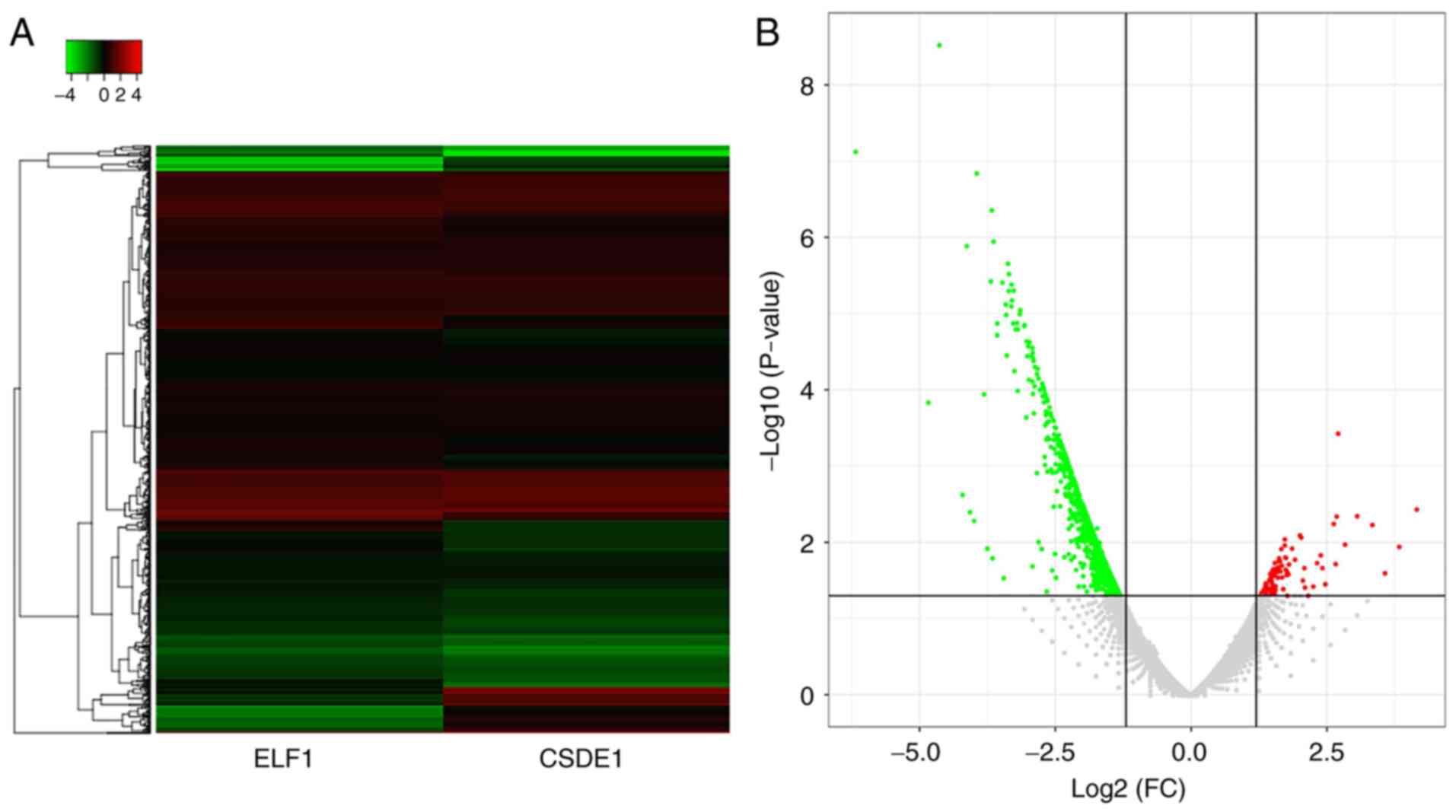 |
Figure 1
(A) Heatmap and (B) volcano plot providing an overview of differential target genes in the ELF1 and CSDE1 chromatin immunoprecipitation sequencing datasets. ELF1, E74-like factor 1; CSDE1, cold-shock domain-containing E1; FC, fold change.
|
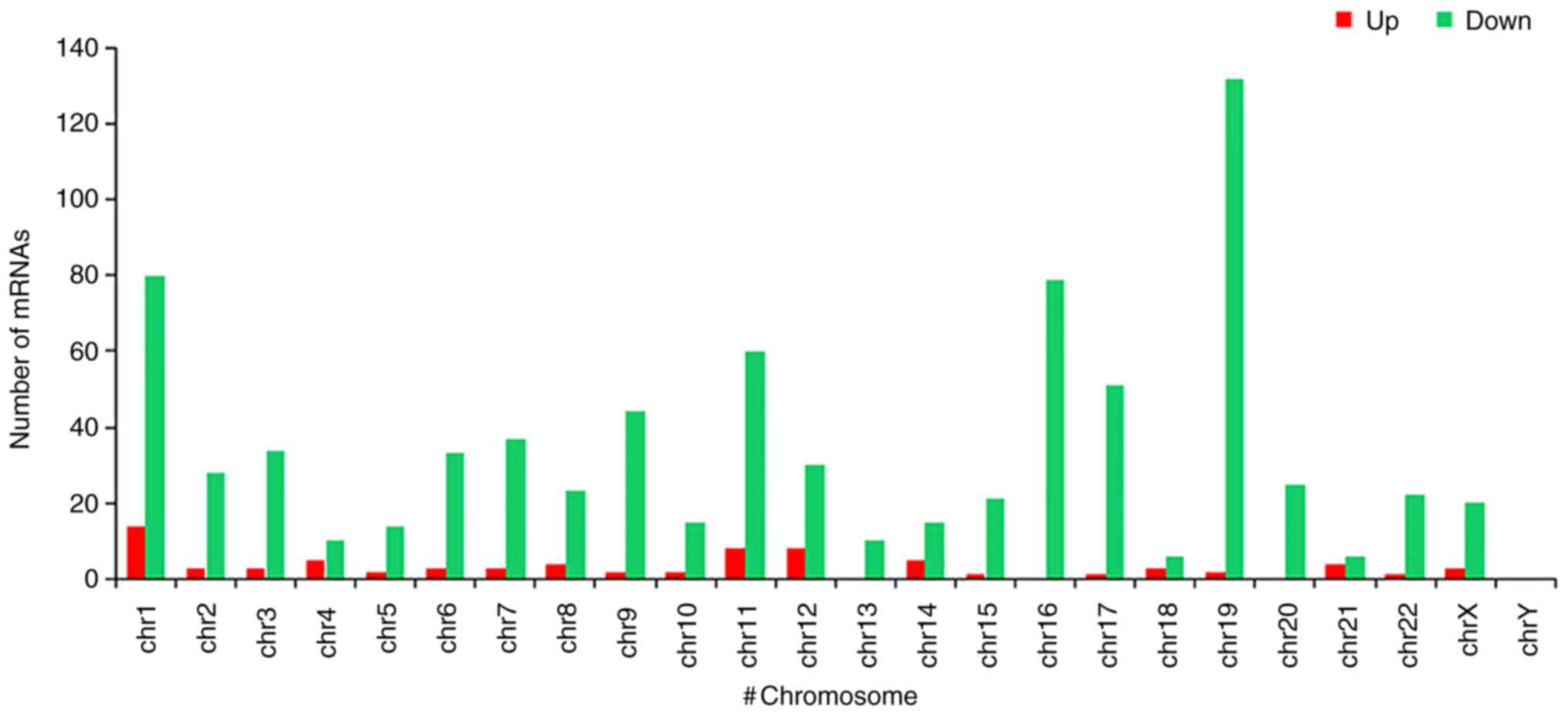 |
Figure 2
Distribution of differential target genes in the genome; Up: Genes with a higher enrichment level in the ELF1 ChIP-seq dataset compared to the CSDE1 ChIP-seq dataset. Down: Genes with a higher enrichment level in the CSDE1 ChIP-seq dataset compared to the ELF1 ChIP-seq dataset. ELF1, E74-like factor 1; CSDE1, cold-shock domain-containing E1; ChIP-seq, chromatin immunoprecipitation sequencing; chr, chromosome.
|
Predicting the functions of differential target genes of ELF1 and CSDE1
The GO and KEGG analyses were performed on ELF1- and CSDE1-associated target genes, respectively. The 95 upregulated and 826 downregulated differential target genes were enriched in 276 and 1,882 GO terms, respectively (Table SII), and were divided into the GO domains cellular component (CC), molecular function (MF) and biological process (BP). Among the ELF1-associated target genes, the two most highly represented terms in CC were ‘cell part’ and ‘cell membrane’, those in MF were ‘molecular transduced activity’ and ‘catalytic activity’, and those in BP were ‘cellular process’ and ‘response to stimulus’ (Fig. 3A). Among the CSDE1-associated target genes, the two most highly represented terms in CC were ‘cell’ and ‘cell part’, in MF, they were ‘binding’ and ‘transcription regulator activity’, and those in BP were ‘cellular process’ and ‘metabolic process’ (Fig. 3B).
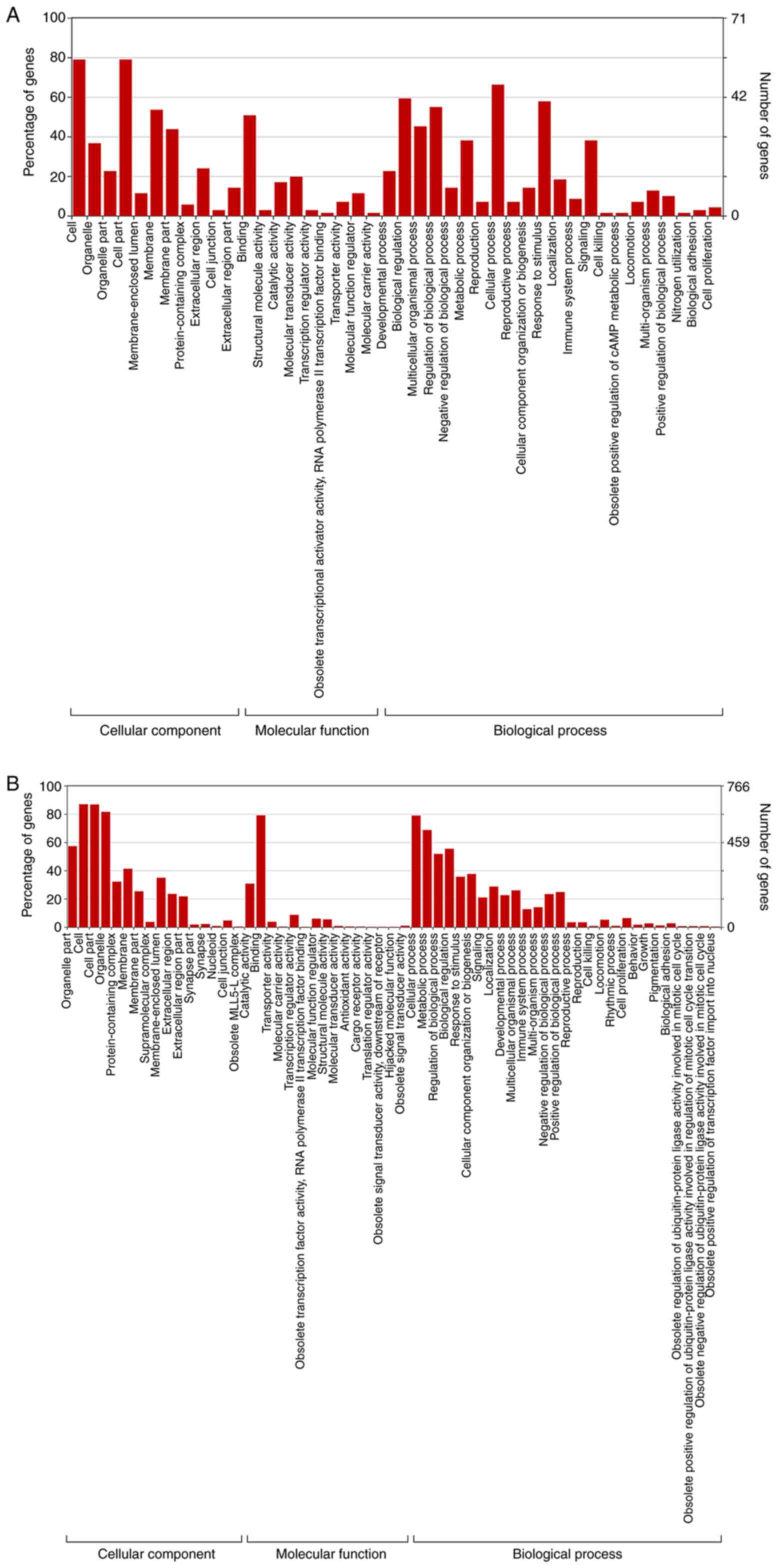 |
Figure 3
GO analysis indicated the potential function of differential target genes which exhibited higher enrichment in the (A) ELF1 ChIP-seq dataset compared to the CSDE1 ChIP-seq dataset. GO, Gene Ontology; ELF1, E74-like factor 1; CSDE1, cold-shock domain-containing E1; ChIP-seq, chromatin immunoprecipitation sequencing. GO analysis indicated the potential function of differential target genes which exhibited higher enrichment in the (B) CSDE1 ChIP-seq dataset compared to the ELF1 ChIP-seq dataset. GO, Gene Ontology; ELF1, E74-like factor 1; CSDE1, cold-shock domain-containing E1; ChIP-seq, chromatin immunoprecipitation sequencing.
|
In addition, the ELF1-and CSDE1-associated target genes were subjected to KEGG pathway enrichment analysis. The results revealed that the ELF1-associated target genes were mainly enriched in the categories ‘olfactory transduction’, ‘chemokine signaling pathway’, ‘carbohydrate digestion and absorption’, and ‘starch and sucrose metabolism’, while CSDE1-associated target genes were mainly enriched in ‘ribosomes’, ‘metabolic pathways’, ‘endocytosis’, ‘oxidative phosphorylation’, and ‘transcriptional misregulation in cancer’ (Fig. 4 and Table SIII).
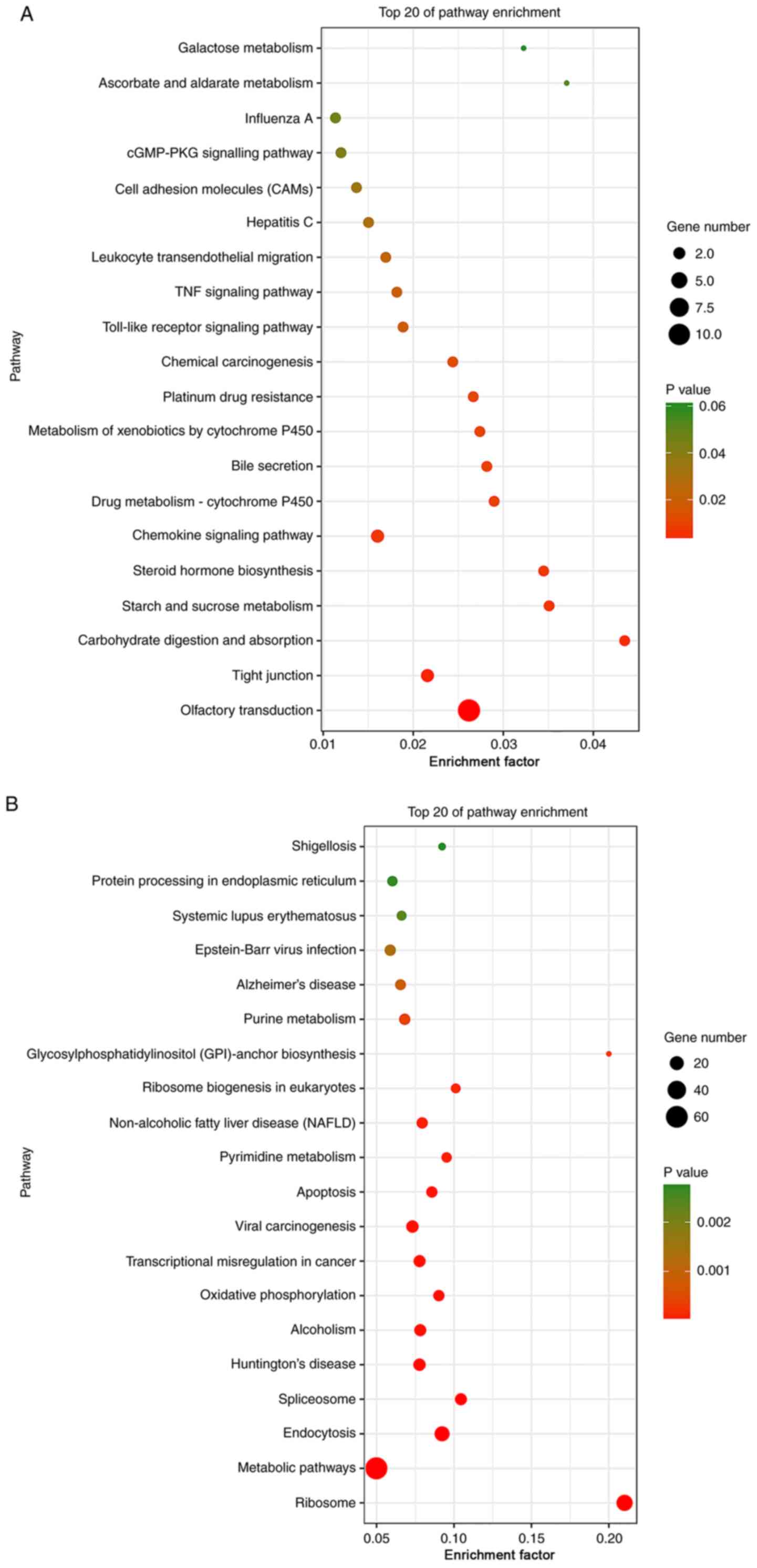 |
Figure 4
KEGG pathway enrichment analysis. (A) Top 20 significantly enriched pathways by the differential target genes in the ELF1 ChIP-seq dataset. KEGG, Kyoto Encyclopedia of Genes and Genomes; ELF1, E74-like factor 1; CSDE1, cold-shock domain-containing E1; ChIP-seq, chromatin immunoprecipitation sequencing; TNF, tumor necrosis factor; cGMP-PKG, cyclic guanosine monophosphate protein kinase G. KEGG pathway enrichment analysis. (B) Top 20 significantly enriched pathways by the differential target genes in the CSDE1 ChIP-seq dataset. KEGG, Kyoto Encyclopedia of Genes and Genomes; ELF1, E74-like factor 1; CSDE1, cold-shock domain-containing E1; ChIP-seq, chromatin immunoprecipitation sequencing; TNF, tumor necrosis factor; cGMP-PKG, cyclic guanosine monophosphate protein kinase G.
|
Regulatory network of differential target genes of ELF1 and CSDE1
PPI networks of the ELF1- and CSDE1-associated target genes were constructed. The results suggested that the ELF1 regulatory network had no obvious key node genes (Fig. 5A). The largest network contained seven proteins, including the chemokines C-X-C motif chemokine ligand (CXCL) 10, CXCL8 and CXCL6 (Fig. 5B). The CSDE1 regulatory network contained six major sub-networks, in which two sub-network genes were abundant. The largest sub-network contained multiple ribosomal proteins and the second-largest sub-network consisted of mitochondrial ribosomal proteins. Mitochondrial ribosomal protein L4 (MRPL4), NADH: ubiquinone oxidoreductase subunit B7 (NDUFB7), small nuclear ribonucleoprotein polypeptide E (SNRPE), as well as ribosomal protein (RP)S26, RPS11 and RPS6, were important nodal genes in each sub-network.
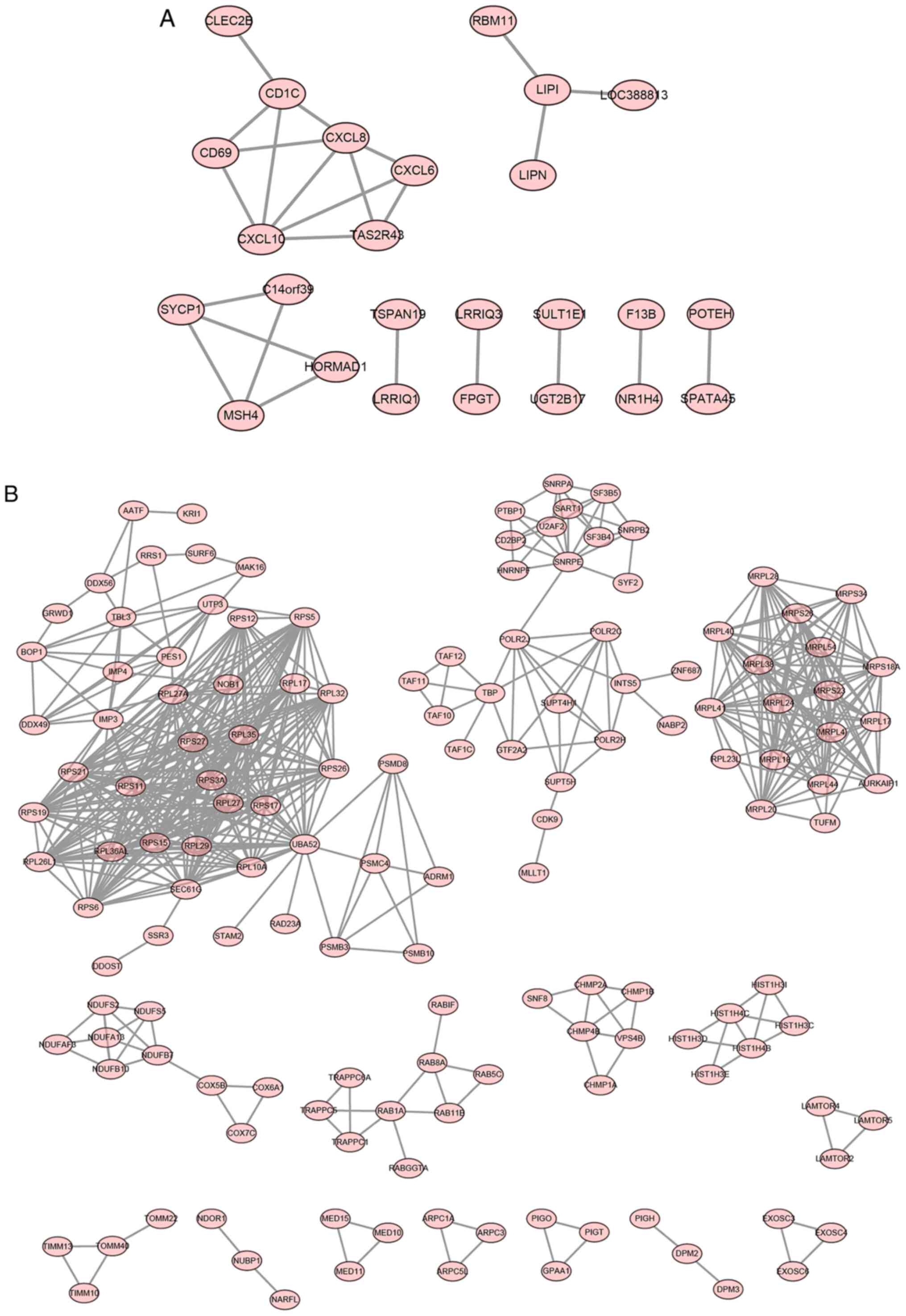 |
Figure 5
Interaction network generated with differential target genes in the (A) ELF1 ChIP-seq dataset and (B) CSDE1 ChIP-seq dataset. ELF1, E74-like factor 1; CSDE1, cold-shock domain-containing E1; ChIP-seq, chromatin immuno-precipitation sequencing.
|
Gene expression analysis
To further analyze the role of ELF1 and CSDE1 in breast cancer and the interaction between ELF1- and CSDE1-associated target genes, RT-qPCR was used to determine the ELF1 and CSDE1 mRNA expression in different breast cancer cell lines. In addition, the mRNA expression levels of ELF1- and CSDE1-associated target genes in MCF-7 cells transfected with LV003-ELF1, LV003-CSDE1 and siRNA fragments-mediated silencing of ELF1 and CSDE1 were also analyzed by RT-qPCR. The ELF1 mRNA expression levels in the breast cancer cell lines MCF-7, BT-20, HS 578T and MDA-MB-468 were lower than those in the normal breast cell line MCF-10A (Fig. 6A). The CSDE1 mRNA expression levels in the breast cancer cell line MCF-7 was the highest and significantly higher than that in the normal breast cell line MCF-10A (Fig. 6A). Therefore, MCF-7 cells were selected for subsequent analysis. The ELF1 and CSDE1 mRNA expression levels in the MCF-7 cells transfected with LV003-ELF1 and LV003-CSDE1 were >2 fold higher than those in the MCF-7 cells transfected with LV003 plasmids (negative control), indicating that overexpression plasmids were successfully transduced into the MCF-7 cells (Fig. 6B and C). CXCL8 and CXCL6 mRNA expression levels in the MCF-7 cells transfected with LV003-ELF1 were significantly lower than those in MCF-7 cells transfected with LV003, indicating that ELF1 overexpression may downregulate CXCL8 and CXCL6 expression levels in the MCF-7 cells (Fig. 6B). MRPL4, NDUFB7, SNRPE, RPS26, RPS11 and RPS6 mRNA expression levels in the MCF-7 cells transfected with LV003-CSDE1 were higher than those in the MCF-7 cells transfected with LV003, indicating that CSDE1 overexpression may induce the mRNA expression of CSDE1-associated target genes (Fig. 6C). By contrast, the ELF1 and CSDE1 mRNA expression levels in the MCF-7 cells transfected with si-ELF1 and si-CSDE1 were lower than those in the MCF-7 cells transfected with siRNA (negative control) and the interference efficiency reached 50% for si-ELF1 and 25% for si-CSDE1, indicating that knockdown of ELF1 and CSDE1 expression levels in MCF-7 cells was successful (Fig. 6D and E). CXCL8 and CXCL6 mRNA expression levels in the MCF-7 cells transfected with si-ELF1 were significantly higher than those in MCF-7 cells transfected with siRNA (Fig. 6D). MRPL4, NDUFB7, SNRPE, RPS26, RPS11 and RPS6 mRNA expression levels in the MCF-7 cells transfected with si-CSDE1 were lower than those in the MCF-7 cells transfected with siRNA (Fig. 6E), indicating that ELF1 knockdown may upregulate CXCL8 and CXCL6 expression levels in the MCF-7 cells and CSDE1 overexpression may suppress the mRNA expression of CSDE1-associated target genes.
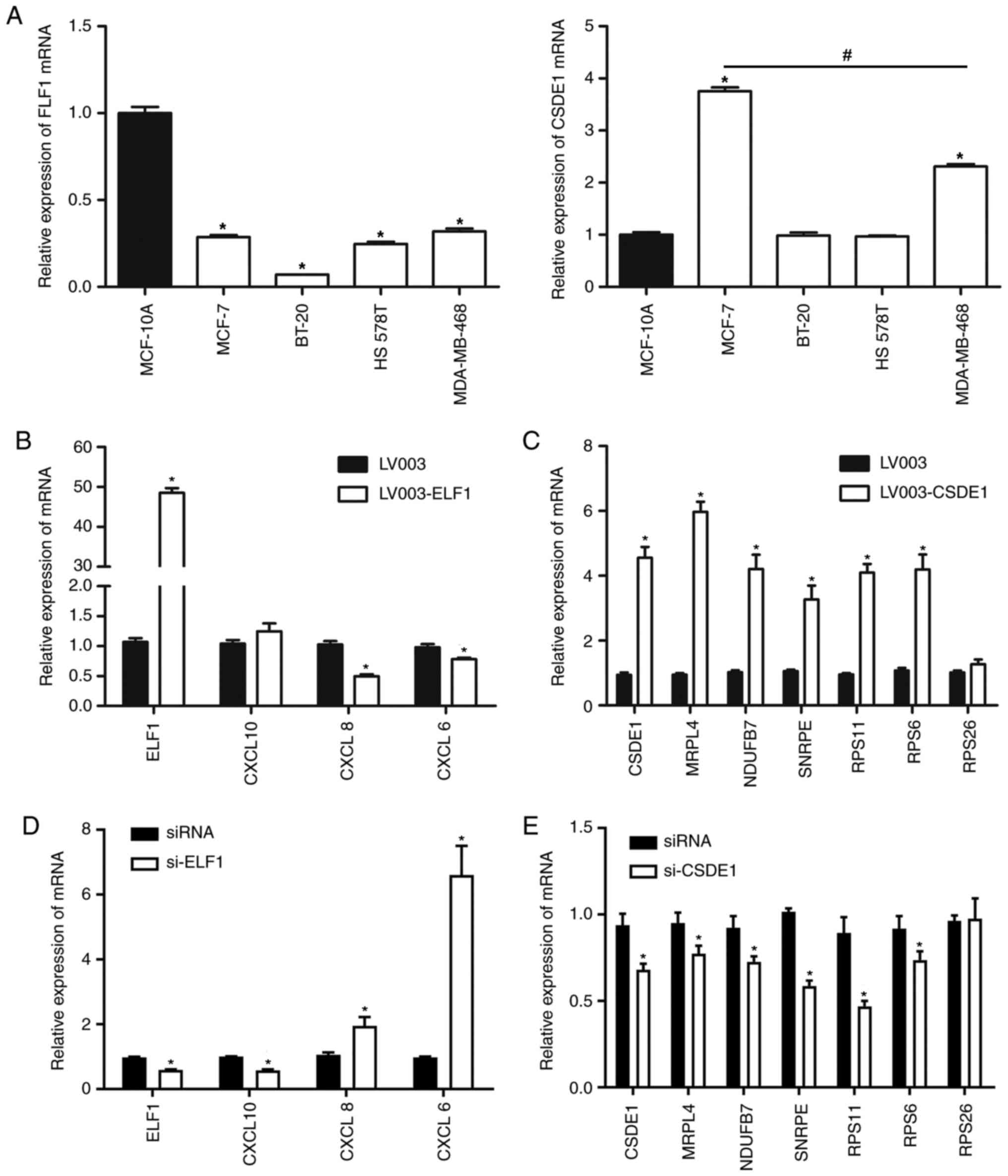 |
Figure 6
Gene expression level analysis in breast cancer cells determined by reverse transcription-quantitative PCR. (A) ELF1 and CDSE1 mRNA expression in MCF-10A, MCF-7, BT-20, HS 578T and MDA-MB-468 cells. (B-E) Expression levels of various genes in MCF-7 subjected to various transfections. (B and D) ELF1, CXCL10, CXCL8 and CXCL6 mRNA expression levels in MCF-7 cells transfected with LV003-ELF1 and si-ELF1, respectively. (C and E) CSDE1, MRPL4, NDUFB7, SNRPE, RPS11, RPS6 and RPS26 mRNA expression levels in MCF-7 cells transfected with LV003-CDSE1 and si-CDSE1, respectively. *P<0.05 vs. the negative control (LV003 and siRNA) and MCF-10A. #P<0.05 vs. MDA-MB-468. CXCL, C-X-C motif chemokine ligand; ELF1, E74-like factor 1; CSDE1, cold-shock domain-containing E1; MRPL4, mitochondrial ribosomal protein L4; NDUFB7, NADH: ubiquinone oxidoreductase subunit B7; SNRPE, small nuclear ribonucleoprotein polypeptide E; RPS26, ribosomal protein S26; siRNA, small interfering RNA; LV003, empty lentivirus vector.
|
Discussion
In the present study, 921 differential target genes were identified to be differentially expressed in the ELF1 and CSDE1 ChIP-seq datasets, of which 95 genes were significantly dominant in the ELF1 ChIP-seq dataset, while 826 were dominant in the CSDE1 ChIP-seq dataset. Bioinformatics analysis suggested that the function and regulatory networks between the ELF1- and CSDE1-associated target genes were different.
Chemokines and their receptors are among the essential mediators of leukocyte and cancer cell migration (16). In addition to promoting leukocyte migration and inducing inflammatory responses, chemokines also have a role in promoting tumor growth, proliferation and metastasis by promoting tumor cell proliferation or neovascularization (17,18). Chemokines also shape the pre-metastatic micro-environment, which favors metastasis, providing navigation to metastatic sites (19), and may serve as a tumor immunotherapy tool to target cytotoxic T-cells containing engineered chemokine receptors and accumulate at metastatic sites (20). Chemokines may be roughly divided into two groups: The first group is associated with inflammatory regulatory functions induced by inflammation, while the other group is associated with maintaining the body's steady state. In the present study, the largest ELF1 regulatory network contained inflammatory chemokines, including CXCL10, CXCL8 and CXCL6, of which CXCL10 and CXCL8 are frequently induced simultaneously in various nervous system inflammations (21,22). CXCL8 is frequently induced in highly invasive breast cancer cells, which is closely linked to tumor metastatic behavior (23). Particularly in breast cancer, epidermal growth factor may induce breast cancer cells to upregulate the CXCL8 secretion level, thereby promoting malignant transformation of tumor cells (24). These results suggest that CXCL8, an inflammatory chemokine, mainly regulates malignant transformation of breast cancer cells. CXCL6, which is located on the same chromosome as CXCL8, was also identified to be upregulated in high-stage breast cancer cells, suggesting that CXCL6 is associated with breast cancer progression. In fact, CXCL6 promotes tumor survival and metastasis in non-small-cell lung cancer and liver cancer (25,26). Compared to CXCL8 and CXCL6, CXCL10 promotes the progression and metastasis of breast cancer (27). After its expression, CXCL10 binds to its receptor CXCR3 and initiates downstream factors to regulate tumor progression (28-30). In the present study, CXCL6, CXCL8 and CXCL10 were important nodes of the regulatory network of ELF1-associated target genes, as indicated by PPI network analysis. The present results also suggested that CXCL8 and CXCL6 mRNA expression levels in MCF-7 cells transfected with LV003-ELF1 were significantly lower than those in MCF-7 cells transfected with LV003. In addition, KEGG analysis indicated that the chemokine signaling pathway is one of the most abundant target gene-enriched networks of ELF1. It may be inferred that the synergy of CXCL8 and CXCL6, which are ELF1-associated target genes, has an important role in the occurrence and development of breast cancer.
In the present study, the largest PPI sub-network of CSDE1-associated target genes comprised RPs, including RPS27, RPS26, RPL35, RPL27 and RPS12. The function of these proteins in breast cancer has yet to be elucidated. Previous studies suggested that RPS27, which is frequently upregulated in tumors, is a tumor-associated factor and may be used as an early diagnostic marker for a variety of tumor types (31). It was also indicated that RPS27 binds to the mouse double minute 2 homologue to maintain the function of P53, suggesting that RPS27 may be involved in the regulation of tumor cell apoptosis (32). The second-largest network of CSDE1-associated target genes contains numerous mitochondrial ribosomal proteins (MRPs) in the center of the PPI network in the present study. The major functions of MRPs include participation in mitochondrial gene transcription and translation, and synthesis and regulation of cell growth (33-36). Since the protein product of the mitochondrial gene is involved in oxidative phosphorylation regulation and adenosine triphosphate (ATP) production, although most of the functions of MRPs have remained to be fully elucidated, it may be deduced that MRPs are closely associated with cell growth, differentiation and development requiring ATP. In addition, the KEGG pathway enrichment analysis revealed that the CSDE1-associated target genes were mainly enriched in ribosomes, metabolic pathways and oxidative phosphorylation, which is consistent with the functions of these networks. The PPI also indicated that MRPL4, NDUFB7, SNRPE, RPS11 and RPS6 are important nodal genes of these small networks. Of these, SNRPE, RPS11 and RPS6 are associated with tumor growth and poor prognosis (37-41). NDUFB7 is a subunit of the multi-subunit NADH: ubiquinone oxidoreductase (complex I), which has NADH dehydrogenase activity and oxidoreductase activity and is involved in ATP synthesis (42). The present results also demonstrated that MRPL4, NDUFB7, SNRPE, RPS26, RPS11 and RPS6 mRNA expression levels in the MCF-7 cells transfected with LV003-CSDE1 were elevated. Interference with CSDE1 resulted in the opposite effect in the MCF-7 cells. It may therefore be inferred that MRPL4, NDUFB7, SNRPE, RPS26, RPS11 and RPS6 are involved in the regulation of breast cancer in terms of ribosomes, metabolic pathways and oxidative phosphorylation.
The protein expression levels of CXCL8, CXCL6, MRPL4, NDUFB7, SNRPE, RPS26, RPS11 and RPS6 in MCF-7 cells require to be further verified. The specific molecular mechanisms of the roles of CXCL8 and CXCL6 in ELF-1 regulating breast cancer require further verification. The specific molecular mechanisms of MRPL4, NDUFB7, SNRPE, RPS26, RPS11 and RPS6 in CSDE1 regulating breast cancer also require further verification to provide a theoretical basis for novel treatments of breast cancer.
In summary, in MCF-7 cells, the ELF1 and CSDE1 target genes and regulatory pathways were observed to be different. ELF1- and CSDE1-associated target genes had different potential functions and signaling pathways. The ELF1 regulatory network mainly regulated chemokine-mediated malignant tumor cells, while the CSDE1 regulatory network mainly regulated oncogene transcription and translation, as well as oxidative phosphorylation. Understanding these regulatory pathways may help devise strategies for personalized breast cancer treatment.
Supplementary Material
Table SI. Differential target genes between GSE105503 and GSE105291 datasets.
Table SII. GO analysis of differential target genes between GSE105503 and GSE105291 datasets.
Table SIII. KEGG analysis of differential target genes between the GSE105503 and GSE105291 datasets.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HD and DP conceived and designed the study. HH, QH, YL, SC, XL and LL participated in the data analysis. HD and DP wrote the manuscript. HH, QH, YL, SC, XL and LL performed the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Malvezzi M, Carioli G, Bertuccio P, Rosso T, Boffetta P, Levi F, La Vecchia C and Negri E: European cancer mortality predictions for the year 2016 with focus on leukaemias. Ann Oncol. 27:725–731. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Gradishar WJ, Anderson BO, Balassanian R, Blair SL, Burstein HJ, Cyr A, Elias AD, Farrar WB, Forero A, Giordano SH, et al: Breast cancer, version 4.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 16:310–320. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chen W, Sun K, Zheng R, Zheng R, Zeng H, Zhang S, Xia C, Yang Z, Li H, Zou X and He J: Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 30:1–12. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Brueffer C, Vallon-Christersson J, Grabau D, Ehinger A, Häkkinen J, Hegardt C, Malina J, Chen Y, Bendahl PO, Manjer J, et al: Clinical value of RNA sequencing-based classifiers for prediction of the five conventional breast cancer biomarkers: A report from the population-based multicenter Sweden cancerome analysis network-breast initiative. JCO Precision Oncol. 2:1–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
O'Regan RM and Nahta R: Targeting forkhead box M1 transcription factor in breast cancer. Biochem Pharmacol. 154:407–413. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Safe S, Abbruzzese J, Abdelrahim M and Hedrick E: Specificity protein transcription factors and cancer: Opportunities for drug development. Cancer Prev Res (Phila). 11:371–382. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Seth A and Watson DK: ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 41:2462–2478. 2005.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Andrews PG, Kennedy MW, Popadiuk CM and Kao KR: Oncogenic activation of the human Pygopus2 promoter by E74-like factor-1. Mol Cancer Res. 6:259–266. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lindquist JA and Mertens PR: Cold shock proteins: From cellular mechanisms to pathophysiology and disease. Cell Commun Signal. 16(63)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Doniger J, Landsman D, Gonda MA and Wistow G: The product of unr, the highly conserved gene upstream of N-ras, contains multiple repeats similar to the cold-shock domain (CSD), a putative DNA-binding motif. New Biol. 4:389–395. 1992.PubMed/NCBI
|
|
12
|
Wurth L, Papasaikas P, Olmeda D, Bley N, Calvo GT, Guerrero S, Cerezo-Wallis D, Martinez-Useros J, García-Fernández M, Hüttelmaier S, et al: UNR/CSDE1 drives a post-transcriptional program to promote melanoma invasion and metastasis. Cancer Cell. 30:694–707. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Arndt S, Unger P, Wacker E, Shimizu T, Heinlin J, Li YF, Thomas HM, Morfill GE, Zimmermann JL, Bosserhoff AK and Karrer S: Cold atmospheric plasma (CAP) changes gene expression of key molecules of the wound healing machinery and improves wound healing in vitro and in vivo. PLoS One. 8(e79325)2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Heidari N, Phanstiel DH, He C, Grubert F, Jahanbani F, Kasowski M, Zhang MQ and Snyder MP: Genome-wide map of regulatory interactions in the human genome. Genome Res. 24:1905–1917. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Pan W, Liang J, Tang H, Fang X, Wang F, Ding Y, Huang H and Zhang H: Differentially expressed microRNA profiles in exosomes from vascular smooth muscle cells associated with coronary artery calcification. Int J Biochem Cell Biol. 118(105645)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang J and Knaut H: Chemokine signaling in development and disease. Development. 141:4199–4205. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zlotnik A: Chemokines and cancer. Int J Cancer. 119:2026–2029. 2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Strieter RM, Belperio JA, Phillips RJ and Keane MP: CXC chemokines in angiogenesis of cancer. Semin Cancer Biol. 14:195–200. 2004.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Atretkhany KN, Drutskaya MS, Nedospasov SA, Grivennikov SI and Kuprash DV: Chemokines, cytokines and exosomes help tumors to shape inflammatory microenvironment. Pharmacol Ther. 168:98–112. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Garetto S, Sardi C, Morone D and Kallikourdis M: Chemokines and T cell trafficking into tumors: Strategies to enhance recruitment of T cells into tumors. Defects in T Cell Trafficking and Resistance to Cancer Immunotherapy pp 163-177, 2016.
|
|
21
|
Moniuszko A, Czupryna P, Pancewicz S, Rutkowski K, Zajkowska O, Swierzbińska R, Grygorczuk S, Kondrusik M, Owłasiuk P and Zajkowska J: Evaluation of CXCL8, CXCL10, CXCL11, CXCL12 and CXCL13 in serum and cerebrospinal fluid of patients with neuroborreliosis. Immunol Lett. 157:45–50. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang C, Wu K, Yu Q, Zhang S, Gao Z, Liu Y, Ni L, Cheng Y, Guan Z, Shi M, et al: CXCL13, CXCL10 and CXCL8 as potential biomarkers for the diagnosis of neurosyphilis patients. Sci Rep. 6(33569)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Brysse A, Mestdagt M, Polette M, Luczka E, Hunziker W, Noël A, Birembaut P, Foidart JM and Gilles C: Regulation of CXCL8/IL-8 expression by zonula occludens-1 in human breast cancer cells. Mol Cancer Res. 10:121–132. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Azenshtein E, Meshel T, Shina S, Barak N, Keydar I and Ben-Baruch A: The angiogenic factors CXCL8 and VEGF in breast cancer: Regulation by an array of pro-malignancy factors. Cancer Lett. 217:73–86. 2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li J, Tang Z, Wang H, Wu W, Zhou F, Ke H, Lu W, Zhang S, Zhang Y, Yang S, et al: CXCL6 promotes non-small cell lung cancer cell survival and metastasis via down-regulation of miR-515-5p. Biomed Pharmacother. 97:1182–1188. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tian H, Huang P, Zhao Z, Tang W and Xia J: HIF-1α plays a role in the chemotactic migration of hepatocarcinoma cells through the modulation of CXCL6 expression. Cell Physiol Biochem. 34:1536–1546. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ejaeidi AA, Craft BS, Puneky LV, Lewis RE and Cruse JM: Hormone receptor-independent CXCL10 production is associated with the regulation of cellular factors linked to breast cancer progression and metastasis. Exp Mol Pathol. 99:163–172. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Mulligan AM, Raitman I, Feeley L, Pinnaduwage D, Nguyen LT, O'Malley FP, Ohashi PS and Andrulis IL: Tumoral lymphocytic infiltration and expression of the chemokine CXCL10 in breast cancers from the Ontario Familial breast cancer registry. Clin Cancer Res. 19:336–346. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bu H, Shu B, Gao F, Liu C, Guan X, Ke C, Cao F, Hinton AO Jr, Xiang H, Yang H, et al: Spinal IFN-γ-induced protein-10 (CXCL10) mediates metastatic breast cancer-induced bone pain by activation of microglia in rat models. Breast Cancer Res Treat. 143:255–263. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Datta D, Flaxenburg JA, Laxmanan S, Geehan C, Grimm M, Waaga-Gasser AM, Briscoe DM and Pal S: Ras-induced modulation of CXCL10 and its receptor splice variant CXCR3-B in MDA-MB-435 and MCF-7 cells: Relevance for the development of human breast cancer. Cancer Res. 66:9509–9518. 2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fernandez-Pol JA: Increased serum level of RPMPS-1/S27 protein in patients with various types of cancer is useful for the early detection, prevention and therapy. Cancer Genomics Proteomics. 9:203–256. 2012.PubMed/NCBI
|
|
32
|
Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X and Prives C: Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 35:316–326. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wang Z, Cotney J and Shadel GS: Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J Biol Chem. 282:12610–12618. 2007.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Miller C, Saada A, Shaul N, Shabtai N, Ben-Shalom E, Shaag A, Hershkovitz E and Elpeleg O: Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann Neurol. 56:734–738. 2004.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yang Y, Cimen H, Han MJ, Shi T, Deng JH, Koc H, Palacios OM, Montier L, Bai Y, Tong Q and Koc EC: NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem. 285:7417–7429. 2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yoo YA, Kim MJ, Park JK, Chung YM, Lee JH, Chi SG, Kim JS and Yoo YD: Mitochondrial ribosomal protein L41 suppresses cell growth in association with p53 and p27Kip1. Mol Cell Biol. 25:6603–6616. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Anchi T, Tamura K, Furihata M, Satake H, Sakoda H, Kawada C, Kamei M, Shimamoto T, Fukuhara H, Fukata S, et al: SNRPE is involved in cell proliferation and progression of high-grade prostate cancer through the regulation of androgen receptor expression. Oncol Lett. 3:264–268. 2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yong WH, Shabihkhani M, Telesca D, Yang S, Tso JL, Menjivar JC, Wei B, Lucey GM, Mareninov S, Chen Z, et al: Ribosomal proteins RPS11 and RPS20, two stress-response markers of glioblastoma stem cells, are novel predictors of poor prognosis in glioblastoma patients. PLoS One. 10(e0141334)2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S, Destefanis G, Delogu S, Zimmermann A, Ericsson J, et al: Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 140:1071–1083. 2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Li G, Shan C, Liu L, Zhou T, Zhou J, Hu X, Chen Y, Cui H and Gao N: Tanshinone IIA inhibits HIF-1α and VEGF expression in breast cancer cells via mTOR/p70S6K/RPS6/4E-BP1 signaling pathway. PLoS One. 10(e0117440)2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang C, Cigliano A, Jiang L, Li X, Fan B, Pilo MG, Liu Y, Gui B, Sini M, Smith JW, et al: 4EBP1/eIF4E and p70S6K/RPS6 axes play critical and distinct roles in hepatocarcinogenesis driven by AKT and N-Ras proto-oncogenes in mice. Hepatology. 61:200–213. 2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Smeitink J and van den Heuvel L: Human mitochondrial complex I in health and disease. Am J Hum Genet. 64:1505–1510. 1999.PubMed/NCBI View Article : Google Scholar
|




















