Introduction
Osteoarthritis (OA) is a complex chronic, severe
degenerative arthropathy involving the synovium of the joint. It
has a significant economic impact on the public health sector and
as the population ages, the number of patients is expected to
increase (1). OA is widely
recognized as an ‘arthralgia syndrome’ in Traditional Chinese
Medicine (TCM). Inflammation is a critical process in the
occurrence and development of OA, as it is able to stimulate and
enhance the catabolism of chondrocytes and lead to joint injury.
One of the primary symptoms of OA is pain, which may be caused and
worsened by inflammation. Inhibition of inflammation is able to
provide an effective treatment for OA by relieving pain and
reducing joint injury (2-4).
Radix Angelicae biseratae is the root of the
plant Angelica biserrata, known as ‘Duhuo’ in China. As a
natural herb, Radix Angelicae biseratae has long been used
to treat ‘arthralgia syndrome’ by eliminating dampness and
relieving pain according to the concepts of TCM (5). Modern pharmacological studies have
demonstrated that Radix Angelicae biseratae has a
significant anti-inflammatory effect for the treatment of OA
(6). However, the chemical
composition of Radix Angelicae biseratae is complex and the
mechanism for treating OA at the molecular level has remained
elusive.
Systematic pharmacology is a subject where the
knowledge of systems biology is applied to pharmacology. The
purpose of this discipline is to clarify how drugs act on the body
through biological systems. Instead of evaluating the effect of a
drug by analyzing a particular protein-drug interaction, systematic
pharmacology suggests that drugs act by forming a network of
interactions (7). Interaction
systems may include drug-protein and protein-protein interactions,
as well as genetic, signaling and physiological interactions
(7). Systematic pharmacology uses
bioinformatics and statistics to integrate and explain the network
of drug interactions, which provides an effective method to
investigate the underlying mechanisms of TCM and other naturally
occurring compounds for the treatment of a number of diseases
(8,9).
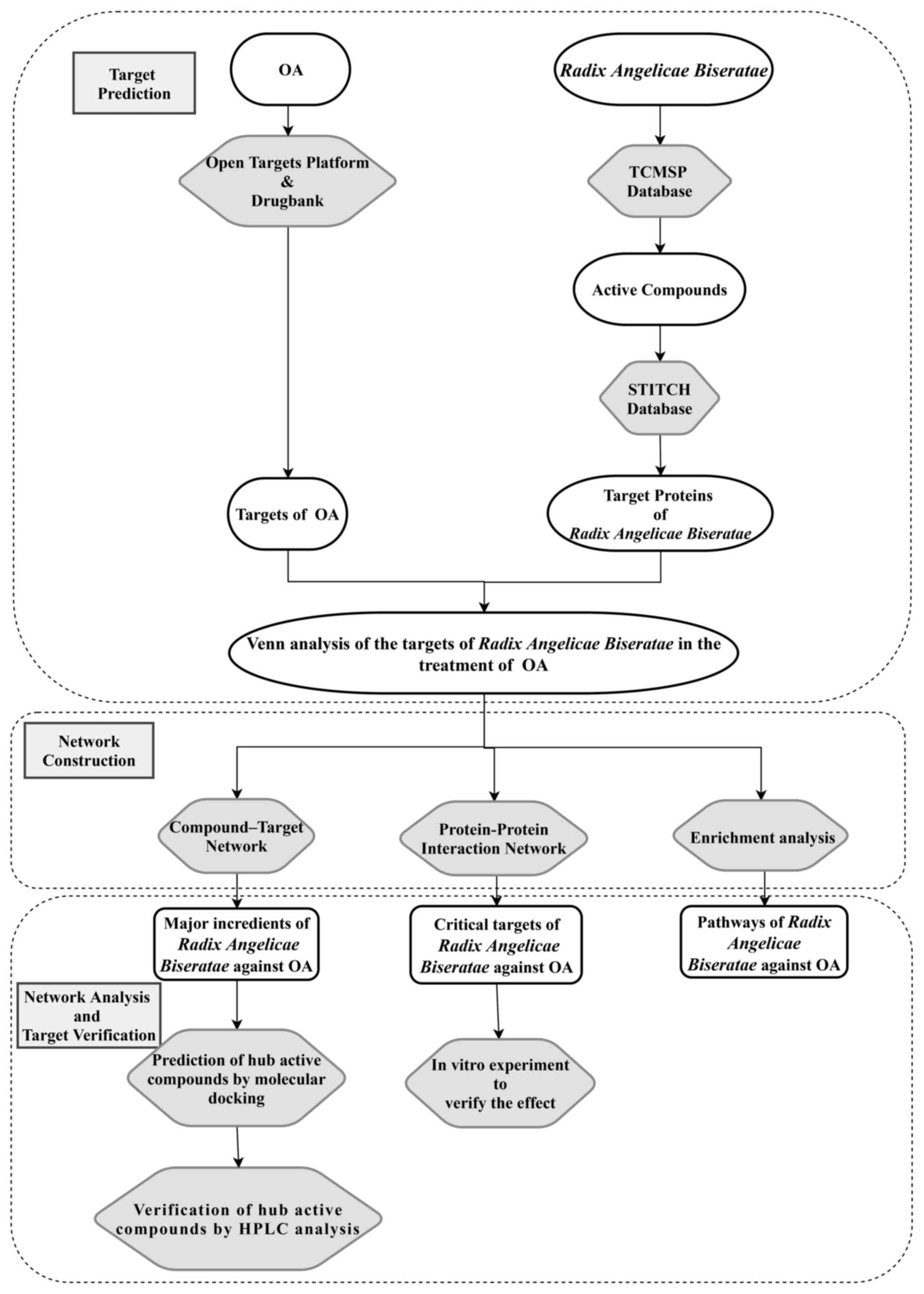
Therefore, in the present study, systematic
pharmacology was used to screen the potential active components of
Radix Angelicae biseratae for the treatment of OA, to
predict and verify its targets and provide a theoretical basis for
drug development and disease treatment. An experimental flowchart
for the present study is presented in Fig. 1.
Materials and methods
Screening for active components
The TCM systems pharmacology (TCMSP) database
(http://lsp.nwu.edu.cn/tcmsp.php) is a
systematic pharmacological database for evaluating the
pharmacokinetic properties of TCM or associated components
(10). It provides data on the
absorption, distribution, metabolism and systemic excretion (ADME)
of drugs with potential biological effects, such as oral
bioavailability (OB) and druglikeness (DL), as well as Caco-2 and
blood-brain barrier permeability. OB is the most critical feature
of oral drugs because it is important in evaluating the
effectiveness of drug distribution through the systemic circulation
(11). DL refers to the similarity
between components and known drugs. In drug development, DL
evaluation helps to identify qualified components and improve the
success rate of candidate drugs (12). Based on the published literature and
preliminary information in the TCMSP database (13), the drug name ‘Radix Angelicae
biseratae’ was entered into the search window in the TCMSP
database. The ingredients with OB ≥30% and DL ≥18% were screened
for further study.
Identification of drug targets
The STITCH (http://stitch.embl.de/) database was used to identify
the potential targets of active components in Radix Angelicae
biseratae (14). STITCH is a
database used to explore and predict the interaction between
components and proteins. Through experiments, databases and
literature evidence, chemicals associated with other chemicals and
proteins were retrieved. In the present study, the active
components were inputted into the STITCH database and human species
were set up to screen the target proteins of Radix Angelicae
biseratae with a confidence score >0.4 for further
analysis.
Identification of disease targets
OA targets were collected from the Open Targets
platform (https://www.targetvalidation.org) (15,16)
and the DrugBank database (https://www.drugbank.ca) (17-19).
The Open Targets platform is a comprehensive and robust data
integration platform for accessing and visualizing potential drug
targets related to diseases. It brings together a variety of data
types designed to help users identify and prioritize targets for
further research (20). The disease
name ‘osteoarthritis’ was entered into the search window in the
Open Targets platform database and target proteins were collected
for further analysis. DrugBank database is a comprehensive online
database that contains information concerning drugs and drug
targets. The DrugBank database was searched with the key word
‘osteoarthritis’ to screen and collect targets for known drugs in
the treatment of OA. Subsequently, the proteins overlapping the
Radix Angelicae biseratae targets were selected for further
study.
Construction of component-target
network
A ‘component-target network’ diagram was constructed
using Cytoscape 3.7.2 (https://cytoscape.org/) software in order to reveal
the major active components and regulatory mechanisms of Radix
Angelicae biseratae in the treatment of OA (21).
Construction of target-target
interaction network
In order to identify the critical proteins among the
target proteins identified as specified above, a ‘target-target
interaction network’ was established using the Search Tool for the
Retrieval of Interacting Genes and proteins (STRING; v11.0;
http://string-db.org/) and visualized with
Cytoscape 3.7.2(22).
Gene functional enrichment
analysis
ClusterProfiler was used to analyze the enrichment
of gene function and signaling pathways. ClusterProfiler is an R
package for gene cluster enrichment analysis, which may be used to
understand the function of genes and the enrichment of pathways
(23). When provided with the
overlapping target gene list, ClusterProfiler uses the Gene
Ontology (GO) database to analyze the biological function and
signaling pathway enrichment of these genes (24,25).
Verification of target proteins by OA
gene expression array
A set of microarray data of gene expression in
cartilage tissue of patients with OA was obtained from the gene
expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/; dataset no.
GSE75181). The dataset contained 48 samples, of which 12
non-control samples and 12 control samples treated with IL-1β were
selected. The gene matrix file of the GSE75181 dataset was
downloaded and it was standardized with the limma package (26), including background correction,
standardization and normalization. The impute package was used to
fill in the missing values (27).
The platform annotation file was used to annotate the probes and
delete those probes (genesymbols) that did not match the gene. For
multiple probes targeting the same gene, the average value of
different probes was used as the expression value of the gene.
Depending on whether the samples were treated with IL-1β or not,
the samples were divided into two groups: The non-control group and
the control group. Regarding the difference in expression, the
P-value and fold change (FC) in expression of the non-control vs.
control samples were calculated using the limma package. Adjusted
P-value (adj.P.Val) <0.05 and |log2FC|>1 were chosen as the
threshold for filtering the significant differentially expressed
mRNA and display the results in the form of volcano plot. Critical
proteins on the volcano plot were then marked.
Prediction of hub target
protein-component docking
The Blind Docking server (https://bio-hpc.ucam.edu/achilles/entry) was used for
molecular docking experiments (28). The crystal structure of active
components was downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov) and the
structure of the hub target proteins was obtained from the protein
databank (PDB; https://rscb.org) (29). BIOVIA DiscoveryStudio®
2016 software (Dassault Systemes) was used to modify the protein
structure for molecular docking. All water molecules and ligands
were removed and the necessary hydrogen atoms were added. The
obtained receptor and ligand structures were uploaded to the Blind
Docking server for docking calculation and the best structures with
the lowest binding energy were used.
In vitro verification Animals
Male 4-week-old Sprague Dawley rats (n=6; weight,
80-100 g) were purchased from Shanghai SLAC Laboratory Animal Co.,
Ltd [Laboratory Animal Use Certificate no. SCXK(SH)2017-0005]. The
present study was approved by the Animal Care and Use Committee of
Fujian University of Traditional Chinese Medicine (Fuzhou, China).
Experiments involving animals complied with the 2006 Edition of the
Guidelines for the Care and Use of Experimental Animals by the
Ministry of Science and Technology, China (30).
Preparation and analysis of Radix
Angelicae biseratae extract
Radix Angelicae biseratae was purchased from
the local Guo Yi Tang Chinese herbal medicine store. The original
herb was identified as Radix Angelicae biseratae by Dr Wen
Xu at the Department of Pharmacology, Fujian University of
Traditional Chinese Medicine (Fujian, China). Radix Angelicae
biseratae (500 g) was crushed into a fine powder and boiled
twice with 4 l of 80% ethanol for 1 h. The ethanolic extract was
collected and filtered. The filtrate was concentrated to 500 ml at
50˚C under reduced pressure and the concentration was 1 g/ml. The
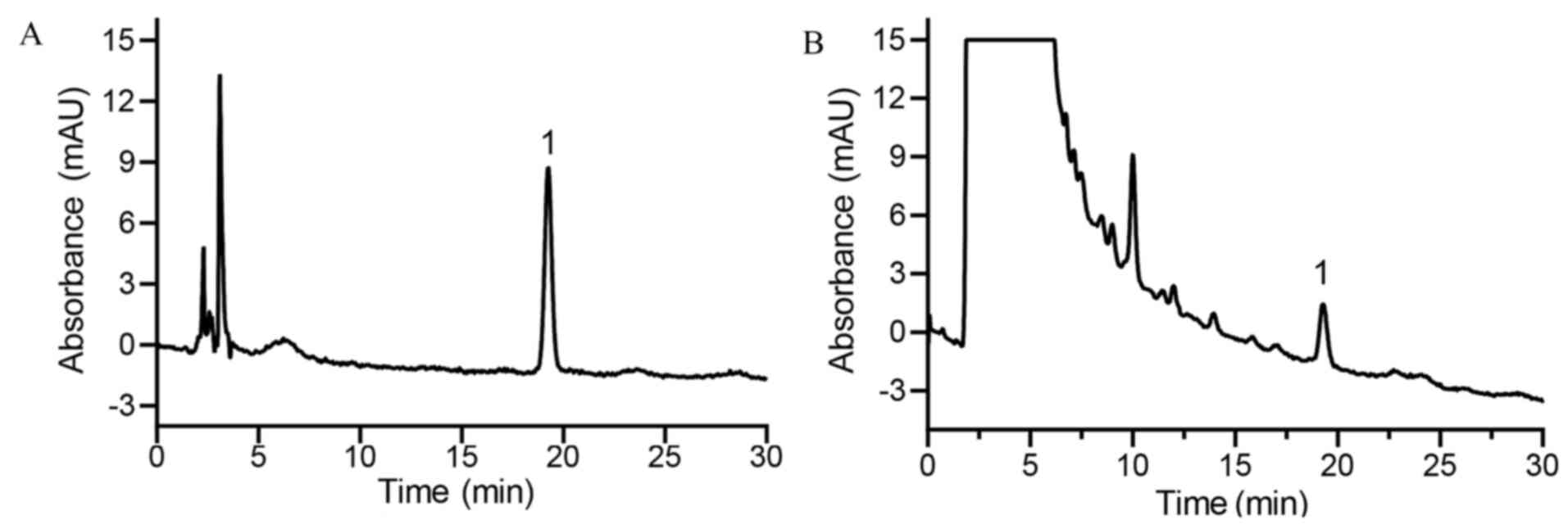
content of sitosterol in the extract was determined by
high-performance liquid chromatography (HPLC) method using an
Agilent 1200 HPLC system (Agilent Technologies, Inc.) and the
Thermo Scientific™ BETASIL™ C18 column (4.6x150 mm, 5 µm; Thermo
Fisher Scientific, Inc.). The column temperature was set at 35˚C
and the injection volume was 10 µl. The mobile phase was 100%
methanol and the flow rate was 1.0 ml/min. Ultraviolet detection
wavelengths were set at 210 nm at 0-30 min.
Chondrocyte observation and
identification
Chondrocytes were isolated from the knee articular
cartilage of 4-week-old Sprague Dawley rats. After the rats were
anesthetized by intraperitoneal injection of 30 mg/kg
pentobarbital, they were sacrificed via rapid decapitation. The
chondrocytes were isolated, cultured and verified, as previously
described (31,32).
Degenerative chondrocyte model
The degenerative chondrocyte model was established
as previously described (33,34).
In brief, the chondrocytes at passage three were exposed to 10
ng/ml IL-1β for 24 h.
Experimental grouping
Cells were divided into the following groups: i)
control group without treatment; ii) degenerative chondrocyte
group; iii) degenerative chondrocytes with 1 µM Radix Angelicae
biseratae extract group; iv) degenerative chondrocytes with 5
µM Radix Angelicae biseratae extract group; and v)
degenerative chondrocytes with 25 µM Radix Angelicae
biseratae extract group. The intervention time for all groups
was 24 h.
Reverse transcription-quantitative PCR
(RT-qPCR)
After 24 h of intervention, total RNA was extracted
from chondrocytes of each group using a TRIzol® kit
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA(1 µg) was reverse
transcribed into cDNA using the PrimeScript™ RT reagent kit (cat.
no. RR0047A; Takara Bio, Inc.) in the PCR amplifier (S1000 Thermal
Cycler; Bio-Rad Laboratories, Inc.) according to the manufacturer's
protocol. The following primer sequences were used:
Prostaglandin-endoperoxide synthase 2 (PTGS2) forward,
5'-AGCACAATAGACGCCCAAGA-3' and reverse,
5'-GGAGTCAAAGCATAGGTCTTCA-3'; β-actin forward,
5'-ACCACTGGCATTGTGATGGA-3' and reverse, 5'-CGCTCGGTCAGGATCTTCT-3'.
β-actin was used to normalize the expression of mRNA. AceQ qPCR
SYBR® Green Master Mix (cat. no. Q111-02; Vazyme Biotech
Co., Ltd.) was used to detect the respective mRNA expression
levels. qPCR was conducted using the 7500 Fast Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) using the
following reaction conditions: Initial denaturation at 95˚C for 3
min, followed by 40 cycles at 95˚C for 10 sec and 60˚C for 30 sec;
dissolution curve at 95˚C for 15 sec, 60˚C for 60 sec and 95˚C for
15 sec. The experiment was performed three times and the gene
expression ratios were expressed in the form of average value and
standard deviation according to the independent measurement results
of the three experiments. The 2-ΔΔCq method (35) was used to evaluate the experimental
results.
Western blot analysis
The expression levels of the target proteins in
chondrocytes of each group were measured by western blot analysis.
Western blotting and semi-quantitative analysis were performed
according to a previously published protocol (36). The following antibodies were used:
PTGS2 antibody (1:1,000 dilution; cat. no. ab52237; Abcam), β-actin
antibody (1:2,000 dilution; cat. no. ab8227; Abcam) and horseradish
peroxidase-conjugated secondary antibodies (1:20,000 dilution; cat.
no. AP132P; Merck KGaA).
Statistical analysis
The experimental data were processed and analyzed by
Graph Pad Prism 8.02 software (GraphPad Software, Inc.). The
Shapiro-Wilk test was used to determine the normality of all groups
of data. If the data exhibited a normal distribution, they were
compared by one-way analysis of variance followed by a least
significant difference or Games Howell post-hoc test; if not, the
Kruskal-Wallis test was used and the Mann-Whitney U test with
Bonferroni's correction was applied as a post hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Components and potential targets for
Radix Angelicae biseratae
The TCMSP database was searched with the key words
‘Radix Angelicae biseratae’ for 99 herbal ingredients. When
applying the threshold specified in the methods section, nine
herbal ingredients were obtained.
These nine components screened from the TCMSP
database were searched in the STITCH database, of which eight
components had 30 target proteins. Among them, ammidin had five
targets, isoimperatorin had seven targets, sitosterol had 14
targets, O-acetyl columbianetin had nine targets, angelol D had two
targets, angelol G had one target, angelicone had five targets and
nodakenin had eight targets. The ADME properties of these eight
components are presented in Table
I.
 | Table IAbsorption, distribution, metabolism
and excretion properties of 8 active components from Radix
Angelicae biseratae. |
Table I
Absorption, distribution, metabolism
and excretion properties of 8 active components from Radix
Angelicae biseratae.
| Mol ID | Molecule name | OB | DL |
|---|
| MOL001941 | Ammidin | 34.55 | 0.28 |
| MOL001942 | Isoimperatorin | 45.46 | 0.27 |
| MOL000358 | Sitosterol | 36.91 | 0.23 |
| MOL003608 |
O-Acetylcolumbianetin | 60.04 | 0.29 |
| MOL004777 | Angelol D | 34.85 | 0.30 |
| MOL004778 | Angelol G | 46.03 | 0.29 |
| MOL004780 | Angelicone | 30.99 | 0.26 |
| MOL004792 | Nodakenin | 57.12 | 0.27 |
Identification of OA-associated
proteins
By searching the Open Targets Platform and DrugBank
database, 20 targets linked to the treatment of OA were screened
(Fig. 2A). As presented in Fig. 2B, a network of pharmacologically
active components and targets was established. Of the eight
components, seven had OA-related targets. Among these seven
components, sitosterol (degree=11) may have an essential role in
the treatment of OA, followed by O-acetylcolumbianetin (degree=7)
and nodakenin (degree=6). All components were linked to PTGS2,
which suggests that Radix Angelicae biseratae may be a
potential inhibitor of PTGS2.
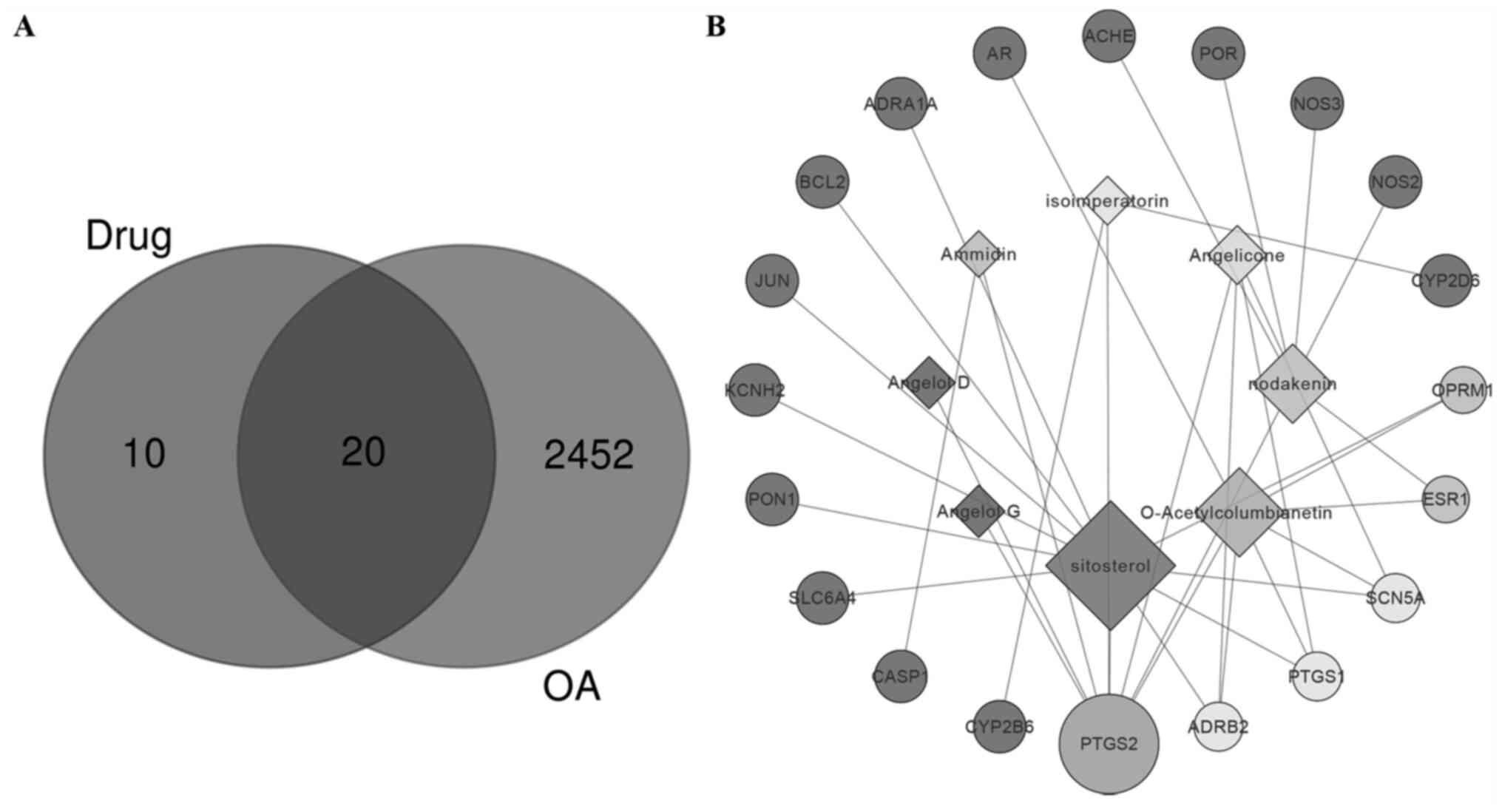 | Figure 2Venn analysis and network analysis of
active components and potential targets of Radix Angelicae
biseratae in the treatment of OA. (A) Venn diagram of target
proteins of Radix Angelicae biseratae and OA-related
proteins based on the Open Targets Platform and Drugbank database.
(B) The compound-target network of Radix Angelicae biseratae
to treat OA, the square nodes represent the compounds, the circular
nodes represent the targets, and the node size is proportional to
the degree. OA, osteoarthritis; PTGS2, prostaglandin-endoperoxide
synthase 2; NOS3, nitric oxide synthase 3; CYP2B6, cytochrome P450
2B6; ACHE, acetylcholinesterase; ESR1, estrogen receptor 1; SLC6A4,
solute carrier family 6 member 4; CYP2D6, cytochrome P450 2D6;
NOS2, nitric oxide synthase 2; OPRM1, opioid receptor µ1; CASP1,
caspase 1; AR, androgen receptor; PTGS1, prostaglandin-endoperoxide
synthase 1; PON1, paraoxonase 1; ADRA1A, adrenoceptor α1A; POR,
cytochrome P450 oxidoreductase; ADRB2, adrenoceptor β2; KCNH2,
potassium voltage-gated channel subfamily H member 2; BCL2, B-cell
lymphoma 2; SCN5A, sodium voltage-gated channel α subunit 5. |
Target-target interaction
analysis
Using Cytoscape 3.7.1, a diagram of the
‘target-target network’ was constructed based on the query results
of the STRING database (Fig. 3A).
In addition, the bar plot was constructed according to the degrees
of connection between the targets (Fig.
3B). The results indicated that nitric oxide synthase 3 (NOS3),
PTGS2 and cytochrome P450 2B6 (CYP2B6) were the hub targets in this
network with high degree values.
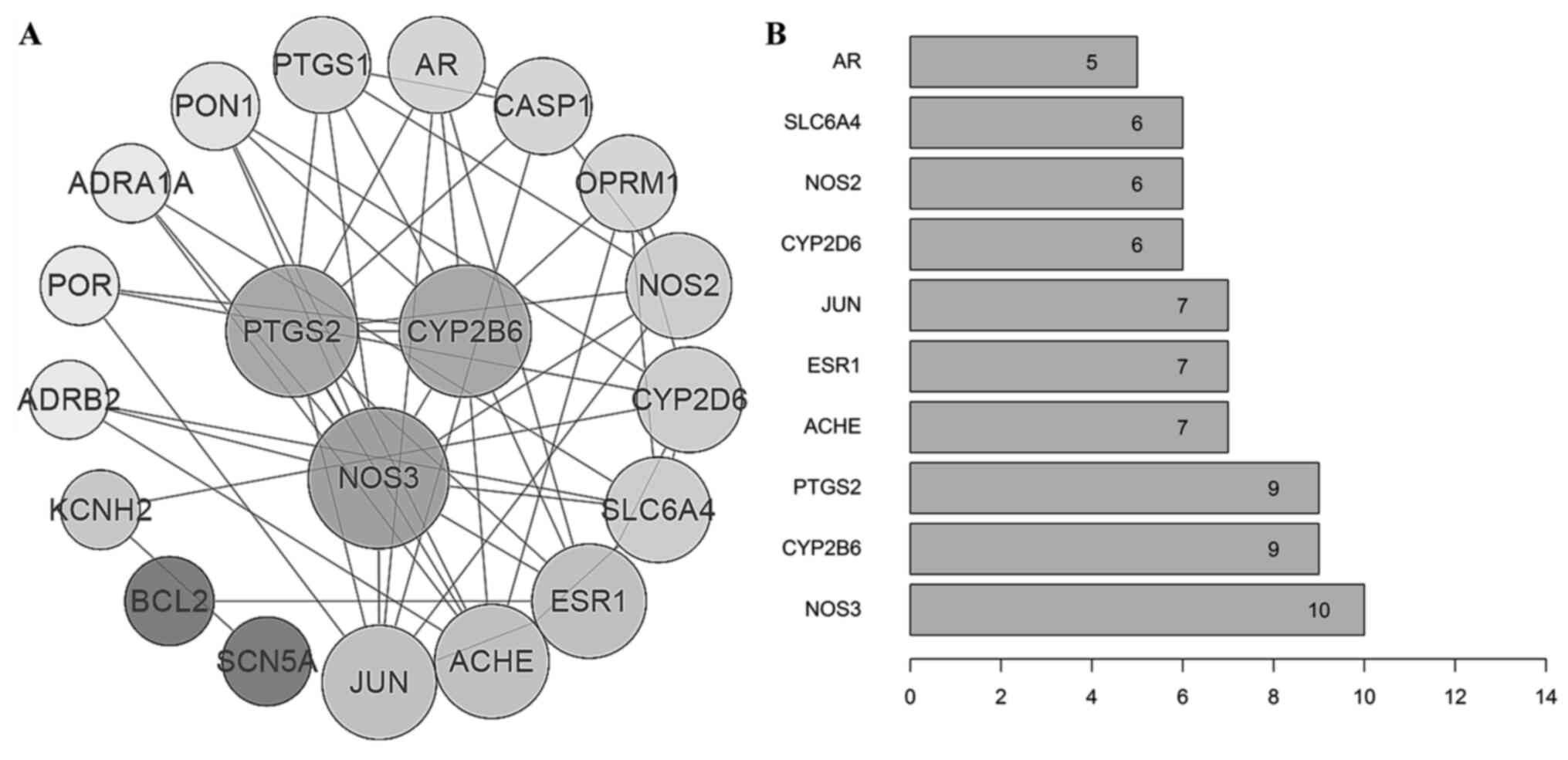 | Figure 3‘Target-target’ interaction network
analysis. (A) Target-target interaction network constructed using
Cytoscape. The sizes of the nodes are proportional to their
degrees. (B) Barplot of the degrees of connection between the
targets. PTGS2, prostaglandin-endoperoxide synthase 2; NOS3, nitric
oxide synthase 3; CYP2B6, cytochrome P450 2B6; JUN, jun
proto-oncogene; ACHE, acetylcholinesterase; ESR1, estrogen receptor
1; SLC6A4, solute carrier family 6 member 4; CYP2D6, cytochrome
P450 2D6; NOS2, nitric oxide synthase 2; OPRM1, opioid receptor µ1;
CASP1, caspase 1; AR, androgen receptor; PTGS1,
prostaglandin-endoperoxide synthase 1; PON1, paraoxonase 1; ADRA1A,
adrenoceptor α1A; POR, cytochrome P450 oxidoreductase; ADRB2,
adrenoceptor β2; KCNH2, potassium voltage-gated channel subfamily H
member 2; BCL2, B-cell lymphoma 2; SCN5A, sodium voltage-gated
channel α subunit 5. |
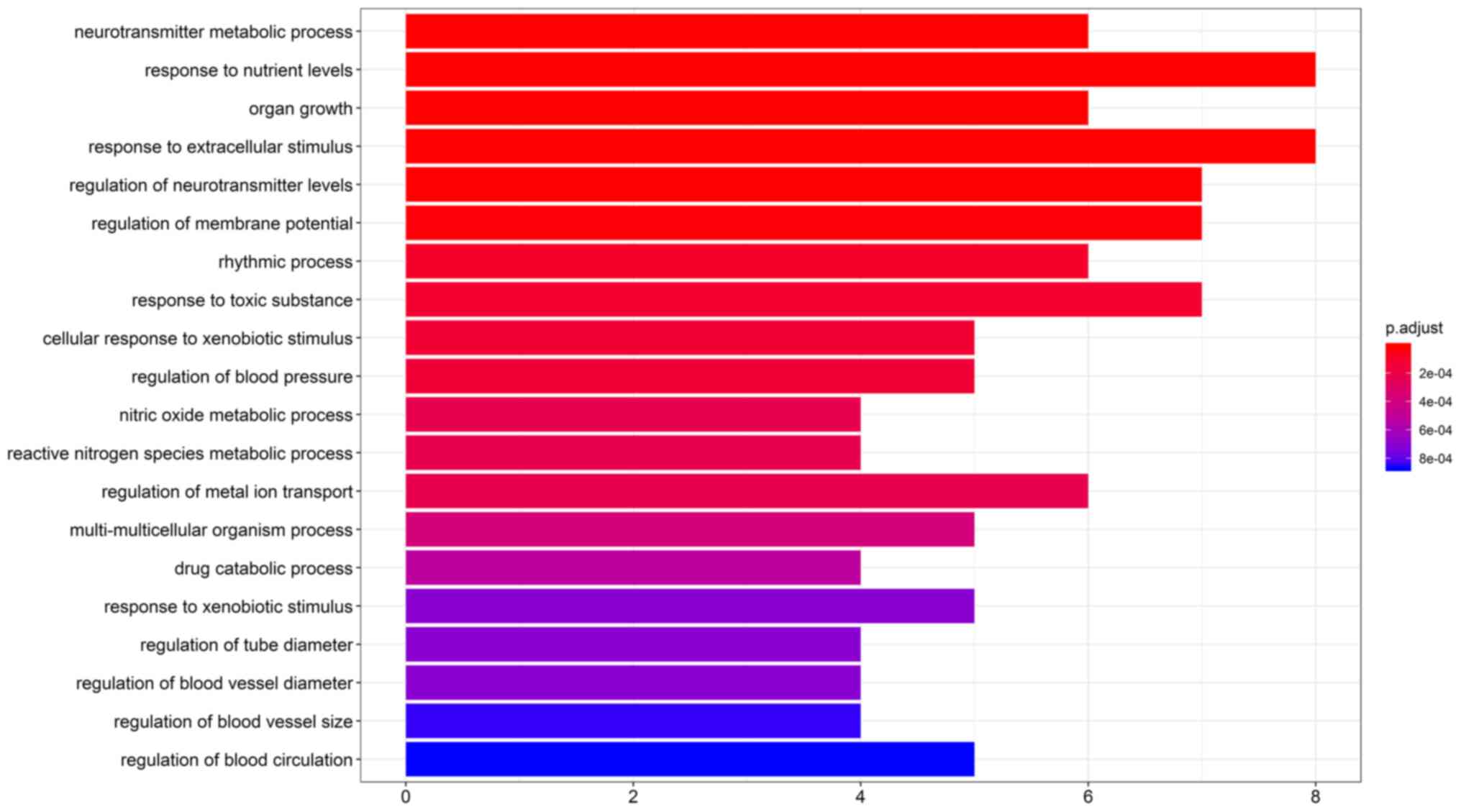
Pathway analysis
The ClusterProfiler package in R was used to perform
GO analysis of the functional characteristics of targets and the
threshold was set as P≤0.01. The results of the GO analysis
suggested that 75 GO terms were enriched in ‘Biological Process’.
The first 20 items with the most significant P-values are provided
in Fig. 4. The neurotransmitter
metabolic process, response to nutrient levels and nitric oxide
metabolic process were among the functional roles of Radix
Angelicae biseratae and this may explain its anti-inflammatory
action.
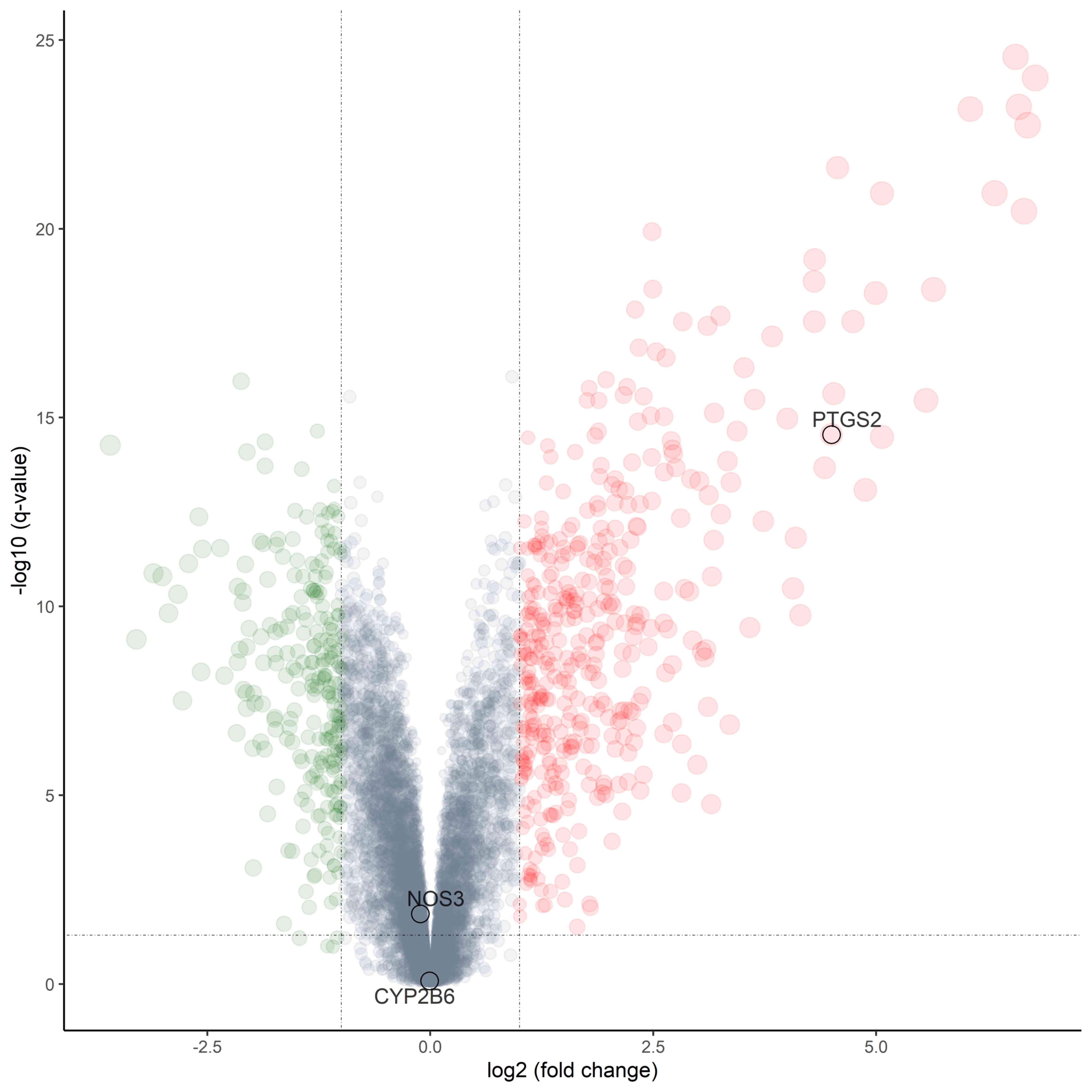
Verification of the target protein in
GSE75181
A total of 665 differentially expressed genes were
identified from two groups of cartilage samples. As presented in
Fig. 5, compared with the hub
target of Radix Angelicae biseratae, PTGS2 was significantly
upregulated in the IL-1β-treated group (adj.P.Val <0.05;
|log2FC|>1).
Docking of hub target proteins and
major components
Docking studies were performed using the Blind
Docking server in the active sites of the two critical targets to
investigate the possible interactions between the active components
and potential targets, namely PTGS2 (PDB ID: 5F19) and NOS3 (PDB
ID: 3NOS). In general, a lower binding energy indicates a stronger
binding between the molecules (37). The conformation with the lowest
binding energy was selected in the component-target complex between
each component and the target and their binding energy is listed in
Table II.
 | Table IILowest binding energy between active
components of Radix Angelicae biseratae and the active sites
of proteins PTGS2 and 3NOS (Protein Databank ID: 5F19 and 3NOS,
respectively) determined using the Blind Docking server
(kcal/mol). |
Table II
Lowest binding energy between active
components of Radix Angelicae biseratae and the active sites
of proteins PTGS2 and 3NOS (Protein Databank ID: 5F19 and 3NOS,
respectively) determined using the Blind Docking server
(kcal/mol).
| Compound | PTGS2 | NOS3 |
|---|
| Sitosterol | -11.76 | -10.67 |
|
O-Acetylcolumbianetin | -9.12 | -8.71 |
| Ammidin | -8.47 | -8.16 |
| Angelicone | -7.71 | -7.91 |
| Angelol D | -8.16 | -7.34 |
| Angelol G | -8.12 | -7.36 |
| Nodakenin | -9.13 | -9.31 |
| Isoimperatorin | -7.62 | -8.19 |
As presented in Table
II, among the active components, a certain conformation of the
complex of sitosterol and hub targets had the highest binding
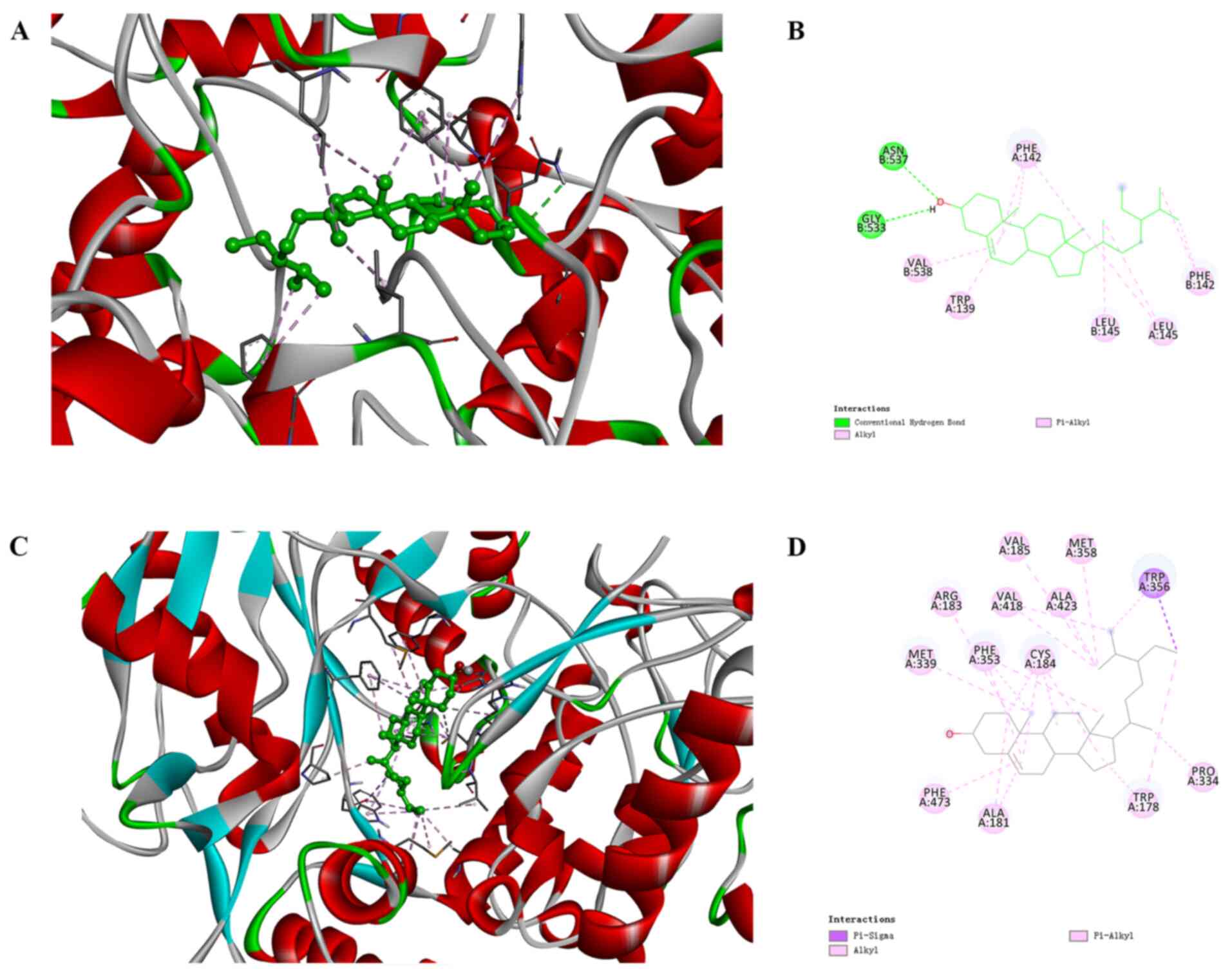
energy. Discovery Studio was used to map the binding pattern of
sitosterol to hub target proteins. The binding pattern of
sitosterol in the active site of PTGS2 is presented as a
three-dimensional model in Fig. 6A
and a two-dimensional diagram is shown in Fig. 6B. Sitosterol was indicated to
interact with the ASN537 residue in the B chain and also, the
GLY533 residue in the B chain has two hydrogen bonds, and
sitosterol has two hydrophobic interactions with the PHE142 and
LEU145 amino acid residues. The binding pattern of sitosterol at
the active site of NOS3 is presented in its three-dimensional model
in Fig. 6C and a two-dimensional
diagram is provided in Fig. 6D.
Sitosterol exhibited an electrostatic interaction in the TRP356
residue on the B chain, and also involved in the alkyl-alkyl
interaction with ALA423, MET358, VAL185, VAL418, CYS184, PHE353,
ARG183, MET339, PHE473, ALA181, TRP178 and PRO334 in the A chain.
These interactions increased the binding affinity between the two
molecules.
Analysis of active components of Radix
Angelicae biseratae extract
HPLC analysis indicated that the Radix Angelicae
biseratae extract contained sitosterol (Fig. 7A and B), which was demonstrated to be a
significant active compound in the treatment of OA that may be
responsible for its anti-inflammatory activity, as determined by
network analysis and molecular docking.
Radix Angelicae biseratae extract
regulates gene and protein expression in the degenerative
chondrocyte model
In order to verify the effect of Radix Angelicae
biseratae extract on inflammatory factors in degenerative
chondrocytes, the rat chondrocytes were treated with 10 ng/ml IL-1β
for 24 h, following which they were treated with Radix Angelicae
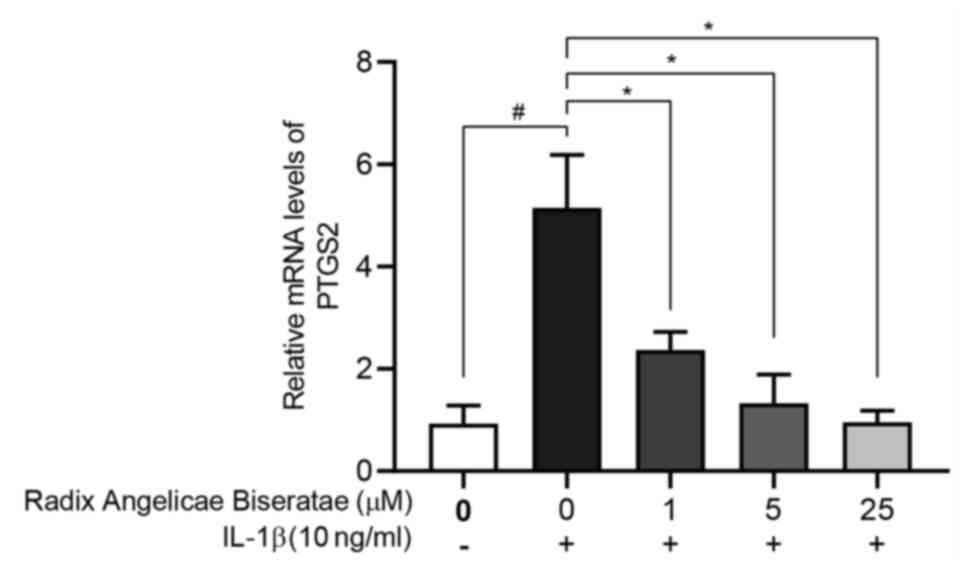
biseratae extract (1, 5 or 25 µM) for 24 h. As presented in
Fig. 8, the expression of PTGS2 in
the IL-1β group was significantly higher compared with that in the
regular group, which was consistent with the results of the gene
expression array. At the same time, Radix Angelicae
biseratae extract significantly decreased the expression of
PTGS2 in degenerative chondrocytes treated with IL-1β (P<0.05),
which indicated that certain active components in Radix
Angelicae biseratae extract were able to inhibit the expression
of PTGS2 in degenerative chondrocytes.
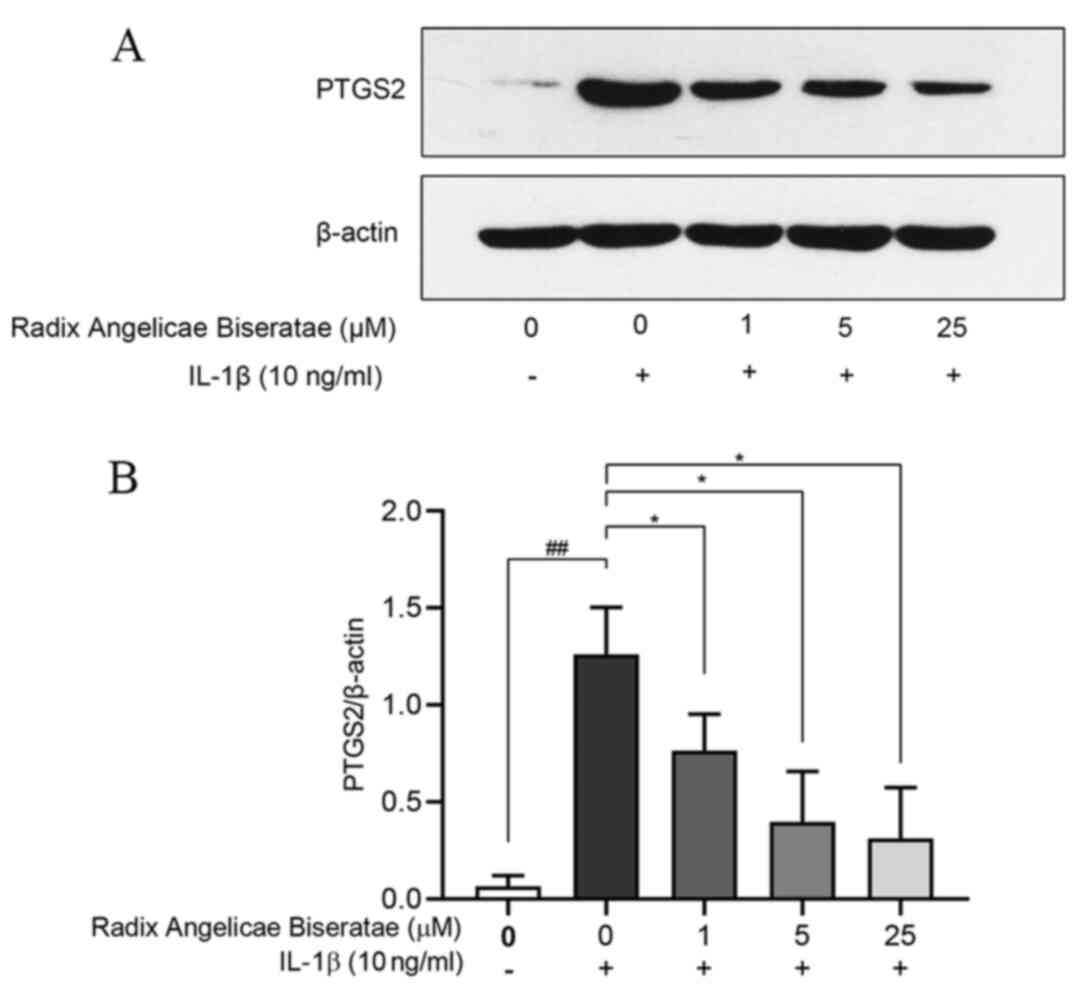
The protein expression of PTGS2 was determined to
verify the effect of Radix Angelicae biseratae on
degenerative chondrocytes (Fig. 9A
and B). Compared with the regular
group, the expression of PTGS2 protein in IL-1β-induced
degenerative chondrocytes was significantly increased (P<0.01).
Compared with that in the degenerative chondrocyte group, the
expression of PTGS2 protein in the Radix Angelicae biseratae
groups (1, 5 and 25 µM) was significantly inhibited
(P<0.05).
Discussion
The systematic pharmacology method has been
developed for determining molecular biological networks and may be
used to discover novel therapeutic effects of drugs from medicinal
plants (8). Therefore, it provides
a systematic method to expand the application of available drug
components in TCM in a variety of complex diseases (9). OA, which is a common disease of
chronic arthritis, is associated with painful symptoms and
significantly affects the patients' quality of life (1,2).
Although Radix Angelicae biseratae has been used as an
effective Chinese herbal medicine for the treatment of OA for
several centuries, its pharmacological mechanism has remained to be
elucidated (5).
In the present study, the active components and
potential targets of Radix Angelicae biseratae in the
treatment of OA were evaluated based on systematic pharmacology,
including ADME system evaluation, network analysis, pathway
analysis and molecular docking. A total of eight active components
were screened and were indicated to interact with 30 different
targets related to OA. According to the analysis of the
components-targets network model, sitosterol had the most
significant number of target connections (degree=11), followed by
O-acetyl columbianetin (degree=7) and nodakenin (degree=6).
Sitosterol has a wide range of biological functions in vivo,
including antitumor, antioxidant, antidiabetic and
anti-inflammatory activity and roles in reducing gallstone activity
(38). Combined with the results of
molecular docking in the present study, it is proposed that
sitosterol is a critical potential active component of Radix
Angelicae biseratae in the treatment of OA.
PTGS2 is the coding gene of cyclooxygenase-2, which
is an essential target of OA anti-inflammatory therapy and one of
the major inflammatory factors involved in articular cartilage
degradation (39). NOS encoded by
the NOS3 gene is also a crucial inflammatory mediator in the
occurrence and development of OA. It can promote the production of
NO in the microenvironment, increase the production of reactive
oxygen species and induce chondrocyte apoptosis (40). CYP2B6 is a member of the cytochrome
P450 superfamily of enzymes that is closely related to drug
metabolism (41). A recent study
demonstrated that CYP2B6 may affect the metabolism of NO under
inflammatory stimulation (42). The
present study indicated that PTGS2, NOS3 and CYP2B6 are potential
targets for Radix Angelicae biseratae in the treatment of
OA, with high degree values in the ‘target-target’ network.
Microarray analysis demonstrated that compared with the non-control
group, PTGS2 was highly expressed in the degenerative chondrocyte
group. RT-qPCR and western blot analysis confirmed that Radix
Angelicae biseratae extract inhibited the expression of PTGS2
in the cartilage degeneration model in vitro, which
indicates that certain active components of Radix Angelicae
biseratae may have a role in the treatment of OA via
anti-inflammatory mechanisms.
Combined with the results of molecular docking, the
present results indicated that sitosterol may be a potential
inhibitor of NOS3 and PTGS2 and may have an anti-inflammatory
effect on OA. The levels of PTGS2 and NOS3 and the effect of
sitosterol on their expression in animal models will be
investigated in future experiments.
The results of the present pathway analysis
indicated that Radix Angelicae biseratae exerted its
pharmacological effects in OA by modulating multiple pathways,
including drug metabolism, inflammation and immune modulation.
In conclusion, sitosterol was demonstrated to be a
critical active component of Radix Angelicae biseratae in
the treatment of OA. Radix Angelicae biseratae extract
reduced the expression of PTGS2 in degenerative chondrocytes, which
may be the underlying mechanism of action of Radix Angelicae
biseratae in reducing inflammation in the treatment of OA.
However, the present study is based on data mining analysis and
in vitro experiments and the results require to be further
verified by in vivo experiments.
Acknowledgements
The authors would like to thank Dr Wen Xu from the
Department of Pharmacology, Fujian University of Traditional
Chinese Medicine (Fuzhou, China), for his help with the
identification of the original herbs.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81774345), the
Natural Science Foundation of Fujian Province (grant no.
2018J01874) and the Health Family Planning Research Talent Training
Project of Fujian Province (grant no. 2017-ZQN-62).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZC and GW conceived and designed the study. ZC, RZ,
CF, JC and JL determined the active components of Radix
Angelicae biseratae and potential targets for the treatment of
OA, downloaded datasets and screened differentially expressed
genes. ZC and RZ performed the experiments and analyzed the data.
ZC ran the statistics and wrote the manuscript. GW reviewed and
edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All animal experimental procedures were approved by
the Animal Care and Use Committee of Fujian University of
Traditional Chinese Medicine (Fuzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lespasio MJ, Piuzzi NS, Husni ME, Muschler
GF, Guarino A and Mont MA: Knee osteoarthritis: A primer. Perm J.
21:16–183. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bonnet CS and Walsh DA: Osteoarthritis,
angiogenesis and inflammation. Rheumatology (Oxford). 44:7–16.
2005.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Shen J, Abu-Amer Y, O'Keefe RJ and
McAlinden A: Inflammation and epigenetic regulation in
osteoarthritis. Connect Tissue Res. 58:49–63. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Urban H and Little CB: The role of fat and
inflammation in the pathogenesis and management of osteoarthritis.
Rheumatology (Oxford). 57 (Suppl_4):iv10–iv21. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ma J, Huang J, Hua S, Zhang Y, Zhang Y, Li
T, Dong L, Gao Q and Fu X: The ethnopharmacology, phytochemistry
and pharmacology of Angelica biserrata: A review. J Ethnopharmacol.
231:152–169. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lyu S, Ji B, Gao W, Chen X, Xie X and Zhou
J: Effects of angelicae pubescentis and loranthi decotion on
repairing knee joint cartilages in rats. J Orthop Surg Res.
12(189)2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hopkins AL: Network pharmacology: The next
paradigm in drug discovery. Nat Chem Biol. 4:682–690.
2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang YF, Huang Y, Ni YH and Xu ZM:
Systematic elucidation of the mechanism of geraniol via network
pharmacology. Drug Des Devel Ther. 13:1069–1075. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yue SJ, Xin LT, Fan YC, Li SJ, Tang YP,
Duan JA, Guan HS and Wang CY: Herb pair Danggui-Honghua: Mechanisms
underlying blood stasis syndrome by system pharmacology approach.
Sci Rep. 7(40318)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ru J, Li P, Wang J, Zhou W, Li B, Huang C,
Li P, Guo Z, Tao W, Yang Y, et al: TCMSP: A database of systems
pharmacology for drug discovery from herbal medicines. J
Cheminform. 6(13)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cabrera-Pérez M and Pham-The H:
Computational modeling of human oral bioavailability: What will be
next? Expert Opin Drug Discov. 13:509–521. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tao W, Xu X, Wang X, Li B, Wang Y, Li Y
and Yang L: Network pharmacology-based prediction of the active
ingredients and potential targets of Chinese herbal Radix Curcumae
formula for application to cardiovascular disease. J
Ethnopharmacol. 145:1–10. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yue SJ, Liu J, Feng WW, Zhang FL, Chen JX,
Xin LT, Peng C, Guan HS, Wang CY and Yan D: System
pharmacology-based dissection of the synergistic mechanism of
huangqi and huanglian for diabetes mellitus. Front Pharmacol.
8(694)2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kuhn M, Szklarczyk D, Franceschini A, von
Mering C, Jensen LJ and Bork P: STITCH 3: Zooming in on
protein-chemical interactions. Nucleic Acids Res. 40 (Database
issue):D876–D880. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Koscielny G, An P, Carvalho-Silva D, Cham
JA, Fumis L, Gasparyan R, Hasan S, Karamanis N, Maguire M, Papa E,
et al: Open Targets: A platform for therapeutic target
identification and validation. Nucleic Acids Res. 45
(D1):D985–D994. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Carvalho-Silva D, Pierleoni A, Pignatelli
M, Ong C, Fumis L, Karamanis N, Carmona M, Faulconbridge A,
Hercules A, McAuley E, et al: Open targets platform: New
developments and updates two years on. Nucleic Acids Res. 47
(D1):D1056–D1065. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wishart DS, Knox C, Guo AC, Shrivastava S,
Hassanali M, Stothard P, Chang Z and Woolsey J: DrugBank: A
comprehensive resource for in silico drug discovery and
exploration. Nucleic Acids Res. 34 (Database issue):D668–D672.
2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wishart DS, Knox C, Guo AC, Cheng D,
Shrivastava S, Tzur D, Gautam B and Hassanali M: DrugBank: A
knowledgebase for drugs, drug actions and drug targets. Nucleic
Acids Res. 36 (Database issue):D901–D906. 2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wishart DS, Feunang YD, Guo AC, Lo EJ,
Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al:
DrugBank 5.0: A major update to the DrugBank database for 2018.
Nucleic Acids Res. 46 (D1):D1074–D1082. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kafkas Ş, Dunham I and McEntyre J:
Literature evidence in open targets-a target validation platform. J
Biomed Semantics. 8(20)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
von Mering C, Jensen LJ, Snel B, Hooper
SD, Krupp M, Foglierini M, Jouffre N, Huynen MA and Bork P: STRING:
Known and predicted protein-protein associations, integrated and
transferred across organisms. Nucleic Acids Res. 33 (Database
issue):D433–D437. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Huntley RP, Harris MA, Alam-Faruque Y,
Blake JA, Carbon S, Dietze H, Dimmer EC, Foulger RE, Hill DP,
Khodiyar VK, et al: A method for increasing expressivity of Gene
Ontology annotations using a compositional approach. BMC
Bioinformatics. 15(155)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Troyanskaya O, Cantor M, Sherlock G, Brown
P, Hastie T, Tibshirani R, Botstein D and Altman RB: Missing value
estimation methods for DNA microarrays. Bioinformatics. 17:520–525.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sanchez-Linares I, Perez-Sanchez H,
Cecilia JM and Garcia JM: High-Throughput parallel blind virtual
screening using BINDSURF. BMC Bioinformatics. 13 Suppl 14 (Suppl
14)(S13)2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Burley SK, Berman HM, Bhikadiya C, Bi C,
Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S,
et al: RCSB Protein Data Bank: Biological macromolecular structures
enabling research and education in fundamental biology,
biomedicine, biotechnology and energy. Nucleic Acids Res. 47
(D1):D464–D474. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Science Mo and China TotPsRo: Guidance
suggestions for the care and use of laboratory animals. 2006.
|
|
31
|
Li X, Du M, Liu X, Wu M, Ye H, Lin J, Chen
W and Wu G: Millimeter wave treatment inhibits NO-induced apoptosis
of chondrocytes through the p38MAPK pathway. Int J Mol Med.
25:393–399. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Li XH, Wu MX, Ye HZ, Chen WL, Lin JM,
Zheng LP and Liu XX: Experimental study on the suppression of
sodium nitroprussiate-induced chondrocyte apoptosis by Tougu
Xiaotong Capsule-containing serum. Chin J Integr Med. 17:436–443.
2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sanchez C, Mathy-Hartert M, Deberg MA,
Ficheux H, Reginster JY and Henrotin YE: Effects of rhein on human
articular chondrocytes in alginate beads. Biochem Pharmacol.
65:377–388. 2003.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wu SQ, Otero M, Unger FM, Goldring MB,
Phrutivorapongkul A, Chiari C, Kolb A, Viernstein H and Toegel S:
Anti-inflammatory activity of an ethanolic Caesalpinia
sappan extract in human chondrocytes and macrophages. J
Ethnopharmacol. 138:364–372. 2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lin J, Wu G, Chen J, Fu C, Hong X, Li L,
Liu X and Wu M: Electroacupuncture inhibits sodium
nitroprusside-mediated chondrocyte apoptosis through the
mitochondrial pathway. Mol Med Rep. 18:4922–4930. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Takamatsu Y, Sugiyama A, Purqon A, Nagao H
and Nishikawa K: Binding free energy calculation and structural
analysis for antigen-antibody complex. J. 832:566–569. 2006.
|
|
38
|
Liao PC, Lai MH, Hsu KP, Kuo YH, Chen J,
Tsai MC, Li CX, Yin XJ, Jeyashoke N and Chao LK: Identification of
β-sitosterol as in vitro anti-inflammatory constituent in moringa
oleifera. J Agric Food Chem. 66:10748–10759. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Fukai A, Kamekura S, Chikazu D, Nakagawa
T, Hirata M, Saito T, Hosaka Y, Ikeda T, Nakamura K, Chung UI and
Kawaguchi H: Lack of a chondroprotective effect of cyclooxygenase 2
inhibition in a surgically induced model of osteoarthritis in mice.
Arthritis Rheum. 64:198–203. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhang LC, Hu Y and Sui C: Lack of
associations between polymorphisms in SOD2 (rs2758331), NOS3
(rs1808593), PPARδ (rs9794 and rs10865710) and the risk of
osteoarthritis in a Chinese Han population: A case-control study.
Chin Med J (Engl). 132:1113–1114. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Crettol S, Déglon JJ, Besson J,
Croquette-Krokkar M, Gothuey I, Hämmig R, Monnat M, Hüttemann H,
Baumann P and Eap CB: Methadone enantiomer plasma levels, CYP2B6,
CYP2C19, and CYP2C9 genotypes, and response to treatment. Clin
Pharmacol Ther. 78:593–604. 2005.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lee CM, Tripathi S and Morgan ET: Nitric
oxide-regulated proteolysis of human CYP2B6 via the
ubiquitin-proteasome system. Free Radic Biol Med. 108:478–486.
2017.PubMed/NCBI View Article : Google Scholar
|