Introduction
Abnormally invasive placenta (AIP) is a pathologic
condition in which the placenta abnormally attaches to and invades
the uterine wall (1). AIP is
regarded as one of the most dangerous conditions associated with
pregnancy. The main complication of AIP is massive hemorrhaging
when attempts are made to remove the placenta manually during
delivery. Massive hemorrhaging may result in multisystem organ
failure, disseminated intravascular coagulation, hysterectomy,
intensive care requirement and even death (2). AIP is a common complication in
obstetrics and it is the leading cause of hysterectomies associated
with caesarean delivery and peripartum hysterectomies (3). A standardized approach with a
comprehensive multidisciplinary care team accustomed to the
management of AIP is essential for favourable maternal and neonatal
outcomes (4-6).
The early diagnosis of pregnant females with AIP will provide more
opportunities to receive multidisciplinary care and good perinatal
outcomes (7). Studies have
indicated that certain placental and fetal hormones, such as human
chorionic gonadotropin, alpha-fetoprotein and pregnancy-associated
plasma protein A, are helpful in the diagnosis of AIP (8,9).
However, none of the placental/fetal proteins have proved useful
for diagnosing AIP earlier than ultrasound and MRI. The major
prenatal diagnostic methods of AIP are still ultrasound and MRI
examination. AIP remains unpreventable and at present, more than
half of all pregnant females with AIP remain undiagnosed after
imaging examinations prior to delivery (10).
Although AIP may be diagnosed through imaging
examinations as well as placental and fetal hormones, the
expression of lncRNAs (long non-coding RNAs) and the roles they
have in AIP have remained elusive. lncRNAs are transcripts no
longer than 200 bp, which were indicated to actively regulate the
expression and function of thousands of protein-coding genes
through various mechanisms (11).
Numerous lncRNAs identified in the human placenta are reported to
be involved in preeclampsia and intrauterine growth restriction due
to their regulation of trophoblast differentiation, proliferation
and migration (12). lncRNA
metastasis-associated adenocarcinoma transcript 1 was even proved
to be involved in AIP and associated with trophoblast-like cell
invasion (13). These studies imply
that lncRNAs may participate in the process of AIP and have great
potential to serve as biomarkers of AIP.
Reliable biomarkers for prediction and diagnosis may
contribute to perinatal outcomes. Thus, it was attempted to seek
potential and valid lncRNAs to serve as biomarkers for the
efficient and precise prediction and diagnosis of AIP. In the
present study, the lncRNA expression profiles of patients with AIP
were determined and 5 randomly selected lncRNAs were validated by
reverse transcription-quantitative (RT-q)PCR.
Materials and methods
Sample collection and ethics
approval
In the present study, placenta tissues from pregnant
females enrolled at Southern Medical University Affiliated Maternal
& Child Health Hospital of Foshan (Foshan, China) between
December 2017 and June 2018 were analysed. A total of 5 pregnant
females who were antenatally suspected of having AIP according to
array-scale and doppler sonography were enrolled in the study. All
of these 5 patients were diagnosed with AIP by expert surgeons
during the operation according to the clinical grading system
(14). Subsequently, the diagnosis
was confirmed through pathological examination according to the
pathological classification criteria (15). Clinical and demographic data on each
of the patients is provided in Table
I. All pregnant females were diagnosed with a scarred uterus
and placenta previa, and they had no hypertension, diabetes or
other obstetric complications. Tissues from each patient of the AIP
group included one sample from the invasive site and another sample
from the adherent noninvasive site of the placenta according to the
operators' instructions. They were divided into the invasive and
control group. In addition, for extended sample validation,
placental tissues were collected from 15 patients with AIP and 15
patients with a normal placenta. The two groups were referred to as
the AIP group and the normal group. After placental delivery, all
of the samples were cut from the surface of the maternal placenta.
They were washed in PBS five times to remove blood cells and were
then transferred into liquid nitrogen within 10 min.
 | Table IClinical details of pregnant
females. |
Table I
Clinical details of pregnant
females.
| No. | Age (years) | Gestational age
(weeks) | Prior caesarean
section | Other uterine
surgery | Hysterectomy | Histological
gradea |
|---|
| 1 | 29 | 36+1 | 1 | 2 (induced
abortion) | No | Percreta |
| 2 | 30 | 34+5 | 1 | 0 | No | Increta |
| 3 | 28 | 35+1 | 1 | 1 (induced
abortion) | No | Increta |
| 4 | 30 | 36+1 | 1 | 0 | No | Increta |
| 5 | 29 | 35+4 | 1 | 0 | No | Accreta |
The Human Ethics Committee of the Foshan Women and
Children's Hospital Affiliated to Southern Medical University
(Foshan, China) approved this study (approval no.
FSFY-MEC-2017-055). Written informed consent was obtained from all
pregnant females for using their specimens and other clinical
information. All of the methods were performed according to the
approved guidelines of the Human Ethics Committee.
RNA extraction, amplification and
hybridization
For microarray analysis, lncRNAs and mRNAs were
detected with the Arraystar Human LncRNA Microarray v4.0
(Arraystar, Inc.). Following the manufacturer's protocol, total RNA
was extracted from frozen samples using TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc.). The RNA quantity and quality were
measured using a NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.).
An Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) and
standard denaturing agarose gel electrophoresis were used to
evaluate RNA integrity. Following the Agilent One-Color
Microarray-Based Gene Expression Analysis protocol (Agilent
Technologies, Inc.), array hybridization and sample labelling were
performed. After removing ribosomal RNA and performing
amplification, 100 ng of total RNA was labelled and hybridized.
Subsequently, the hybridized arrays were washed, fixed and scanned
on the Agilent DNA Microarray Scanner. The expression data were
stored in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo) under the
accession number GSE126552.
RT-qPCR validation
In the present study, original validation (samples
were divided into invasive group and control group, and all sample
were collected from 5 patients with AIP) and extended validation
(samples were divided into AIP group and normal group. Samples of
AIP group were collected from 15 patients with AIP, samples of the
normal group were collected from 15 patients without AIP) was
conducted to validate the results of microarray through RT-qPCR.
For RT-qPCR validation, total RNA was extracted with TRIzol reagent
from the remaining portion of tissues not used in the lncRNA
microarray. Subsequently, first-strand complementary DNAs were
generated using SuperScript™ III Reverse Transcriptase
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. The RT-qPCR process was performed
using a ViiA-7 RT-PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and 2X PCR master mix (Arraystar, Inc.). The
relative expression levels of lncRNAs were quantified using the
2-ΔΔCq method (16) and
were normalized to β-actin expression. The primers for RT-qPCR were
designed according to the lncRNA sequences in NONCODE (http://www.noncode.org) and primer sequences were
synthesized and purified by Yingjun Co. The primer sequences are
listed in Table SI.
Bioinformatics analysis of RNA scanned
data
Raw data were extracted from the acquired array
images using Agilent Feature Extraction software 11.0.1.1.
Subsequently, GeneSpring GX v12.1 software (Agilent Technologies,
Inc.) was used to normalize quantiles of raw data. lncRNAs and
mRNAs with flags in ‘Present’ or ‘Marginal’ (‘All Targets Value’)
in at least 5 out of 10 samples were chosen for further data
analysis. Dysregulated lncRNAs and mRNAs between two groups with a
fold change >1.5 were filtered based on their P-value/false
discovery rate (<0.05).
Gene Ontology (GO) and Kyoto Encyclopaedia of Genes
and Genomes (KEGG) pathway analyses were applied to predict the
roles of the dysregulated mRNAs by determining GO terms and
biological pathways, respectively. In-house scripts of R3.5.0 were
used to perform hierarchical clustering and to perform combined
analysis. First, http://bioconductor.org/biocLite.R was sourced to
install the clusterProfiler package and load org.Hs.eg.db.
Subsequently, the function of enrichGO and enrichKEGG was used to
obtain the results.
Coexpression analysis of the dysregulated lncRNAs
subgroup was also performed to identify coregulated lncRNA-mRNA
pairs. The subgroups of dysregulated RNA coregulation pairs that
were dysregulated included large intervening noncoding RNAs
(lincRNAs), antisense lncRNAs and their paired dysregulated
mRNAs.
Statistical analysis
In present study, all subsequent analysis was
conducted using R 3.5.0 (https://www.r-project.org/). Proportions or mean ±
standard error were used to present all variables when appropriate.
A Mann-Whitney U test was used to compare differences in RT-qPCR
results between invasive group and control group and a Mann-Whitney
U test was used to compare differences in RT-qPCR results between
the AIP group and normal group. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression profile of lncRNAs and
mRNAs
To explore the lncRNA and mRNA expression profiles
in AIP, 5 pairs of placenta tissues of patients with AIP were
screened and divided into two groups: The invasive group and the
control group. In total, 20,730 mRNAs and 40,173 lncRNAs were
detected by microarray analysis. Among them, 329 lncRNAs were
significantly differentially expressed (fold change >1.5,
P<0.05). Of the dysregulated lncRNAs, 101 transcripts were
upregulated, while 228 transcripts were downregulated in the
invasive group. The top dysregulated lncRNAs in the invasive group
were leucine-rich repeat-containing 38 among the upregulated
lncRNAs and ELMO domain-containing 1 among the downregulated
lncRNAs. Furthermore, a total of 179 dysregulated mRNAs (fold
change >1.5, P<0.05) were identified. Of the dysregulated
mRNAs in the invasive group, 95 transcripts were upregulated and 84
transcripts were downregulated. The most significantly dysregulated
mRNAs in the invasive group were AF003625.3 (upregulated) and
G037111 (downregulated). Hierarchical clustering was performed for
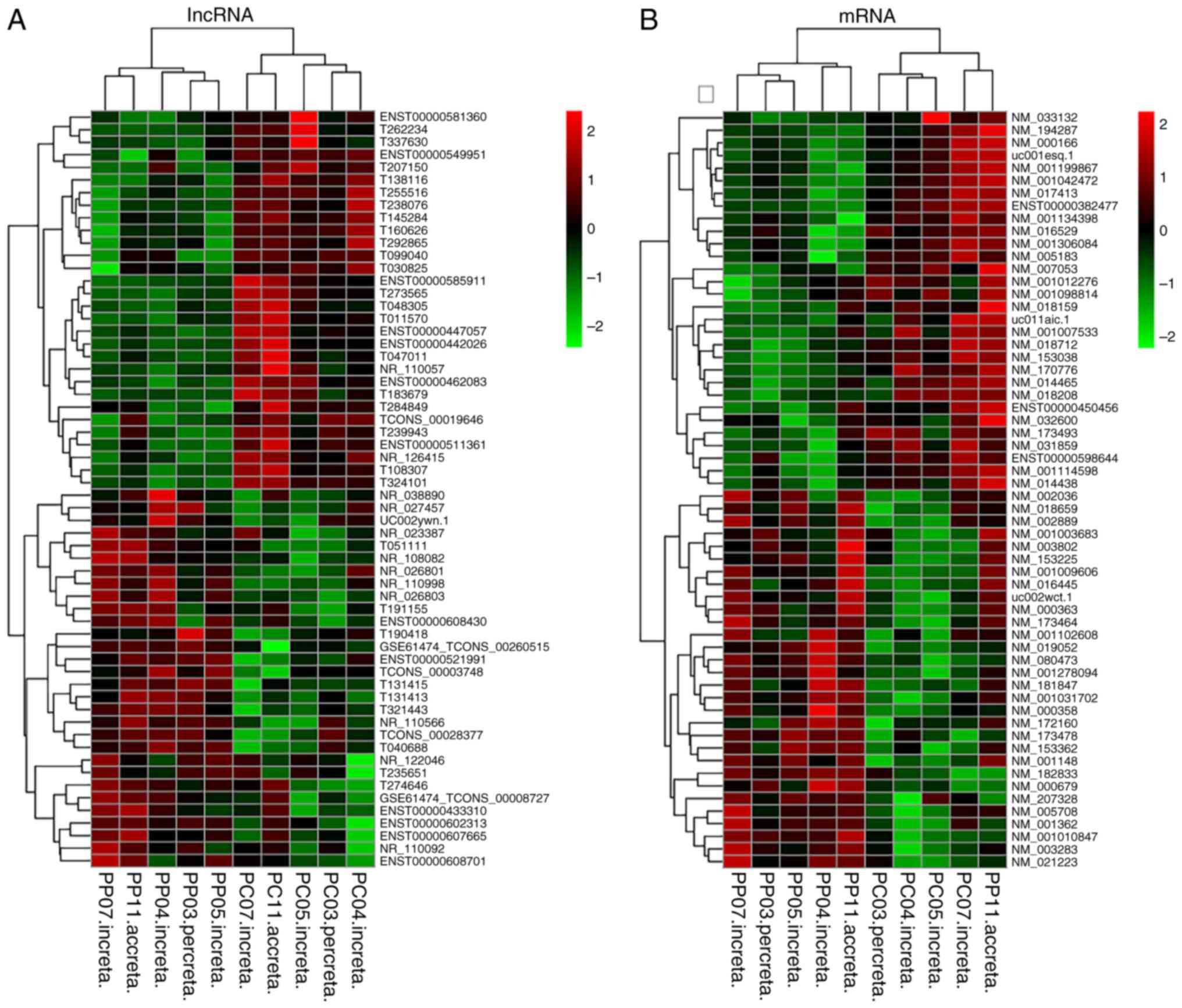
inter-group comparisons of differential gene expression patterns
(Fig. 1A and B).
Expression signatures of the
dysregulated lncRNAs and coexpression analysis of subgroup
To further understand lncRNA expression patterns in
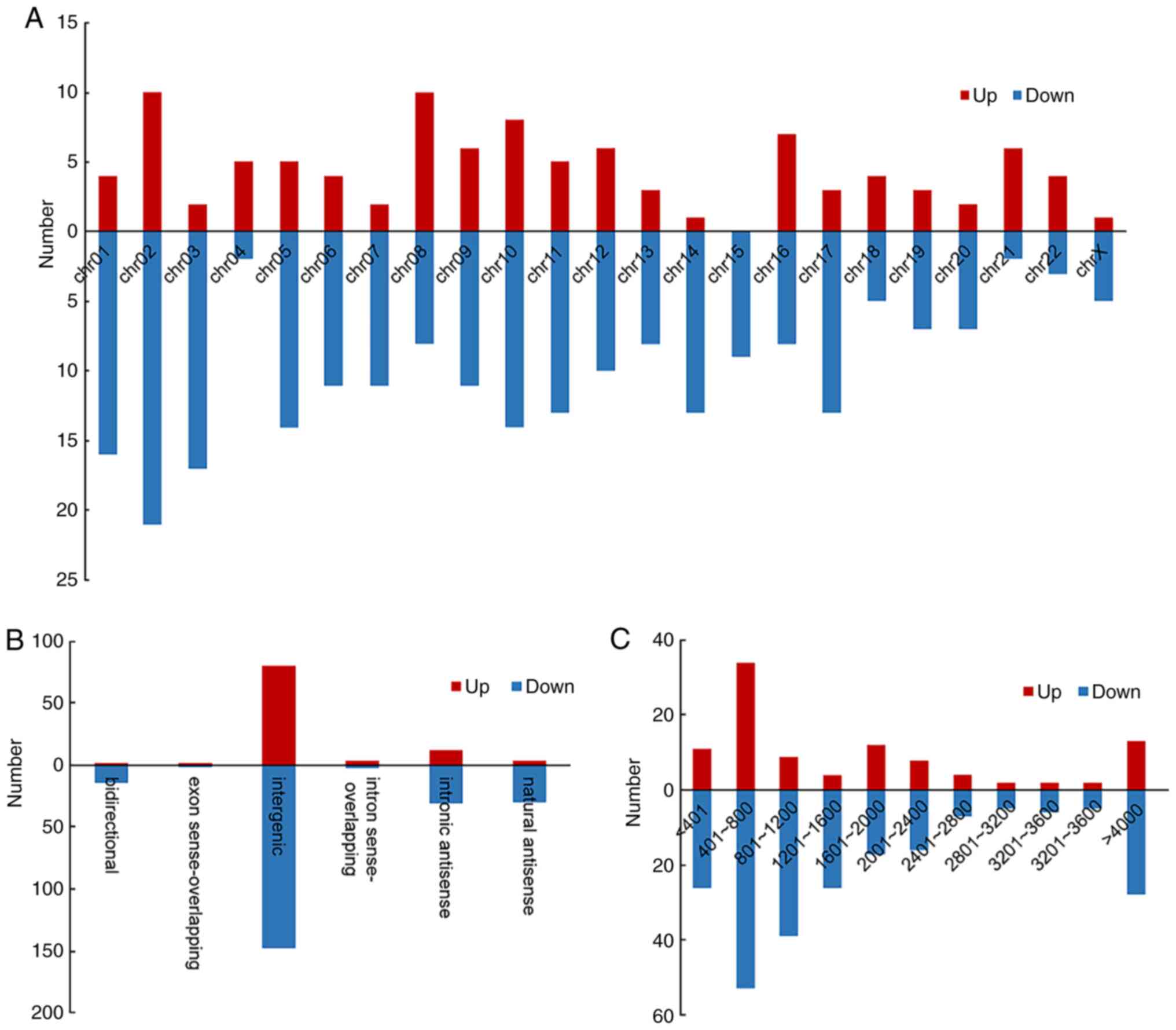
AIPs, the length, chromosome and classification distribution of the
dysregulated lncRNAs were also analysed (Fig. 2). lncRNAs were divided into six
categories according to their association with protein-coding
genes. The categories were bidirectional, intergenic, exon
sense-overlapping, intron sense-overlapping, natural antisense and
intronic antisense. The statistical results indicated that the
majority of dysregulated lncRNAs were intergenic (72.15%; Fig. 2B) and were 400-800 nt in length
(69.30%; Fig. 2C). The chromosome
distribution indicated that chromosome 2 (9.42%) and 10 (6.69%)
contained most of the differentially expressed lncRNAs (Fig. 2A). The expression profile of the
lncRNAs revealed dysregulated lncRNAs that may take part in
AIP.
A coexpression analysis was used to further
determine the potential functions of differentially expressed
lncRNA subgroup in AIP. A total of 10 pairs of lincRNA-mRNA and 2
pairs of antisense lncRNA-mRNA (distance <300 kb) were indicated
to be coregulated transcripts (Table
II).
 | Table IICoexpression analysis of dysregulated
lncRNAs subgroup. |
Table II
Coexpression analysis of dysregulated
lncRNAs subgroup.
| Gene symbol | Fold change-
lncRNAs | Genome
relationship | Nearby gene
symbol | Nearby protein
name | Fold
change-mRNAs |
|---|
| CTD-2587H24.10 | -2.73297 | Upstream | TNNI3 | Troponin I type 3
(cardiac) | +1.88938 |
| CTD-2587H24.10 | -2.73297 | Upstream | TNNT1 | Troponin T type 1
(skeletal, slow) | +1.983701 |
| VTRNA2-1 | -1.53625 | Downstream | TGFBI | Transforming growth
factor beta induced | +1.983147 |
| G008916 | +1.677219 | Upstream | BAMBI | BMP and activin
membrane-bound inhibitor | +1.557582 |
| G023160 | +1.567031 | Downstream | ZIC5 | Zic family member
5 | -2.799566 |
| G037467 | -1.617928 | Downstream | ACAA2 | Acetyl-CoA
acyltransferase 2 | -1.680578 |
| G045620 | -1.640229 | Upstream | PCDP1 | Primary ciliary
dyskinesia 1 homolog | -2.004573 |
| G052555 | +1.570389 | Upstream | ADARB1 | Adenosine
deaminase, RNA-specific, B1 | +1.525046 |
| G052559 | -1.515549 | Downstream | ADARB1 | Adenosine
deaminase, RNA-specific, B1 | +1.525046 |
| G052938 | -1.686997 | Downstream | GGT2 |
Gamma-glutamyltransferase 2 | -1.921371 |
| RP11-247A12.1 | -1.716386 | Natural
antisense | CRAT | Carnitine
O-acetyltransferase | -1.592573 |
| AK057317 | -1.758716 | Intronic
antisense | ASIC2 | Acid sensing ion
channel subunit 2 | +1.618979 |
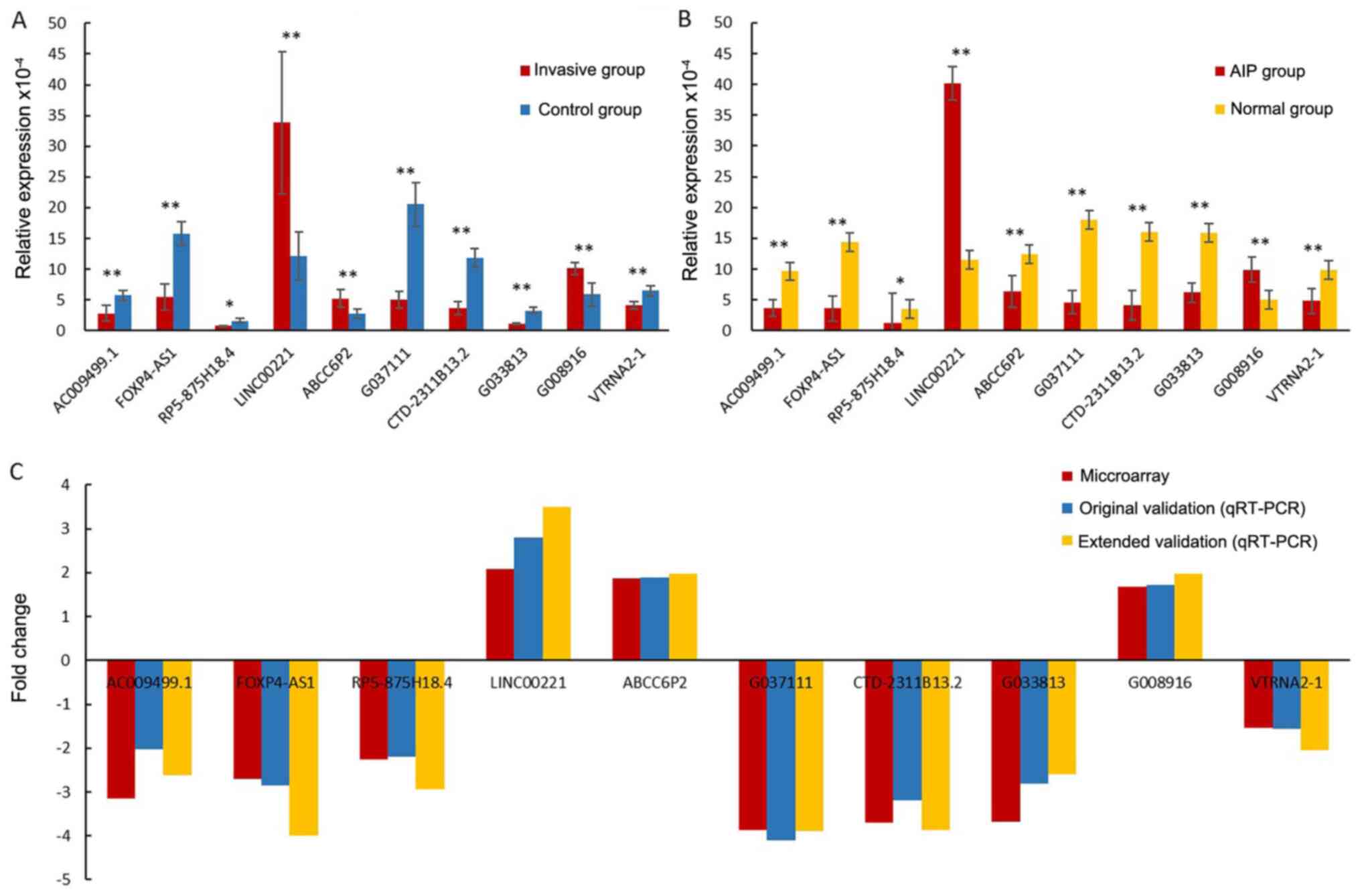
RT-qPCR results
To independently validate the dysregulated lncRNAs
detected by the RNA-microarray, 10 aberrantly expressed lncRNAs
[AC009499.1, FOXP4 antisense RNA 1 (FOXP4-AS1), RP5-875H18.4, long
intergenic non-protein coding RNA 221 (LINC00221), ATP binding
cassette subfamily C member 6 pseudogene 2 (ABCC6P2), G037111,
CTD-2311B13.2, G033813, G008916 and vault RNA 2-1 (VTRNA2-1)] were
randomly selected. Their expression was validated by RT-qPCR. The
validation results demonstrated that AC009499.1, FOXP4-AS1,
RP5-875H18.4, G037111, CTD-2311B13.2, G033813 and VTRNA2-1 were
downregulated, while LINC00221, ABCC6P2 and G008916 were
upregulated in the invasive group and AIP group vs. the control and
normal group (P<0.05; Fig. 3A
and B). In addition, the fold
changes of the 10 lncRNAs in both original and extended validation
were close to the results of the microarray (Fig. 3C). In summary, the RT-qPCR results
exhibited good consistency with the RNA-microarray data, indicating
that to determine lncRNA changes, RNA-microarray methodology has
good reliability and reproducibility.
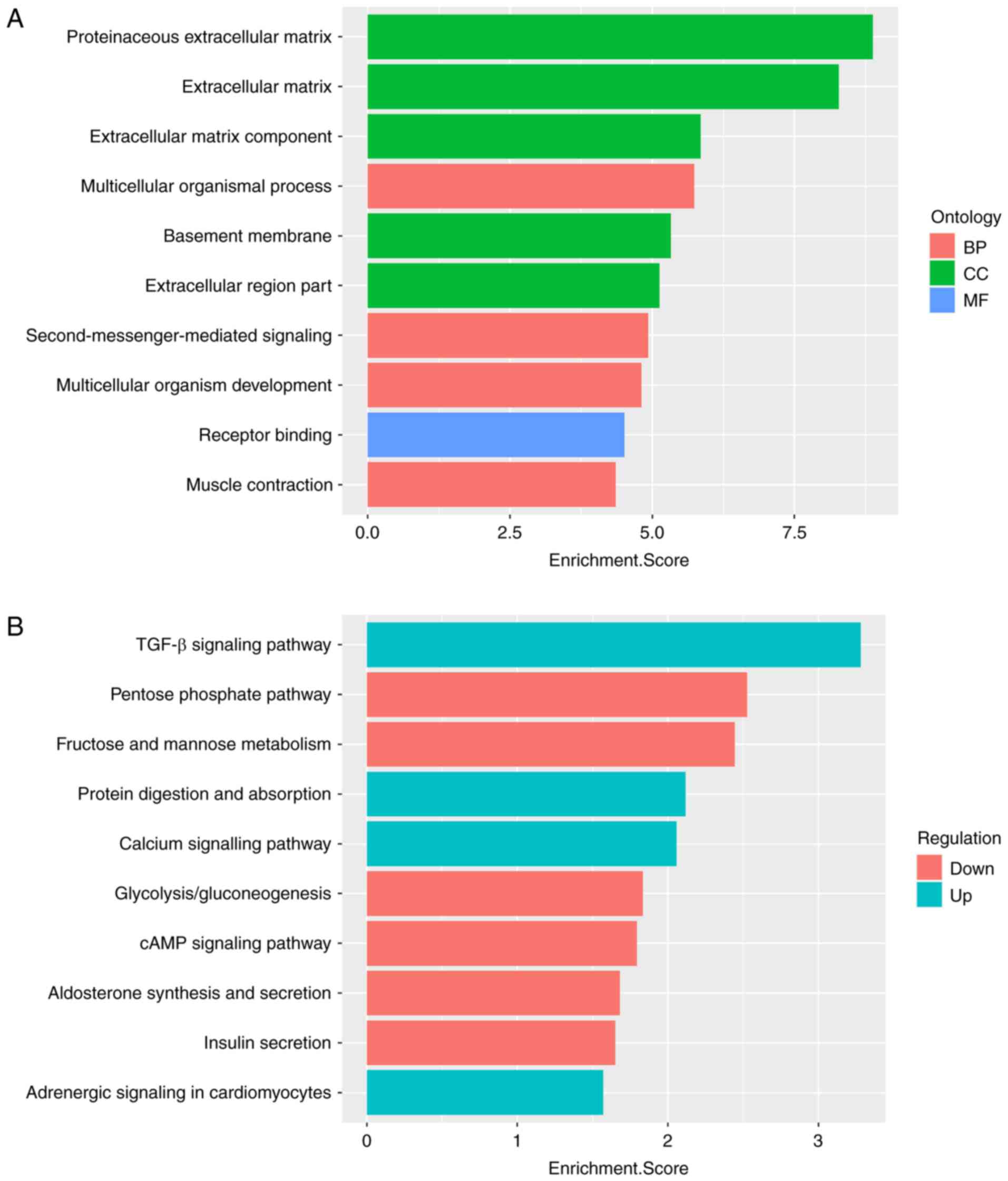
Functional enrichment analysis
GO and KEGG pathway analyses were performed to
identify the functional roles of differentially expressed mRNAs in
AIPs. GO analysis was performed in three domains: Biological
process (BP), cellular component (CC) and molecular function (MF).
The results suggested that all 10 of the most significant functions
were enriched with upregulated mRNAs. As presented in Fig. 4A, in the BP domain, the upregulated
genes were most enriched in the terms proteinaceous extracellular
matrix (GO:0005578), extracellular matrix (ECM; GO:0031012) and ECM
component (GO:0044420). For CC and MF, the upregulated genes were
most enriched in multicellular organismal process (GO:0032501) and
receptor binding (GO:0005102), respectively.
Pathway analysis was performed by mapping genes to
KEGG pathways. The 10 most significantly enriched pathways
corresponding to dysregulated mRNAs are listed in Fig. 4B. The most enriched pathways were
the TGF-β signaling pathway (KEGG ID: hsa04350) for upregulated
mRNAs and the pentose phosphate pathway (KEGG ID: hsa00030) for
downregulated mRNAs.
Discussion
The present study provided a comprehensive analysis
of lncRNA and mRNA expression profiles in AIP. In invasive placenta
tissues, a total of 329 lncRNAs (101 upregulated; 228
downregulated) and 179 mRNAs (95 upregulated; 84 downregulated)
were differentially expressed compared with adherent noninvasive
tissues. Most of the dysregulated lncRNAs were from intergenic
regions (72.15%). The RT-qPCR results are consistent with the
results of the microarray assays, proving the reliability of the
microarray results.
GO analysis indicated that the top three functions
of all dysregulated genes were enriched in proteinaceous ECM, ECM
and ECM components. Physiologically, ECM provides biochemical and
structural support to surrounding cells (17) and undergoes vigorous reorganization
in the process of placenta implantation (18). ECM components are critical for
trophoblast invasion (19). Heparan
sulfate proteoglycans, an ample ECM component of the placenta, may
decentralize the ECM and promote trophoblast invasion to the
myometrium when they degrade (19).
Other ECM components, such as decorin and biglycan, were reported
to be involved in placental invasion (20). In addition, ECM elasticity may
direct cellular differentiation, including the
epithelial-mesenchymal transition (EMT), and studies have indicated
that trophoblast EMT has an important role in AIP (21,22).
MMPs in the ECM are able remodel the ECM to facilitate EMT,
promoting cell specification during embryonic and placental
development (23,24). Furthermore, placental basement
membrane proteins also participate in trophoblast invasion; they
constitute >80% of ECM proteins in the placental basal plate and
are indispensable for effective cytotrophoblast invasion (25). As the results of the present
functional enrichment analysis indicate, the basement membrane had
the fifth-highest enrichment score, which is consistent with the
above results. The ECM is derived from a multicellular organismal
process, which has important functions in cell adhesion,
differentiation and cell-to-cell communication (26). These different roles may be the
reason why the category of multicellular organismal process has the
fourth-highest enrichment score. KEGG pathway analysis for
dysregulated mRNAs revealed that the top enriched pathway was the
TGF-β signaling pathway. According to previous studies, numerous
cellular processes, including EMT (27-29),
ECM neogenesis and placenta formation are regulated by the TGF-β
signaling pathway (30). The
results of the KEGG analysis of the present study were consistent
with those of the GO enrichment analysis. Furthermore, Duzyj et
al (31) indicated that the
endoglin-TGF-β pathway system is abnormally expressed in the
invasive site of the placenta, which increases the invasiveness of
placental cells. While no previous lncRNA studies have been
performed on the regulation of AIP, to the best of our knowledge,
the present study provided clues that dysregulated lncRNAs may
regulate the process of ECM neogenesis and placenta formation to
facilitate the process of AIP through the TGF-β signaling
pathway.
Previous research has demonstrated that lncRNAs are
able to regulate the expression of nearby mRNAs (32). A coexpression analysis performed in
the present study suggested that the expression of 10 lncRNAs and 2
antisense lncRNAs was correlated with the expression of nearby
mRNAs. Although the corresponding functions of these mRNAs in AIP
remain elusive, based on previous studies and GO analysis, it was
indicated that they are mainly involved in muscle contraction
regulation [troponin I1 (TNNI1), troponin I3 (TNNI3)] (33), TGF-β signaling [BMP and activin
membrane-bound inhibitor (BAMBI) and TGF-β-induced (TGFBI)]
(34,35), cell proliferation, migration,
aggressiveness [ZIC5, acetyl-CoA acyltransferase 2 (ACAA2)]
(36-39),
multicellular organism function [adenosine deaminase RNA specific
B1 (ADARB1), acid sensing ion channel subunit 2 (ASIC2)], amino
acid regulation [gamma-glutamyltransferase 2 (GGT2)] and fat
metabolism [carnitine O-acetyltransferase (CRAT)]. Of note, BAMBI
and TGFBI, as the target genes of the TGF-β pathway (34), are closely related to the ECM and
invasive placental cells, and their nearby lncRNAs G008916 and
VTRNA2-1 were upregulated and downregulated, respectively.
According to the results of the subgroup and the GO and KEGG
analyses, it is indicated that BAMBI and TGFBI have important roles
in AIP through TGF-β pathway to promote placental invasion.
The present study has certain strengths and
limitations. The present study was the first to explore the lncRNAs
expression profile in patients with AIP. The present study
identified and validated certain dysregulated lncRNAs in AIP.
However, there are certain limitations to this study. One major
limitation was that no normal group was used for comparison with
the invasive group and control group when performing lncRNA
microarray scanning, as both of them were sampled from patients
with AIP. Using a normal group with samples from patients without
AIP may make the comparison more complete. In order to remedy this,
extended sample validation was performed and the results of the
extended qPCR validation were consistent with the original sample
validation and microarray data.
In summary, the dysregulated mRNAs were mainly
enriched in functional terms associated with the extracellular
matrix and in the TGF-β signaling pathway. lncRNAs G008916 and
VTRNA2-1 corresponded to the nearby mRNAs BAMBI and TGFBI, which
were both involved in the TGF-β pathway and were strictly
associated with the ECM and the invasion of placental cells. These
lncRNAs have great potential to serve as biomarkers of AIP.
Supplementary Material
Sequences of primers designed for
quantitative PCR validation of candidate lncRNAs.
Acknowledgements
The authors thank the obstetricians Dr Yan Wang and
Dr Meng Zeng (all, Department of Obstetrics, Foshan Women and
Children's Hospital Affiliated to Southern Medical University) for
helping with the collection of the placenta tissues.
Funding
No funding was received.
Availability of data and materials
The expression data were stored in the Gene
Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo; accession no.
GSE126552).
Authors' contributions
ZL, SY, SW, HM and HZ designed the experiments. ZL
decided the final experimental scheme. SY collected and analysed
the data. SW and HM interpreted the data. HZ analysed the data and
drafted the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The Human Ethics Committee of the Foshan Women and
Children's Hospital Affiliated to Southern Medical University
approved the present study (approval no. FSFY-MEC-2017-055).
Written informed consent was obtained from all pregnant females for
using their specimens and clinical information.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Baldwin HJ, Patterson JA, Nippita TA,
Torvaldsen S, Ibiebele I, Simpson JM and Ford JB: Antecedents of
abnormally invasive placenta in primiparous women: Risk Associated
with gynecologic procedures. Obstet Gynecol. 131:227–233.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Baldwin HJ, Patterson JA, Nippita TA,
Torvaldsen S, Ibiebele I, Simpson JM and Ford JB: Maternal and
neonatal outcomes following abnormally invasive placenta: A
population-based record linkage study. Acta Obstet Gynecol Scand.
96:1373–1381. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Iacovelli A, Liberati M, Khalil A,
Timor-Trisch I, Leombroni M, Buca D, Milani M, Flacco ME, Manzoli
L, Fanfani F, et al: Risk factors for abnormally invasive placenta:
A systematic review and meta-analysis. J Matern Fetal Neonatal Med.
33:471–481. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Nieto-Calvache AJ, López-Girón MC,
Quintero-Santacruz M, Bryon AM, Burgos-Luna JM, Echavarría-David
MP, López L, Macia-Mejia C and Benavides-Calvache JP: A systematic
multidisciplinary initiative may reduce the need for blood products
in patients with abnormally invasive placenta. J Matern Fetal
Neonatal Med: 1-7, 2020.
|
|
5
|
Erfani H, Fox KA, Clark SL, Rac M, Rocky
Hui SK, Rezaei A, Aalipour S, Shamshirsaz AA, Nassr AA, Salmanian
B, et al: Maternal outcomes in unexpected placenta accreta spectrum
disorders: Single-center experience with a multidisciplinary team.
Am J Obstet Gynecol. 221:337.e1–337.e5. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Collins SL, Alemdar B, van Beekhuizen HJ,
Bertholdt C, Braun T, Calda P, Delorme P, Duvekot JJ, Gronbeck L,
Kayem G, et al: Evidence-based guidelines for the management of
abnormally invasive placenta: Recommendations from the
international society for abnormally invasive placenta. Am J Obstet
Gynecol. 220:511–526. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Buca D, Liberati M, Cali G, Forlani F,
Caisutti C, Flacco ME, Manzoli L, Familiari A, Scambia G and
D'Antonio F: Influence of prenatal diagnosis of abnormally invasive
placenta on maternal outcome: Systematic review and meta-analysis.
Ultrasound Obstet Gynecol. 52:304–309. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Shainker SA, Silver RM, Modest AM, Hacker
MR, Hecht JL, Salahuddin S, Dillon ST, Ciampa EJ, Dalton ME, Otu
HH, et al: Placenta accreta spectrum: Biomarker discovery using
plasma proteomics. Am J Obstet Gynecol. 223:433.e1–433.e14.
2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Al-Khan A, Youssef YH, Feldman KM, Illsley
NP, Remache Y, Alvarez-Perez J, Mannion C, Alvarez M and Zamudio S:
Biomarkers of abnormally invasive placenta. Placenta. 91:37–42.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jauniaux E, Bhide A, Kennedy A, Woodward
P, Hubinont C and Collins S: FIGO Placenta Accreta Diagnosis and
Management Expert Consensus Panel. FIGO consensus guidelines on
placenta accreta spectrum disorders: Prenatal diagnosis and
screening. Int J Gynaecol Obstet. 140:274–280. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kopp F and Mendell J: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
McAninch D, Roberts CT and Bianco-Miotto
T: Mechanistic insight into long noncoding RNAs and the placenta.
Int J Mol Sci. 18(1371)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tseng JJ, Hsieh YT, Hsu SL and Chou MM:
Metastasis associated lung adenocarcinoma transcript 1 is
up-regulated in placenta previa increta/percreta and strongly
associated with trophoblast-like cell invasion in vitro. Mol Hum
Reprod. 15:725–731. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jauniaux E, Chantraine F, Silver RM and
Langhoff-Roos J: FIGO Placenta Accreta Diagnosis and Management
Expert Consensus Panel. FIGO consensus guidelines on placenta
accreta spectrum disorders: Epidemiology. Int J Gynaecol Obstet.
140:265–273. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jauniaux E and Ayres-de-Campos D: FIGO
Placenta Accreta Diagnosis and Management Expert Consensus Panel.
FIGO consensus guidelines on placenta accreta spectrum disorders:
Introduction. Int J Gynaecol Obstet. 140:261–264. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mazzei M, Vascellari M, Zanardello C,
Melchiotti E, Vannini S, Forzan M, Marchetti V, Albanese F and
Abramo F: Quantitative real time polymerase chain reaction
(qRT-PCR) and RNAscope in situ hybridization (RNA-ISH) as effective
tools to diagnose feline herpesvirus-1-associated dermatitis. Vet
Dermatol. 30:491–e147. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Michel G, Tonon T, Scornet D, Cock JM and
Kloareg B: The cell wall polysaccharide metabolism of the brown
alga Ectocarpus siliculosus. Insights into the evolution of
extracellular matrix polysaccharides in Eukaryotes. New Phytol.
188:82–97. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Graubner FR, Boos A, Aslan S, Kücükaslan I
and Kowalewski MP: Uterine and placental distribution of selected
extracellular matrix (ECM) components in the dog. Reproduction.
155:403–421. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Haimov-Kochman R, Friedmann Y, Prus D,
Goldman-Wohl DS, Greenfield C, Anteby EY, Aviv A, Vlodavsky I and
Yagel S: Localization of heparanase in normal and pathological
human placenta. Mol Hum Reprod. 8:566–573. 2002.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Borbely AU, Daher S, Ishigai MM, Mattar R,
Sun SY, Knöfler M, Bevilacqua E and Oliveira SF: Decorin and
biglycan immunolocalization in non-villous structures of healthy
and pathological human placentas. Histopathology. 64:616–625.
2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
DaSilva-Arnold SC, Zamudio S, Al-Khan A,
Alvarez-Perez J, Mannion C, Koenig C, Luke D, Perez AM, Petroff M,
Alvarez M and Illsley NP: Human trophoblast epithelial-mesenchymal
transition in abnormally invasive placenta. Biol Reprod.
99:409–421. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Hiden U, Eyth CP, Majali-Martinez A,
Desoye G, Tam-Amersdorfer C, Huppertz B and Ghaffari Tabrizi-Wizsy
N: Expression of matrix metalloproteinase 12 is highly specific for
non-proliferating invasive trophoblasts in the first trimester and
temporally regulated by oxygen-dependent mechanisms including
HIF-1A. Histochem Cell Biol. 149:31–42. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Nguyen AH, Wang Y, White DE, Platt MO and
McDevitt TC: MMP-mediated mesenchymal morphogenesis of pluripotent
stem cell aggregates stimulated by gelatin methacrylate
microparticle incorporation. Biomaterials. 76:66–75.
2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kuo CY, Guo T, Cabrera-Luque J,
Arumugasaamy N, Bracaglia L, Garcia-Vivas A, Santoro M, Baker H,
Fisher J and Kim P: Placental basement membrane proteins are
required for effective cytotrophoblast invasion in a
three-dimensional bioprinted placenta model. J Biomed Mater Res A.
106:1476–1487. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Abedin M and King N: Diverse evolutionary
paths to cell adhesion. Trends Cell Biol. 20:734–742.
2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Miyazono K, Katsuno Y, Koinuma D, Ehata S
and Morikawa M: Intracellular and extracellular TGF-β signaling in
cancer: Some recent topics. Front Med. 12:387–411. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Huang TW, Li ST, Fang KM and Young TH:
Hyaluronan antagonizes the differentiation effect of TGF-β1 on
nasal epithelial cells through down-regulation of TGF-β type I
receptor. Artif Cells Nanomed Biotechnol. 46 (Suppl 3):S254–S263.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Rocha MR, Barcellos-de-Souza P,
Sousa-Squiavinato ACM, Fernandes PV, de Oliveira IM, Boroni M and
Morgado-Diaz JA: Annexin A2 overexpression associates with
colorectal cancer invasiveness and TGF-β induced epithelial
mesenchymal transition via Src/ANXA2/STAT3. Sci Rep.
8(11285)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Prossler J, Chen Q, Chamley L and James
JL: The relationship between TGFβ, low oxygen and the outgrowth of
extravillous trophoblasts from anchoring villi during the first
trimester of pregnancy. Cytokine. 68:9–15. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Duzyj CM, Buhimschi IA, Laky CA, Cozzini
G, Zhao G, Wehrum M and Buhimschi CS: Extravillous trophoblast
invasion in placenta accreta is associated with differential local
expression of angiogenic and growth factors: A cross-sectional
study. BJOG. 125:1441–1448. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sheng JJ and Jin JP: TNNI1, TNNI2 and
TNNI3: Evolution, regulation, and protein structure-function
relationships. Gene. 576:385–394. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ge J, Apicella M, Mills JA, Garcon L,
French DL, Weiss MJ, Bessler M and Mason PJ: Dysregulation of the
transforming growth factor β pathway in induced pluripotent stem
cells generated from patients with diamond blackfan anemia. PLoS
One. 10(e0134878)2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Guo SK, Shen MF, Yao HW and Liu YS:
Enhanced expression of TGFBI promotes the proliferation and
migration of glioma cells. Cell Physiol Biochem. 49:1097–1109.
2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Li GF, Li L, Yao ZQ and Zhuang SJ:
Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the
proliferation and migration of glioma cells. Biochem Biophys Res
Commun. 499:765–771. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Nyholm MK, Wu SF, Dorsky RI and Grinblat
Y: The zebrafish zic2a-zic5 gene pair acts downstream of canonical
Wnt signaling to control cell proliferation in the developing
tectum. Development. 134:735–746. 2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Satow R, Nakamura T, Kato C, Endo M,
Tamura M, Batori R, Tomura S, Murayama Y and Fukami K: ZIC5 drives
melanoma aggressiveness by PDGFD-mediated activation of FAK and
STAT3. Cancer Res. 77:366–377. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yang Y, Fang X, Yang R, Yu H, Jiang P, Sun
B and Zhao Z: MiR-152 regulates apoptosis and triglyceride
production in MECs via targeting ACAA2 and HSD17B12 genes. Sci Rep.
8(417)2018.PubMed/NCBI View Article : Google Scholar
|