|
1
|
Loeser RF, Goldring SR, Scanzello CR and
Goldring MB: Osteoarthritis: A disease of the joint as an organ.
Arthritis Rheum. 64:1697–1707. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sagar DR, Ashraf S, Xu L, Burston JJ,
Menhinick MR, Poulter CL, Bennett AJ, Walsh DA and Chapman V:
Osteoprotegerin reduces the development of pain behaviour and joint
pathology in a model of osteoarthritis. Ann Rheum Dis.
73:1558–1565. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Glynjones S, Palmer AJ, Agricola R, Price
AJ, Vincent TL, Weinans H and Carr AJ: Osteoarthritis. Lancet.
386:376–387. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Vos T, Allen C, Arora M, Barber RM, Bhutta
ZA, Brown A, Carter A, Casey DC, Charlson FJ, Chen AZ and
Coggeshall M: Global, regional, and national incidence, prevalence,
and years lived with disability for 310 diseases and injuries,
1990-2015: A systematic analysis for the global burden of disease
study 2015. Lancet. 388:1545–1602. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hashimoto S, Ochs RL, Komiya S and Lotz M:
Linkage of chondrocyte apoptosis and cartilage degradation in human
osteoarthritis. Arthritis Rheum. 41:1632–1638. 1998.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kaneko S, Satoh T, Chiba J, Ju C, Inoue K
and Kagawa J: Interleukin-6 and interleukin-8 levels in serum and
synovial fluid of patients with osteoarthritis. Cytokines Cell Mol
Ther. 6:71–79. 2000.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Roach HI, Yamada N, Cheung KS, Tilley S,
Clarke NM, Oreffo RO, Kokubun S and Bronner F: Association between
the abnormal expression of matrix-degrading enzymes by human
osteoarthritic chondrocytes and demethylation of specific CpG sites
in the promoter regions. Arthritis Rheum. 52:3110–3124.
2005.PubMed/NCBI View Article : Google Scholar
|
|
8
|
O'neill TW, Parkes MJ, Maricar N,
Marjanovic EJ, Hodgson R, Gait AD, Cootes TF, Hutchinson CE and
Felson DT: Synovial tissue volume: A treatment target in knee
osteoarthritis (OA). Ann Rheum Dis. 75:84–90. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hackinger S, Trajanoska K, Styrkarsdottir
U, Zengini E, Steinberg J, Ritchie GRS, Hatzikotoulas K, Gilly A,
Evangelou E, Kemp JP, et al: Evaluation of shared genetic aetiology
between osteoarthritis and bone mineral density identifies SMAD3 as
a novel osteoarthritis risk locus. Hum Mol Genet. 26:3850–3858.
2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Koike M, Nojiri H, Ozawa Y, Watanabe K,
Muramatsu Y, Kaneko H, Morikawa D, Kobayashi K, Saita Y, Sasho T,
et al: Mechanical overloading causes mitochondrial superoxide and
SOD2 imbalance in chondrocytes resulting in cartilage degeneration.
Sci Rep. 5(11722)2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Muñoz-Valle JF, Oregón-Romero E,
Rangel-Villalobos H, Martínez-Bonilla GE, Castañeda-Saucedo E,
Salgado-Goytia L, Leyva-Vázquez MA, Illades-Aguiar B,
Alarcón-Romero Ldel C, Espinoza-Rojo M and Parra-Rojas I: High
expression of TNF alpha is associated with- 308 and- 238 TNF alpha
polymorphisms in knee osteoarthritis. Clin Exp Med. 14:61–67.
2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jiang Y, Hu C, Yu S, Yan J, Peng H, Ouyang
HW and Tuan RS: Cartilage stem/progenitor cells are activated in
osteoarthritis via interleukin-1β/nerve growth factor signaling.
Arthritis Res Ther. 17(327)2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vuolteenaho K, Koskinen-Kolasa A, Laavola
M, Nieminen R, Moilanen T and Moilanen E: High synovial fluid
interleukin-6 levels are associated with increased matrix
metalloproteinase levels and severe radiographic changes in
osteoarthritis patients. Osteoarthritis Cartilage. 25 (Suppl
1):S92–S93. 2017.
|
|
14
|
Guns LA, Kvasnytsia M, Kerckhofs G,
Vandooren J, Martens E, Opdenakker G, Lories RJ and Cailotto F:
Suramin protects against osteoarthritis by increasing tissue
inhibitor of matrix metalloproteinase-3 and glycosaminoglycans in
the articular cartilage. Osteoarthritis Cartilage. 25:S145–S146.
2017.
|
|
15
|
van Helvoort EM, Popov-Celeketic J,
Coeleveld K, Tryfonidou MA, Lafeber FP and Mastbergen SC: Effects
of the human IL4-10 fusion protein in the canine groove model of
osteoarthritis. Osteoarthritis Cartilage. 25(S434)2017.
|
|
16
|
Ratnayake M, Plöger F, Santibanez-Koref M
and Loughlin J: Human chondrocytes respond discordantly to the
protein encoded by the osteoarthritis susceptibility gene GDF5.
PLoS One. 9(e86590)2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Nepple JJ, Thomason KM, An TW,
Harris-Hayes M and Clohisy JC: What is the utility of biomarkers
for assessing the pathophysiology of hip osteoarthritis? A
systematic review. Clin Orthop Relat Res. 473:1683–1701.
2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sliz E, Taipale M, Welling M, Skarp S,
Alaraudanjoki V, Ignatius J, Ruddock L, Nissi R and Männikkö M:
TUFT1, a novel candidate gene for metatarsophalangeal
osteoarthritis, plays a role in chondrogenesis on a calcium-related
pathway. PLoS One. 12(e0175474)2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Carvalho BS and Irizarry RA: A framework
for oligonucleotide microarray preprocessing. Bioinformatics.
26:2363–2367. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Troyanskaya O, Cantor M, Sherlock G, Brown
P, Hastie T, Tibshirani R, Botstein D and Altman RB: Missing value
estimation methods for DNA microarrays. Bioinformatics. 17:520–525.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Rao Y, Lee Y, Jarjoura D, Ruppert AS, Liu
CG, Hsu JC and Hagan JP: A comparison of normalization techniques
for microRNA microarray data. Stat Appl Genet Mol Biol.
7(Article22)2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kang DD, Sibille E, Kaminski N and Tseng
GC: MetaQC: Objective quality control and inclusion/exclusion
criteria for genomic meta-analysis. Nucleic Acids Res.
40(e15)2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
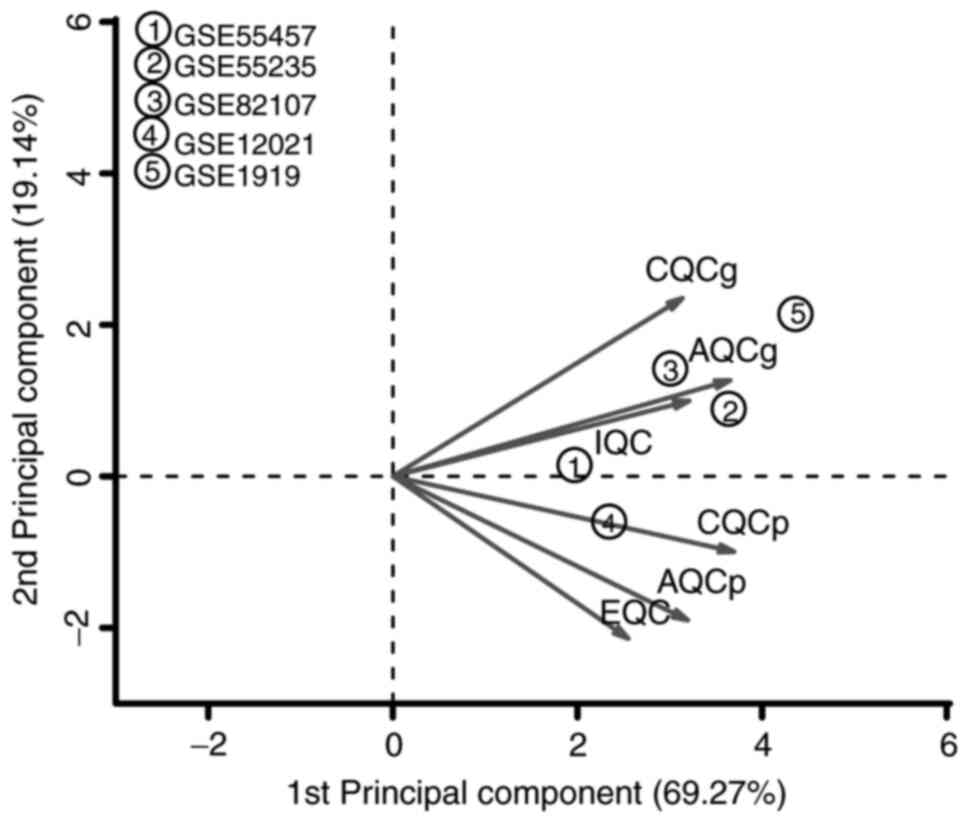
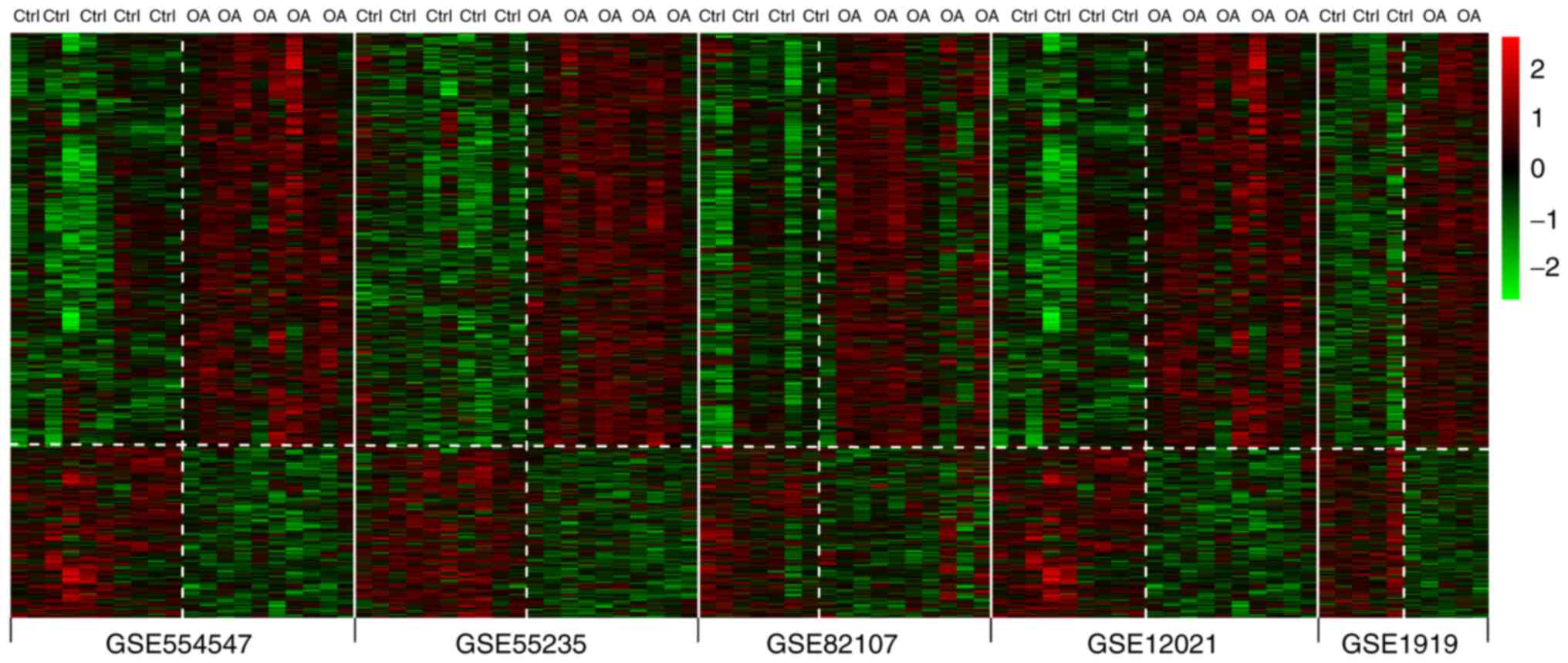
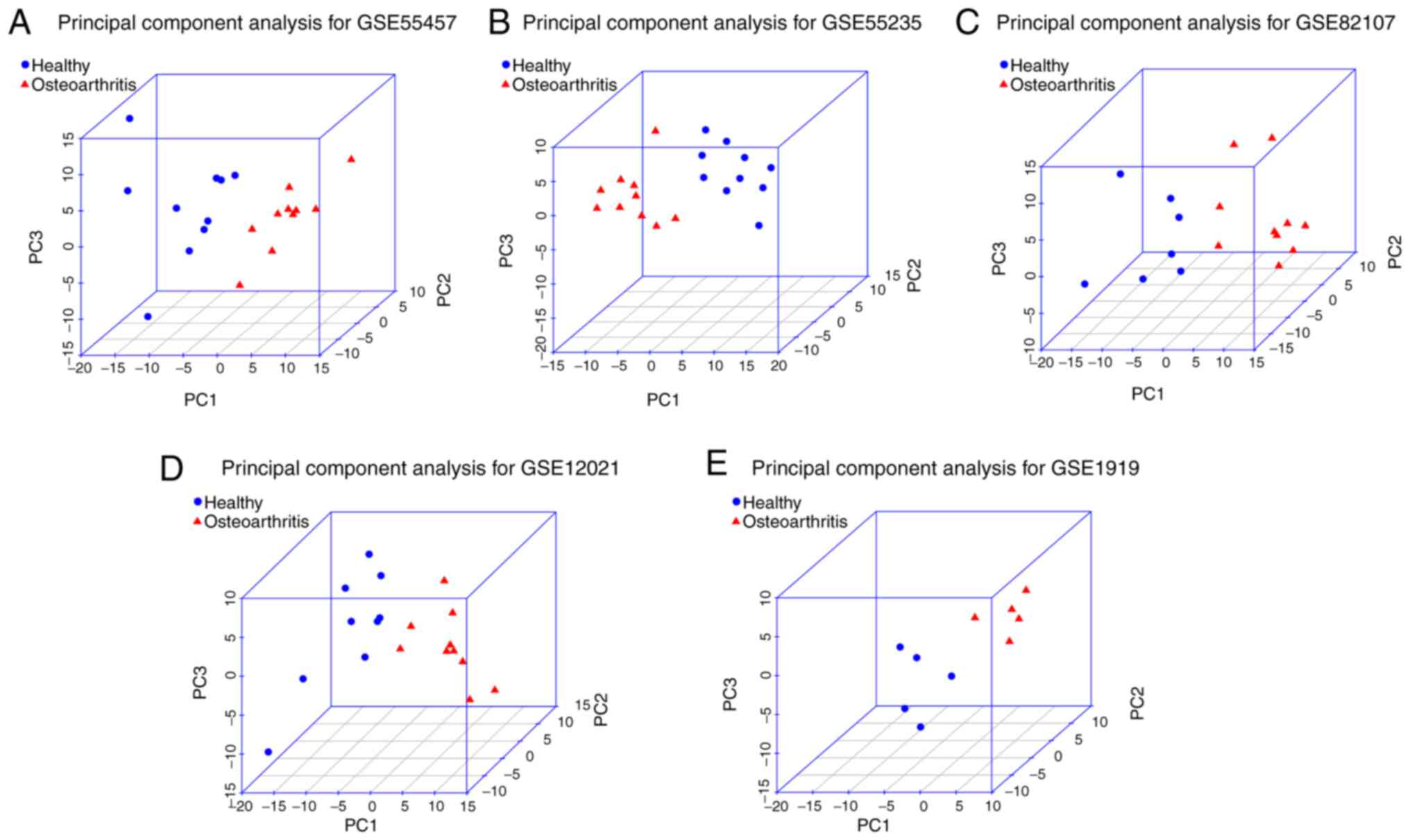
Demšar U, Harris P, Brunsdon C,
Fotheringham AS and McLoone S: Principal component analysis on
spatial data: An overview. Ann Assoc Am Geogr. 103:106–128.
2013.
|
|
24
|
Wang X, Kang DD, Shen K, Song C, Lu S,
Chang LC, Liao SG, Huo Z, Tang S, Ding Y, et al: An R package suite
for microarray meta-analysis in quality control, differentially
expressed gene analysis and pathway enrichment detection.
Bioinformatics. 28:2534–2536. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
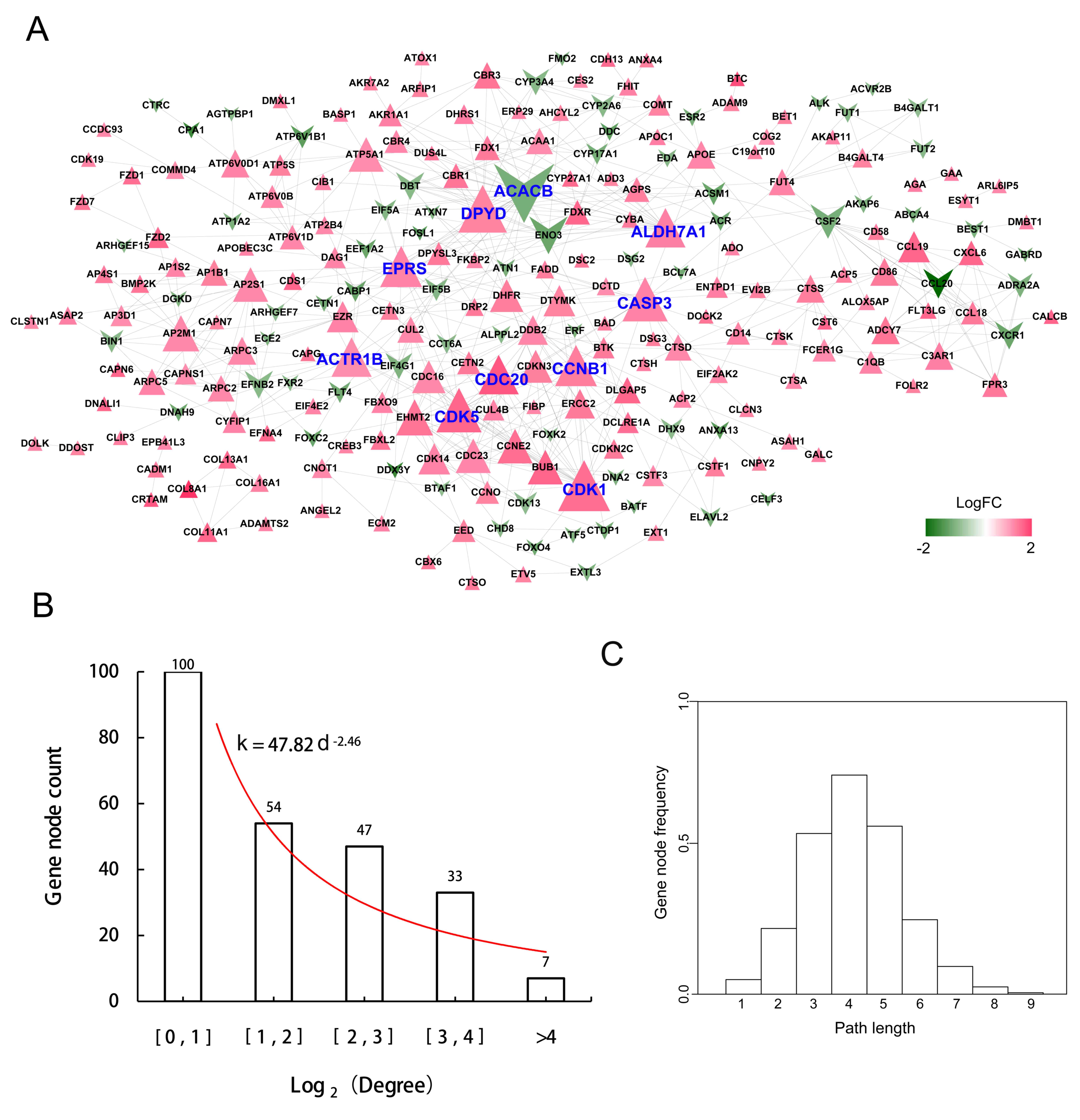
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res. 43
(Database Issue):D447–D452. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jeong H, Mason SP, Barabási AL and Oltvai
ZN: Lethality and centrality in protein networks. Nature.
411:41–42. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Alizadeh E, Lyons S, Castle J and Prasad
A: Measuring systematic changes in invasive cancer cell shape using
Zernike moments. Integr Biol (Camb). 8:1183–1193. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hayami T, Pickarski M, Zhuo Y, Wesolowski
GA, Rodan GA and Duong LT: Characterization of articular cartilage
and subchondral bone changes in the rat anterior cruciate ligament
transection and meniscectomized models of osteoarthritis. Bone.
38:234–243. 2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Réka A: Scale-free networks in cell
biology. J Cell Sci. 118:4947–4957. 2005.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lei X, Wu FX, Tian J and Zhao J: ABC and
IFC: Modules detection method for PPI network. Biomed Res Int.
2014(968173)2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Qasba PK, Ramakrishnan B and Boeggeman E:
Structure and function of beta-1,4-galactosyltransferase. Curr Drug
Targets. 9:292–309. 2008.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Al-Obaide MA, Alobydi H, Abdelsalam AG,
Zhang R and Srivenugopal KS: Multifaceted roles of 5'-regulatory
region of the cancer associated gene B4GALT1 and its comparison
with the gene family. Int J Oncoly. 47:1393–1404. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liu W, Cui Z, Wang Y, Zhu X, Fan J, Bao G,
Qiu J and Xu D: Elevated expression of β1,4-galactosyltransferase-I
in cartilage and synovial tissue of patients with osteoarthritis.
Inflammation. 35:647–655. 2012.
|
|
37
|
Sun Y, Franklin AM, Mauerhan DR and Hanley
EN: biological effects of phosphocitrate on osteoarthritic
articular chondrocytes. Open Rheumatol J. 11:62–74. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Park EH, Walker SE, Lee JM, Rothenburg S,
Lorsch JR and Hinnebusch AG: Multiple elements in the eIF4G1
N-terminus promote assembly of eIF4G1•PABP mRNPs in vivo. EMBO J.
30:302–316. 2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Okazaki Y, Suzuki A, Sawada T,
Ohtake-Yamanaka M, Inoue T, Hasebe T, Yamada R and Yamamoto K:
Identification of citrullinated eukaryotic translation initiation
factor 4G1 as novel autoantigen in rheumatoid arthritis. Biochem
Biophys Res Commun. 341:94–100. 2006.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Coldwell MJ, Cowan JL, Vlasak M, Mead A,
Willett M, Perry LS and Morley SJ: Phosphorylation of eIF4GII and
4E-BP1 in response to nocodazole treatment: A reappraisal of
translation initiation during mitosis. Cell Cycle. 12:3615–3628.
2013.PubMed/NCBI View
Article : Google Scholar
|
|
41
|
Hu W, Zhang W, Li F, Guo F and Chen A:
miR-139 is up-regulated in osteoarthritis and inhibits chondrocyte
proliferation and migration possibly via suppressing EIF4G2 and
IGF1R. Biochem Biophys Res Commun. 474:296–302. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhang X, Yuan Z and Cui S: Identifying
candidate genes involved in osteoarthritis through bioinformatics
analysis. Clin Exp Rheumatol. 34:282–290. 2016.PubMed/NCBI
|