Introduction
Peripheral nerve injury (PNI) poses a significant
challenge in the medical field worldwide, resulting in issues such
as the economic loss of the patient's family and society, physical
disability and mental shock (1-4).
Symptoms caused by various open and closed peripheral nerve
injuries include neuropathic palsy, motor dysfunction, muscle
atrophy and neuropathic pain (5-8).
Pain reduction in patients with PNI is an important area of focus
in medicine. Previous studies have demonstrated that Schwann cells
serve an important role in promoting axonal regeneration, myelin
repair and functional recovery in neuroregeneration (9-11).
Schwann cells have been the focus of research on
neural regeneration due to their ability to promote axon
regeneration (12-16).
After PNI, Schwann cells turn into an ‘activated state’ and acquire
the capacity to migrate, proliferate and secrete soluble mediators
that control Wallerian degeneration and axonal regeneration
(17-20).
Previous studies have demonstrated that activated Schwann cells
(ASCs) can be extracted from pre-injured peripheral nerves
(21,22). In brief, the sciatic nerve was
exposed through a dorsal the incision under general anesthesia and
ligated to allow pre-degeneration to take place. One week later,
the distal segment of the pre-degenerated sciatic nerve was removed
in order to isolate activated Schwann cells. Furthermore, current
research has focused on combining ASCs with various biomaterials to
repair nerve damage (23). In 2012,
Zhou et al (24) used human
autologous activated Schwann cells to repair spinal cord injury and
achieved notable results. Besides, ASCs also be combined with some
scaffold as one of the promising methods to treat spinal cord
injury (25). A previous study
revealed that the survival of Schwann cells was enhanced by
exposure to perfluorotributylamine through the promotion of sciatic
nerve regeneration (26).
DNA methylation, which directs transcriptional
silencing via heterochromatin formation, is a genetic marker for
the growth and development of a number of eukaryotes (27-29).
As one of the earliest epigenetic modifications to be recognized,
DNA methylation has been observed in numerous organisms (30,31).
Methylation of eukaryotic DNA typically occurs at position 5 of
cytosine to produce 5-methylcytosine (32). A previous study reported that PNI
led to DNA methylome remodeling in rat dorsal root ganglion
(33). These observations suggest
that the biological impact of DNA methylation should not be
underestimated. In our previous studies, a genome-wide methylation
analysis and isobaric tags for relative and absolute quantitation
(iTRAQ)-based proteomics profiling were performed to compare normal
Schwann cells (NSCs) and ASCs (34,35).
However, to the best of our knowledge, no studies have compared the
functional changes associated with adhesion and proliferation
between NSCs and ASCs.
In the present study, both NSCs and ASCs were
isolated from the sciatic nerve of Wistar rats and purified using
differential adhesion methods. Subsequently, several assays were
performed, including proliferation and adhesion assays, reverse
transcription-quantitative (RT-q)PCR, bioinformatic analysis and
methylated DNA immunoprecipitation-sequencing (MeDIP-seq)
analysis.
Materials and methods
Ethics
All animal handling experimental protocols and
procedures were approved by the Animal Care and Use Committee of
Tianjin Medical University General Hospital (Tianjin, China).
Wistar rats (100±10 g) were provided by the Laboratory Animal
Center of the Chinese People's Liberation Army General Hospital
(Beijing, China; Animal license no. SCXK2012-0066).
Preparation of Schwann cells
Schwann cells were isolated from the proximal
section of the sciatic nerves of Wistar rats, as previously
described (36,37). All cells were cultured with a
culture medium contain Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1%
Antibiotic-Antimycotic (AA) in 5% CO2 at 37˚C. A total
of 18 Wistar female rats (4 weeks, 100±10 g) were acclimatized for
≥2 weeks under a humidity- and temperature-controlled environment
with a 12-h light/dark cycle before surgery. All rats had free
access to food and water, and the health and behavior of rats were
monitored once per day. Before surgery, all rats were deeply
anesthetized using 2% isoflurane to minimize suffering.
Subsequently, each rat received unilateral ligation of the sciatic
nerve. One week after surgery, all rats were euthanized using 5%
isoflurane until respiration ceased and death ensued. There were
two groups in the present study: Group A, ASCs from the ligated
sciatic nerves of all rats (n=18); and Group B, NSCs from the
untreated sciatic nerves of all rats (n=18).
The euthanasia protocols and procedures of Tianjin
Medical University General Hospital are based on and consistent
with the Institutional Animal Care and Use Committee of the
University of Iowa (38).
Additionally, 5% isoflurane was used for sacrifice of the rats as
previously described (39-41).
No other physical method was used as the second sacrifice method.
Two different methods were used in the present study to confirm
death: i) Heartbeat, heartbeat was assessed for 10 min after
euthanasia, and a lack of electrical activity of the heart as
determined by electrocardiogram; ii) respiratory pattern, the
respiratory pattern was assessed for 10 min after euthanasia, and a
lack of breathing for >10 min.
Characterization of Schwann cells
For immunofluorescence labeling, Schwann cells from
the different groups were fixed in 4% paraformaldehyde (ChemCruz™
Biochemicals; Santa Cruz Biotechnology, Inc.) at room temperature
for 15 min. Subsequently, the cells were permeabilized with 0.1%
Triton X-100 (Sigma-Aldrich; Merck KGaA) at room temperature for 10
min and incubated with 5% normal goat serum (Cell Signaling
Technology, Inc.) at room temperature for 1 h. A primary rabbit
anti-rat S100 (1:200; cat. no. ab52642; Abcam) antibody was added
to the cells and incubated at 37˚C for 2 h. Fluorescein
isothiocyanate-conjugated goat anti-rabbit IgG (1:500; cat. no.
111-545-003; Jackson ImmunoResearch Laboratories, Inc.) was added
as a secondary antibody. The nucleus was stained with DAPI
(Sigma-Aldrich; Merck KGaA) at room temperature for 10 min.
Finally, cells were observed under an inverted fluorescence
microscope (magnification, x20; Leica Microsystems GmbH). All
images were imported into Zeiss confocal system (v3.0) for further
analysis and statistics.
Proliferation and adhesion assays
A WST-8 assay was used for the second passages cells
in order to investigate the cell proliferation according to
previous publications (42-44).
Briefly, Schwann cells were seeded into 96-well plates at a density
of 1x103 cells/well, the Cell Counting Kit-8 (Dojindo
Molecular Technologies, Inc.) solution was added to the culture
medium, and cells were incubated at 37˚C for 1.5 h. The absorbance
was measured at 450 nm.
For cell adhesion assays, Schwann cells were seeded
into six-well plates (cat. no. SRP3186; Sigma-Aldrich; Merck KGaA)
coated with vitronectin at a density of 3x105
cells/well. After 45 min of incubation at 37˚C, cells were washed
five times with PBS (Gibco; Thermo Fisher Scientific, Inc.) and
fixed in 4% paraformaldehyde at room temperature for 15 min.
Adherent cells were stained with crystal violet (1% in
ddH2O; Sigma-Aldrich; Merck KGaA) at room temperature
for 10 min. Six fields of view/well (magnification, x200) were
randomly selected and the number of cells adherent to the bottom of
plates was counted manually with a counting chamber (Hausser
Scientific Co.) under the light scope (Zeiss AG) (45,46).
RT-qPCR
Total RNA was extracted from Schwann cells using
trizol reagent (Beijing Solarbio Science & Technology Co.,
Ltd.). Total RNA (1 µg/sample) was reverse transcribed (5 min at
20˚C, 20 min at 46˚C, 1 min at 95˚C and held at 4˚C) using a
Reverse Transcription kit (Takara Bio, Inc.). qPCR was performed
with the SYBR Green PCR Master Mix (Applied Biosystems; Thermo
Fisher Scientific, Inc.) on a CFX96 Touch™ Deep Well Real-Time PCR
Detection system (Bio-Rad Laboratories, Inc.) according to previous
publications (47,48). Briefly, the thermal program
consisted of 2 min at 95˚C, then 40 cycles of amplifications, 5 sec
at 95˚C for denaturation, 5 sec at 65˚C for annealing, and 5 sec at
95˚C for extension. Each sample was analyzed in triplicate. The
data were then calculated using the 2-∆∆Cq formula with
reference to the basal controls. β-actin was used as an internal
control. The expression levels of nine genes, namely vinculin
(Vcl), BCAR1 scaffold protein (Bcar1), laminin
subunit γ1 (Lamc1), collagen type V α3 chain
(Col5a3), collagen type XVIII α1 chain (Col18a1),
ezrin (Ezr), integrin subunit β6 (Itgb6), collagen
type III α1 chain (Col3a1) and Stat5a, were evaluated
in both NSCs and ASCs. Primer3 (v0.4.0) software was used for all
PCR primer design. All primers are listed in Table I.
 | Table IDetails of the primers used for
quantitative PCR. |
Table I
Details of the primers used for
quantitative PCR.
| Gene | NCBI accession
no. | Product length | Forward primer,
5'-3' | Reverse primer,
5'-3' | Annealing
temperature, ˚C |
|---|
| β-actin | NM_031144.3 | 85 |
AGCGTGGCTACAGCTTCACC | AAGTCTAGGGCAAC
ATAGCACAGC | 57.5 |
| Ezr | NM_019357.1 | 190 | TGGACGACCGTAACGAGGA
GAAG | CTGATCTGCCGCAGCGT
CTTATAC | 58.6 |
| Itgb6 | XM_017591686.1 | 132 | GCTCATCGGTGTCGTGCTA
CTG | CCTCGGTACAGCGGATTG
GTTC | 58.2 |
| Lamc1 | NM_053966.2 | 173 | AACGAGGTGAATGGCAT
GTTGAGG | TGGCTGAGGAGGCTGCT
GAC | 58.6 |
| Col3a1 | NM_032085.1 | 177 | GACACGCTGGTGCTCAAG
GAC | GTTCGCCTGAAGGACCTCG
TTG | 58.6 |
| Vcl | XM_006251639.3 | 162 | AAGGCAAGATTGAGCAGG
CACAG | CACGGTCACACTTGGCGA
GAAG | 58.8 |
| Col5a3 | XM_017595884.1 | 86 | TCAGGTGACCACAGGC
ACTCTATC | TTGATGGTGGCTGCTGTTG
TCTG | 58.7 |
| Stat5a | NM_017064.1 | 102 | GACCATCATCAGCGAGCA
GCAG | TACTCCATGACGCAGCAGT
TGTTC | 58.3 |
| Col18a1 | NM_053489.2 | 153 | GAGTCAACAGTTCCTAC
GCACCAG |
TGCCTGCATCGCCAACACTG | 58.3 |
| Bcar1 | XM_006255629.3 | 82 |
AGCACACGCAGCAGCCAATC | CCACAGCAACTTCCAGCTC
CAG | 58.6 |
MeDIP-seq
MeDIP-seq was used to detect each sample according
to the protocol of a previous study (Annoroad Genomics) (49). Briefly, 5 µg DNA from each group
(with two duplicates) was sonicated (20 kHz) at 4˚C for 10 min to
produce DNA fragments (100-500 bp). Adapter-ligated DNA was
immunoprecipitated at room temperature for 2 h using an
anti-5-methylcytosine monoclonal antibody (1:1,000; cat. no.
ab214727; Abcam). Subsequently, RT-qPCR analysis was performed to
verify the specificity of the immunoprecipitated fragments. A
200-300 bp DNA fragment was purified using a DNA Clean and
Concentrator-5 column (D4003; Zymo Research Corp.). The amplicon
quality and quantity were assessed using a 2100 analyzer DNA 1000
chip (Agilent Technologies, Inc.). MeDIP library was sequenced on
an Illumina HiSeq 2000 Sequencing system (Illumina, Inc.).
Bioinformatics analysis
The adhesion-associated differential methylation
regions (DMRs) between NSCs and ASCs were identified (mean
difference, ≥20; P<0.05). All genes containing DMRs were used
for Kyoto Encyclopedia of Genes and Genomes (KEGG) functional
enrichment analysis and Gene Ontology (GO) analysis using the
Database for Annotation, Visualization and Integrated Discovery
(https://david.ncifcrf.gov/list.jsp) (50). The biological processes (BPs),
cellular components (CCs) and molecular functions (MFs) of
differentially expressed genes were evaluated in detail in GO
analysis, while KEGG pathway analysis was used to investigate the
systemic functional, chemical and genomic information of
differentially methylated genes. P<0.001 was used to denote the
significance of GO and KEGG pathway enrichment in the
differentially methylated genes (mean difference ≥20,
P<0.001).
Protein-protein interaction (PPI)
network analysis
The information of the interaction of proteins, and
neighborhood, gene fusions were provided using the Search Tool for
the Retrieval of Interacting Genes (STRING) database (a publicly
available database; http://string-db.org/) (51). In the present study, the input gene
sets were gene modules and the species was Rattus
norvegicus. To further explore the potential relevance of the
differentially methylated genes in Schwann cells, an evidence
threshold >0.9 was set as the cut-off value. The core genes were
selected by the interaction with other genes and further verified
by RTq-PCR.
Statistical analysis
GraphPad Prism statistical software (version 8.0.2;
GraphPad Software, Inc.) was used to perform data analysis.
Statistical differences between two groups were analyzed using
paired Student's t-test. All data are presented as the mean ± SEM.
All experiments were repeated in triplicate. P<0.05 was
considered to indicate a statistically significant difference.
Results
ASCs exhibit a stronger proliferative
capacity than NSCs
To investigate whether there were proliferation
differences between ASCs and NSCs, a cell proliferation assay was
performed. Both groups exhibited a sustained proliferation rate.
ASCs exhibited a significantly higher proliferation activity after
4, 6 and 7 days compared with NSCs. However, no marked
morphological differences were observed between the two groups when
observed under an optical microscope (Fig. 1A). Furthermore, adhesion experiments
were used to further verify whether adhesion of Schwann cells
changed after PNI. The number of cells adherent to the bottom of
the plates was counted. ASCs exhibited a significantly increased
capacity of adhesion compared with NSCs (Fig. 1B). Additionally, both NSCs and ASCs
were positive for S100 protein as revealed by immunocytochemical
staining under a fluorescence microscope (Fig. 1C).
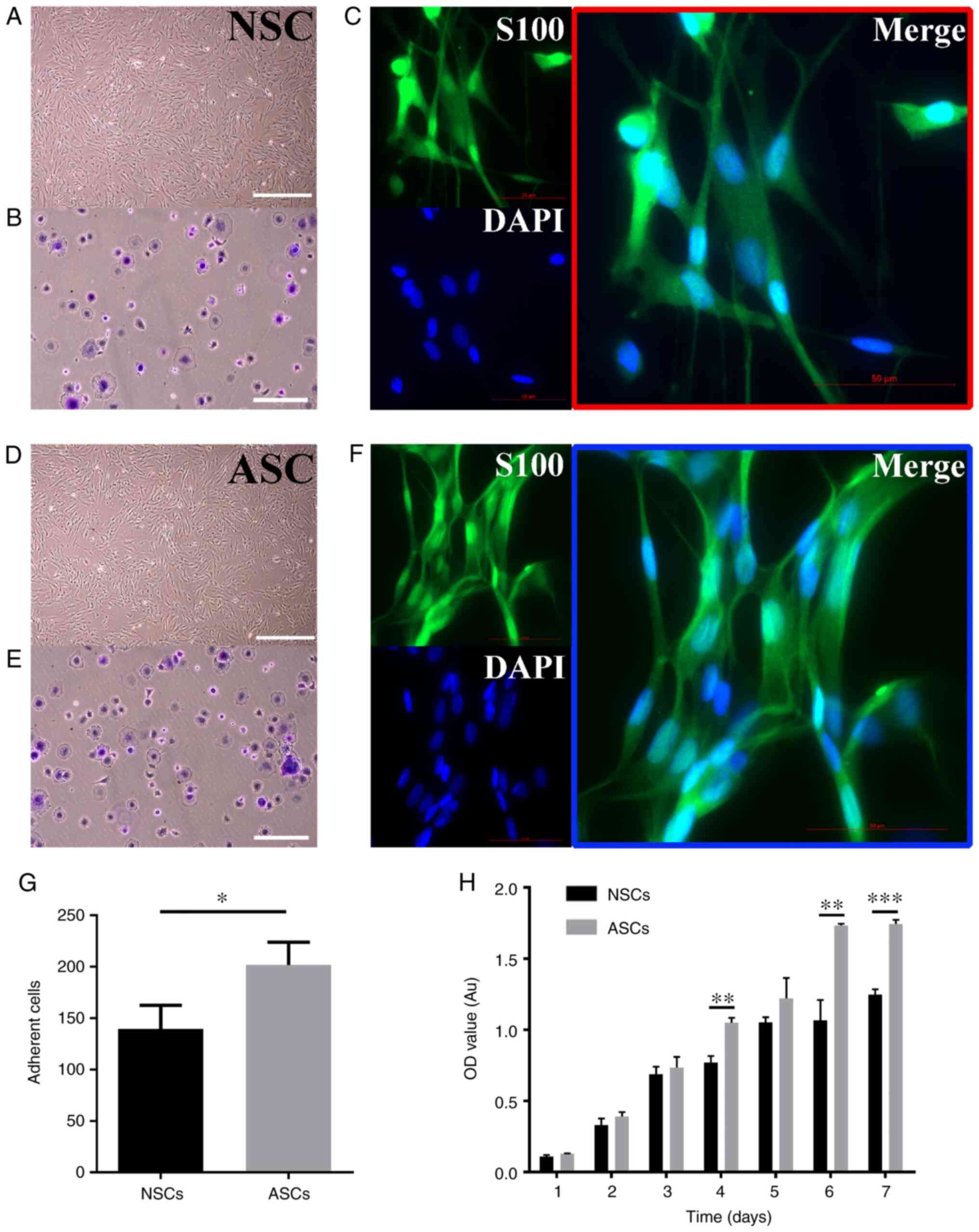 | Figure 1Culture and identification of Schwann
cells. (A, D) The shape of NSCs and ASCs under an optical
microscope, all cells were arranged in a long spindle shape and
nucleus was oblong or ovoid. Scale bar: 200 μm (B, E, G) Adhesion
assay of Schwann cells. Scale bar: 100 μm. (C, F) Immunofluorescent
staining images of s100, DAPI, and Merge of NSCs and ASCs. (H)
Proliferation assay of ASCs and NSCs. *P<0.05;
**P<0.01; ***P<0.001. Scale bar, 50 µm.
NSCs, normal schwann cells; ASCs, activated schwann cells; OD,
optical density; Au, absorbance units. |
Identification of adhesion-associated
DMRs after PNI
Specific experimental procedures for MeDIP-seq are
shown in Fig. 2A. A total of 429
differentially methylated genes associated with Schwann cell
adhesion were identified. After PNI, DMRs were classified into
seven major groups according to the genomic location, including
distal intergenic (56.6%), intron (29.3%), exon (7.0%), promoter
(2.8%), 3'-untranslated region (UTR; 1.2%), 5'-UTR (0.5%) and
downstream (2.6%) (Fig. 2B). The
chromosomal distribution of these DMRs after PNI is shown in
Fig. 2C. In terms of chromosomal
distribution, chromosome 2 had the greatest number of
differentially methylated genes. Among all 429 differentially
methylated genes, nine genes were selected according to the gene
interaction with other genes, including: Vcl, Bcar1, Lamc1,
Col5a3, Col18a1, Ezr, Itgb6, Col3a1 and Stat5a. All of
these nine aberrantly expressed genes are shown in a heat map in
Fig. 2D. For the heat map of this
study, the expression of Bcar1 and Lamc1 exhibited a
high level in ASCs compare with the NSCs. Besides, the expression
of Vcl, Col5a3, Col18a1, Ezr, Itgb6, Col3a1 and
Stat5a exhibited a low level in ASCs compare with the
NSCs.
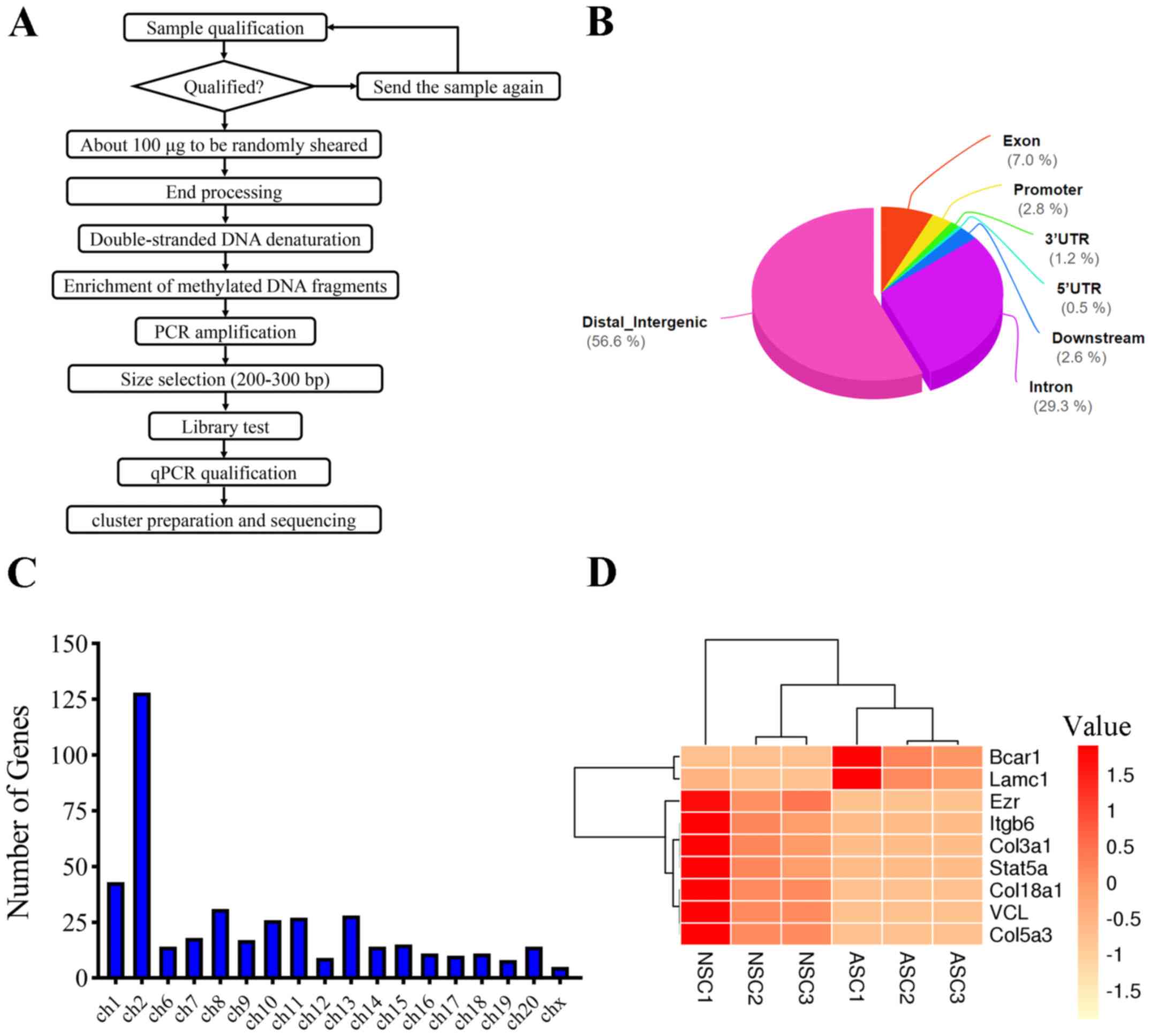 | Figure 2Expression signatures of differential
methylation genes after PNI. (A) Concise experimental procedure for
the Methylated DNA immunoprecipitation-sequencing. (B) Differential
methylation genes were classified according their genomic
architecture after PNI. (C) Chromosome distribution showed the
numbers of regulated genes located at different chromosomes. (D)
Heat map of nine aberrantly expressed genes. Vcl, vinculin;
Bcar1, BCAR1 scaffold protein; Lamc1, laminin subunit
γ1; Col5a3, collagen type V α3 chain; Col18a1,
collagen type XVIII α1 chain; Ezr, ezrin; Itgb6,
integrin subunit β6; Col3a1, collagen type III α1 chain;
MeDIP-seq, methylated DNA immunoprecipitation-sequencing; PNI,
peripheral nerve injury; ch, chromosome; NSC, normal Schwann cell;
ASC, activated Schwann cell; qPCR, quantitative PCR. |
Analysis of functional categories of
differentially methylated genes
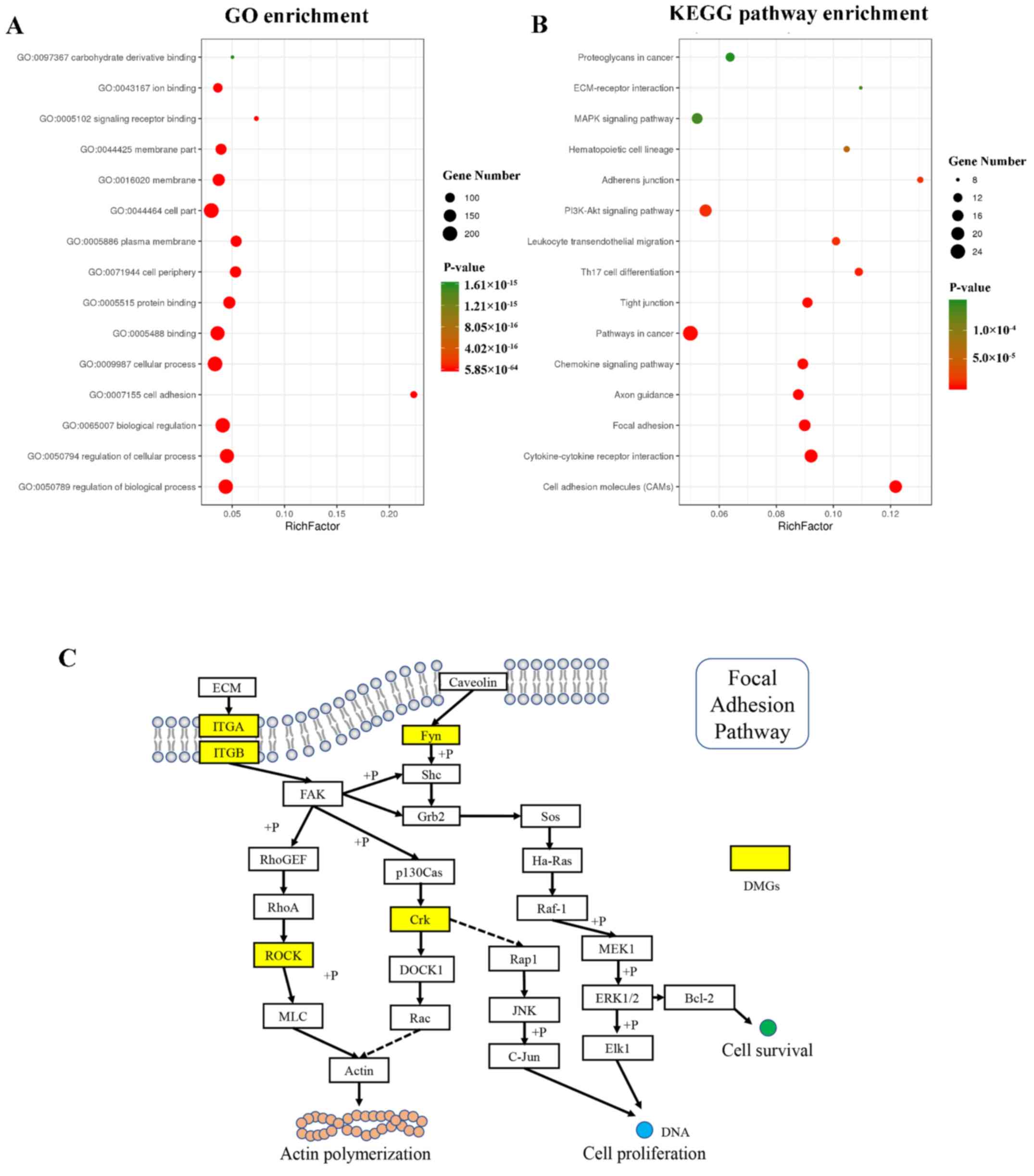
The results of GO analysis are shown in Fig. 3A. The differentially methylated
genes were significantly enriched in BPs, including ‘regulation of
biological process’ (GO:0050789), ‘regulation of cellular process’
(GO:0050794), ‘biological regulation’ (GO:0065007), ‘cell adhesion’
(GO:0007155) and ‘cellular process’ (GO:0009987). The enriched CCs
included ‘cell periphery’ (GO:0071944), ‘plasma membrane’
(GO:0005886), ‘cell part’ (GO:0044464), ‘membrane’ (GO:0016020) and
‘membrane part’ (GO:0044425). The enriched MFs included ‘binding’
(GO:0005488), ‘protein binding’ (GO:0005515), ‘signaling receptor
binding’ (GO:0005102), ‘ion binding’ (GO:0043167) and ‘carbohydrate
derivative binding’ (GO:0097367).
KEGG pathway analysis results are shown in Fig. 3B. Of all adhesion-associated
differentially methylated genes, the 15 most enriched pathways were
selected. According to the KEGG pathway analysis results,
differentially methylated genes were significantly enriched in
‘Cell adhesion molecules’, ‘Cytokine-cytokine receptor
interaction’, ‘Focal adhesion’, ‘Pathways in cancer’, ‘Axon
guidance’, ‘Chemokine signaling pathway’, ‘Tight junction’, ‘Th17
cell differentiation’, ‘Leukocyte transendothelial migration’,
‘PI3K-Akt signaling pathway’, ‘Adherens junction’, ‘Hematopoietic
cell lineage’, ‘ECM-receptor interaction’, ‘MAPK signaling pathway’
and ‘Proteoglycans in cancer’. The differentially methylated genes
in ASCs compared with NSCs involved in the ‘Focal adhesion’ pathway
are shown in Fig. 3C.
ITGA/ITGB, ROCK, Fyn and
Crk were the differential methylated genes detected in
MeDIP-seq, suggesting that these genes may be associated with actin
polymerization, cell proliferation and survival through the focal
adhesion pathway.
PPI network analysis
PPI network analysis was performed using STRING. The
PPI network of differentially methylated genes is shown in Fig. 4A. A total of 294 nodes and 205
interaction pairs were included in the network. Some proteins
involved in the ‘Focal adhesion’ pathway, such as Lamc1, Lamc2,
Itga11, Bcar1, Fyn, Rock2, Flt3, Kdr and Pgf, were central nodes in
the present network. Some proteins involved in the ‘Cell adhesion
molecules’ pathway, such as Sdc3, Cd276, Ptprc, Cd86, Lcam1 and
Alcam, were also central nodes in the network. The top 10
high-degree hub nodes included Fyn, App, Ptpn11, Jak2, Cxcl13, Crk,
Ctnnb1, Efna1, Kitlg and Epha7 (Fig.
4B). Among these proteins, Fyn was the node with the highest
degree.
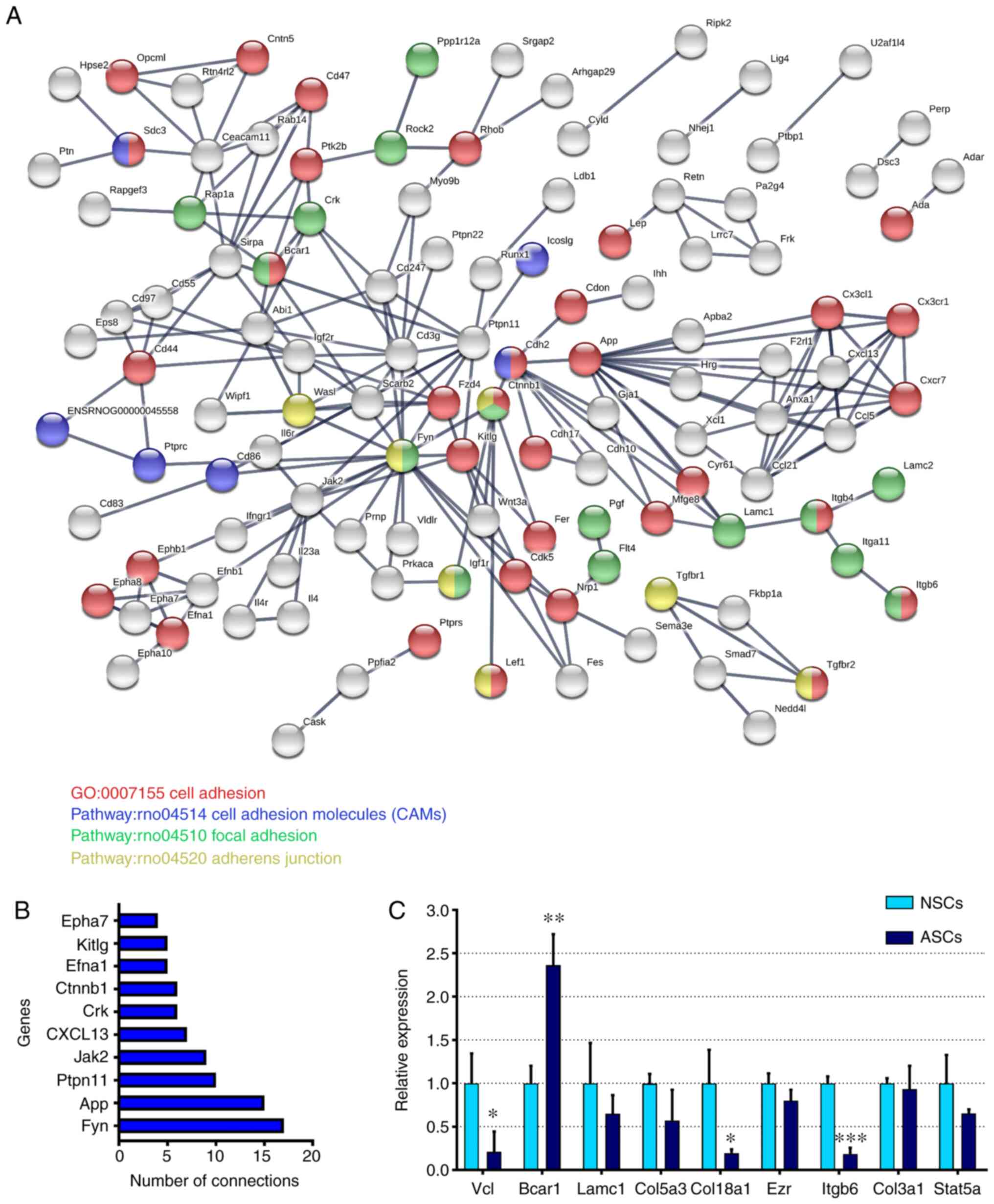 | Figure 4Bioinformatics analysis and
verification by RTq-PCR. (A) PPI network analysis of
adhesion-associated differentially expressed genes in schwann
cells. (B) The top 10 high-degree hub nodes in the PPI network. (C)
Expression levels of adhesion-associated genes verified via reverse
transcription-quantitative PCR. Values are expressed as the mean ±
SEM. *P<0.05; **P<0.01;
***P<0.001 vs. NSCs. PPI, protein-protein
interaction; NSCs, normal Schwann cells; ASCs, activated Schwann
cells; GO, Gene Ontology; Vcl, vinculin; Bcar1, BCAR1 scaffold
protein; Lamc1, laminin subunit γ1; Col5a3, collagen type V α3
chain; Col18a1, collagen type XVIII α1 chain; Ezr, ezrin; Itgb6,
integrin subunit β6; Col3a1, collagen type III α1 chain. |
Validation of differentially expressed
genes via RT-qPCR
To further investigate the changes in cell adhesion,
RT-qPCR was performed to explore several known adhesion-associated
genes (Vcl, Bcar1, Lamc1, Col5a3, Col18a1, Ezr, Itgb6,
Col3a1 and Stat5a) in NSCs and ASCs. Among these genes,
Bcar1 expression was significantly upregulated, while the
expression levels of Vcl, Col18a1 and Itgb6
were significantly downregulated in ASCs compared with in NSCs
(Fig. 4C). The present results
indicated that these genes may cause changes in Schwann cell
adhesion after PNI.
Discussion
DNA methylation, histone modification and non-coding
RNA regulation have been studied in previous epigenetic research
(31,52-54).
In our previous studies, genome-wide methylation analyses and
iTRAQ-based proteomics profiling of Schwann cells after PNI were
performed (23,35). The Schwann cell phenotype induced by
DNA methylation is known to change after PNI, but this phenomenon
requires further investigation (55-57).
In the present study, a unilateral sciatic nerve injury model was
established using the proximal region of the sciatic nerve in
Wistar rats. A previous study reported differences in the
expression of immunomodulatory genes between the proximal and
distal segments after PNI (58).
Future studies should investigate DNA methylation changes between
the proximal and distal segments after PNI. The present study
identified and characterized Schwann cells, and revealed that the
proliferative and adhesive abilities of ASCs were stronger than
those of NSCs. Additionally, MeDIP-seq was used to identify
epigenetic changes between NSCs and ASCs. A total of 429
adhesion-associated differentially methylated genes and 15 closely
associated signaling pathways were identified. After performing GO,
KEGG signaling pathway and PPI network analyses, the genes of heat
map were verified by RT-qPCR and were determined to be potential
regulators of rat Schwann cell proliferation and adhesion.
The present study investigated epigenetic changes
that occurred after nerve injury. Among the adhesion-associated
differentially methylated genes identified in the present study,
several important genes have been implicated in cell adhesion or
proliferation, including Bcar1, Col5a3, Col18a1, Ezr, Col3a1
and Stat5a (59-61).
Notably, Bcar1 was one of the differentially
methylated genes identified in the present study. Bcar1 is a vital
scaffolding protein that modulates numerous essential cellular
processes (62,63). It belongs to the Crk-associated
substrate family of scaffolding proteins (64). There is increasing evidence that
Bcar1 serves a crucial role in cell-extracellular matrix
adhesion, tissue homeostasis and pathogenesis of Kaposi's
sarcoma-associated herpesvirus (65). Furthermore, the lack of Bcar1
in some cells may lead to altered integrin signaling, which is
reflected by aberrant basal membrane adhesion (63). This is consistent with the
expression of changes in core genes that affect adhesion in Schwann
cells after PNI, as shown in the present study.
In the KEGG analysis in the current study, the
differentially methylated genes were significantly enriched in
several pathways, including ‘cell adhesion molecules’,
‘cytokine-cytokine receptor interaction’ and ‘focal adhesion’
pathways. It has been reported that the focal adhesion pathway is
regulated by microRNA (miR)-29s, while the Lamc1 gene is a
candidate target of miR-29s regulation (66). This indicates that Lamc1 may
regulate the focal adhesion pathway indirectly through miR-29s.
Additionally, a previous study demonstrated that the presence of
Lamc1 may promote the migration without affecting the
proliferative activity of prostate cancer cells (66). After PPI network analysis of the
differentially methylated genes, the top 10 high-degree hub nodes
included Fyn, Efna1, Jak2, Vav3, Flt4, Epha7, Crk, Kitlg, Ctnnb1
and Ptpn11.
Although the current study revealed numerous
epigenetic changes after PNI in Wistar rats, some limitations were
noted. First, even though it has been reported that rats are useful
animal models for experimental studies of peripheral neuropathology
and repair, future experiments should focus on large animals and
primate models (67,68). Second, the present study used
unilateral ligation of one sciatic nerve, while the other nerve
served as the control. However, it is unclear whether unilateral
ligation of one sciatic nerve may affect the other nerve. A
previous study reported that the recoveries observed using an upper
limb model were faster, and the time required for function
restoration was shorter, compared with the model of sciatic nerve
injury (69). Additionally, the
sciatic nerve cannot represent all the peripheral nerves. Third, it
should be noted that the current study only examined the adhesion
and proliferation of Schwann cells. Schwann cells have numerous
additional functions that occur before and after PNI, including
cell migration, cell secretion and axonal regeneration, which
require further research.
In conclusion, the present results provided novel
information concerning basic Schwann cell adhesion. However, in
vitro culture, amplification and purification of Schwann cells
are complex processes. It is challenging to obtain Schwann cells in
sufficient numbers, with substantial purity and biological
activity, no immune rejection and limited proliferation
characteristics (70).
Proliferation, adhesion and migration may markedly change with the
epigenetic changes that occur in ASCs. Therefore, Schwann cell
transplantation in peripheral nerve repair requires to overcome
numerous difficulties (71). The
present study performed bioinformatics analyses of DNA methylation
patterns associated with Schwann cell adhesion and proliferation. A
total of 429 differentially methylated genes were identified in
ASCs compared with in NSCs. Among these genes, Vcl, Bcar1,
Col18a1 and Itgb6 may affect cell adhesion after PNI.
The screened genes and pathways suggested potential candidates for
further study of epigenetic mechanisms associated with Schwann
cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
State Key Program of National Natural Science Foundation of China
(grant no. 81930070), the State General Program National Natural
Science Foundation of China (grant no. 81371957), the International
Cooperation Program of National Natural Science Foundation of China
(grant no. 81620108018), the Key Program Sponsored by the Tianjin
Science and Technology Committee of China (grant nos. 14ZCZDSY00044
and 13RCGFSY19000), Key Projects for Science and Technology Support
(the Tianjin Key Research and Development Plan, grant no.
19YFZCSY0060), China Scholarship Council and Tianjin Research
Innovation Project for Postgraduate Students (grant no.
2019YJSB109), and Postgraduate Innovation Fund of ‘13th Five-Year
comprehensive investment’ of Tianjin Medical University. (grant no.
YJSCX201903).
Availability of data and materials
For all datasets from (STRING) database (a publicly
available database; http://string-db.org/), data are available from the
authors upon reasonable request and with permission of STRING. The
other datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
SZ, GS and JF designed the study. GS analyzed the
data and wrote the manuscript. XZha, BF, SL, YH and ZW collected
and analyzed all data. XZho and SF acquired funding, made
substantial contributions to the study design and gave final
approval of the version to be published. All authors read and
approved the final manuscript. All the above authors critically
revised the manuscript for important intellectual content and made
substantial contributions to conception and design.
Ethics approval and consent to
participate
All animal procedures were approved by the Animal
Ethics Committee of Tianjin Medical University General Hospital
(Tianjin, China) in January 2019 (approval no. IRB2019-WZ-004). All
experimental procedures described were in accordance with the Guide
for the Care and Use of Laboratory Animals (38).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ghosh M, Tuesta LM, Puentes R, Patel S,
Melendez K, El Maarouf A, Rutishauser U and Pearse DD: Extensive
cell migration, axon regeneration, and improved function with
polysialic acid-modified Schwann cells after spinal cord injury.
Glia. 60:979–992. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Parrinello S, Napoli I, Ribeiro S,
Wingfield Digby P, Fedorova M, Parkinson DB, Doddrell RD, Nakayama
M, Adams RH and Lloyd AC: EphB signaling directs peripheral nerve
regeneration through Sox2-dependent Schwann cell sorting. Cell.
143:145–155. 2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Stassart RM, Fledrich R, Velanac V,
Brinkmann BG, Schwab MH, Meijer D, Sereda MW and Nave KA: A role
for Schwann cell-derived neuregulin-1 in remyelination. Nat
Neurosci. 16:48–54. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Scheib J and Höke A: Advances in
peripheral nerve regeneration. Nat Rev Neuro. 9:668–676.
2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Johnson S, Ayling H, Sharma M and Goebel
A: External noninvasive peripheral nerve stimulation treatment of
neuropathic pain: A prospective audit. Neuromodulation. 18:384–391.
2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jessen KR and Mirsky R: The success and
failure of the schwann cell response to nerve injury. Front Cell
Neurosci. 13(33)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Pereira JA, Lebrun-Julien F and Suter U:
Molecular mechanisms regulating myelination in the peripheral
nervous system. Trends Neurosci. 35:123–134. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Esposito E and Cuzzocrea S: Anti-TNF
therapy in the injured spinal cord. Trends Pharmacol Sci.
32:107–115. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang H, Shao Z, Zhu Y, Shi L, Li Z, Hou
R, Zhang C and Yao D: Toll-like receptor 4 (TLR4) expression
affects Schwann cell behavior in vitro. Sci Rep.
8(11179)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhu H, Xue C, Yao M, Wang H, Zhang P, Qian
T, Zhou S, Li S, Yu B, Wang Y and Gu X: miR-129 controls axonal
regeneration via regulating insulin-like growth factor-1 in
peripheral nerve injury. Cell Death Dis. 9(720)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Staser K, Yang FC and Clapp DW: Mast cells
and the neurofibroma microenvironment. Blood. 116:157–164.
2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Painter MW, Brosius Lutz A, Cheng YC,
Latremoliere A, Duong K, Miller CM, Posada S, Cobos EJ, Zhang AX,
Wagers AJ, et al: Diminished Schwann cell repair responses underlie
age-associated impaired axonal regeneration. Neuron. 83:331–343.
2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lopez-Verrilli MA, Picou F and Court FA:
Schwann cell-derived exosomes enhance axonal regeneration in the
peripheral nervous system. Glia. 61:1795–1806. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li A, Hokugo A, Yalom A, Berns EJ,
Stephanopoulos N, McClendon MT, Segovia LA, Spigelman I, Stupp SI
and Jarrahy R: A bioengineered peripheral nerve construct using
aligned peptide amphiphile nanofibers. Biomaterials. 35:8780–8790.
2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Afshari FT, Kwok JC, White L and Fawcett
JW: Schwann cell migration is integrin-dependent and inhibited by
astrocyte-produced aggrecan. Glia. 58:857–869. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Huang H, Mao G, Chen L and Liu A: Progress
and challenges with clinical cell therapy in neurorestoratology. J
Neurorestoratol. 3:91–95. 2015.
|
|
17
|
Campana WM: Schwann cells: Activated
peripheral glia and their role in neuropathic pain. Brain Behav
Immun. 21:522–527. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Guertin AD, Zhang DP, Mak KS, Alberta JA
and Kim HA: Microanatomy of axon/glial signaling during Wallerian
degeneration. J Neurosci. 25:3478–3487. 2005.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Weis J, Claeys KG, Roos A, Azzedine H,
Katona I, Schröder JM and Senderek J: Towards a functional
pathology of hereditary neuropathies. Acta Neuropathol.
133:493–515. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Boilly B, Faulkner S, Jobling P and
Hondermarck H: Nerve dependence: From regeneration to cancer.
Cancer Cell. 31:342–354. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Akassoglou K, Yu WM, Akpinar P and
Strickland S: Fibrin inhibits peripheral nerve remyelination by
regulating Schwann cell differentiation. Neuron. 33:861–875.
2002.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Keilhoff G, Fansa H, Schneider W and Wolf
G: In vivo predegeneration of peripheral nerves: An effective
technique to obtain activated Schwann cells for nerve conduits. J
Neurosci Methods. 89:17–24. 1999.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shi GD, Zhang XL, Cheng X, Wang X, Fan BY,
Liu S, Hao Y, Wei ZJ, Zhou XH and Feng SQ: Abnormal DNA methylation
in thoracic spinal cord tissue following transection injury. Med
Sci Monit. 24:8878–8890. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhou XH, Ning GZ, Feng SQ, Kong XH, Chen
JT, Zheng YF, Ban DX, Liu T, Li H and Wang P: Transplantation of
autologous activated Schwann cells in the treatment of spinal cord
injury: Six cases, more than five years of follow-up. Cell
Transplantat. 21 (Suppl 1):S39–S47. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhou X, Shi G, Fan B, Cheng X, Zhang X,
Wang X, Liu S, Hao Y, Wei Z, Wang L and Feng S: Polycaprolactone
electrospun fiber scaffold loaded with iPSCs-NSCs and ASCs as a
novel tissue engineering scaffold for the treatment of spinal cord
injury. Int J Nanomedicine. 13:6265–6277. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ma T, Zhu L, Yang Y, Quan X, Huang L, Liu
Z, Sun Z, Zhu S, Huang J and Luo Z: Enhanced in vivo survival of
Schwann cells by a synthetic oxygen carrier promotes sciatic nerve
regeneration and functional recovery. J Tissue Eng Regen Med.
12:e177–e189. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bender J: DNA methylation and epigenetics.
Annu Rev Plant Biol. 55:41–68. 2004.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lev Maor G, Yearim A and Ast G: The
alternative role of DNA methylation in splicing regulation. Trends
Genet. 31:274–280. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Filipponi D, Muller J, Emelyanov A and
Bulavin DV: Wip1 controls global heterochromatin silencing via
ATM/BRCA1-dependent DNA methylation. Cancer Cell. 24:528–541.
2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Garriga J, Laumet G, Chen SR, Zhang Y,
Madzo J, Issa JJ, Pan HL and Jelinek J: Nerve injury-induced
chronic pain is associated with persistent DNA methylation
reprogramming in dorsal root ganglion. J Neurosci. 38:6090–6101.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Shi G, Zhou X, Wang X, Zhang X, Zhang P
and Feng S: Signatures of altered DNA methylation gene expression
after central and peripheral nerve injury. J Cell Physiol.
235:5171–5181. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Finnegan EJ, Genger RK, Peacock WJ and
Dennis ES: DNA METHYLATION IN PLANTS. Annu Rev Plant Physiol Plant
Mol Biol. 49:223–247. 1998.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gölzenleuchter M, Kanwar R, Zaibak M, Al
Saiegh F, Hartung T, Klukas J, Smalley RL, Cunningham JM, Figueroa
ME, Schroth GP, et al: Plasticity of DNA methylation in a nerve
injury model of pain. Epigenetics. 10:200–212. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhou XH, Lin W, Ren YM, Liu S, Fan BY, Wei
ZJ, Shi GD, Cheng X, Hao Y and Feng SQ: Comparison of DNA
methylation in schwann cells before and after peripheral nerve
injury in rats. Biomed Res Int. 2017(5393268)2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Shi GD, Cheng X, Zhou XH, Fan BY, Ren YM,
Lin W, Zhang XL, Liu S, Hao Y, Wei ZJ and Feng SQ: iTRAQ-based
proteomics profiling of Schwann cells before and after peripheral
nerve injury. Iran J Basic Med Sci. 21:832–841. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Bennett GJ and Xie YK: A peripheral
mononeuropathy in rat that produces disorders of pain sensation
like those seen in man. Pain. 33:87–107. 1988.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Michot B, Deumens R and Hermans E:
Immunohistochemical comparison of astrocytic mGluR5 upregulation in
infraorbital nerve-versus sciatic nerve-ligated rat. Neurosci Lett.
653:113–119. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
National Research Council Committee for
the Update of the Guide for the C and Use of Laboratory A: The
National Academies Collection: Reports funded by National
Institutes of Health. In: Guide for the Care and Use of Laboratory
Animals National Academies Press (US) Copyright©. 2011,
National Academy of Sciences., Washington (DC), 2011.
|
|
39
|
Chisholm JM and Pang DS: Assessment of
carbon dioxide, carbon dioxide/oxygen, isoflurane and pentobarbital
killing methods in adult female Sprague-Dawley rats. PLoS One.
11(e0162639)2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Hickman DL and Johnson SW: Evaluation of
the aesthetics of physical methods of euthanasia of anesthetized
rats. J Am Assoc Lab Anim Sci. 50:695–701. 2011.PubMed/NCBI
|
|
41
|
Stutler SA, Johnson EW, Still KR,
Schaeffer DJ, Hess RA and Arfsten DP: Effect of method of
euthanasia on sperm motility of mature Sprague-Dawley rats. J Am
Assoc Lab Anim Sci. 46:13–20. 2007.PubMed/NCBI
|
|
42
|
Yang M and Zhou H: Grass carp transforming
growth factor-beta 1 (TGF-beta 1): Molecular cloning, tissue
distribution and immunobiological activity in teleost peripheral
blood lymphocytes. Mol Immunol. 45:1792–1798. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhao H, Zhang Y, Sun J, Zhan C and Zhao L:
Raltitrexed inhibits HepG2 cell proliferation via G0/G1 cell cycle
arrest. Oncol Res. 23:237–248. 2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wang X, Niu Z, Jia Y, Cui M, Han L, Zhang
Y, Liu Z, Bi D and Liu S: Ubenimex inhibits cell proliferation,
migration and invasion by inhibiting the expression of APN and
inducing autophagic cell death in prostate cancer cells. Oncol Rep.
35:2121–2130. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Xiao G, Cheng H, Cao H, Chen K, Tu Y, Yu
S, Jiao H, Yang S, Im HJ, Chen D, et al: Critical role of
filamin-binding LIM protein 1 (FBLP-1)/migfilin in regulation of
bone remodeling. J Biol Chem. 287:21450–21460. 2012.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Liu D, Zhang Y, Li X, Li J, Yang S, Xing
X, Fan G, Yokota H and Zhang P: eIF2α signaling regulates ischemic
osteonecrosis through endoplasmic reticulum stress. Sci Rep.
7(5062)2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Liu Q, Zhu Y, Qi J, Amadio PC, Moran SL,
Gingery A and Zhao C: Isolation and characterization of turkey bone
marrow-derived mesenchymal stem cells. J Orthop Res. 37:1419–1428.
2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Li N, Ye M, Li Y, Yan Z, Butcher LM, Sun
J, Han X, Chen Q, Zhang X and Wang J: Whole genome DNA methylation
analysis based on high throughput sequencing technology. Methods.
52:203–212. 2010.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Huang da W, Sherman BT, Stephens R,
Baseler MW, Lane HC and Lempicki RA: DAVID gene ID conversion tool.
Bioinformation. 2:428–430. 2008.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Petersen SC, Luo R, Liebscher I, Giera S,
Jeong SJ, Mogha A, Ghidinelli M, Feltri ML, Schöneberg T, Piao X
and Monk KR: The adhesion GPCR GPR126 has distinct,
domain-dependent functions in Schwann cell development mediated by
interaction with laminin-211. Neuron. 85:755–769. 2015.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Jessen KR and Arthur-Farraj P: Repair
Schwann cell update: Adaptive reprogramming, EMT, and stemness in
regenerating nerves. Glia. 67:421–437. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhao X, Chen C, Wei Y, Zhao G, Liu L, Wang
C, Zhang J and Kong X: Novel mutations of COL4A3, COL4A4, and
COL4A5 genes in Chinese patients with Alport Syndrome using next
generation sequence technique. Mol Genet Genomic Med.
7(e653)2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Assinck P, Duncan GJ, Hilton BJ, Plemel JR
and Tetzlaff W: Cell transplantation therapy for spinal cord
injury. Nat Neurosci. 20:637–647. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Li L, Xiong WC and Mei L: Neuromuscular
junction formation, aging, and disorders. Annu Rev Physiol.
80:159–188. 2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Adams D, Koike H, Slama M and Coelho T:
Hereditary transthyretin amyloidosis: A model of medical progress
for a fatal disease. Nat Rev Neurol. 15:387–404. 2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Chernov AV, Dolkas J, Hoang K, Angert M,
Srikrishna G, Vogl T, Baranovskaya S, Strongin AY and Shubayev VI:
The calcium-binding proteins S100A8 and S100A9 initiate the early
inflammatory program in injured peripheral nerves. J Biol Chem.
290:11771–11784. 2015.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Ratushnyy AY and Buravkova LB: Expression
of focal adhesion genes in mesenchymal stem cells under simulated
microgravity. Dokl Biochem Biophys. 477:354–356. 2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Chen Y, Teng L, Liu W, Cao Y, Ding D, Wang
W, Chen H, Li C and An R: Identification of biological targets of
therapeutic intervention for clear cell renal cell carcinoma based
on bioinformatics approach. Cancer Cell Int. 16(16)2016.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Tabur S, Oztuzcu S, Oguz E, Demiryürek S,
Dagli H, Alasehirli B, Ozkaya M and Demiryürek AT: Evidence for
elevated (LIMK2 and CFL1) and suppressed (ICAM1, EZR, MAP2K2, and
NOS3) gene expressions in metabolic syndrome. Endocrine.
53:465–470. 2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Fromont G, Vallancien G, Validire P,
Levillain P and Cussenot O: BCAR1 expression in prostate cancer:
Association with 16q23 LOH status, tumor progression and EGFR/KAI1
staining. Prostate. 67:268–273. 2007.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Camacho Leal MD, Costamagna A, Tassone B,
Saoncella S, Simoni M, Natalini D, Dadone A, Sciortino M, Turco E,
Defilippi P, et al: Conditional ablation of p130Cas/BCAR1 adaptor
protein impairs epidermal homeostasis by altering cell adhesion and
differentiation. Cell Commun Signal. 16(73)2018.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Camacho Leal Mdel P, Sciortino M, Tornillo
G, Colombo S, Defilippi P and Cabodi S: p130Cas/BCAR1 scaffold
protein in tissue homeostasis and pathogenesis. Gene. 562:1–7.
2015.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Barrett A, Pellet-Many C, Zachary IC,
Evans IM and Frankel P: p130Cas: A key signalling node in health
and disease. Cell Signal. 25:766–777. 2013.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Nishikawa R, Goto Y, Kojima S, Enokida H,
Chiyomaru T, Kinoshita T, Sakamoto S, Fuse M, Nakagawa M, Naya Y,
et al: Tumor-suppressive microRNA-29s inhibit cancer cell migration
and invasion via targeting LAMC1 in prostate cancer. Int J Oncol.
45:401–410. 2014.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Zhang P, Kou Y, Yin X, Wang Y, Zhang H and
Jiang B: The experimental research of nerve fibers compensation
amplification innervation of ulnar nerve and musculocutaneous nerve
in rhesus monkeys. Artif Cells Blood Substit Immobil Biotechnol.
39:39–43. 2011.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Hu J, Zhu QT, Liu XL, Xu YB and Zhu JK:
Repair of extended peripheral nerve lesions in rhesus monkeys using
acellular allogenic nerve grafts implanted with autologous
mesenchymal stem cells. Exp Neurol. 204:658–666. 2007.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Bontioti EN, Kanje M and Dahlin LB:
Regeneration and functional recovery in the upper extremity of rats
after various types of nerve injuries. J Peripher Nerv Syst.
8:159–168. 2003.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Zhu J, Qin J, Shen Z, Kretlow JD, Wang X,
Liu Z and Jin Y: Dispase rapidly and effectively purifies Schwann
cells from newborn mice and adult rats. Neural Regen Res.
7:256–260. 2012.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Wang G, Ma Z, Cao L, Yan G, Wang Y, Jin Y,
Shen H, Zhang Y, Xu X, Chen X and Shen Z: A novel method for
obtaining highly enriched Schwann cell populations from mature
monkey nerves based on in vitro pre-degeneration. Mol Med
Rep. 16:6600–6607. 2017.PubMed/NCBI View Article : Google Scholar
|