Introduction
Preeclampsia (PE) is medical condition that begins
after week 20 of pregnancy, with hypertension and proteinuria as
the main clinical manifestations (1). In the United States, the prevalence of
preeclampsia has increased from 2.4% in 1980 to 3.8% in 2010 in all
pregnancies over the past three decades (2,3).
Several studies have reported that PE pathogenesis is associated
with insufficient recasting of the uterine spiral artery,
inflammatory immune overactivation, vascular endothelial cell
damage, genetic factors and nutritional deficiencies (4). Other theories include the weakening of
extravillous trophoblast (EVT) invasion, leading to shallow
placental implantation (5).
IFN-γ is produced by decidual natural killer (NK)
cells and can induce apoptosis of primary human trophoblasts
(6). Previous studies have revealed
that IFN-γ increases EVT apoptosis, reduces active protease and
insulin growth factor receptor-2 levels and inhibits EVT invasion
(7). Additionally, IFN-γ can reduce
abnormal expression of MMP1, MMP3 and MMP9 in decidual cells, and
reverses the effects of TNF-α on EVT during decidual invasion
(8). Therefore, IFN-γ can avoid
excessive EVT invasion and reduce PE.
IFN-γ regulates the expression of different genes by
activating the JAK/STAT pathway (9). Previous studies on STAT1-deficient
fibrosarcoma bone cancer cells observed that STAT1 phosphorylation
at tyrosine (Tyr)701 and serine (Ser)727 residues are essential for
achieving transcriptional regulation (10). In HTR8/SVneo cells, IFN-γ can
activate STAT1 at Ser1727 and Tyr701 residues and reduce basic
leucine zipper transcriptional factor ATF-like 2 (BATF2) expression
to reduce cell invasion, thus inhibiting JUN. By regulating the
expression of JUN, cell invasion is diminished. Moreover, STAT1 and
BATF2 function dependently or independently of each other (11).
Suppressor of cytokine signaling (SOCS) is an
important negative regulatory protein in Janus kinase (JAK)/STAT
activator pathways, and also inhibits the signaling pathways of
various cytokines (12). Previous
studies have discovered seven SOCS molecules (SOCS1-7) and
cytokine-inducible src homology 2 (SH-2) proteins, and cytokines
and growth factors can produce most of these molecules (13). By regulating the Tyr phosphorylation
levels of STAT protein, SOCS may negatively regulate the signaling
pathways (14). Furthermore, the
interaction of SOCS molecules and other signaling pathways may be
key to maintain cytokine balance and fetal immune tolerance
(15). Numerous cytokines,
including the T-helper (Th) 1 cytokines IFN-γ and IL-2, and the Th2
cytokine TNF-α, could rapidly induce SOCS (16,17),
and in turn, inhibit cytokine signal transduction.
The present study aimed to investigate
IFN-γ-mediated activation of JAK/STAT1 in HTR-8/SVneo cells and its
effects on cell viability, migration, invasion and apoptosis.
Additionally, the present study examined how SOCS1 feedback
regulated JAK/STAT1 and affected EVT invasion. Immortalized human
chorionic trophoblast cell HTR-8/SVneo cells were used instead of
EVTs.
Materials and methods
Tissue collection
Between January 2018 and June 2018, 30 patients with
PE (age range, 21.0-42.0 years; mean age, 31.80±5.91 years) were
selected for obstetric examinations and deliveries in The Second
Hospital of Shanxi Medical University (Taiyuan, China). The Ethics
Committee of The Second Hospital of Shanxi Medical University
approved the research protocol, and written informed consent was
obtained from each patient. The criteria to diagnose PE was a
systolic blood pressure of ≥140 mmHg, a diastolic blood pressure of
≥90 mmHg and 0.3 g protein in urine samples from females that were
≥20 weeks pregnant and had previously healthy blood pressure and
24-h urine samples. In total, 10 healthy pregnant women (age range,
17.0-36.0 years; mean age, 28.90±6.12 years) who gave birth in the
Second Hospital of Shanxi Medical University at the same time were
selected as a control group. The gestational weeks of pregnant
women were 34-39 weeks. Both groups of pregnant women (PE and
control) underwent single primary labor, and none of these women
had a history of repeated abortions or in vitro
fertilization. The exclusion criteria were as follows: Patients
that presented complications such as pregnancy with hypertension,
gestational diabetes, pregnancy with diabetes and pregnancy with
heart disease. A total of 3 min after delivery of the placenta,
~1x1x1 cm of placental tissue was collected, which was frozen and
stored at -80˚C for further use.
Cell culture
HTR-8/SVneo cells (American Type Culture Collection)
were cultured in RPMI-1640 medium (Wuhan Boster Biological
Technology, Ltd.) supplemented with 10% FBS (Biological
Industries), 100 mg/ml streptomycin and 100 U/ml penicillin. Cells
were cultured in a humid atmosphere of 5% CO2 and 37˚C.
Cells were extracted after reaching 80% confluence. Drug
intervention cells were cultured in RPMI-1640 (Sigma-Aldrich; Merck
KGaA) supplemented with 100 ng/ml IFN-γ, while control cells were
cultured in conventional RPMI-1640 in a humid atmosphere of 5%
CO2 and 37˚C for 48 h. For inhibition experiments, cells
were subjected to fluorescein amidites-labeled small interfering
RNA (siRNA) transfection and cultured in RPMI-1640 medium for 48 h.
Cells were collected for analysis 24 h after transfection.
MTT assay
HTR-8/SVneo cells in the logarithmic growth phase
were plated in 96-well plates with 100 µl cell suspension in each
well, and a blank group, with 100 µl sterile PBS added to the wells
surrounding the cells was also established. Cells were cultured at
37˚C overnight. Cells were treated with 0, 0.1, 1, 10, 100 and
1,000 ng/ml IFN-γ and were cultured at 37˚C and 5% CO2
saturated humidity for 48 h. A total 10 µl MTT solution was then
added to each well and incubated at 37˚C for 4 h. The media was
aspirated, followed by addition of 150 µl DMSO and agitation for 10
min at 37˚C. The absorbance at 568 nm in each well was determined
using a microplate reader. All experiments were repeated three
times.
Wound healing assay
HTR-8/SVneo cells were seeded in 6-well plates with
2 ml cell suspension/well (2x105 cells/ml) and cultured
overnight at 37˚C. The cells were then treated with 0 and 100 ng/ml
IFN-γ (Sigma-Aldrich; Merck KGaA) at 37˚C for 48 h. Once cells
reached 90% confluence, a scratch was made using a pipette tip to
create a 1-mm-wide scratch, at the same time, cell culture medium
was replaced with FBS-free medium. After 0 and 24 h, following
three washes with PBS, images were captured using an Olympus IX51
Inverted fluorescence microscope (Olympus Corporation) at x100
magnification. The control and experimental group scratch widths
were measured using ImageJ 1.42 (National Institutes of Health) and
normalized to their values at 0 h. The migration distance is
defined as follows: Migration distance (%) = (At 0 h-At 24 h)/At 0
h x100%. All experiments were repeated three times.
Cell apoptosis assay
Cell apoptosis analysis was performed to detect the
percentage of early and late apoptotic cells using the Annexin
V-APC apoptosis analysis kit (cat. no. AO2001-11A-G; Tianjin
Sungene Biotech Co., Ltd.) were used to measure the
apoptotic-inducing ability of IFN-γ. In total, ~4x105
cells were seeded on a 6-well plate, cultured at 37˚C overnight,
treated with complete medium containing 0 and 100 ng/ml IFN-γ at
37˚C for 48 h and digested with 0.25% trypsin without EDTA to
terminate digestion. Cells were subsequently collected, washed
twice with PBS and centrifuged at 300 x g for 3 min at 37˚C. A
total of 500 µl binding buffer was added and cells were
resuspended. Following the addition of 5 µl APC, 5 µl 7-AAD was
added to the mix and left to react at room temperature for 5 min in
the dark. A CytoFLEX flow cytometer and CytExpert software v2.3
(Beckman Coulter, Inc.) were used for apoptosis analysis. All
experiments were repeated three times.
Invasion assay
Cells were treated with 0 and 100 ng/ml IFN-γ
complete medium at 37˚C for 48 h, and cells (2x105/ml)
were then seeded into 24-well plates (Corning, Inc.). Transwell
chambers (5 µm pore size, Corning, Inc.) with Matrigel (cat. no.
356234; Corning, Inc.) were used in the cell invasion assay.
Matrigel was melted at 4˚C 1 day in advance, which was used to
precoat the Transwell chambers at 37˚C for 30 min. The upper
inserts (containing 200 µl cell suspension) were filled with
serum-free medium at a concentration of 2x105 cells/ml,
while the medium (800 µl) in the lower chamber contained 10% FBS
and RPMI-1640 complete medium supplemented with 1% penicillin and
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.).
Following 24 h incubation, the chambers were carefully washed with
PBS. Cells were fixed with 4% paraformaldehyde for 30 min at room
temperature and stained with 0.5% crystal violet at room
temperature for 20 min. The non-migratory cells were then removed,
the membrane was imaged with an Olympus IX51 inverted light
microscope (Olympus Corporation) at x200 magnification and the cell
numbers was measured randomly from five visual fields per well. All
experiments were repeated three times.
Western blotting
RIPA lysis buffer (Beyotime Institute of
Biotechnology) and loading buffer (1 µg/µl, 50 µg) were used to
extract total protein from the placenta. Protein concentration was
determined using a bicinchoninic acid Protein Assay kit (Pierce;
Thermo Fisher Scientific, Inc.). Total proteins (20 µg) were loaded
per lane and separated using 12% SDS-PAGE, and transferred to a
PVDF membrane (EMD Millipore) overnight at 4˚C. The membrane was
blocked with 10% skim milk in TBS-0.1% Tween (TBS-T) for 2 h at
room temperature, followed by three TBS-T washes. Membranes were
incubated overnight at 4˚C with the following primary antibodies:
Murine anti-STAT1 (dilution 1:2,000; cat. no. 66545-1-Ig;
Proteintech Group, Inc.), rabbit anti-phosphorylated (p)-STAT1
(dilution 1:1,000; cat. no. 9177T; Cell Signaling Technology, Inc.)
and murine anti-JAK1 (dilution 1:2,000; cat. no. 66466-1-Ig;
Proteintech Group, Inc.) in 10% milk; rabbit anti-p-JAK1 (dilution
1:1,000; cat. no. 74129S; Cell Signaling Technology, Inc.), mouse
anti-PDGFRA (dilution 1:500; cat. no. sc-398206; Santa Cruz
Biotechnology, Inc.), rabbit anti-cleaved caspase3 (dilution
1:1,000; cat. no. 19677-1-AP; Proteintech Group, Inc.), rabbit
anti-caspase3 (dilution 1:1,000; cat. no. 19677-1-AP; Proteintech
Group, Inc.), rabbit anti-Ezrin (EZR; dilution 1:2,000; cat. no.
ab75840; Abcam), rabbit anti-GAPDH (dilution 1:1,000; cat. no.
AB-P-R 001; Hangzhou Xianzhi Biotechnology Co., Ltd.) and goat
anti-SOCS1 (dilution 1:250; cat. no. ab9870; Abcam). Membranes were
then washed three times with TBST and incubated with horseradish
peroxidase-conjugated secondary antibodies (dilution 1:50,000; cat.
nos. BA1051 and BA1054; Wuhan Boster Biological Technology, Ltd.)
for 2 h at room temperature. An enhanced chemiluminescence
detection kit (Applygen Technologies, Inc.) was used to detect
protein signals. An X-ray film was pressed into the developing
solution, a fixing solution was used at room temperature for 5 min
and the film was washed. Glyko BandScan 5.0 software (ProZyme,
Inc.; Agilent Biotechnologies, Inc.) was used to analyze film gray
values. All experiments were repeated three times.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was extracted using TRIzol®
reagent (cat. no. 15596-026; Thermo Fisher Scientific, Inc.), and
the RNA pellet was resuspended in Millipore water and temporarily
stored at -80˚C. NanoDrop was used to assess RNA quantity and
purity. RNA (1 µg) was reverse transcribed into cDNA using a Maxima
H Minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific,
Inc.). The temperature protocol for reverse transcription was as
follows: 25˚C for 5 min, 50˚C for 15 min, 85˚C for 5 min and 4˚C 10
min. cDNA (20 ng) was used to amplify the reference gene (GAPDH) or
target genes (SOCS1, STAT1 and JAK1). The efficiency of
primer-amplified amplicons was ~100%. The following primer pairs
were used for the qPCR: GAPDH forward, 5'-GGATTTGGTCGTATTGGGCG-3'
and reverse, 5'-GGATTTGGTCGTATTGGGCG-3'; SOCS1 forward,
5'-CGACACGCACTTCCGCACATT-3' and reverse,
5'-TGGGTCCCGAGGCCATCTTCAC-3'; STAT1 forward,
5'-TGAACTTACCCAGAATGCC-3' and reverse, 5'-TCTTTCCACCACAAACGAG-3';
and JAK1 forward, 5'-CCTGCCGTGCCCACCTAACT-3' and reverse,
5'-GCTTGTCCGATTGGATGGTT-3'. The reaction mixture consisted of 4 µl
cDNA, 0.4 µl (10 µM) forward and reverse primers, 4.8 µl
RNase/DNase-free water, 10 µl SYBR-Green buffer (Vazyme Biotech
Co., Ltd.) and 0.4 µl 50X ROX (Vazyme Biotech Co., Ltd). The final
total reaction volume was 20 µl. ABI QuantStudio 6 Pro systems
V2.4.3 (Thermo Fisher Scientific, Inc.) was used for quantitative
analysis of mRNA levels. The following thermocycling conditions
were used for the qPCR: Initial denaturation at 95˚C for 2 min,
followed by 40 cycles of 95˚C for 30 sec and annealing/extension at
60˚C for 60 sec. Gene expression was calculated using the
2-ΔΔCq method (18), and
all experiments were repeated three times.
SOCS1 siRNA transient
transfection
siRNA against SOCS1 and NC siRNA were all produced
by Shanghai GenePharma Co., Ltd. Cells were extracted at the
logarithmic growth phase, and the cell density was adjusted to
2x105 cells/ml via the addition of RPMI-1640 complete
medium. Cells were seeded in 6-well plates with 2 ml cell
suspension/well and cultured overnight at 37˚C. The following
sequences were used: SOCS1 siRNA, 5'-UCGCCCUUAGCGUGAAGAUTT-3' and
NC siRNA, 5'-UUCUCCGAACGUGUCACGUTT-3'. Cells were transfected with
fluorescein amidites-labeled siRNA and cultured in a 37˚C incubator
for 24 h and then subjected to the following analysis. A total of
10 µl siRNA was diluted in 100 µl serum-free Opti-MEM (Gibco;
Thermo Fisher Scientific, Inc.). The mixture was gently mixed with
a pipette tip and left to stand for 5 min at room temperature.
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was gently mixed before use. A total of 5 µl
Lipofectamine 2000 was diluted in 100 µl Opti-MEM and left to stand
for 5 min at room temperature. Lipofectamine and siRNA dilutions
were mixed gently and left to stand for a further 20 min at room
temperature. A total of 200 µl Lipofectamine and siRNA solution was
added to each culture well plate and the plate was gently agitated
in order for the solution to mix. The cells were cultured in a 37˚C
CO2 incubator. After 6 h, the mixture was aspirated and
replaced with standard medium. Cells were cultivated in a 5%
CO2 incubator at 37˚C for 24 h, and photomicrographs
were captured using the Olympus IX51 inverted fluorescence
microscope (Olympus Corporation) at x100 magnification.
Hematoxylin & eosin (H&E)
staining
The placenta tissue was fixed in 4% paraformaldehyde
for 24 h at room temperature and the sections (thickness, 5 µm)
were stained with hematoxylin (37˚C for 5 min) and eosin (37˚C for
1 min) using a HE Staining kit (Beijing Solarbio Science &
Technology Co., Ltd.). For each placental sample, two randomly
selected fields of view were captured at x100 or 400 magnification
using the Olympus IX51 inverted light microscope (Olympus
Corporation).
ELISA
Placental tissue was washed with cold PBS (0.01 M;
pH=7.2-7.4) to remove blood. The clean tissue was cut into small
pieces and homogenized in PBS on ice. The placenta homogenate was
collected and centrifuged at 5,000 x g for 5 min at 4˚C.
Supernatants were tested using ELISA kits for IL-10 (cat. no.
E-EL-H0103c; Elabscience, Inc.) and IFN-γ (cat. no. E-EL-H0108c;
Elabscience, Inc.) according to the manufacturer's protocol.
Immunohistochemistry
Placental tissue was fixed in 4% formalin overnight
and embedded in paraffin at room temperature. Placental sections
(thickness, 5 µm) placental sections were cut and SOCS1 was applied
using the 3,3'-diaminobenzidine staining method (cat. no. PV-6000;
OriGene Technologies, Inc.). Following deparaffinization using
xylene followed by a descending ethanol gradient, the antigen
retrieval was performed using EDTA buffer (1 mM EDTA; pH 8.0) in a
high-pressure steamer at 37˚C for 15 min. Samples were incubated
with 3% peroxidase and protein blocking solution at 37˚C for 10 min
and then incubated with an anti-SOCS1 antibody (dilution 1:500;
cat. no. ab9870; Abcam) overnight at 4˚C. The following day,
samples were incubated with secondary antibody using the Universal
SP kit (mouse/rabbit streptavidin-biotin method detection system,
ready for use; cat. no. SP-9000; ZSGB-BIO; OriGene Technologies,
Inc.) at 37˚C for 30 min. The sections were then washed and
incubated with chromogen (liquid diaminobenzidine and peroxide
buffer; dilution 1:200) at 37˚C for 5 min until a reaction was
observed. The slides were counterstained with hematoxylin at 37˚C
for 5 min to provide nuclear and morphological details and
fixation. For each placental sample, two randomly selected fields
of view were captured at x100 or 400 magnification using the
Olympus IX51 inverted light microscope (Olympus Corporation).
Statistical analysis
Each experiment was repeated three times. Data are
presented as the mean ± SD. GraphPad Prism 5 (GraphPad Software,
Inc.) was used for statistical analysis. An unpaired t-test was
used for comparison between two groups. A one-way ANOVA was
performed followed by Dunnett's test to compare differences between
>3 groups. A Spearman's rank correlation coefficient was used
for correlation analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
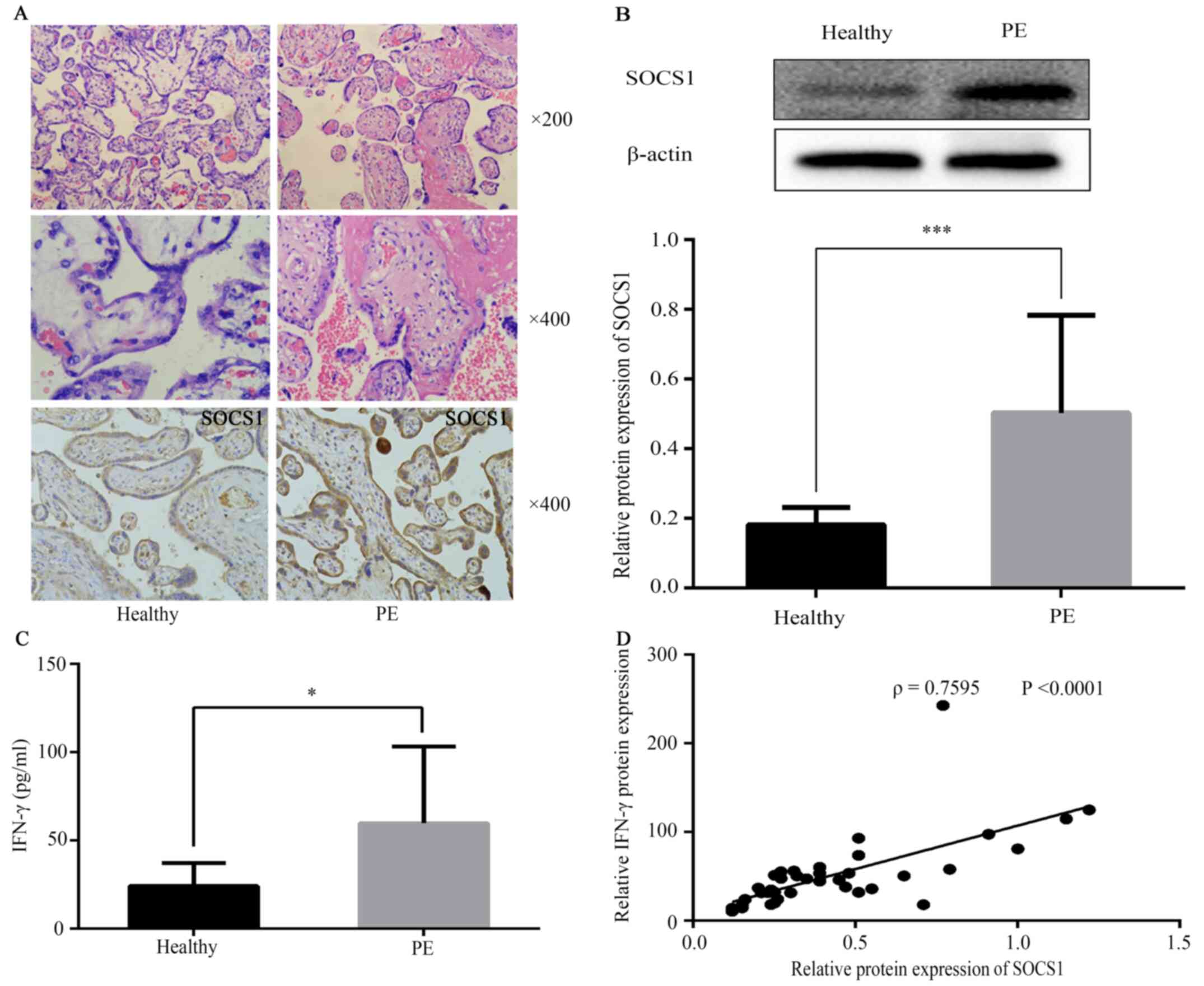
Placental tissue pathology
In the control group, the placental villi were rich
in blood vessels, with a clear structure and fewer cytotrophoblast
cells compared with the PE group (Fig.
1A). By contrast, in the PE group, the number of placental
villi was reduced, the structure was irregular and atrophic, with
parts of the villus cellulose necrotic. The placental villi
trophoblast nodules were increased, and most of the villi were
immature (Fig. 1A).
Expression levels of SOCS1 and IFN-γ
in villi and outer villous trophoblasts
SOCS1 in patients with PE demonstrated weak
syncytial staining with some nuclei and cytotrophoblasts strongly
stained. Immunostaining results are presented in Fig. 1A. SOCS1 protein expression in PE
placental tissue was measured using western blot analysis (Fig. 1B), which indicated that SOCS1
protein expression was increased in the PE group (Fig. 1B; 0.50±0.28). Compared with the
control group, tissue expression levels of IFN-γ were increased in
the PE group (Fig. 1C). The
correlation between SOCS1 expression and IFN-γ was evaluated, and
it was found that SOCS1 expression was strongly, positively
correlated with IFN-γ in healthy placental and PE groups (Fig. 1D).
IFN-γ activates JAK/STAT, inhibits
HTR8/SVneo cell viability, migration and invasion and promotes
apoptosis
MTT experiments were performed to detect the cell
viability of HTR-8/SVneo cells. The results showed a dose-dependent
inhibitory effect of IFN-γ on the cell viability of HTR-8/SVneo
cells (Fig. 2A). The data
demonstrated a significant reduction in the number of live cells
treated with IFN-γ (100 ng/ml) compared with the untreated control
group (Fig. 2A), therefore 100
ng/ml was selected as the concentration for the subsequent
analysis. Furthermore, the migratory and invasive abilities were
significantly reduced (Fig. 2B-E),
while apoptosis (Fig. 2F and
G) was significantly increased in
HTR-8/SVneo cells treated with 100 ng/ml IFN-γ compared with
untreated cells.
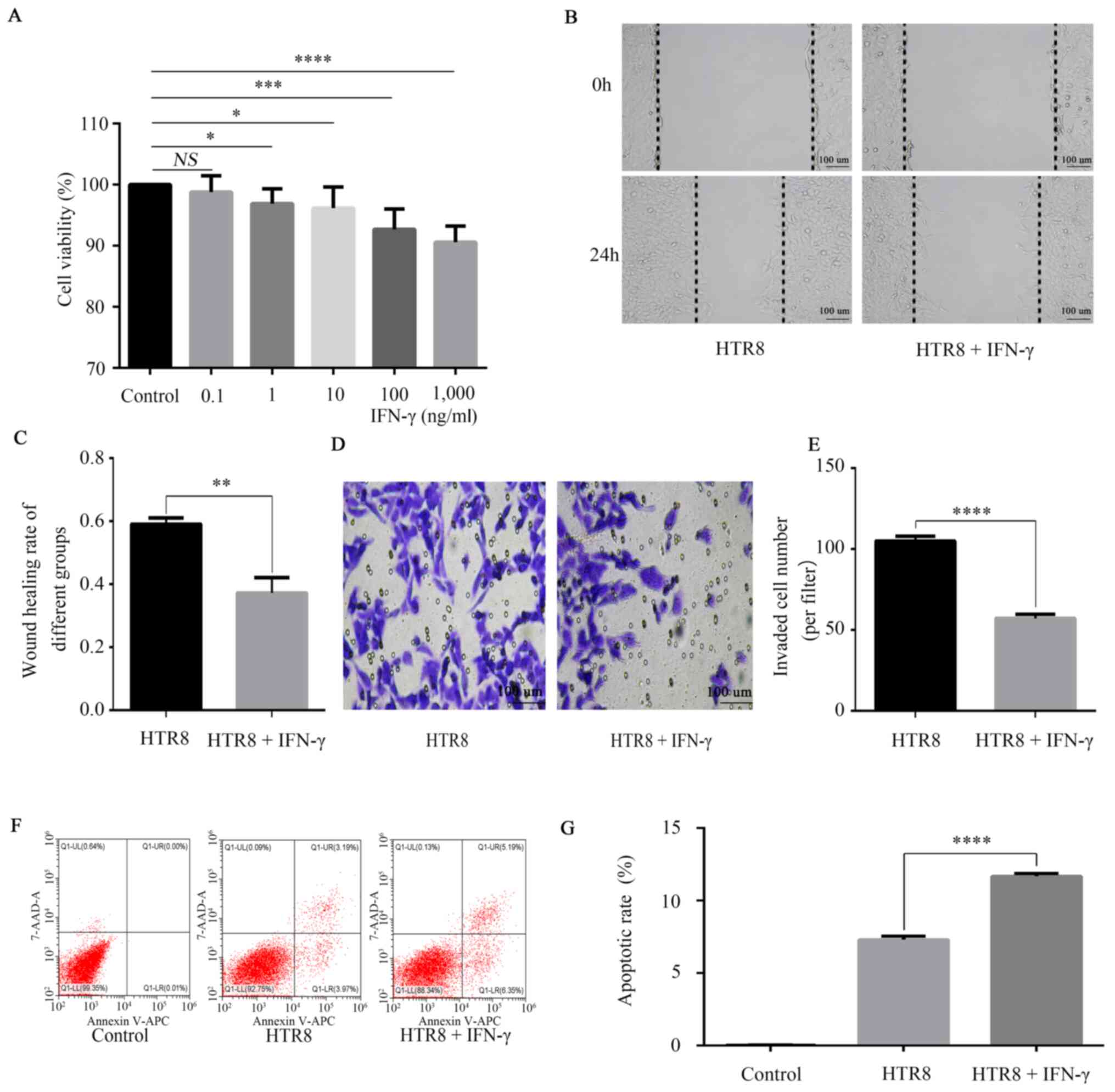 | Figure 2Effects of IFN-γ on cell viability,
migration, migration and apoptosis of HTR8/SVneo cells. (A)
HTR8/SVneo cells were treated with 0, 0.1, 1, 10, 100 and 1,000
ng/ml IFN-γ. MTT assay was conducted to detect cell viability, with
the absorbance measured at a wavelength of 568 nm. HTR8/SVneo cells
were treated with 0 and 100 ng/ml IFN-γ for 48 h. NS, not
significant; *P<0.05; ***P<0.001;
****P<0.0001. (B) Wound healing results indicated
that (C) IFN-γ decreased HTR8/SVneo migration.
**P<0.01. (D) Transwell assay results demonstrated
that (E) IFN-γ decreased HTR8/SVneo invasion.
****P<0.0001. (F) Flow cytometry findings suggested
that (G) IFN-γ increased the apoptotic rate.
****P<0.0001. Data are presented as the mean ± SD of
three independent experiments. IFN-γ, interferon-γ. |
SOCS1 may be involved in IFN-γ
mediated reduction of HTR8/SVneo cell invasion
RT-qPCR results demonstrated that compared with
untreated controls, HTR-8/SVneo cells treated with IFN-γ (100
ng/ml) for 48 h had significantly increased mRNA expression levels
of SOCS1 (Fig. 3A). Additionally, a
significant increase in SOCS1 protein expression was observed
(Fig. 3D and E). RT-qPCR results also indicated that,
with GAPDH was used as a reference, JAK and STAT1 mRNA expression
levels were significantly increased compared with the untreated
control group (Fig. 3B and C). Western blot analysis found that p-JAK
and p-STAT1 expression levels were significantly increased compared
with the control group, while JAK and STAT1 expression levels were
not significantly altered (Fig. 3H
and I). In the IFN-γ treatment
group, cell migration and invasion-related markers PDGFRA and EZR
were significantly reduced compared with the control group, and the
apoptosis-related gene cleaved caspase3 expression was
significantly increased (Fig. 3F
and G).
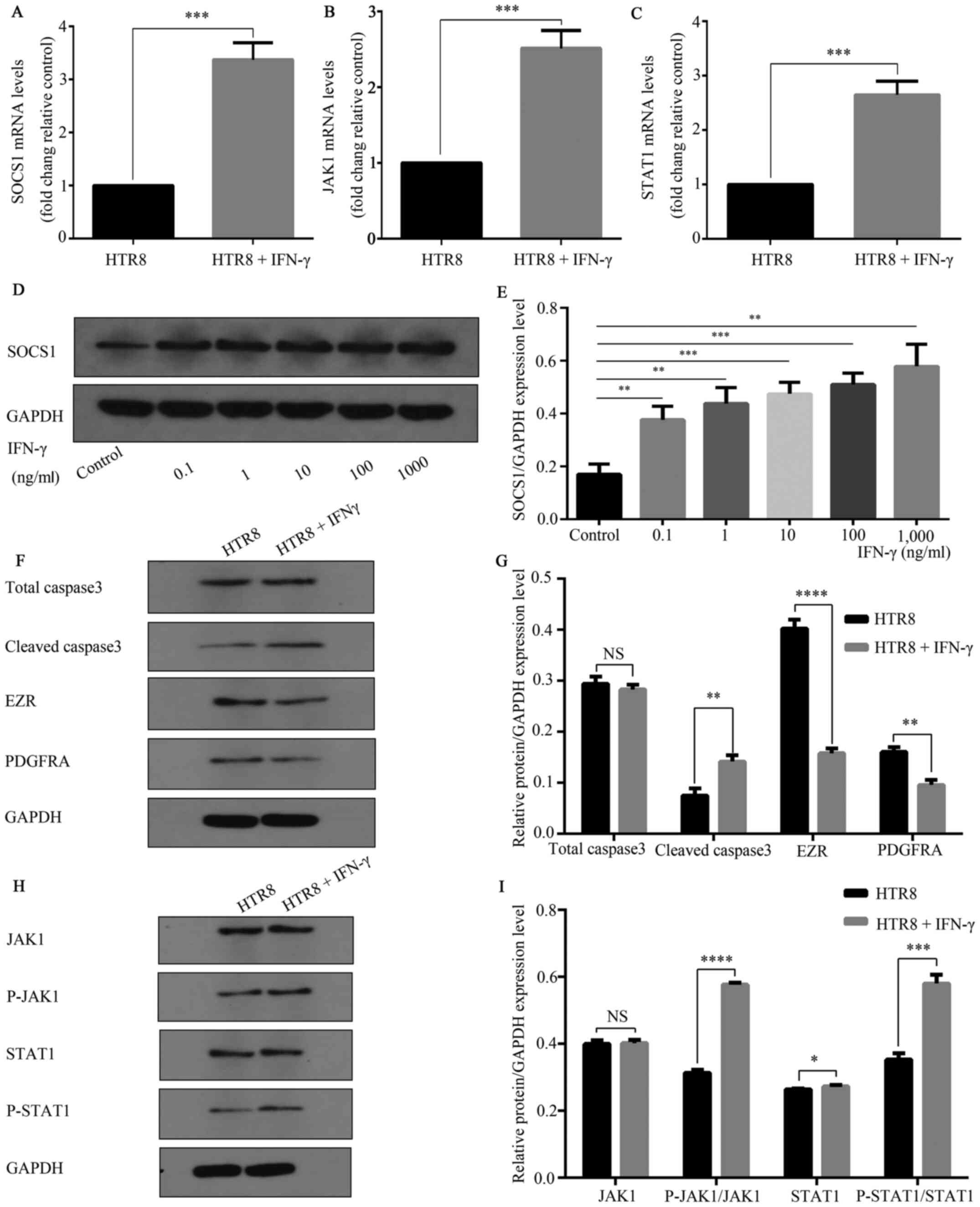 | Figure 3IFN-γ activates JAK1/STAT1 and
increases SOCS1 expression. HTR8/SVneo cells were treated in 0 and
100 ng/ml complete medium for 48 h. Total cellular RNA was
extracted. IFN-γ increased the mRNA expression levels of (A) SOCS1,
***P=0.0002; (B) JAK1, ***P=0.0004; and (C)
STAT1, ***P=0.0003 mRNA. (D) HTR8/SVNeo cells were
treated with 0, 0.1, 1, 10, 100 and 1,000 ng/ml and analyzed using
western blotting. (E) Different concentrations of IFN-γ increased
the expression of SOCS1 protein. **P<0.01;
***P<0.001. (F) Caspase3, EZR and PDGFRA are markers
of apoptosis, migration and invasion, respectively. (G) IFN-γ (100
ng/ml) reduced the expression levels of total caspase3 (NS, not
significant), cleaved caspase3 **P=0.01; EZR,
****P<0.0001; and PDGFRA, **P=0.01. (H)
Western blotting results suggested that 100 ng/ml activated
JAK/STAT1. (I) p-JAK1, ****P<0.0001; and p-STAT,
***P=0.0003, expression levels were increased, while
JAK1 (NS, not significant) and STAT1 (*P=0.0434)
expression levels did not significantly change. Data are presented
as the mean ± SD of three independent experiments. SOCS1,
suppressor of cytokine signaling 1; PDGFRA, platelet-derived growth
factor receptor A; EZR, Ezrin; p-, phosphorylated; JAK, Janus
kinase. |
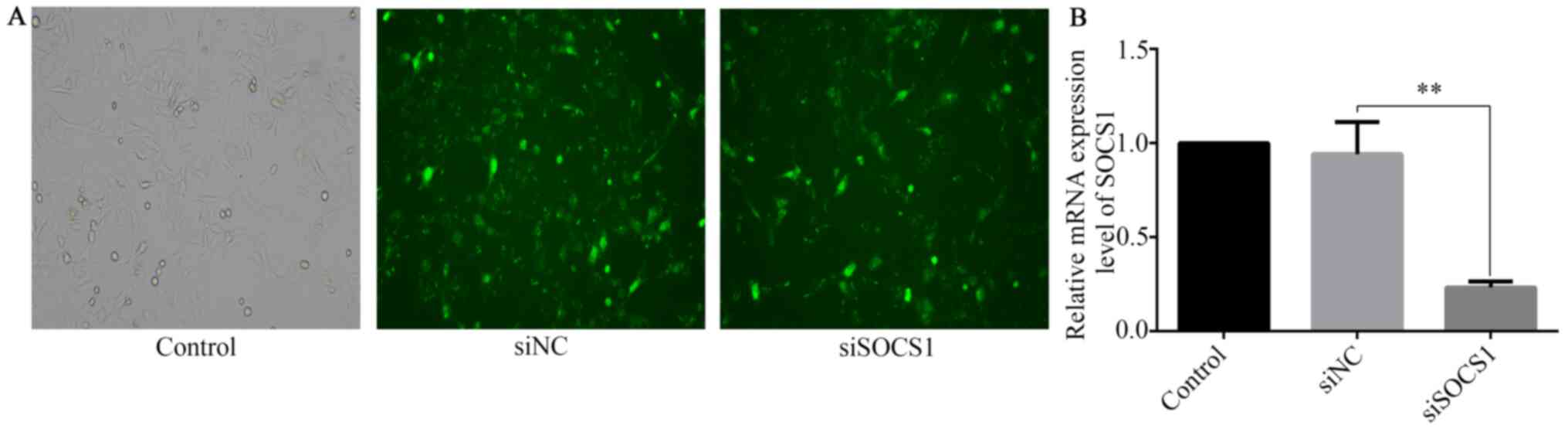
SOCS1 silencing further inhibits
IFN-γ-mediated HTR8/SVneo cell invasion
SOCS1 siRNA was transfected into HTR8/SVneo cells to
investigate the role of SOCS1 in the reduction of IFN-γ-mediated
invasion. RT-qPCR and fluorescence microscopy were used to assess
SOCS1 silencing at the transcript level. Compared with cells
transfected with siRNA negative control (siNC), the expression of
SOCS1 in SOCS1 siRNA-transfected cells was significantly reduced
(Fig. 4A and B).
The results demonstrated that IFN-γ-treated SOCS1
siRNA-transfected cells had a significantly lower migratory
capacity (Fig. 5A and B). The data showed that the invasion of
siSOCS1-transfected cells was significantly reduced in comparison
with the siNC group (Fig. 5C).
Moreover, the invasion of cells transfected with SOCS1 siRNA and
treated with IFN-γ was further reduced compared with cells
transfected with siNC and treated with IFN-γ (Fig. 5C and D). In addition, IFN-γ-treated SOCS1
siRNA-transfected cells significantly increased apoptosis compared
with that in the IFN-γ-treated siNC-transfected group (Fig. 5E and F). Concurrently, RT-qPCR (Fig. 6A) and western blotting (Fig. 6B-E) results demonstrated that
knockdown of SOCS1 reduced SOCS1 expression in HTR8+ siSOCS1 group
compared with that in the HTR + NC group, but IFN-γ intervention
increased SOCS1 expression in HTR8 + IFN-γ + siSOCS1 group compared
with that in the HTR + siSOCS1 group. Compared with the siNC group,
western blot analysis indicated that EZR and PDGFRA expressions,
which is related to cell invasion and migration respectively
(19,20), were reduced in the siSOCS1 group;
whilst the expression of cell apoptosis marker cleaved caspase3 was
increased in the siSOCS1 group (Fig.
6B and C). Furthermore, western
blot results showed that EZR and PDGFRA expressions were reduced in
the IFN-γ + siSOCS1 group compared with the IFN-γ + siNC group,
whilst the expression of cell apoptosis marker cleaved caspase3 was
increased in the IFN-γ and siSOCS1 group compared with that in the
IFN-γ and siNC group (Fig. 6D and
E). These results indicated that
IFN-γ-treated SOCS1 siRNA-transfected cells had a significantly
lower migratory capacity and increased apoptosis.
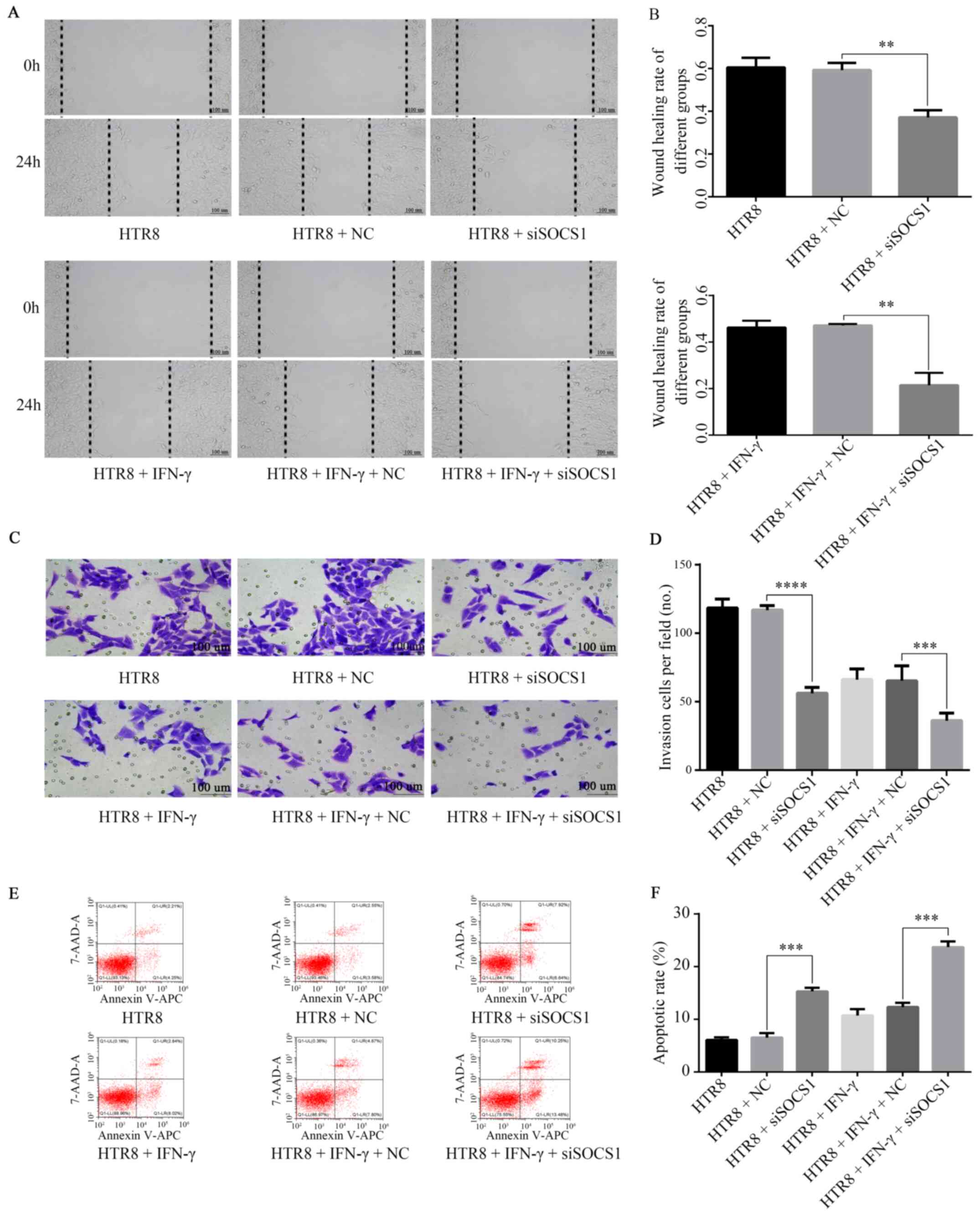 | Figure 5Role of SOCS1 in IFN-γ-mediated
reduction of HTR8/SVneo cell invasion. (A) siSOCS1 was transfected
into HTR8/SVneo cells, resulting in decreased cell migration. (B)
Following co-treatment with 100 ng/ml IFN-γ and siSOCS1, cell
migration was further reduced. **P=0.01. (C) siSOCS1 was
transfected into HTR8/SVneo cells, resulting in decreased invasive
abilities. (D) Following co-treatment with 100 ng/ml IFN-γ and
siSOCS1, cell invasion were further reduced.
***P=0.0007, ****P<0.0001. (E) Flow
cytometry results found that (F) in siSOCS1-transfected cells, the
apoptotic rate increased. ***P=0.0001. After treatment
with 100 ng/ml IFN-γ, the apoptotic rate was further increased.
***P=0.0001. SOCS1, suppressor of cytokine signaling 1;
siSOCS1, siRNA targeting SOCS1; NC, negative control. |
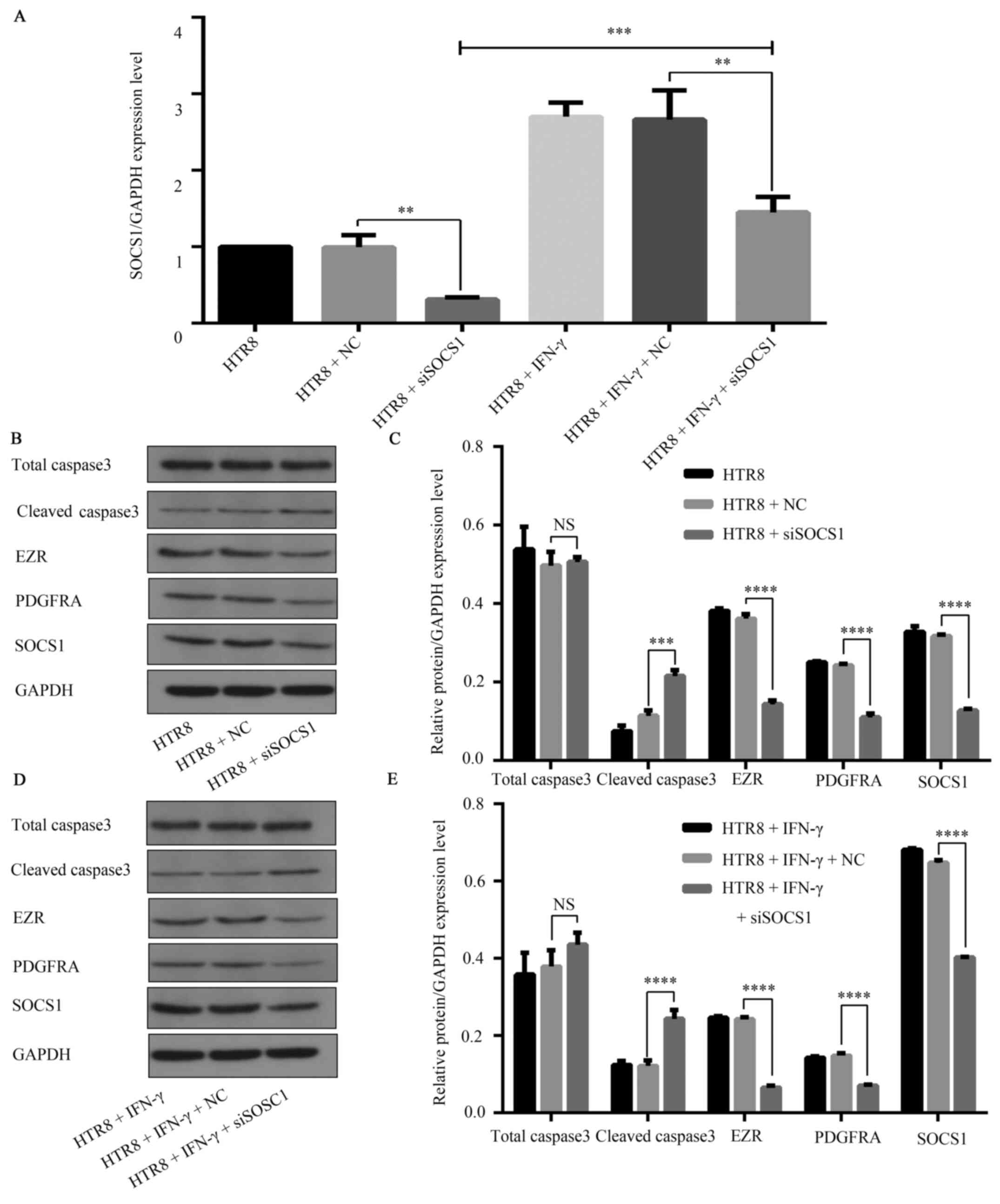 | Figure 6SOCS1 feedback regulation of the
JAK1/STAT1 signaling pathway in IFN-γ-activated HTR8/SVneo cells.
(A) siSOCS1 was transfected into HTR8/SVneo cells, resulting in
decreased SOCS1 mRNA (**P=0.0018) levels. Upon
co-treatment with 100 ng/ml IFN-γ, the mRNA levels of SOCS1
decreased compared with the NC group. **P=0.0083. The
mRNA levels increased compared with the HTR8 + siSOCS1 group.
***P=0.0007. (B) Representative western blotting images.
(C) In siSOCS1-transfected cells, the expression of the apoptosis
marker cleaved caspase3 increased (***P=0.0002), while
expression levels of the migration and invasion markers EZR
(****P<0.0001) and PDGFRA, as well as
(****P<0.0001) SOCS1 (****P<0.0001)
were decreased. (D) Representative western blotting images. (E) In
HTR8/SVneo cells transfected with siSOCS1 and 100 ng/ml IFN-γ,
Caspase3 expression was increased (****P<0.0001),
while EZR and PDGFRA expression levels were further decreased
(****P<0.0001). SOCS1 expression was lower compared
with the NC group in drug intervention
(****P<0.0001), but increased compared with siSOCS1
without drug intervention. SOCS1, suppressor of cytokine signaling
1; siSOCS1, small interfering RNA targeting SOCS1; PDGFRA,
platelet-derived growth factor receptor A; NC, negative control;
EZR, Ezrin; JAK, Janus kinase. |
Discussion
Placental trophoblast cell differentiation is a
complex process (21). Some of the
trophoblast cells differentiate into trophoblastic cells, which
have a high degree of infiltration ability and assists in
implantation (22). Following
implantation, trophoblast cells are further separated into villous
trophoblastic and EVTs. During placental implantation, EVTs migrate
into the uterus and change the blood vessels. EVT enters the lumen
of the uterine spiral arterioles and gradually replaces the smooth
muscle cells and endothelial cells of the vascular wall (23,24).
This transforms the artery from a high-resistance low-volume blood
vessel to a low-resistance high-volume blood vessel to increase the
blood flow of the placenta and ensure the normal exchange of
materials between mother and fetus (23,24).
The present study investigated the relationship between IFN-γ and
trophoblast invasion using HTR-8/SVneo cells as the in vitro
model due to its highly similar physiological phenotype with EVTs
in early pregnancy (25,26).
In the present study, treatment of HTR-8/SVneo cells
with IFN-γ reduced the invasion of HTR-8/SVneo cells, while cell
viability was not affected. Laganà et al (27) reported that endothelial progenitor
cells were significantly lower, whereas NK cells were significantly
higher in a PE group compared with uncomplicated pregnancies during
the first trimester. NK cells in the maternal endometrium produce
high amounts of IFN-γ (28).
Previous studies (29-32)
have aimed to identify precise and unique biochemical markers in
the serum to predict PE; however, there has been limited success
(33). In early pregnancy, high
levels of IFN-γ are beneficial for pregnancy (34), but excessive IFN-γ can inhibit cell
reproduction and metabolic activities, as well as induce apoptosis.
Simultaneously, during the second to third trimester of pregnancy,
IFN-γ secretion decreases, regulating the number and invasiveness
of trophoblast cells (35). There
have been reports of persistently high levels of IFN-γ secretion in
the plasma of patients with PE (36,37),
and the present clinical data also demonstrated that patients with
PE secrete higher levels of IFN-γ in the placenta. Pinheiro et
al (38) reported that elevated
levels of pro-inflammatory cytokines in the maternal circulation
with a deviation in the IL-8/IL-6 axis towards IFN-γ may drive the
cytokine network in women with PE towards an excessive systemic
inflammatory state. Collectively, these studies suggest that IFN-γ
is required during implantation, but levels above or below the
threshold may be harmful to pregnant women.
Previous studies have reported that IFN-γ can
activate JAK/STAT1 and innate and adaptive immune responses, as
well as promote apoptosis. For example, it was revealed that IFN-γ
binds to its receptor and then activates JAK1/2(39). Subsequently, cytoplasmic STAT1 is
recruited to the receptor complex and phosphorylated at Tyr701 and
Ser727(40). Following
phosphorylation, STAT1 forms a homodimer and translocates to the
nucleus where it binds to the conserved DNA sequence in the
promoter region of the downstream target gene and activates its
transcription (9). Previous studies
have shown that IFN-γ could activate the JAK/STAT pathway in
HTR-8/SVneo cells (41,42). STAT1 is phosphorylated at Ser727 and
Tyr701 residues (43). In the
present study, knockdown of STAT1 expression results in significant
increase of HTR8/SVneo cell invasion. Moreover, clinical samples
and cytological experiments indicated that IFN-γ could activate
JAK/STAT, inhibit HTR8/SVneo cell migration and invasion, promote
apoptosis, increase expression levels of p-JAK, p-STAT1 and
caspase3 and decrease expression levels of PDGFRA and EZR.
SOCS is a newly-discovered cytokine-induced protein
family composed of an N-terminal variable region, central SH-2
region and C-terminal SOCS box (44). SOCS can inhibit signal transduction
by inhibiting JAK/STAT pathways and regulating the signaling of
cytokine factors, such as INF-γ, prolactin and growth hormones
(45,46). Among the family, SOCS1 potently
suppresses cytokine actions by inhibiting JAK kinase activity
(47). While SOCS expression levels
are low under physiological conditions, these are upregulated in
response to cytokine stimulation in a number of immune and
inflammatory processes (48).
Overexpression of SOCS1 in keratinocyte clones abrogates
IFN-γ-induced expression of numerous pro-inflammatory genes and the
release of related chemokines via blocking the JAK/STAT pathway
(49). In addition, SOCS1 inhibits
JAK2 kinase activity by binding the catalytic site of JAK2, with
its kinase-inhibitory region acting as a pseudo-substrate of the
enzyme (49). Skjesol et al
(50) were the first to demonstrate
that SOCS1 is a potent inhibitor of IFN-γ-mediated JAK/STAT
signaling in teleost fish. Previous studies (51,52)
have also reported that SOCS1 is one of the most effective IFN-γ
signaling inhibitors (53).
In the present study, SOCS1 was upregulated in
patients with PE, and HTR-8/SVneo cells treated with IFN-γ also had
increased SOCS1 expression. However, the role of SOCS1 in the
regulation of trophoblast invasion remains to be elucidated.
Therefore, the present study investigated how SOCS1 in IFN-γ led to
reduced HTR-8/SVneo invasion. SOCS1 siRNA was transfected in
HTR-8/SVneo cells and the effects on cell invasion with or without
IFN-γ treatment were examined. Knockdown of SOCS1 further reduced
the invasion and migration of HTR-8/SVneo cells, reduced the
expression levels of PDGFRA and EZR, increased apoptosis and
enhanced the expression of caspase3. Therefore, the current
findings indicated that SOCS1 may negatively regulate JAK/STAT1 and
affect HTR-8 SVneo invasiveness (Fig.
7).
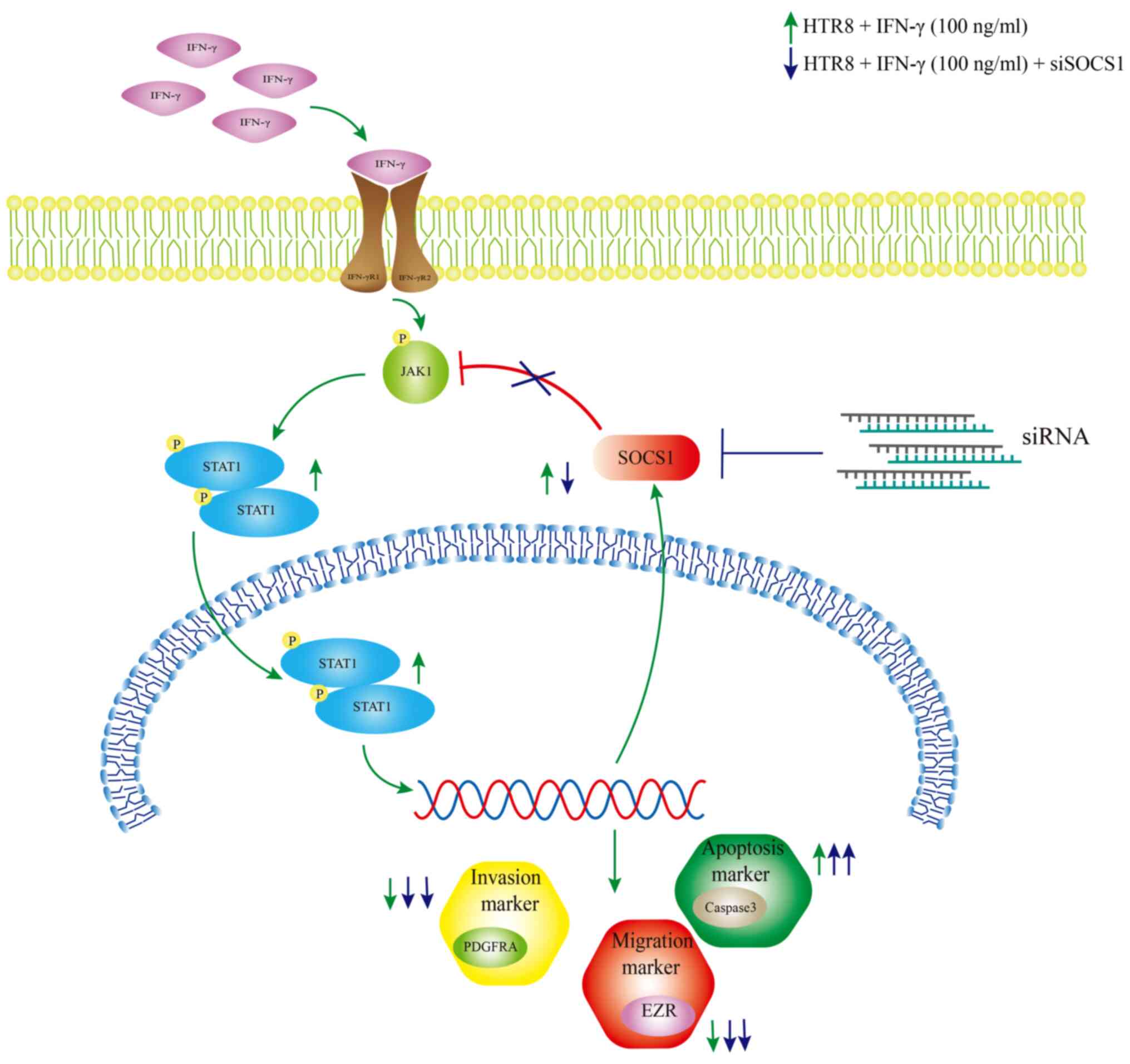 | Figure 7Schematic effect of SOCS1 feedback
regulation of IFN-γ-activated JAK1/STAT1 in HTR8/SVneo cells,
resulting in reduced cell invasion. IFN-γ increased the
phosphorylation levels of JAK1 and STAT1 in HTR8/SVneo cells,
leading to a decrease in cell invasion and migration, an increase
in apoptosis rate, decrease in PDGFRA, EZR and caspase3 protein
expression levels and an increase in SOCS1 protein expression.
Following knockdown of SOCS1, the cell invasive and migratory
abilities were further reduced, the apoptotic rate was increased,
PDGFRA and EZR protein expression levels were reduced and cleaved
caspase3 protein expression was increased. SOCS1, suppressor of
cytokine signaling 1; PDGFRA, platelet-derived growth factor
receptor A; EZR, Ezrin; p, phosphate; JAK, Janus kinase. |
There are limitations to the current study. For
instance, the present report is only a preliminary investigation
into the role of IFN-γ in PE, and additional studies are required
to further illustrate the functions and underlying mechanism of
IFN-γ in PE. For example, increasing evidence suggests that
microRNAs (miRNAs), a type of non-coding small RNA that consists of
20-26 nucleotides and regulates gene expression by targeting the
3'-untranslated region, are involved in various pregnancy-related
disorders, including PE and fetal growth restriction (53,54).
Thus, would be worth investigating whether miRNAs are in involved
in the regulation of the IFN-γ/SOCS1/JAK/STAT1 feedback loop.
Moreover, additional siRNAs for SOCS1 could be used to exclude
off-target effects in future studies. Finally, the present study
only conducted experiments on HTR-8/SVneo cells and clinical tissue
samples. Since HTR-8/SVneo is different from primary EVTs, the
pathogenesis of IFN-γ and PE requires further research.
In conclusion, IFN-γ reduced the invasion of
HTR-8/SVneo cells by activating JAK/STAT1, leading to an increase
in SOCS1 expression, which negatively regulated JAK/STAT1 and
eliminated the pro-inflammatory effects of IFN-γ, thus forming a
feedback loop. These findings could explain the higher expression
of IFN-γ and SOCS1 in the placental tissue of patients with PE
compared with the healthy control group.
Acknowledgements
Not applicable.
Funding
This work was generously sponsored by Shanxi
Provincial Science and Technology Department of the Shanxi Province
Applied Basic Research Program (grant no. 201801D121322) and
Beijing Municipal Administration of Hospitals Clinical Medicine
Development of Special Funding-YangFan Project (grant no.
ZYLX201713).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL and CL conceived and designed the present study.
HL and WW performed the experiments and analyzed the data. HL and
CL interpreted the data and wrote the manuscript. All authors read
and approved the final manuscript and agree to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Second Hospital of Shanxi Medical University.
Written informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Burton GJ, Redman CW, Roberts JM and
Moffett A: Pre-eclampsia: Pathophysiology and clinical
implications. BMJ. 366(l2381)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bujold E, Chaillet N and Kingdom J:
Placental growth factor testing for suspected pre-eclampsia.
Lancet. 393:1775–1776. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Nakashima A, Yamanaka-Tatematsu M, Fujita
N, Koizumi K, Shima T, Yoshida T, Nikaido T, Okamoto A, Yoshimori T
and Saito S: Impaired autophagy by soluble endoglin, under
physiological hypoxia in early pregnant period, is involved in poor
placentation in preeclampsia. Autophagy. 9:303–316. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ananth Cande V, Keyes Katherine M and
Wapner Ronald J: Pre-eclampsia rates in the United States,
1980-2010: Age-period-cohort analysis. BMJ.
347(f6564)2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Stevens W, Shih T, Incerti D, Ton TGN, Lee
HC, Peneva D, Macones GA, Sibai BM and Jena AB: Short-term costs of
preeclampsia to the United States health care system. Am J Obstet
Gynecol. 217:237–248.e16. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Murphy SP, Tayade C, Ashkar AA, Hatta K,
Zhang J and Croy BA: Interferon gamma in successful pregnancies.
Biol Reprod. 80:848–859. 2009.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lash GE, Otun HA, Innes BA, Kirkley M, De
Oliveira L, Searle RF, Robson SC and Bulmer JN: Interferon-gamma
inhibits extravillous trophoblast cell invasion by a mechanism that
involves both changes in apoptosis and protease levels. FASEB J.
20:2512–2518. 2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lockwood CJ, Basar M, Kayisli UA,
Guzeloglu-Kayisli O, Murk W, Wang J, De Paz N, Shapiro JP, Masch
RJ, Semerci N, et al: Interferon-gamma protects first-trimester
decidual cells against aberrant matrix metalloproteinases 1, 3, and
9 expression in preeclampsia. Am J Pathol. 184:2549–2559.
2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Midgley AC, Morris G, Phillips AO and
Steadman R: 17β-estradiol ameliorates age-associated loss of
fibroblast function by attenuating IFN-γ/STAT1-dependent miR-7
upregulation. Aging Cell. 15:531–541. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Stark GR: How cells respond to interferons
revisited: From early history to current complexity. Cytokine
Growth Factor Rev. 18:419–423. 2007.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Verma S, Pal R and Gupta SK: Decrease in
invasion of HTR-8/SVneo trophoblastic cells by interferon gamma
involves cross-communication of STAT1 and BATF2 that regulates the
expression of JUN. Cell Adh Migr. 12:432–446. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Nita-Lazar M, Banerjee A, Feng C and Vasta
GR: Galectins regulate the inflammatory response in airway
epithelial cells exposed to microbial neuraminidase by modulating
the expression of SOCS1 and RIG1. Mol Immunol. 68:194–202.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yoshimura A, Naka T and Kubo M: SOCS
proteins, cytokine signalling and immune regulation. Nat Rev
Immunol. 7:454–465. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Seif F, Khoshmirsafa M, Aazami H,
Mohsenzadegan M, Sedighi G and Bahar M: The role of JAK-STAT
signaling pathway and its regulators in the fate of T helper cells.
Cell Commun Signal. 15(23)2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Poehlmann TG, Busch S, Mussil B, Winzer H,
Weinert J, Mebes I, Schaumann A, Fitzgerald JS and Markert UR: The
possible role of the Jak/STAT pathway in lymphocytes at the
fetomaternal interface. Chem Immunol Allergy. 89:26–35.
2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li Y, Chu N, Rostami A and Zhang GX:
Dendritic cells transduced with SOCS-3 exhibit a tolerogenic/DC2
phenotype that directs type 2 Th cell differentiation in vitro and
in vivo. J Immunol. 177:1679–1688. 2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Endo TA, Masuhara M, Yokouchi M, Suzuki R,
Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H,
et al: A new protein containing an SH2 domain that inhibits JAK
kinases. Nature. 387:921–924. 1997.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gerashchenko TS, Zolotaryova SY, Kiselev
AM, Tashireva LA, Novikov NM, Krakhmal NV, Cherdyntseva NV,
Zavyalova MV, Perelmuter VM and Denisov EV: The activity of KIF14,
Mieap, and EZR in a new type of the invasive component,
torpedo-like structures, predetermines the metastatic potential of
breast cancer. Cancers (Basel). 12(1909)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sun T, Yin L and Kuang H: miR-181a/b-5p
regulates human umbilical vein endothelial cell angiogenesis by
targeting PDGFRA. Cell Biochem Funct. 38:222–230. 2020.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Turco MY, Gardner L, Kay RG, Hamilton RS,
Prater M, Hollinshead MS, McWhinnie A, Esposito L, Fernando R,
Skelton H, et al: Trophoblast organoids as a model for
maternal-fetal interactions during human placentation. Nature.
564:263–267. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Choudhury RH, Dunk CE, Lye SJ, Aplin JD,
Harris LK and Jones RL: Extravillous trophoblast and endothelial
cell crosstalk mediates leukocyte infiltration to the early
remodeling decidual spiral arteriole wall. J Immunol.
198:4115–4128. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Knöfler M, Haider S, Saleh L, Pollheimer
J, Gamage TKJB and James J: Human placenta and trophoblast
development: Key molecular mechanisms and model systems. Cell Mol
Life Sci. 76:3479–3496. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang Y and Zhao S: Placental Blood
Circulation. In: Vascular Biology of the Placenta. San Rafael (ed).
Chapter 2. Morgan & Claypool Life Sciences, pp3-11, 2010.
|
|
25
|
Chen Y, Zhang Y, Deng Q, Shan N, Peng W,
Luo X, Zhang H, Baker PN, Tong C and Qi H: Wnt5a inhibited human
trophoblast cell line HTR8/SVneo invasion: Implications for early
placentation and preeclampsia. J Matern Fetal Neonatal Med.
29:3532–3538. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Malik A, Pal R and Gupta SK:
Interdependence of JAK-STAT and MAPK signaling pathways during
EGF-mediated HTR-8/SVneo cell invasion. PLoS One.
12(e0178269)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Laganà AS, Giordano D, Loddo S, Zoccali G,
Vitale SG, Santamaria A, Buemi M and D'Anna R: Decreased
endothelial progenitor cells (EPCs) and increased Natural Killer
(NK) cells in peripheral blood as possible early markers of
preeclampsia: A case-control analysis. Arch Gynecol Obstet.
295:867–872. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Guleria I and Pollard JW: The trophoblast
is a component of the innate immune system during pregnancy. Nat
Med. 6:589–593. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Szarka A, Rigó J Jr, Lázár L, Beko G and
Molvarec A: Circulating cytokines, chemokines and adhesion
molecules in normal pregnancy and preeclampsia determined by
multiplex suspension array. BMC Immunol. 11(59)2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Nicolaides KH, Bindra R, Turan OM, Chefetz
I, Sammar M, Meiri H, Tal J and Cuckle HS: A novel approach to
first-trimester screening for early pre-eclampsia combining serum
PP-13 and Doppler ultrasound. Ultrasound Obstet Gynecol. 27:13–7.
2006.PubMed/NCBI View
Article : Google Scholar
|
|
31
|
Molvarec A, Rigó J Jr, Lázár L, Balogh K,
Makó V, Cervenak L, Mézes M and Prohászka Z: Increased serum
heat-shock protein 70 levels reflect systemic inflammation,
oxidative stress and hepatocellular injury in preeclampsia. Cell
Stress Chaperones. 14:151–159. 2009.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Molvarec A, Kalabay L, Derzsy Z, Szarka A,
Halmos A, Stenczer B, Arnaud P, Karádi I, Prohászka Z and Rigó J
Jr: Preeclampsia is associated with decreased serum alpha(2)-HS
glycoprotein (fetuin-A) concentration. Hypertens Res. 32:665–669.
2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Laganà AS, Favilli A, Triolo O, Granese R
and Gerli S: Early serum markers of pre-eclampsia: Are we stepping
forward? J Matern Fetal Neonatal Med. 29:3019–3023. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Laird SM, Tuckerman EM, Cork BA, Linjawi
S, Blakemore AI and Li TC: A review of immune cells and molecules
in women with recurrent miscarriage. Hum Reprod Update. 9:163–174.
2003.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Banerjee S, Smallwood A, Moorhead J,
Chambers AE, Papageorghiou A, Campbell S and Nicolaides K:
Placental expression of interferon-gamma (IFN-gamma) and its
receptor IFN-gamma R2 fail to switch from early hypoxic to late
normotensive development in preeclampsia. J Clin Endocrinol Metab.
90:944–952. 2005.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Santner-Nanan B, Peek MJ, Khanam R,
Richarts L, Zhu E, Fazekas de St Groth B and Nanan R: Systemic
increase in the ratio between Foxp3+ and IL-17-producing
CD4+ T cells in healthy pregnancy but not in
preeclampsia. J Immunol. 183:7023–7030. 2009.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Laresgoiti-Servitje E, Gómez-López N and
Olson DM: An immunological insight into the origins of
pre-eclampsia. Hum Reprod Update. 16:510–524. 2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Pinheiro MB, Martins-Filho OA, Mota AP,
Alpoim PN, Godoi LC, Silveira AC, Teixeira-Carvalho A, Gomes KB and
Dusse LM: Severe preeclampsia goes along with a cytokine network
disturbance towards a systemic inflammatory state. Cytokine.
62:165–173. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Aota K, Yamanoi T, Kani K and Azuma M:
Cepharanthine inhibits IFN-gamma-induced CXCL10 by suppressing the
JAK2/STAT1 signal pathway in human salivary gland ductal cells.
Inflammation. 41:50–58. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Coombs MR, Harrison ME and Hoskin DW:
Apigenin inhibits the inducible expression of programmed death
ligand 1 by human and mouse mammary carcinoma cells. Cancer Lett.
380:424–433. 2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Verma S, Kang AK, Pal R and Gupta SK: BST2
regulates interferon gamma-dependent decrease in invasion of
HTR-8/SVneo cells via STAT1 and AKT signaling pathways and
expression of E-cadherin. Cell Adh Migr. 14:24–41. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Qin S, Zhang Y, Zhang J, Tian F, Sun L, He
X, Ma X, Zhang J, Liu XR, Zeng W and Lin Y: SPRY4 regulates
trophoblast proliferation and apoptosis via regulating
IFN-γ-induced STAT1 expression and activation in recurrent
miscarriage. Am J Reprod Immunol. 83(e13234)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Nguyen H, Ramana CV, Bayes J and Stark GR:
Roles of phosphatidylinositol 3-kinase in
interferon-gamma-dependent phosphorylation of STAT1 on serine 727
and activation of gene expression. J Biol Chem. 276:33361–33368.
2001.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Yu CR, Mahdi RM, Ebong S, Vistica BP, Gery
I and Egwuagu CE: Suppressor of cytokine signaling 3 regulates
proliferation and activation of T-helper cells. J Biol Chem.
278:29752–29759. 2003.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Bachmann J, Raue A, Schilling M, Böhm ME,
Kreutz C, Kaschek D, Busch H, Gretz N, Lehmann WD, Timmer J and
Klingmüller U: Division of labor by dual feedback regulators
controls JAK2/STAT5 signaling over broad ligand range. Mol Syst
Biol. 7(516)2011.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lindemann C, Hackmann O, Delic S, Schmidt
N, Reifenberger G and Riemenschneider MJ: SOCS3 promoter
methylation is mutually exclusive to EGFR amplification in gliomas
and promotes glioma cell invasion through STAT3 and FAK activation.
Acta Neuropathol. 122:241–251. 2011.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Yasukawa H, Nagata T, Oba T and Imaizumi
T: SOCS3: A novel therapeutic target for cardioprotection. JAKSTAT.
1:234–240. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Liang Y, Xu WD, Peng H, Pan HF and Ye DQ:
SOCS signaling in autoimmune diseases: Molecular mechanisms and
therapeutic implications. Eur J Immunol. 44:1265–1275.
2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Doti N, Scognamiglio PL, Madonna S,
Scarponi C, Ruvo M, Perretta G, Albanesi C and Marasco D: New
mimetic peptides of the kinase-inhibitory region (KIR) of SOCS1
through focused peptide libraries. Biochem J. 443:231–240.
2012.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Skjesol A, Liebe T, Iliev DB, Thomassen
EI, Tollersrud LG, Sobhkhez M, Lindenskov Joensen L, Secombes CJ
and Jørgensen JB: Functional conservation of suppressors of
cytokine signaling proteins between teleosts and mammals: Atlantic
salmon SOCS1 binds to JAK/STAT family members and suppresses type I
and II IFN signaling. Dev Comp Immunol. 45:177–189. 2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Alexander WS, Starr R, Fenner JE, Scott
CL, Handman E, Sprigg NS, Corbin JE, Cornish AL, Darwiche R,
Owczarek CM, et al: SOCS1 is a critical inhibitor of interferon
gamma signaling and prevents the potentially fatal neonatal actions
of this cytokine. Cell. 98:597–608. 1999.PubMed/NCBI View Article : Google Scholar
|
|
52
|
DiGiandomenico A, Wylezinski LS and
Hawiger J: Intracellular delivery of a cell-penetrating SOCS1 that
targets IFN-gamma signaling. Sci Signal. 2(ra37)2009.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Laganà AS, Vitale SG, Sapia F, Valenti G,
Corrado F, Padula F, Rapisarda AMC and D'Anna R: miRNA expression
for early diagnosis of preeclampsia onset: Hope or hype? J Matern
Fetal Neonatal Med. 31:817–821. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Chiofalo B, Laganà AS, Vaiarelli A, La
Rosa VL, Rossetti D, Palmara V, Valenti G, Rapisarda AMC, Granese
R, Sapia F, et al: Do miRNAs play a role in fetal growth
restriction? A fresh look to a busy corner. Biomed Res Int.
2017(6073167)2017.PubMed/NCBI View Article : Google Scholar
|