Introduction
Glucocorticoids (GCs), which have anti-inflammatory
and immune-suppressing properties, are commonly used in the
treatment of a number of adult dyspnea syndromes (such as atypical
pneumonia), asthma, autoimmune diseases and cardiopulmonary-related
inflammation (1,2). However, high doses or prolonged use of
GCs can cause many side effects (3-6).
For muscle tissue, the main side effect caused by GCs is steroid
myopathy, which is characterized by muscle weakness and atrophy
(1,7,8).
Glucocorticoid-induced skeletal muscle atrophy (GIMA) increases the
disease risk for patients, leading to weakened immunity, increased
infection rates, limited mobility, fracture and even paralysis
(9,10). Severe systemic muscle atrophy can
lead to swallowing disorders, breathing difficulty and other
secondary life-threatening complications (5,11). The
diseases induced by skeletal muscle atrophy seriously affect the
quality of life and prognosis of patients. At present, the
pathogenesis of GIMA is not fully understood, but some researchers
believe that inhibition of protein anabolism or stimulation of
protein catabolism is responsible for skeletal muscle atrophy
(12,13), GCs can downregulate myogenic
regulatory factors, thereby inhibiting proliferation and
differentiation processes (1,10). GCs
can activate skeletal muscle proteolysis through mechanisms such as
the muscle-specific ubiquitin proteasome system, lysosomal system
and calcium-dependent protease system (1). In addition, GCs induce muscular
atrophy by affecting the production of growth factors such as
insulin-like growth factor (10).
Therefore, researchers are also aiming to explore the possibility
of treating GIMA by blocking the mechanism of GC-induced muscular
atrophy. For example, Bodine et al (14) demonstrated that the IGF1/PI3K/Akt
pathway is sufficient to induce Myotube hypertrophy by activating
the protein synthesis pathway. Stitt et al (15) indicated that inhibiting the
activation of the muscular atrophy pathway can inhibit the
upregulation of muscle RING finger protein 1 (MuRF1) and muscle
atrophy F-box (MAFbx) induced by glucocorticoid. Despite recent
significant advances in the understanding of the mechanism of GIMA
pathogenesis, the mechanism by which GCs are expressed through
atrophy-related genes, particularly long non-coding RNAs (lncRNAs),
has not been fully described.
MicroRNAs (miRNAs) and lncRNAs are both non-coding
RNAs; however, more attention has been paid to the role of miRNAs
in skeletal muscle development (11,16-19).
miRNAs are a class of small, non-coding RNAs that are ~22
nucleotides in length. The mechanism of miRNA regulation of muscle
atrophy mainly involves muscle protein metabolism, muscle
regeneration, angiogenesis and muscle cell apoptosis (20). miRNAs that are expressed
specifically in muscles can alter the diseases that affect muscles
(17,21-25).
For example, in dexamethasone (DEX)-mediated muscular atrophy,
muscle-specific miR-1 is induced and heat shock protein 70 (HSP70)
levels are reduced (26). In
addition, miR-23a is inhibited in diabetes and DEX-induced muscular
atrophy (27). LncRNAs are a novel
class of non-coding RNAs that are >200 nucleotides in length
(28,29). Muscle-specific lncRNAs are important
regulators of muscle proliferation, differentiation and atrophy
(26,28-30).
Long intergenic non-protein coding RNA muscle differentiation 1
(linc-MD1) regulates the expression of mastermind-like
transcriptional coactivator 1 and myocyte enhancer factor 2C during
muscle differentiation (31). The
lncRNA muscle anabolic regulator 1 (MAR1) acts as a miR-487b sponge
to regulate Wnt5a protein, resulting in muscle differentiation and
regeneration (12). Given that
aberrant gene expression underlies muscle atrophy, it is important
to understand how lncRNAs regulate gene expression in response to
diverse stresses or diseases that contribute to muscle atrophy
(10,28,32).
A growing number of studies have demonstrated that
the genetic hierarchies and transcriptional networks involved in
myogenesis include lncRNA molecules (31,33-36).
However, few studies have provided a comprehensive perspective on
the regulation of miRNAs and lncRNAs in GC-induced skeletal muscle
atrophy. Therefore, the aim of the current study was to investigate
the expression of regulatory miRNAs and lncRNAs in GC-induced
muscular atrophy within C2C12 cells. DEX is the most effective
synthetic GC and conferred anti-inflammatory GC activity compared
to natural cortisol and corticosterone (37). DEX has the potential to promote
protein degradation and is considered an effective drug to induce
muscle atrophy in vivo and in vitro (38-40).
In the present study, the expression of selected miRNAs and lncRNAs
that exhibited expression profiles similar to those previously
reported (16,18,19,26,30,31,36),
between control and DEX-treated C2C12 cells was investigated, and
miRNAs and lncRNAs that were differentially expressed between
normal (control) and atrophy conditions were identified. The
results from the present study may provide candidate miRNAs and
lncRNAs that may lead to a better understanding of the molecular
pathways by which GCs regulate skeletal muscular atrophy.
Materials and methods
Cell culture
The mouse myoblast C2C12 cell line was purchased
from the Stem Cell Bank of the Type Culture Collection of the
Chinese Academy of Sciences and cultured at 37˚C in 5%
CO2 and high glucose DMEM (cat. no. 12100046; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% (v/v) FBS
(cat. no. SH30070.03; HyClone; Cytiva). For myotube
differentiation, C2C12 myoblasts were incubated at 37˚C in 12-well
plates with 2% horse serum (cat. no. SH30074.03; HyClone; Cytiva)
for 72 h, according to previous studies (8,41).
DEX-induced muscle atrophy cell
model
C2C12 cells were cultured to 70-80% confluence with
high glucose DMEM (cat. no. 12100046; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% (v/v) FBS (cat. no.
SH30070.03; HyClone; Cytiva), before being digested with trypsin.
In total, 5x104 cells were then seeded into the a
12-well plate, until they reached 80-90% confluence, following
which they were differentiated by incubation in high glucose DMEM
containing 2% horse serum (cat. no. SH30074.03; HyClone; Cytiva)
(28). DEX-induced atrophy was
performed by treating cells on the 3rd day of differentiation with
50, 100 and 200 µM DEX (Sigma-Aldrich; Merck KGaA) dissolved in
ethanol for 12, 24 and 48 h. Control cells were incubated with
0.03% (v/v) ethanol (control) for 48 h. The cell medium was
exchanged every 24 h (42). All
incubations were performed at 37˚C.
RNA extraction and
reverse-transcription quantitative (RT-q) PCR
DEX-treated C2C12 and control cell samples were
washed with phosphate-buffered saline before lysis in
TRIzol® reagent (cat. no. 15596018; Thermo Fisher
Scientific, Inc.), and total RNA was extracted according to the
manufacturer's instructions. Total RNA was reverse transcribed into
complementary DNA (cDNA) at 37˚C for 15 min and 85˚C for 5 sec
using PrimeScript™ RT Master mix (cat. no. RR 036A; Takara Bio,
Inc.) for mRNA and lncRNA detection. At the same time, total RNA
was reverse transcribed into cDNA using the Mir X™ miRNA First
Strand Synthesis kit (cat. no. 638313; Takara Bio, Inc.) to detect
miRNA. All the RNAs (mRNA, lncRNAs and miRNAs) were measured using
TB Green™ Premix Ex Taq™ II (cat. no. RR820A; Takara Bio, Inc.).
The PCR reaction was completed on a StepOnePlus Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.)
according to the following program: Denaturation at 95˚C for 30
sec, followed by 40 cycles of denaturation at 95˚C for 5 sec,
annealing at 55˚C for 30 sec and extension at 72˚C for 30 sec. The
PCR for detecting miRNAs was performed as follows: Denaturation at
95˚C for 10 sec, followed by 40 cycles of denaturation at 95˚C for
5 sec, and annealing and extension at 60˚C for 30 sec. The
2-ΔΔCq method was used to calculate the relative fold
change among biological groups using 18S as an internal normalizer
for mRNA and lncRNA, and U6 as an internal normalizer for miRNA
(43). The list of primers and
sequences is provided in Tables I
and II.
 | Table IPrimers for miRNAs. |
Table I
Primers for miRNAs.
| miRNA | Sequence
(5'-3') |
|---|
| miR-133a |
TTTGGTCCCCTTCAACCAGC |
| miR-133b |
GGTCCCCTTCAACCAGCTA |
| miR-206-3p |
GGAATGTAAGGAAGTGTGTGG |
| miR-18a |
GCCATCTAGTGCAGATAGAAAA |
| miR-186 |
GAATTCTCCTTTTGGGCTAAAA |
| miR-1a-3p |
TGGAATGTAAAGAAGUATGTAT |
| miR-23a-3p |
ATCACATTGCCAGGGATTTCC |
| miR-27b |
TTCACAGTGGCTAAGTTCTGC |
| miR-29b-3p |
TAGCACCATTTGAAATCAGTGTT |
| U6 forward |
GGAACGATACAGAGAAGATTAGC |
| U6 reverse |
TGGAACGCTTCACGAATTTGCG |
 | Table IIPrimers for mRNAs and lncRNAs. |
Table II
Primers for mRNAs and lncRNAs.
| Gene | Forward primer
(5'-3') | Reverse primer
(5'-3') |
|---|
| Atrogin-1 |
GAGTGGCATCGCCCAAAAGA |
TCTGGAGAAGTTCCCGTATAAGT |
| MURF1 |
GTGTGAGGTGCCTACTTGCTC |
GCTCAGTCTTCTGTCCTTGGA |
| Atrolnc-1 |
CAGCTGCCTACACCTGAAGA |
AGGGCTCGCAGATTACACC |
| Dum |
CACAAAGACAGGCAGACAGAC |
TACCAAGCAGGTTCCTACGG |
| MAR1 |
CCAAAGGACTGTCTTGGAACA |
AACAGCACTGAGCAGGGAC |
| linc-MD1 |
AGTGATTGAGGTGGACAGAAGG |
CCCATTGAGGAGCATAGAACC |
| Myolinc |
CGGTGCTATGGTTCTGATCG |
TATGTGGGAAATACAGGGACA |
| lncMyoD |
ACCCAAGGCAAGAAAAGTAGCA |
ACTCACGAGTCAGCGGCAGAAC |
| lnc-mg |
CTGCATCACGGAAGGAGATA |
AACAATCCATCCTCATTGGC |
| 18S |
GTAACCCGTTGAACCCCATT |
CCATCCAATCGGTAGTAGCG |
Myotube area measurements
Myotube area was quantified by analyzing the area of
myotubes covering the culture area. Images were acquired at a
magnification of x400 using an optical electron microscope (Thermo
Fisher Scientific, Inc.). The myotube area was measured using
ImageJ (v1.44P; National Institutes of Health) software from
randomly selected areas of the myotubes in the control group from
three different wells and three different DEX treatments. A total
of 40 myotubes were measured in each well.
Western blot analysis
Western blot analysis was performed to determine
protein levels of MURF1 and Atrogin-1 in myotubes. The C2C12 cells
were lysed in RIPA buffer containing protease inhibitor (Beyotime
Institute of Biotechnology) and phenylmethylsulfonyl fluoride to
extract total proteins. A total of 20 µg of protein, measured using
the bicinchoninic acid Protein Assay Kit (cat. no. P0010S; Beyotime
Institute of Biotechnology) was separated using 10-12% SDS-PAGE.
The protein was transferred to a PVDF membrane and blocked by 5%
skimmed milk powder at room temperature for 1 h. MURF1 (1:1,000;
cat. no. ab77577; Abcam), Atrogin-1 (1:2,000; cat. no. ab168372;
Abcam) or tubulin-targeted primary antibody (1:5,000; cat. no.
AC021; ABclonal Biotech Co., Ltd.) was added and incubated
overnight at 4˚C. After washing the membrane five times, the
membrane was incubated with horseradish peroxidase (HRP)-conjugated
secondary antibody Goat Anti-Rabbit IgG (1:10,000; cat. no. as014;
ABclonal Biotech, Co., Ltd.) at room temperature for 1 h. After
washing the membrane five times, Immobilon western chemilum HRP
substrate (cat. no. WBKlS0100; EMD Millipore) by 5% non-fat
powdered milk was used for detection, and a Tanon series automatic
chemiluminescence imaging analysis system (Tanon-V8 Pro; Tanon
Sciences and Technology Co., Ltd.) was used for scanning the image.
Tubulin was used as the internal control, and the relative
expression of the target protein band was compared with the
internal control.
Statistical analysis
GraphPad Prism 5.0 (GraphPad Software, Inc.) was
used to perform statistical analyses. The experimental data were
represented by the mean ± SD from triplicate data, and the
Student's t-test was applied for the comparison of two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
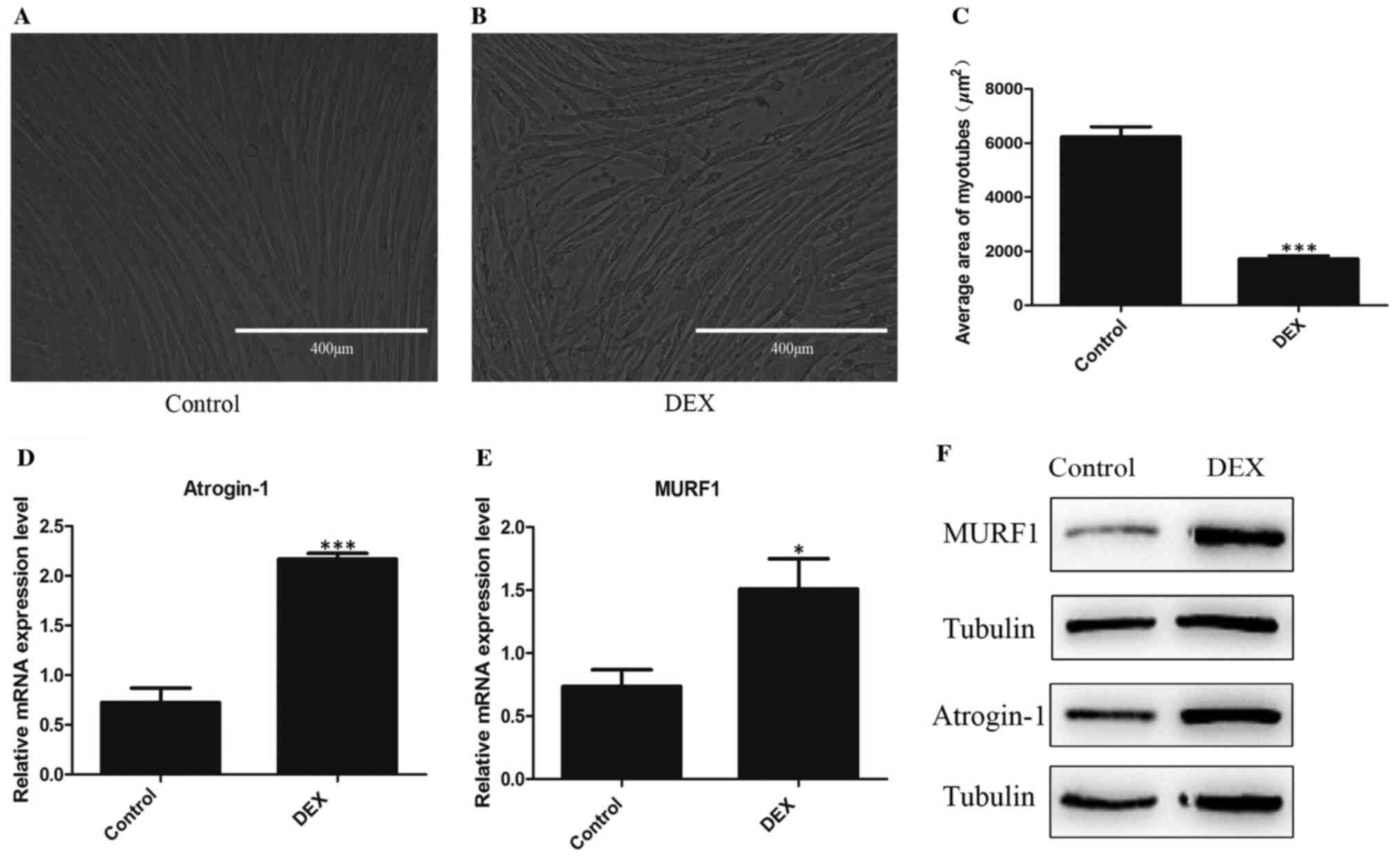
Establishment of the DEX-induced
muscle atrophy cell model
To establish an appropriate in vitro model,
preliminary experiments were conducted by treating cells on day 3
of differentiation with 50, 100 and 200 µM DEX dissolved in ethanol
for 12, 24 and 48 h. Under the treatment conditions aforementioned,
compared with other doses and time points, the mean area of C2C12
myotubes and the protein expression levels of MURF1 and Atrogin-1
in C2C12 after treatment with 100 µM DEX for 48 h were
significantly different, with the mean area of C2C12 myotubes
reduced decreased and the expression of MURF1 and Atrogin-1
increased compared with those in control. Therefore, in the present
study, the concentration of 100 µM DEX and the time point of 48 h
were used to construct the muscular atrophy model (data not shown).
Cells were treated on the 3rd day of differentiation with 100 µM
DEX dissolved in ethanol for 48 h to construct the muscular atrophy
model. The results of optical microscopy images indicated that the
average area of C2C12 myotubes in control group was higher compared
with the DEX group (Fig. 1A-C).
RT-qPCR and western blot analysis revealed that the transcription
and protein expression of MURF1 and Atrogin-1 in DEX-treated
myotubes were increased (Fig. 1D
and E), suggesting that GC-induced
muscle atrophy of C2C12 cells was successfully established.
miRNA expression patterns in atrophic
C2C12 cells
RT-qPCR was used to determine the expression of
multiple miRNAs that have previously been revealed to be associated
with muscle development in DEX-treated and control C2C12 cells.
miRNAs that were previously reported in the literature were
identified and nine miRNAs [miR-133a (19), miR-133b (18,44),
miR-206(45), miR-18a (46), miR-186(47), miR-1a-3p (21,48),
miR-23a-3p (27,49), miR-27b (50), miR-29b-3p (16)] were selected for examination in the
present study, which exhibited expression similar to that
previously reported. The results indicated that the expression of
miR-133a, miR-133b and miR-206 was significantly increased in the
DEX-treated group compared with the control group (Fig. 2A-C), while the abundance of the
other six miRNAs (miR-1a-3p, miR-186, miR-18a, miR-23a-3p, miR-27b
and miR-29b-3p) showed no significant change between DEX-treated
group and the control group (Fig.
2C-I).
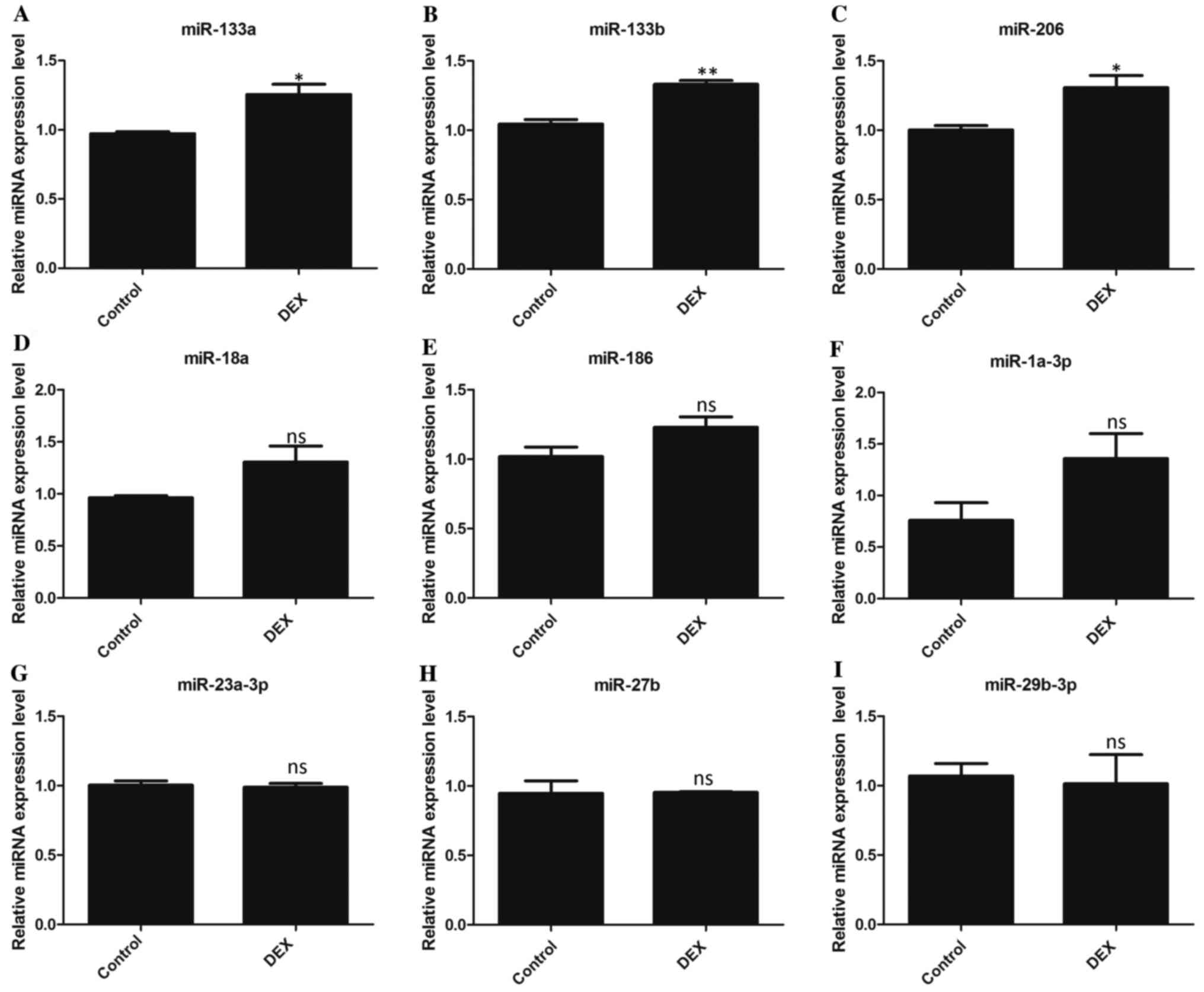 | Figure 2miRNA expression patterns in atrophic
C2C12 cells. The expression levels of miRNAs (A) miR-133a, (B)
miR-133b, (C) miR-206, (D) miR-18a, (E) miR-186, (F) miR-1a-3p, (G)
miR-23a-3p, (H) miR-27b and (I) miR-29b-3p were detected in control
C2C12 cells and DEX-induced muscle atrophy C2C12 cells. All data
are presented as mean ± SD. *P<0.05 or
**P<0.01 vs. control; n=3 per group. microRNA, miRNA
or miR; DEX, dexamethasone; ns, not significant. |
LncRNA expression patterns in atrophic
C2C12 cells
RT-qPCR was used to detect the expression of
multiple lncRNAs related to muscle development and determine if the
expression differed between DEX-treated and control C2C12 cells.
The results revealed that the expression of four lncRNAs
(Atrolnc-1, Dum, MAR1 and linc-MD1) significantly increased in the
DEX-treated group compared with the control group (Fig. 3A-D). However, the expression of
Myolinc in the DEX group was significantly decreased (Fig. 3E). LncMyoD and lnc-mg showed no
statistical difference between the two groups (Fig. 3F, G).
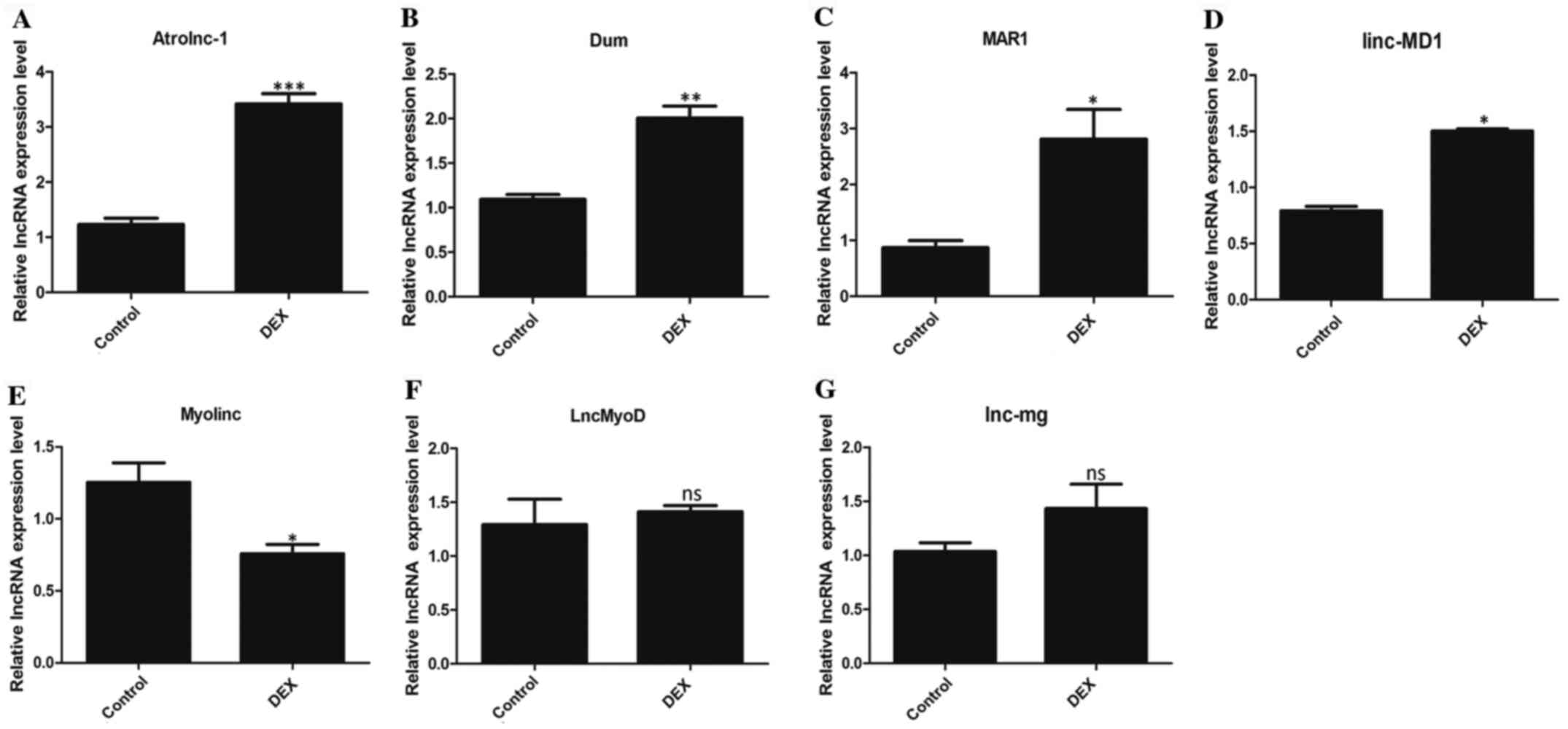 | Figure 3LncRNA expression patterns in
atrophic C2C12 cells. The expression levels of lncRNAs (A)
Atrolnc-1, (B) Dum, (C) MAR1, (D) linc-MD1, (E) Myolinc, (F)
LncMyoD and (G) lnc-mg were detected in control C2C12 cells and
DEX-induced muscle atrophy C2C12 cells. All data are presented as
mean ± SD. *P<0.05, **P<0.01 or
***P<0.001 vs. control; n=3 per group. LncRNA, long
non-coding RNA; DEX, dexamethasone; ns, not significant; linc-MD1,
long intergenic non-protein coding RNA muscle differentiation
1. |
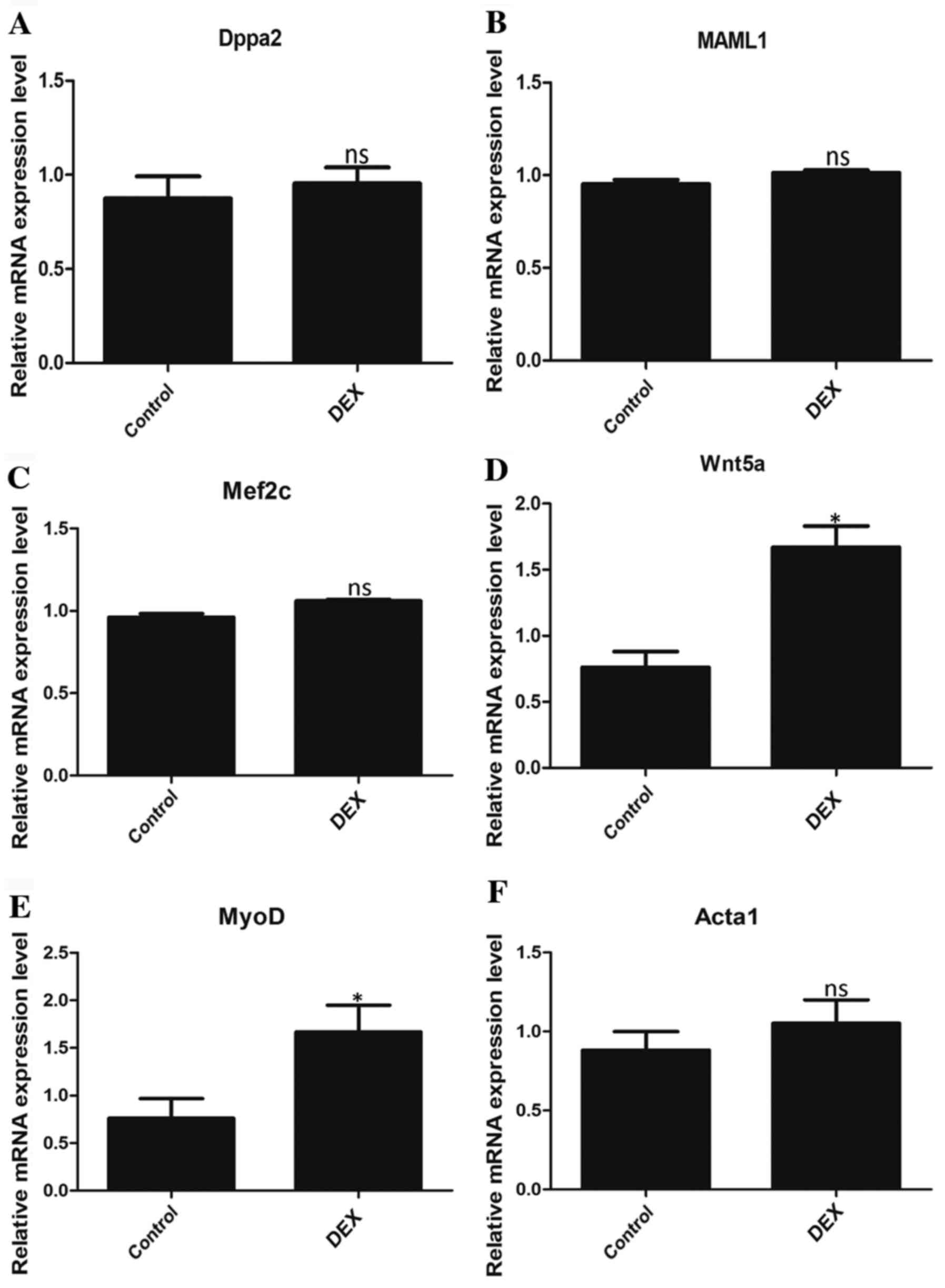
mRNA expression levels of the
downstream targets of differentially expressed lncRNAs
To further determine whether the differentially
expressed lncRNAs identified are involved in the regulation of
DEX-induced muscular atrophy, PCR was performed to determine the
mRNA expression of the downstream targets of Atrolnc-1, Dum, MAR1,
linc-MD1 and Myolinc. The relative mRNA expression levels of Wnt5a
(Fig. 4D) and MyoD (Fig. 4E), which are downstream targets of
MAR1 and Myolinc, respectively, were significantly higher in the
DEX group compared with the control group. However, the expression
levels of Dppa2, MAML1, Mef2c and Acta1 were not significantly
different between the two groups (Fig.
4A-C and F).
Discussion
In clinical practice, the cause of secondary
muscular atrophy due to hormone use is often unclear to doctors and
patients. Patients often refuse treatments that may cause
side-effects such as muscular atrophy and muscle weakness, where
development of these symptoms may reduce a patients' trust in their
doctor and affect their clinical treatment outcome (51,52).
Although GC mechanisms of action have been fully described, the
mechanisms of non-coding RNAs in GIMA yet to be fully understood.
The current study attempted to elucidate the regulatory mechanism
of non-coding RNAs in GC-induced skeletal muscle atrophy to provide
the groundwork for the development of drugs and treatment programs
for the prevention and treatment of muscular atrophy in clinical
practice. Non-coding RNAs were examined, which have been previously
demonstrated to be involved in other muscle atrophy models. The
current study aimed to identify whether non-coding RNAs were
associated with the DEX model.
In skeletal muscle, non-coding RNAs serve multiple
roles in muscle development and regeneration, such as in the
regulation of genes involved in myogenesis, proliferation, and
muscle fiber-type conversion (25,26,31-36).
Among them, miRNA has been revealed to serve a role in regulating
muscle atrophy by being associated with muscle protein metabolism,
muscle regeneration, angiogenesis and muscle cell apoptosis
(21). miR-206 inhibits the
expression of Pax7 and Histone deacetylase 4to promote the
differentiation of muscle satellite cells for the purpose of muscle
regeneration (46). miR-27b
promotes the differentiation of myogenic satellite cells and
promotes muscle regeneration by targeting myogenic regulatory
inhibitory proteins (47). The
current study indicated that the expression of miR-133a, miR-133b
and miR-206 were upregulated in the DEX group. These results were
consistent with previous studies that demonstrated increased
expression of miR-133a and miR-206 in animal models of muscular
dystrophy and in the serum of effected patients (30,48,53).
Myogenic regulatory factors include MyoD, MyoG, Myf5 and MRF4,
which servean important role in the regulation of myogenic
differentiation after proliferation of minisatellite cells
(54). In the present study, C2C12
cells treated with DEX were indicated to exhibit increased levels
of miR-133a, miR-133b and miR-206 and of MyoD and Wnt5. This may be
the result of DEX activating a compensatory mechanism for muscle
atrophy, or MyoD may servea role in this mechanism as it may
contribute toother functional mechanisms that are not yet fully
understood. For example, it may be suggested that MyoD promotes
muscle atrophy under the action or involvement of an unknown molec
ule, such as c-Myc. It is well known that c-Myc is a representative
lncRNA that promotes the development and function of muscles
(55,56). However, Eischen et al
(57) revealedthat c-Myc inhibits
Beclin 1 and Bax by inhibiting the anti-apoptotic protein Bcl-2,
whichenhances autophagy and apoptosis, respectively, leading to
musclewastage (58). In addition,
Amirouche et al (45)
indicated that overexpression of miR-206 can promote the expression
of MyoD, and miR-206 can act as a multi-effect modulator to
regulate muscle atrophy that is caused by Duchenne muscular
dystrophy by targeting a variety of key miRNAs. It may be
speculated that miR-206 may participate in the compensatory effect
of muscle atrophy under DEX-induced conditions via unknown
mechanisms, and this should be examined in future studies. The
expression of miR-18a, miR-186, miR-1a-3p, miR-23a-3p, miR-27b and
miR-29b-3p exhibited no significant change, suggesting that their
role in DEX-induced skeletal muscle atrophy may be minimal. In
muscle atrophy under pathological conditions, the factors that
cause miRNA changes are more complicated (59). Studies have demonstrated that
changes in the activity of the hypothalamus-pituitary-adrenal axis
in patients receiving glucocorticoid therapy may also affect muscle
and miRNA expression (60,61). The current study only investigated
the changes in miRNA expression at the cellular level in
vitro; therefore, it is necessary to further verify the
mechanism of the aforementioned miRNAs in hormone-induced muscular
atrophy in vivo.
In a previous study, a total of 2,922 lncRNAs and
581 circular RNAs exhibited differential regulation during C2C12
differentiation, suggesting that they may be involved in muscle
development (41). Among the
lncRNAs (nine miRNAs and seven lncRNAs) detected in the current
study, only some may be associated with the DEX-induced muscular
atrophy. In the current study, a number of lncRNAs, including
Atrolnc-1, Dum, MAR1, linc-MD1 and Myolinc, were revealed to be
differentially expressed in control and atrophic myotubes. A large
number of studies have demonstrated that lncRNAs can inhibit or
activate gene expression by regulating gene transcription, mRNA
stability, pre-mRNA splicing, protein translation and protein
stability (62,63). LncRNA MAR1 acts as a miR-487b sponge
to regulate Wnt5a protein expression, resulting in the promotion
ofmuscle differentiation and regeneration (36). Myolinc recruits TDP-43 to the
promoters of Filamin-A-interacting protein 1 and muscle marker
genes (such as MyoD) to regulate myogenic regulatory networks
(26). Additionally, lncRNAs can
act as ‘sponges’ for miRNAs by pairing with and titrating them off
their mRNA targets (64). In C2C12
cells, MAR1 has been indicated to promote myogenic differentiation
by acting as a sponge for miR-487b, thereby regulating the
expression of Wnt5a, which serves an important role in muscle
regeneration (36). The present
study also demonstrated that MAR1 expression in C2C12 cells treated
with DEX was higher compared with the control group, and the
expression of the downstream target gene Wnt5a was also increased,
indicating a compensatory increase in MAR1 expression. Zhang et
al (36) reported that the
lncRNA MAR1 was significantly downregulated in mouse gastrocnemius
muscle under senescence and mechanical unloading conditions during
muscular atrophy. LncRNAs may change dynamically during muscular
atrophy, or the same lncRNAs involved in muscular development may
respond in different ways to different stimuli (65,66).
linc-MD1 and Dum have been indicated to regulate the expression of
MYHC and servean important role in the differentiation and
development of skeletal muscle cells (30,31).
Therefore, increased expression of linc-MD1 and Dum may be a
compensatory response to atrophy. Inhibition of MURF1 and Atrogin-1
expression has been indicated to inhibit muscle loss and reduce
muscle atrophy (36). In the
current study, the expression of Atrolnc-1 in the DEX-induced
atrophy model was significantly increased compared with the control
cells. These results are consistent with studies that revealed that
Atrolnc-1 significantly enhanced atrophic muscles in mouse models
of chronic kidney disease, starvation and cancer (30,49).
Atrolnc-1 interacts with A20 binding inhibitors of NF-κB-1 to
promote its activation, leading to increased expression of
MURF1(30). The high expression of
Atrolnc-1 and MURF1 observed in the present study further indicates
that muscular atrophy occurred in DEX-treated C2C12 cells.
It has been suggested that lncRNAs and miRNAs may
mutually restrict muscle development in muscular atrophy model
(33,67). The results of the present study
demonstrated that the expression of miR-133a and miR-133b in
DEX-treated C2C12 cells increased compared with the control cells,
which is consistent with the ability of linc-MD1 to modulate
expression. Legnini et al (34) indicated that in normal skeletal
muscle cells, increased expression of miR-133a led to the cleavage
of linc-MD1 to form miR-133b. Therefore, it can be speculated that,
under the action of DEX, linc-MD1 may be upregulated and exhibit a
compensatory effect via increased expression of miR-133a and
miR-133b. However, the expression of the downstream mRNAs MAML1 and
Mef2c were not indicated to be significantly different between the
DEX-treated C2C12 and control cells, so may serve a role in this
mechanism.
In conclusion, DEX increased catabolism in skeletal
muscle cells and elevated the expression of key genes for muscular
atrophy, which suggested the successful establishment of the
muscular atrophy model in C2C12 cells. Recent studies have
demonstrated that some important miRNAs and lncRNAs may be involved
in regulating the mechanism of action behind muscular atrophy
(16,33,44,67);
however, this remains to be further explored. The results of the
present study provide a novel perspective for studies on miRNAs and
lncRNAs in GC-induced muscular atrophy, and suggest that they may
be used as potential diagnostic tools. Further studies are required
to improve the understanding ofthe role of non-coding RNAs in
GC-induced muscle atrophy.
Acknowledgements
Not applicable.
Funding
The current study was supported by grants from the
Natural Science Foundation of Guangdong Province (grant no.
2018A030313591), the Science and Technology Planning Project of
Guangdong Province (grant no. 2017ZC0333), the Science and
Technology Planning Project of Haizhu District (grant no. 2018-87)
and the Science Foundation of Guangdong Second Provincial General
Hospital (grant nos. YQ2018-010, YN2017-002 and YN2017-003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RC conceived and designed the experiments. YL, HS,
SZ, SL and YS performed the experiments analyzed the data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Schakman O, Kalista S, Barbé C, Loumaye A
and Thissen JP: Glucocorticoid-induced skeletal muscle atrophy. Int
J Biochem Cell Biol. 45:2163–2172. 2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Rhen T and Cidlowski JA: Antiinflammatory
action of glucocorticoids-new mechanisms for old drugs. N Engl J
Med. 353:1711–1723. 2005.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rauch A, Seitz S, Baschant U, Schilling
AF, Illing A, Stride B, Kirilov M, Mandic V, Takacz A,
Schmidt-Ullrich R, et al: Glucocorticoids suppress bone formation
by attenuating osteoblast differentiation via the monomeric
glucocorticoid receptor. Cell Metab. 11:517–531. 2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ma K, Mallidis C, Bhasin S, Mahabadi V,
Artaza J, Gonzalez-Cadavid N, Arias J and Salehian B:
Glucocorticoid-induced skeletal muscle atrophy is associated with
upregulation of myostatin gene expression. Am J Physiol Endocrinol
Metab. 285:E363–E371. 2003.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Waddell DS, Baehr LM, van den Brandt J,
Johnsen SA, Reichardt HM, Furlow JD and Bodine SC: The
glucocorticoid receptor and FOXO1 synergistically activate the
skeletal muscle atrophy-associated MuRF1 gene. Am J Physiol
Endocrinol Metab. 295:E785–E797. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Braun TP, Szumowski M, Levasseur PR,
Grossberg AJ, Zhu X, Agarwal A and Marks DL: Muscle atrophy in
response to cytotoxic chemotherapy is dependent on intact
glucocorticoid signaling in skeletal muscle. PLoS One.
9(e106489)2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Braun TP and Marks DL: The regulation of
muscle mass by endogenous glucocorticoids. Front Physiol.
6(12)2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Shimizu N, Yoshikawa N, Ito N, Maruyama T,
Suzuki Y, Takeda S, Nakae J, Tagata Y, Nishitani S, Takehana K, et
al: Crosstalk between glucocorticoid receptor and nutritional
sensor mTOR in skeletal muscle. Cell Metab. 13:170–182.
2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bodine SC and Furlow JD: Glucocorticoids
and skeletal muscle. Adv Exp Med Biol. 872:145–176. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Schakman O, Gilson H, Kalista S and
Thissen JP: Mechanisms of muscle atrophy induced by
glucocorticoids. Horm Res. 72 (Suppl 1):S36–S41. 2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhao SQ, Xu SQ, Cheng J, Cao XL, Zhang Y,
Zhou WP, Huang YJ, Wang J and Hu XM: Anti-inflammatory effect of
external use of escin on cutaneous inflammation: Possible
involvement of glucocorticoids receptor. Chin J Nat Med.
16:105–112. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zheng B, Ohkawa S, Li H, Roberts-Wilson TK
and Price SR: FOXO3a mediates signaling crosstalk that coordinates
ubiquitin and atrogin-1/MAFbx expression during
glucocorticoid-induced skeletal muscle atrophy. FASEB J.
24:2660–2669. 2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Watson ML, Baehr LM, Reichardt HM,
Tuckermann JP, Bodine SC and Furlow JD: A cell-autonomous role for
the glucocorticoid receptor in skeletal muscle atrophy induced by
systemic glucocorticoid exposure. Am J Physiol Endocrinol Metab.
302:E1210–E1220. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bodine SC, Latres E, Baumhueter S, Lai VK,
Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K,
et al: Identification of ubiquitin ligases required for skeletal
muscle atrophy. Science. 294:1704–1708. 2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Stitt TN, Drujan D, Clarke BA, Panaro F,
Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD and Glass DJ: The
IGF-1/PI3K/Akt pathway prevents expression of muscle
atrophy-induced ubiquitin ligases by inhibiting FOXO transcription
factors. Mol Cell. 14:395–403. 2004.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li J, Chan MC, Yu Y, Bei Y, Chen P, Zhou
Q, Cheng L, Chen L, Ziegler O, Rowe GC, et al: miR-29b contributes
to multiple types of muscle atrophy. Nat Commun.
8(15201)2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Horak M, Novak J and Bienertova-Vasku J:
Muscle-specific microRNAs in skeletal muscle development. Dev Biol.
410:1–13. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chen JF, Mandel EM, Thomson JM, Wu Q,
Callis TE, Hammond SM, Conlon FL and Wang DZ: The role of
microRNA-1 and microRNA-133 in skeletal muscle proliferation and
differentiation. Nat Genet. 38:228–233. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
McCarthy JJ and Esser KA: MicroRNA-1 and
microRNA-133a expression are decreased during skeletal muscle
hypertrophy. J Appl Physiol (1985). 102:306–313. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Soares RJ, Cagnin S, Chemello F,
Silvestrin M, Musaro A, De Pitta C, Lanfranchi G and Sandri M:
Involvement of microRNAs in the regulation of muscle wasting during
catabolic conditions. J Biol Chem. 289:21909–21925. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Walden TB, Timmons JA, Keller P,
Nedergaard J and Cannon B: Distinct expression of muscle-specific
microRNAs (myomirs) in brown adipocytes. J Cell Physiol.
218:444–449. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ivey KN and Srivastava D: microRNAs as
developmental regulators. Cold Spring Harb Perspect Biol.
7(a008144)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lei Z, Sluijter JP and van Mil A: MicroRNA
therapeutics for cardiac regeneration. Mini Rev Med Chem.
15:441–451. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Shen H, Liu T, Fu L, Zhao S, Fan B, Cao J
and Li X: Identification of microRNAs involved in
dexamethasone-induced muscle atrophy. Mol Cell Biochem.
381:105–113. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Militello G, Hosen MR, Ponomareva Y,
Gellert P, Weirick T, John D, Hindi SM, Mamchaoui K, Mouly V,
Döring C, et al: A novel long non-coding RNA myolinc regulates
myogenesis through TDP-43 and Filip1. J Mol Cell Biol. 10:102–117.
2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Xiong W, Jiang YX, Ai YQ, Liu S, Wu XR,
Cui JG, Qin JY, Liu Y, Xia YX, Ju YH, et al: Microarray analysis of
long non-coding RNA expression profile associated with
5-fluorouracil-based chemoradiation resistance in colorectal cancer
cells. Asian Pac J Cancer Prev. 16:3395–3402. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen R, Jiang T, She Y, Xie S, Zhou S, Li
C, Ou J and Liu Y: Comprehensive analysis of lncRNAs and mRNAs with
associated co-expression and ceRNA networks in C2C12 myoblasts and
myotubes. Gene. 647:164–173. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Boltaña S, Valenzuela-Miranda D, Aguilar
A, Mackenzie S and Gallardo-Escárate C: Long noncoding RNAs
(lncRNAs) dynamics evidence immunomodulation during ISAV-Infected
Atlantic salmon (Salmo salar). Sci Rep. 6(22698)2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sun L, Si M, Liu X, Choi JM, Wang Y,
Thomas SS, Peng H and Hu Z: Long-noncoding RNA Atrolnc-1 promotes
muscle wasting in mice with chronic kidney disease. J Cachexia
Sarcopenia Muscle. 9:962–974. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Li Y, Meng X, Li G, Zhou Q and Xiao J:
Noncoding RNAs in muscle atrophy. Adv Exp Med Biol. 1088:249–266.
2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang ZK, Li J, Guan D, Liang C, Zhuo Z,
Liu J, Lu A, Zhang G and Zhang BT: Long noncoding RNA lncMUMA
reverses established skeletal muscle atrophy following mechanical
unloading. Mol Ther. 26:2669–2680. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Legnini I, Morlando M, Mangiavacchi A,
Fatica A and Bozzoni I: A feedforward regulatory loop between HuR
and the long noncoding RNA linc-MD1 controls early phases of
myogenesis. Mol Cell. 53:506–514. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wang L, Zhao Y, Bao X, Zhu X, Kwok YK, Sun
K, Chen X, Huang Y, Jauch R, Esteban MA, et al: LncRNA Dum
interacts with Dnmts to regulate Dppa2 expression during myogenic
differentiation and muscle regeneration. Cell Res. 25:335–350.
2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang ZK, Li J, Guan D, Liang C, Zhuo Z,
Liu J, Lu A, Zhang G and Zhang BT: A newly identified lncRNA MAR1
acts as a miR-487b sponge to promote skeletal muscle
differentiation and regeneration. J Cachexia Sarcopenia Muscle.
9:613–626. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fappi A, Neves JC, Sanches LN, Massaroto
E, Silva PV, Sikusawa GY, Brandão TPC, Chadi G and Zanoteli E:
Skeletal muscle response to deflazacort, dexamethasone and
methylprednisolone. Cells. 8(406)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Troncoso R, Paredes F, Parra V, Gatica D,
Vásquez-Trincado C, Quiroga C, Bravo-Sagua R, López-Crisosto C,
Rodriguez AE, Oyarzún AP, et al: Dexamethasone-induced autophagy
mediates muscle atrophy through mitochondrial clearance. Cell
Cycle. 13:2281–2295. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Becker DE: Basic and clinical pharmacology
of glucocorticosteroids. Anesth Prog. 60:25–32. 2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Son YH, Jang EJ, Kim YW and Lee JH:
Sulforaphane prevents dexamethasone-induced muscle atrophy via
regulation of the Akt/Foxo1 axis in C2C12 myotubes. Biomed
Pharmacother. 95:1486–1492. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chen R, Jiang T, Lei S, She Y, Shi H, Zhou
S, Ou J and Liu Y: Expression of circular RNAs during C2C12
myoblast differentiation and prediction of coding potential based
on the number of open reading frames and N6-methyladenosine motifs.
Cell Cycle. 17:1832–1845. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Massaccesi L, Goi G, Tringali C, Barassi
A, Venerando B and Papini N: Dexamethasone-induced skeletal muscle
atrophy increases O-GlcNAcylation in C2C12 cells. J Cell Biochem.
117:1833–1842. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Townley-Tilson WD, Callis TE and Wang D:
MicroRNAs 1, 133, and 206: Critical factors of skeletal and cardiac
muscle development, function, and disease. Int J Biochem Cell Biol.
42:1252–1255. 2010.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Amirouche A, Jahnke VE, Lunde JA, Koulmann
N, Freyssenet DG and Jasmin BJ: Muscle-specific microRNA-206
targets multiple components in dystrophic skeletal muscle
representing beneficial adaptations. Am J Physiol Cell Physiol.
312:C209–C221. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Liu C, Wang M, Chen M, Zhang K, Gu L, Li
Q, Yu Z, Li N and Meng Q: miR-18a induces myotubes atrophy by
down-regulating IgfI. Int J Biochem Cell Biol. 90:145–154.
2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Antoniou A, Mastroyiannopoulos NP, Uney JB
and Phylactou LA: miR-186 inhibits muscle cell differentiation
through myogenin regulation. J Biol Chem. 289:3923–3935.
2014.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lei S, She Y, Zeng J, Chen R, Zhou S and
Shi H: Expression patterns of regulatory lncRNAs and miRNAs in
muscular atrophy models induced by starvation in vitro and in vivo.
Mol Med Rep. 20:4175–4185. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Mercatelli N, Fittipaldi S, De Paola E,
Dimauro I, Paronetto MP, Jackson MJ and Caporossi D: MiR-23-TrxR1
as a novel molecular axis in skeletal muscle differentiation. Sci
Rep. 7(7219)2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Hou L, Xu J, Jiao Y, Li H, Pan Z, Duan J,
Gu T, Hu C and Wang C: MiR-27b promotes muscle development by
inhibiting MDFI expression. Cell Physiol Biochem. 46:2271–2283.
2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Oray M, Abu Samra K, Ebrahimiadib N, Meese
H and Foster CS: Long-term side effects of glucocorticoids. Expert
Opin Drug Saf. 15:457–465. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Stout A, Friedly J and Standaert CJ:
Systemic absorption and side effects of locally injected
glucocorticoids. PM R. 11:409–419. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Matsuzaka Y, Kishi S, Aoki Y, Komaki H,
Oya Y, Takeda S and Hashido K: Three novel serum biomarkers, miR-1,
miR-133a, and miR-206 for Limb-girdle muscular dystrophy,
Facioscapulohumeral muscular dystrophy, and becker muscular
dystrophy. Environ Health Prev Med. 19:452–458. 2014.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Li G, Li QS, Li WB, Wei J, Chang WK, Chen
Z, Qiao HY, Jia YW, Tian JH and Liang BS: miRNA targeted signaling
pathway in the early stage of denervated fast and slow muscle
atrophy. Neural Regen Res. 11:1293–1303. 2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Luo W, Chen J, Li L, Ren X, Cheng T, Lu S,
Lawal RA, Nie Q, Zhang X and Hanotte O: c-Myc inhibits myoblast
differentiation and promotes myoblast proliferation and muscle
fibre hypertrophy by regulating the expression of its target genes,
miRNAs and lincRNAs. Cell Death Differ. 26:426–442. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lin CH, Jackson AL, Guo J, Linsley PS and
Eisenman RN: Myc-regulated microRNAs attenuate embryonic stem cell
differentiation. EMBO J. 28:3157–3170. 2009.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Eischen CM, Packham G, Nip J, Fee BE,
Hiebert SW, Zambetti GP and Cleveland JL: Bcl-2 is an apoptotic
target suppressed by both c-Myc and E2F-1. Oncogene. 20:6983–6993.
2001.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Alessio E, Buson L, Chemello F, Peggion C,
Grespi F, Martini P, Massimino ML, Pacchioni B, Millino C, Romualdi
C, et al: Single cell analysis reveals the involvement of the long
non-coding RNA Pvt1 in the modulation of muscle atrophy and
mitochondrial network. Nucleic Acids Res. 47:1653–1670.
2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
van de Worp WR, Theys J, van Helvoort A
and Langen RC: Regulation of muscle atrophy by microRNAs:
‘AtromiRs’ as potential target in cachexia. Curr Opin Clin Nutr
Metab Care. 21:423–429. 2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Hildebrandt T, Shope S, Varangis E, Klein
D, Pfaff DW and Yehuda R: Exercise reinforcement, stress, and
β-endorphins: An initial examination of exercise in
anabolic-androgenic steroid dependence. Drug Alcohol Depend.
139:86–92. 2014.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Ng TP, Lu Y, Choo RW, Tan CT, Nyunt MS,
Gao Q, Mok EW and Larbi A: Dysregulated homeostatic pathways in
sarcopenia among frail older adults. Aging Cell.
17(e12842)2018.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Zhu M, Liu J, Xiao J, Yang L, Cai M, Shen
H, Chen X, Ma Y, Hu S, Wang Z, et al: Lnc-mg is a long non-coding
RNA that promotes myogenesis. Nat Commun. 8(14718)2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Devaux Y, Zangrando J, Schroen B, Creemers
EE, Pedrazzini T, Chang CP, Dorn GW II, Thum T and Heymans S:
Cardiolinc network. Long noncoding RNAs in cardiac development and
ageing. Nat Rev Cardiol. 12:415–425. 2015.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Ebert MS, Neilson JR and Sharp PA:
MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian
cells. Nat Methods. 4:721–726. 2007.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Cichewicz MA, Kiran M, Przanowska RK,
Sobierajska E, Shibata Y and Dutta A: MUNC, an enhancer RNA
upstream from the MYOD gene, induces a subgroup of myogenic
transcripts in trans independently of MyoD. Mol Cell Biol.
38:e00655–17. 2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Mueller AC, Cichewicz MA, Dey BK, Layer R,
Reon BJ, Gagan JR and Dutta A: MUNC, a long noncoding RNA that
facilitates the function of MyoD in skeletal myogenesis. Mol Cell
Biol. 35:498–513. 2015.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Li Z, Cai B, Abdalla BA, Zhu X, Zheng M,
Han P, Nie Q and Zhang X: LncIRS1 controls muscle atrophy via
sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J
Cachexia Sarcopenia Muscle. 10:391–410. 2019.PubMed/NCBI View Article : Google Scholar
|