Introduction
Ovarian cancer (OC) is the most common lethal
gynecological malignancy and the fifth leading cause of
cancer-related mortality in women worldwide (1). The majority of patients with OC are
diagnosed at an advanced stage, due to the lack of specific
symptoms at the early stages of the disease (2). Cancer antigen 125 (CA125) is one of
the main biomarkers of OC used widely in the clinical setting
(3). Serum CA125 detection in OC is
a valuable indicator of prognosis, survival time and stage
(4,5). However, false-positive results upon
serum CA125 detection may adversely affect women who are screened,
both psychologically and in terms of unnecessary surgical
intervention (6).
CA125 is a membrane-associated mucin-type
glycoprotein encoded by the mucin 16 cell surface associated
(MUC16) gene. Elevated MUC16 expression was reported to promote
proliferation, migration and chemoresistance of OC cells (7-10),
and to be associated with poor prognosis of the patients (11). These findings indicated the roles of
MUC16 upregulation in OC and explained why CA125 can be used as the
biomarker of OC. CA125 is generally present in normal ovarian
epithelia, endometrium and decidua (12). However, its levels may increase in
benign gynecological diseases and abdominal disorders, as well as
in malignant diseases, including OC (13,14).
Upregulation of CA125 under both benign and malignant conditions
suggested that these conditions may share certain common factors,
such as the inflammatory microenvironment (IME), which may be
involved in MUC16 and CA125 regulation. It has been reported that
malignant ascites from OC enhanced MUC16 expression and stimulated
the release of CA125 in human peritoneal mesothelial cells, with
stimulating factors unknown (15).
IME has been found to be involved in the development and
progression of tumors, including OC (16,17).
Alterations of inflammatory cytokines, such as interleukin (IL)-6,
IL-8 and tumor necrosis factor (TNF)-α in the IME play important
roles in this process (18). For
example, elevated expression of IL-6 was observed in the serum,
ascitic fluid and tumor tissues from patients with OC (19,20).
Upregulation of the expression of IL-6 and IL-6 receptor (IL-6R)
have been reported to contribute to the proliferation, migration
and chemotherapy resistance of OC cells, and may be associated with
poor prognosis of patients with OC (21-23).
TNF-α facilitates tumor progression through promoting the
expression of cytokines and matrix metalloproteinases (MMPs), as
well as angiogenesis in OC (24,25).
In addition, it has been reported that TNF-α and interferon (IFN)-γ
stimulate the expression of MUC16 in breast and endometrial cancer,
as well as OC (26). Moreover,
IFN-γ and IL-8 were reported to induce MUC16 expression in human
ocular surface epithelial cells (27). These findings suggested that
inflammatory cytokines in the IME may contribute to the increase of
MUC16 expression levels in OC; however, the underlying mechanisms
remain to be elucidated.
In the present study, OC cells were treated with
inflammation-associated factors, including lipopolysaccharides
(LPS), IL-6, IL-8 and TNF-α, and the expression of MUC16 was
investigated. The aim was to determine the effect of
inflammation-associated factors on MUC16 expression in OC cells and
CA125 concentration. Moreover, the effect of the activation of the
canonical downstream signaling pathway of each
inflammation-associated factor in the regulation of MUC16 and the
role of nuclear factor (NF)-κB in this process were investigated,
in order to determine whether the IME contributes to the level of
CA125 in OC, and whether it should be taken into consideration in
OC diagnosis.
Materials and methods
Cell lines and culture
Four human OC cell lines (OVCAR3, HEY, A2780 and
SKOV3) were purchased from the American Type Culture Collection.
The OVCAR3, HEY and SKOV3 cell lines were maintained in RPMI-1640
medium (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin (Thermo Fisher Scientific, Inc.). A2780 cells were
cultured in DMEM (Thermo Fisher Scientific, Inc.) supplemented with
10% FBS (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and
100 µg/ml streptomycin (Thermo Fisher Scientific, Inc.). All cells
were cultured in a humidified incubator with 95% air and 5%
CO2 at 37˚C. Cells were routinely passaged and used when
they were in the logarithmic growth phase.
Transient transfection
To overexpress or knock down NF-κB in HEY cells,
transient transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Briefly, 1x106 cells were transfected with 5 µg
pENTER-H-NF-κB plasmid expressing human NF-κB (Vigene Biosciences,
Inc.) for overexpression. Furthermore, a total of 5x105
cells were transfected with 100 pmol siRNAs targeting NF-κB for
downregulation. Three siRNAs (siRNA-1,
5'-TTGCTAGAACATGCTATAACATG-3'; siRNA-2,
5'-ACGATTGCAACATCTCTAAGAAT-3'; siRNA-3,
5'-AAGCAATTAAACAAGTTTGTAAT-3') and non-targeting negative control
(5'-TTCTCCGAACGTGTCACGTT-3') synthesized by Shanghai GenePharma
Co., Ltd. were transfected. At 48 h post-transfection, the cells
were used for subsequent assays.
Treatment with inflammation-associated
factors
A total of 1x106 HEY cells were first
treated with 10, 50 and 100 ng/ml LPS (Sigma-Aldrich; Merk KGaA),
IL-6 (PeproTech, Inc.) or IL-8 (PeproTech, Inc.), or 2.5, 10 and 25
ng/ml of TNF-α (PeproTech, Inc.) for 24, 48 and 72 h at 37˚C, with
1X PBS used as a control. The lowest concentration and shortest
stimulation time of inflammation-associated factors resulting in a
statistically significant change in MUC16 mRNA expression levels
were selected as the optimal concentration and duration,
respectively. HEY cells were treated with LPS, IL-6, IL-8 or TNF-α
at the optimal concentration and for the optimal duration. For
co-treatment,HEY cells were treated with 10 ng/ml LPS combined with
500 nM Toll-like receptor 4 (TLR4) antagonist VIPER (Novus
Biologicals, LLC) for 48 h, 50 ng/ml IL-6 was combined with 10 µM
membrane glycoprotein 130 (gp130) inhibitor SC144 (Selleck
Chemicals) for 24 h, 50 ng/ml IL-8 was combined with 10 µM CXCR2
antagonist SB225002 (Selleck Chemicals) for 48 h or 2.5 ng/ml TNF-α
was combined with 100 nM of its inhibitor GSK2982772 (Selleck
Chemicals) for 24 h.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was extracted from cell lines using
TRIzol® reagent (cat. no. 15596; Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. cDNA was synthesized using a cDNA synthesis kit (cat.
no. 12594; Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. RT-qPCR was performed with
SYBR-Green Master Mix (cat. no. K0223; Thermo Fisher Scientific,
Inc.) using the ABI 7300 platform (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The primer sequences used were as
follows: TLR4 forward, 5'-CCGCTTTCACTTCCTCTCAC-3' and reverse,
5'-CATCCTGGCATCATCCTCAC-3'; IL-6R forward, 5'-GGTGCGAAAGGATGAAAG-3'
and reverse, 5'-TAGGATTACAGGCGTGAG-3'; gp130 forward,
5'-GAAAGGCTGCTTGGGTTC-3' and reverse, 5'-GCTCTGGCTTCGTATCTG-3';
CXCR2 forward, 5'-GGGCACACTTCCACTACTCTC-3' and reverse,
5'-GAACGTGGCCTCCTCTACTTC-3'; TNF receptor superfamily member 1A
(TNFRSF1A) forward, 5'-GCCGCCTACTTGGTGCTAAC-3' and reverse,
5'-CGTCCCTCATCCTCGCAAAC-3'; TNF receptor superfamily member 1B
(TNFRSF1B) forward, 5'-TGAGGCTGGGAAATCGTTTG-3' and reverse,
5'-GCTTTGTCGTTGGCTTGTTG-3'; JNK1 forward,
5'-GCATCTCAACTCTGTCATAG-3' and reverse, 5'-CAGCAGGATTAGCATAGAAC-3';
p38 forward, 5'-AAGGAAGGAGGCAGACTGATG-3' and reverse,
5'-GCTGTGGATGGTGAGGATTTG-3'; extracellular signal-regulated kinase
ERK2 forward, 5'-TGGGTCAGAAACAAATGG-3' and reverse,
5'-TGCTCTACACGCATAAAC-3'; NF-κB forward, 5'-GAATGGCTCGTCTGTAGTG-3'
and reverse, 5'-TGGTATCTGTGCTCCTCTC-3'; MUC16 forward,
5'-GCAGACAGCAGAGACTATC-3' and reverse, 5'-CTGGACTTCCCAACCATTC-3';
and GAPDH forward, 5'-AATCCCATCACCATCTTC-3' and reverse,
5'-AGGCTGTTGTCATACTTC-3'. RT-qPCR reactions were conducted
according to the following thermocycling parameters: 95˚C for 10
min; 40 cycles at 95˚C for 15 sec and 60˚C for 45 sec. Primer
specificity was assessed by melt-curve analysis. Relative mRNA
quantification was calculated using the 2-ΔΔCq method
with GAPDH used as an internal control gene (28).
Western blot analysis
Cell lysates of HEY cells were prepared with RIPA
lysis buffer (cat. no. 89901; Thermo Fisher Scientific, Inc.) and
centrifuged at 111 x g for 15 min at 4˚C. The protein concentration
was measured using a BCA protein assay kit (cat. no. 23250; Thermo
Fisher Scientific, Inc.) and the lysate was stored at -80˚C for
further experiments. Equal amounts of protein (30 µg) were loaded
in each lane, separated by 10% SDS-PAGE and transferred onto
nitrocellulose membranes (EMD Millipore) by semi-dry
electrophoretic transfer method for 30 min (25 V). Subsequently,
the membranes were blocked using 5% non-fat milk at room
temperature for 1 h and incubated for 2 h at room temperature with
primary antibodies against MUC16 (dilution, 1:500; cat. no.
ab110640), TLR4 (dilution, 1:500; cat. no. ab13556), TNFR-I
(dilution, 1:1,000; cat. no. ab19139), TNFR-II (dilution, 1:10,000;
cat. no. ab109322), IL-6R (dilution, 1:200; cat. no. ab128008),
gp130 (dilution, 1:500; cat. no. ab87969), CXCR2 (dilution, 1:500;
cat. no. ab14935), NF-κB/p65 (dilution, 1:1,000; cat. no. ab16502)
(all purchased from Abcam); JNK (dilution, 1:1,000; cat. no. 9252),
p-JNK (dilution, 1:1,000; cat. no. 9251S), p38 (dilution, 1:1,000;
cat. no. 8690), p-p38 (dilution, 1:1,000; cat. no. 9211), ERK
(dilution, 1:1,000; cat. no. 4695), p-ERK (dilution, 1:1,000; cat.
no. 4370) and GAPDH (dilution, 1:2,000; cat. no. 5174) (all
purchased from Cell Signaling Technology, Inc.). GAPDH was used as
an internal protein loading control. The membranes were washed with
TBS-0.1% Tween-20 and incubated with the corresponding secondary
antibodies for 1 h at 37˚C. The horseradish peroxidase
(HRP)-labeled donkey anti-goat IgG (dilution, 1:1,000; cat. no.
A0181), HRP-labeled goat anti-rabbit IgG (dilution, 1:1,000; cat.
no. A0208) and HRP-labeled goat anti-mouse IgG (dilution, 1:1,000;
cat. no. A0216) secondary antibodies were purchased from Beyotime
Institute of Biotechnology. The proteins were visualized by
enhanced chemiluminescence (cat. no. WBKLS0100; EMD Millipore)
according to the manufacturer's instructions, and the densitometric
analyses of the bands were performed by ChemiDoc™ XRS + image
analyzer (Bio-Rad Laboratories, Inc.).
ELISA
Cell culture supernatants were collected from cells
treated as mentioned in Treatment with inflammation-associated
factors. CA125 levels were measured using a commercial ELISA
kit (cat. no. XY-E10325; Shanghai Xinyu Biological Technology Co.,
Ltd.) according to the manufacturer's protocols. The absorbance was
measured at 450 nm on a microplate absorbance reader (MR-960;
Perlong Medical Equipment Co., Ltd.). The concentration of CA125
was quantified by corresponding standard curves.
Bioinformatics analysis
To determine the potential binding sites of NF-κB,
the promoter sequence of human MUC16 (chr19:8981139-8983842)
obtained from the University of California, Santa Cruz database
(https://genome.ucsc.edu/) was sent to the Consit
database (http://consite.genereg.net/) with an
85% Transcription Factor score as the cutoff value. Then, PCR
primers amplifying the retrieved potential binding sites was send
for synthesis and used for chromatin immunoprecipitation (ChIP)
analysis.
ChIP assay
Immunoprecipitation assays were performed with a
commercial CUT&Tag kit (cat. no. S602; Vazyme Biotech Co.,
Ltd.) and p65 antibody (cat. no. 10745-1-APNF-κB; ProteinTech
Group, Inc.) according to the manufacturers' instructions. HEY
cells were harvested using 0.25% Trypsin and washed using washing
buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl and 500 mM
spermidine. Cells were resuspended in antibody buffer containing 2
mM EDTA, 0.1% BSA (cat. no. E661003; Sangon Biotech Co., Ltd.) and
0.5% digitonin (Vazyme Biotech Co., Ltd.) for p65 antibody (1 µg
per immunoprecipitation) incubation at room temperature. IgG
antibody (cat. no. AP162-KC; Sigma-Aldrich) was used as the
negative control. After DNA collection and purification by the
phenol and chloroform method, qPCR was performed with SYBR-Green
Master Mix (cat. no. K0223; Thermo Fisher Scientific, Inc.) with
the ABI 7300 platform (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Primers of two potential binding sites were
used: P65-binding site primers (p65B) forward,
5'-ACCTCCACCTCCTGGGTTC-3' and reverse,
5'-GGTGGGTGGATTACTTGAAGTC-3'; NF-κB-binding site primers forward,
5'-GTCGCCCAGGCTGAAGTG-3' and reverse, 5'-CTGCTGGGCGTGGTGTCT-3'; and
negative site primers (NS) forward, 5'-AGGAGACGCAGCTTAGAACC-3' and
reverse, 5'-TTCAACTTTCCAGCCTCCA-3'. The following thermocycling
conditions were used for the qPCR: Initial denaturation at 95˚C for
30 sec and 40 cycles of 95˚C for 5 sec and 60˚C for 10 sec. The
enrichment of the binding site was calculated using the
2-ΔΔCq method with NS used as internal control.
Statistical analysis
All data were analyzed using SPSS 20.0 software (IBM
Corp.). The measurement values are presented as the mean ± SEM from
at least triplicates. One-way ANOVA followed by Tukey's post hoc
test was used for multiple comparisons. P<0.05 was considered to
indicate a statistically significant difference.
Results
Basal MUC16 mRNA expression levels in
four OC cell lines
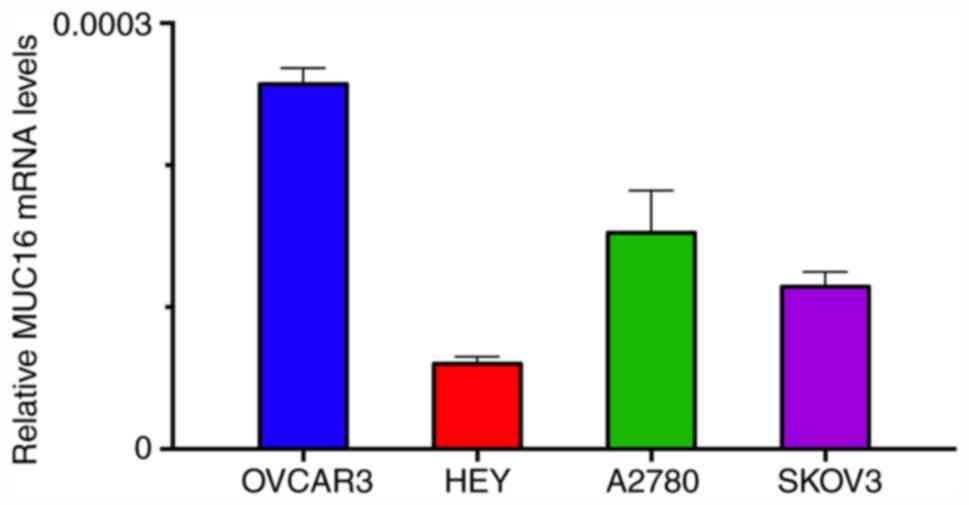
First, the expression levels of MUC16 were assessed
in four different human OC cell lines (OVCAR3, HEY, A2780 and
SKOV3) by RT-qPCR to select the appropriate tumor cells for further
analysis. Among these tumor cells, HEY cells exhibited the lowest
and OVCAR3 the highest levels of MUC16 expression (Fig. 1). Therefore, HEY cells were selected
for further experiments.
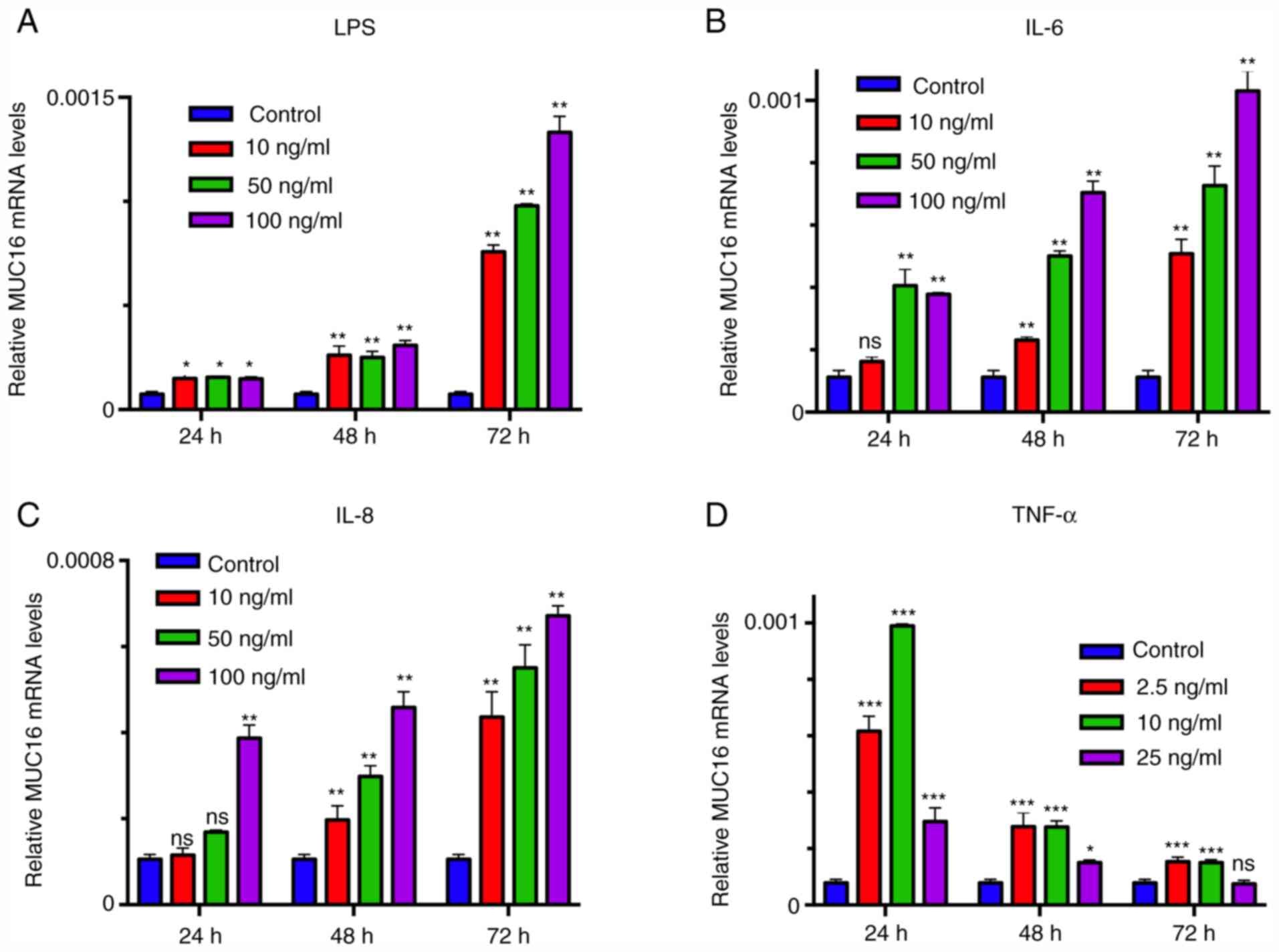
Screening the optimal concentration
and duration of inflammation-associated factor stimulation
To select the optimal concentration and duration of
inflammation-associated factor treatment, HEY cells were treated
with different concentrations of LPS, IL-6, IL-8 or TNF-α for 24,
48 and 72 h and MUC16 mRNA expression was analyzed. The results
demonstrated that the mRNA expression levels of MUC16 were
increased in tumor cells treated with inflammation-associated
factors in a dose and time-dependent (Fig. 2). The highest level of MUC16
expression was observed in cells treated with 100 ng/ml LPS, IL-6
and IL-8 for 72 h respectively, while lower expression levels were
observed in cells treated with lower concentrations or shorter
durations (Fig. 2). For TNF-α
administration, 10 ng/ml incubation for 24 h induced the highest
MUC16 expression while increasing TNF-α concentration and longer
duration induced lower MUC16 expression at 24 h (Fig. 2). Therefore, these concentrations of
inflammation-associated factors and respective treatment durations
were used in the following experiments.
Inflammation-associated factors induce
the expression of MUC16 in OC cells
To investigate whether the inflammation-associated
factors LPS, IL-6, IL-8 and TNF-α affect the expression of MUC16 in
OC cells, the mRNA expression levels of MUC16 were assessed in OC
cells treated with inflammation-associated factors. RT-qPCR
analysis revealed that the mRNA expression levels of MUC16 were
significantly increased in OC cells treated with LPS, IL-6, IL-8 or
TNF-α compared with those in untreated cells (Fig. 3A-D). When the corresponding receptor
antagonists were added, the upregulation of MUC16 was inhibited
(Fig. 3A-D). In addition, the
protein expression levels of MUC16 were detected in tumor cells
treated with inflammation-associated factors. Western blot analysis
demonstrated that the protein expression levels of MUC16 were also
increased following treatment with the aforementioned
inflammation-associated factors (Fig.
3E-H). In addition, the increase induced by treatment with
inflammation-associated factors was inhibited by their
corresponding receptor antagonists (Fig. 3E-H). These results suggested that
LPS, IL-6, IL-8 and TNF-α induced MUC16 expression in HEY cells.
Moreover, the levels of CA125 in the cell culture supernatant were
also investigated. ELISA demonstrated that a higher level of CA125
was observed in the supernatant from tumor cells treated with
inflammation-associated factors compared with that from control
cells (Fig. 3I-L). When cells were
treated with both receptor antagonists and inflammation-associated
factors simultaneously, CA125 levels were decreased to a lower
level compared with the control group, suggesting that receptor
antagonists inhibited the activation of MUC16 expression by
inflammation-associated factors (Fig.
3I-L). Collectively, these data indicated that
inflammation-associated factors increased MUC16 expression and the
level of CA125 in OC cells.
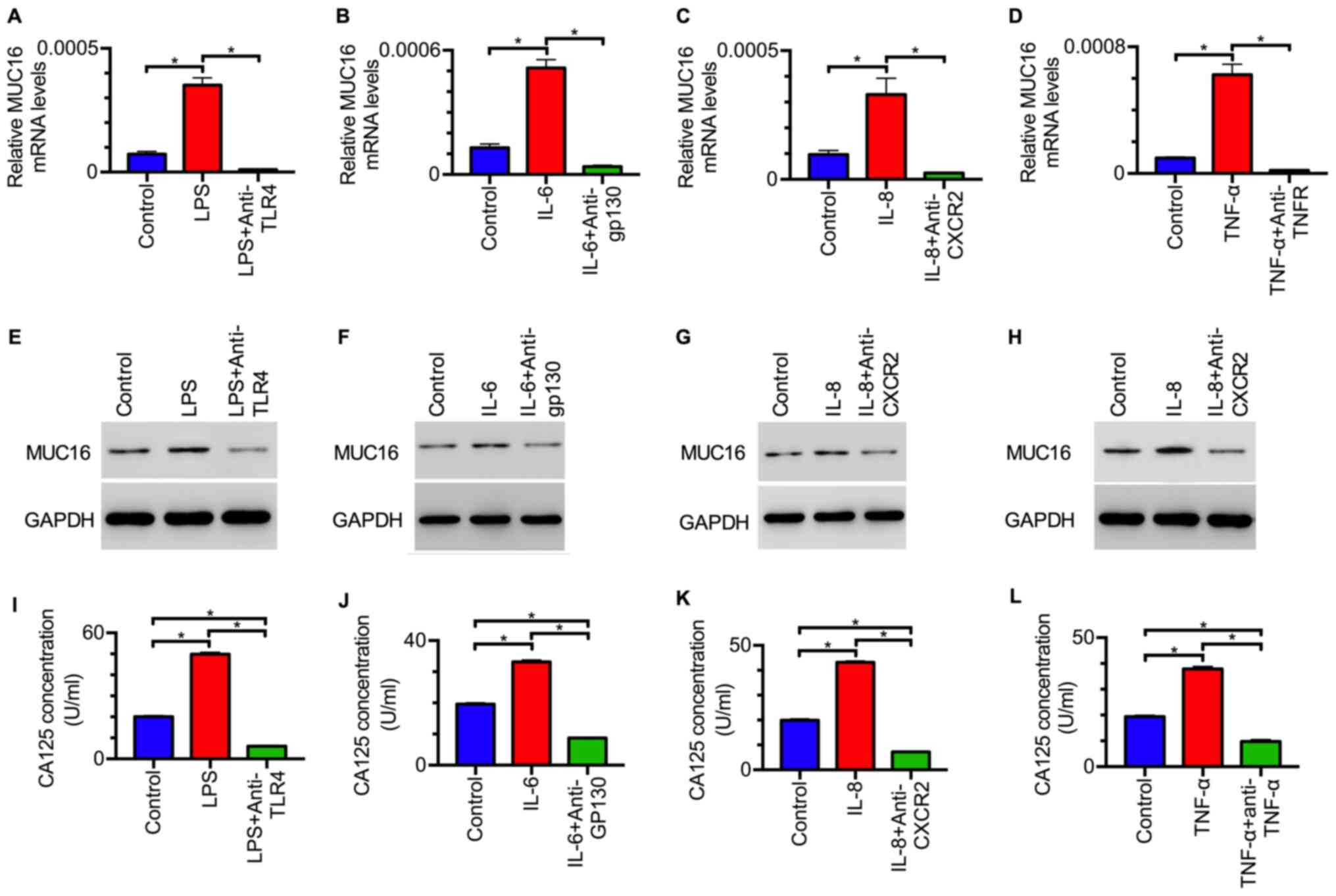 | Figure 3Inflammation-associated factors
induce MUC16 expression in ovarian cancer cells. mRNA expression
levels of MUC16 in HEY cells treated with (A) LPS or LPS +
anti-TLR4 for 48 h, (B) IL-6 or IL-6 + anti-gp130 for 24 h, (C) 50
ng/ml IL-8 or IL-8 + anti-CXCR2 for 48 h and (D) 2.5 ng/ml TNF-α or
TNF-α + anti-TNF-α for 24 h. GAPDH served as an internal control in
reverse transcription-quantitative PCR analysis. Data are presented
as the mean ± SEM of three independent experiments.
*P<0.05 as indicated. Western blot analysis of MUC16
protein expression levels in HEY cells treated with (E) LPS or LPS
+ anti-TLR4, (F) IL-6 or IL-6 + anti-gp130, (G) IL-8 or IL-8 +
anti-CXCR2 and (H) TNF-α or TNF-α + anti-TNF-α. Images are
representative of three independent experiments. Levels of CA125 in
the supernatant of HEY cells treated with (I) LPS or LPS +
anti-TLR4, (J) IL-6 or IL-6 + anti-gp130, (K) IL-8 or IL-8 +
anti-CXCR2 and (L) TNF-α or TNF-α + anti-TNF-α, as determined by
ELISA. Data are presented as the mean ± SEM of three independent
experiments. *P<0.05 as indicated. LPS,
lipopolysaccharides; IL, interleukin; TNF, tumor necrosis factor;
TLR, Toll-like receptor; MUC16, mucin 16 cell surface
associated. |
Inflammation-associated factors
activate downstream signals in OC cells
To elucidate the mechanisms underlying the
regulation of MUC16 expression by inflammation-associated factors,
the activation status of the downstream signaling molecules of each
inflammation-associated factor was investigated. The molecules of
the MAPK signaling pathway, including JNK, p38, ERK and NF-κB, were
selected for investigation, as the MAPK signaling pathways are
activated by all four factors (29-31).
RT-qPCR analysis revealed that treatment with the
inflammation-associated factors increased the mRNA expression
levels of JNK, p38, ERK and
NF-κB in tumor cells, and the addition of the
respective receptor inhibitors lowered the mRNA expression levels
of these genes (Fig. 4A-D). In
addition, the protein expression levels and phosphorylation status
of these molecules were also investigated via western blot
analysis. The results demonstrated that the expression of receptors
including TLR4, IL-6R, CXCR2, TNFRI and TNFRII and the levels of
p-JNK1/2, p-p38, p-ERK1/2 and NF-κB were upregulated by
inflammation-associated factors, and phosphorylation was inhibited
by their receptor inhibitors, whereas the protein expression levels
of JNK1/2, p-38 and ERK1/2 exhibited no marked changes in
expression when compared with the control group for all four
mediators (Fig. 4E-H). The
upregulation of receptor (TLR4, IL-6R, CXCR2, TNFRI and TNFRII)
expression and phosphorylation levels of JNK1/2, p-38 and ERK1/2
were inhibited by inflammation-associated factor receptor
inhibitors, suggesting that these inflammatory mediators activated
downstream signaling cascades in HEY cells (Fig. 4E-H). Taken together, these data
suggested that LPS, IL-6, IL-8 and TNF-α induced the expression and
activation of molecules, including JNK, p38, ERK, NF-κB, in
downstream signaling cascades.
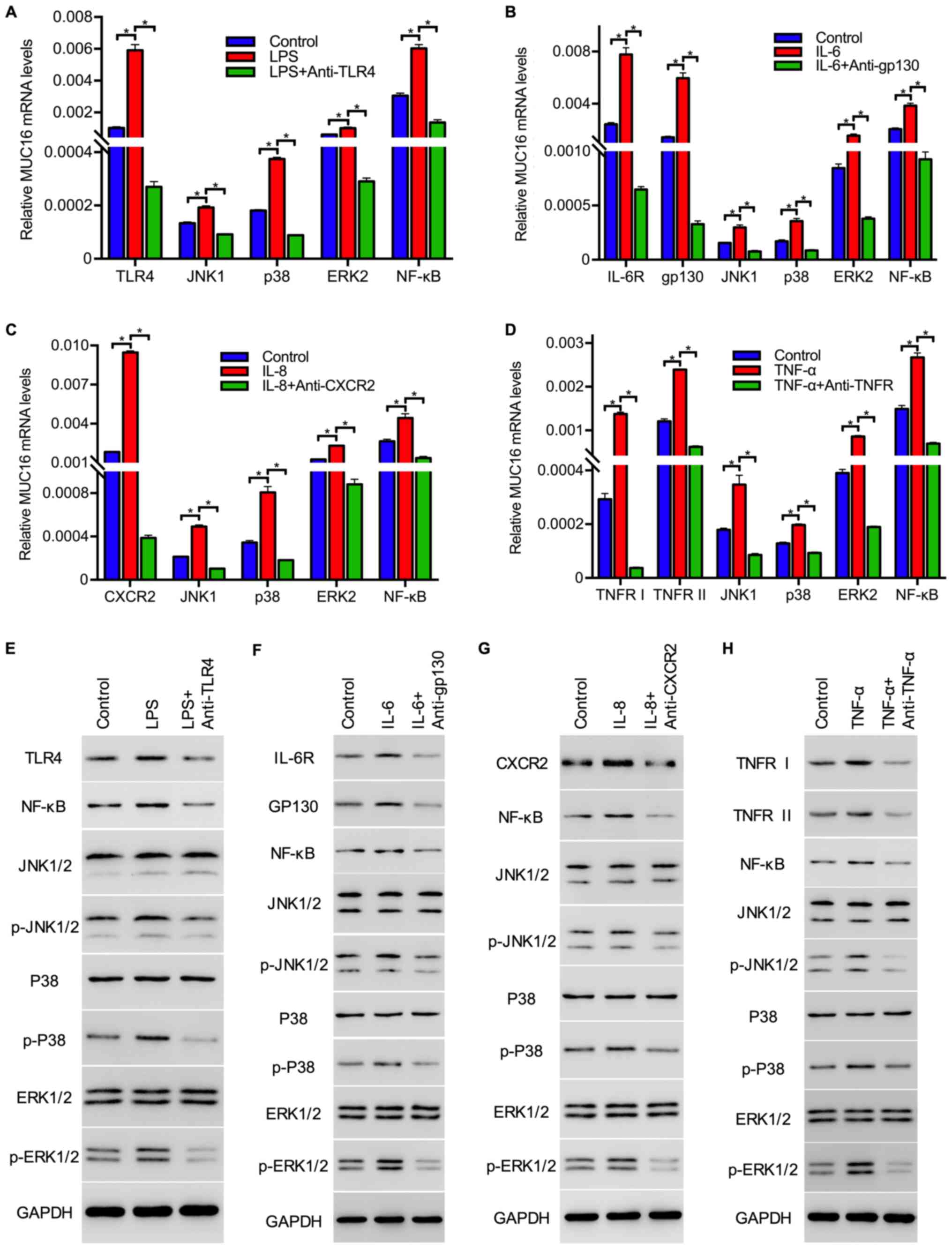 | Figure 4Inflammation-associated factors
stimulate the expression and activation of cell proliferation
signals in ovarian cancer cells. Expression of TLR4, JNK1, p38,
ERK2 and NF-κB in HEY cells treated without or (A) with LPS or LPS
+ anti-TLR4, (B) IL-6 or IL-6 + anti-gp130, (C) IL-8 or IL-8 +
anti-CXCR2 and (D) TNF-α or TNF-α + anti-TNF-α as detected by
reverse transcription-quantitative PCR analysis. GAPDH served as an
internal control. Data are presented as the mean ± SEM of three
independent experiments. *P<0.05 as indicated.
Expression and phosphorylation levels of inflammation-associated
factor receptors, molecules in the MAPK signaling pathway and NF-κB
in HEY cells treated with (E) LPS or LPS + anti-TLR4, (F) IL-6 or
IL-6 + anti-gp130, (G) IL-8 or IL-8 + anti-CXCR2 and (H) TNF-α or
TNF-α + anti-TNF-α, as evaluated by western blotting. NF-κB,
nuclear factor-κB; LPS, lipopolysaccharides; IL, interleukin; TNF,
tumor necrosis factor; TLR, Toll-like receptor; MAPK,
mitogen-activated protein kinase; IL-6R, IL-6 receptor; TNFR, TNF
receptor; gp130, membrane glycoprotein 130. |
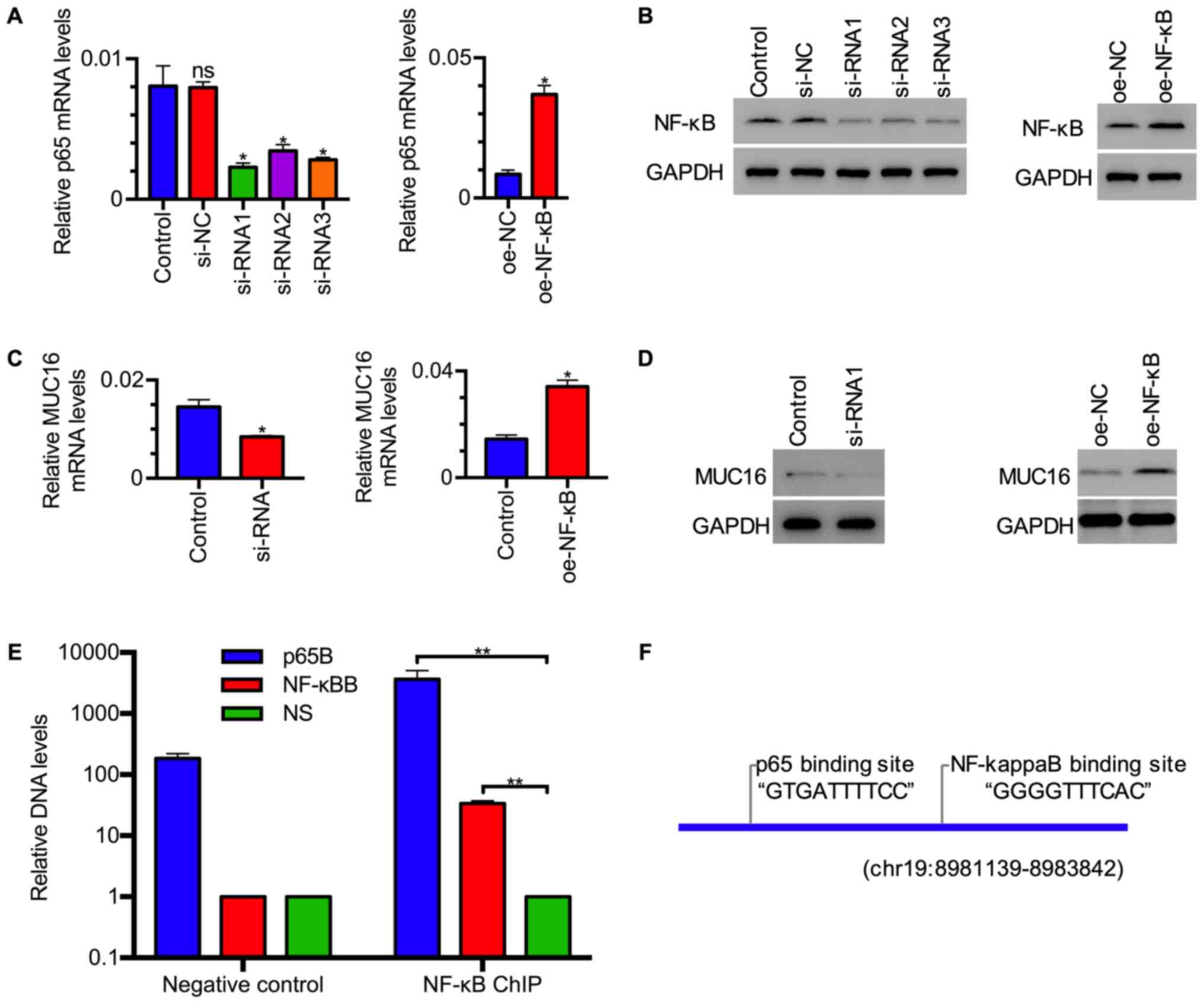
NF-κB/p65 enhances MUC16 expression by
binding to its promoter
Having established that inflammation-associated
factor treatment upregulated MUC16 and NF-κB expression in HEY
cells, the present study sought to investigate whether NF-κB, a
canonical transcription factor, mediated the upregulation of MUC16
by inflammation-associated factors. To this end, transfection of
plasmid expressing NF-κB or siRNAs targeting NF-κB into HEY cells
was performed to upregulate or downregulate NF-κB expression,
respectively (Fig. 5A and B), and then MUC16 expression was assessed.
RT-qPCR analysis revealed that the mRNA expression levels of MUC16
were increased in tumor cells with NF-κB overexpression and
decreased in tumor cells with NF-κB knockdown (Fig. 5C). Western blot analysis revealed
that the protein expression levels of MUC16 exhibited a similar
expression pattern as its mRNA in tumor cells with NF-κB
overexpression or knockdown (Fig.
5D). Moreover, ChIP assay was performed using NF-κB antibody to
investigate how NF-κB regulates MUC16 gene expression. Quantitative
analysis revealed that DNA levels amplified by p65B and NF-κB
primers were higher compared with those by NS primers in samples
immunoprecipitated by p65 antibody, while p65B primers amplified
more DNA in p65 ChIP compared with negative control (Fig. 5E). These data revealed that two
sites from the MUC16 promoter, identified as potential
NF-κB-binding sites by bioinformatics analysis (Fig. 5F), were enriched by ChIP assay,
suggesting that NF-κB binds to these sites on the MUC16 promoter.
These data indicated that NF-κB may activate MUC16 transcription by
binding to its promoter.
Discussion
MUC16, one of the main biomarkers of OC, is involved
in OC development and metastasis, and has been found to be
associated with poor prognosis (7,8).
However, little is known on the association between MUC16
expression and inflammation-associated factors. In the present
study, it was observed that the inflammation-associated factors
LPS, IL-6, IL-8 and TNF-α increased the expression levels of MUC16
and enhanced CA125 release in OC. Moreover, it was demonstrated
that NF-κB mediated regulation of MUC16 via directly binding to the
promoter of MUC16. The finding on the upregulation of MUC16 by
TNF-α was consistent with the observations reported by Morgado
et al (26) who indicated
that TNF-α and IFN-γ stimulated MUC16 expression in OC cells via
NF-κB activation. The present study demonstrated that NF-κB
mediated not only the TNF-α regulation of MUC16, but also LPS, IL-6
and IL-8 regulation, suggesting that NF-κB may be one of the main
transcriptional factors regulating MUC16 expression in OC. These
findings indicated that inflammatory factors regulated MUC16
expression in OC cells and NF-κB may have a role in this
process.
The finding that inflammatory factors regulated
MUC16 expression in OC may improve the understanding of how
inflammation contributes to OC. Inflammation is a hallmark of
cancer that contributes to the occurrence and development of
various tumors, including OC (16,32).
An increasing number of studies have uncovered the role of
inflammation in the initiation and progression of OC, with the
proinflammatory cytokine IL-6 established as a key immunoregulatory
cytokine (31,33,34).
IL-6 was reported to enhance the migratory ability of tumor cells
via increasing MMP9 expression (23). Furthermore, together with IL-8, IL-6
also markedly promoted the proliferation of OC cells in a time- and
dose-dependent manner (35). TNF-α,
another important inflammatory factor, was found to promote tumor
cell migration by upregulating CXCR4 via NF-κB activation in OC
cells (36). Upregulation of MUC16
expression by these inflammation-associated factors in the present
study elucidated another mechanism underlying the effects of
inflammatory factors on OC.
In addition, inflammatory factor-mediated regulation
of MUC16 may explain the high false-positive rate of CA125 in OC
diagnosis. Inflammation is involved in diverse biological and
pathological processes, including non-malignant diseases and
tumors. For example, some patients with endometriosis have been
found to have elevated serum and peritoneal fluid IL-6 levels
(37,38), while an IL-6/TNF-α-based model has
been reported as a potential predictor of chronic endometritis
(39). Some inflammatory and
autoimmune diseases, including rheumatoid arthritis, systemic lupus
erythematosus, Crohn's disease and asthma, have also been
associated with an increased serum IL-6 level (40). It was also reported that 8, or a
combination of IFN-α with TNF-α/IL-17, increased the expression of
MUC16 in human ocular surface epithelial cells (27). Moreover, anti-inflammatory agents,
such as dexamethasone, were reported to upregulate MUC16 in human
corneal epithelial cells (41),
suggesting that some inflammatory factors exert different roles to
IL-6, IL-8 and TNF-α in MUC16 regulation. These findings indicated
that inflammation may increase MUC16 expression in non-malignant
diseases and suggested that other factors should be taken into
consideration together with CA125 in OC diagnosis.
Notably, it was observed that the expression of the
receptors of inflammation-associated factors was upregulated in HEY
cells when treated with their ligands. This type of positive
feed-forward loop has also been reported in hepatocytes (42) and bronchial epithelial cells
(43), while IL-6 treatment
decreased IL-6R expression in primary monocytes (42) and NK92 cells (an IL-2-dependent
natural killer cell line) (44).
These contradictory findings suggested that inflammatory signaling
pathways display cell-specific regulation, which requires further
investigation. In addition, unlike other inflammatory signaling
molecules, JNK1/2, p38 and ERK1/2 protein expression levels in HEY
cells exhibited no synchronous elevation with their mRNA expression
levels when treated with inflammatory factors, which may be
attributed to complex biochemical processes, such as time and space
interval between mRNA transcription and protein translation,
post-translational modification (45), and regulation of inflammatory
signaling networks. More sophisticated experiments involving these
aspects should be performed to elucidate the differences between
mRNA and protein expression levels.
In conclusion, the present study demonstrated that
inflammatory factors, including LPS, IL-6, IL-8 and TNF-α,
upregulated the expression levels of MUC16 in OC cells via NF-κB.
These findings may improve the understanding of the molecular
mechanisms underlying the regulation of MUC16 expression and
uncover the association between inflammation and MUC16 expression
in OC. In addition, these findings suggested that inflammatory
factors may represent promising targets for OC therapy or
diagnosis, along with MUC16/CA125, although further investigation
is required to verify the association of inflammation with MUC16
expression and serum CA125 concentration.
Acknowledgements
Not applicable.
Funding
The present study was supported by funds from the
National Natural Science Foundation of China (grant no. 81874104)
and the Shanghai Municipal Commission of Science and Technology
(grant no. 18411964100).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL, LLi, ZC and XX were responsible for the research
conception and design, analysis and interpretation of the data, the
statistical analysis and manuscript drafting. NL, QL, ZC and XX
contributed to data acquisition, analysis and interpretation of the
data, and critical revision of the manuscript for important
intellectual content. LLiu and DC performed the systematic search
of the literature, and contributed to the acquisition, analysis and
interpretation of the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388.
2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Aithal A, Rauth S, Kshirsagar P, Shah A,
Lakshmanan I, Junker WM, Jain M, Ponnusamy MP and Batra SK: MUC16
as a novel target for cancer therapy. Expert Opin Ther Targets.
22:675–686. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Baek MH, Lee SW, Park JY, Rhim CC, Kim DY,
Suh DS, Kim JH, Kim YM, Kim YT and Nam JH: Preoperative predictive
factors for complete cytoreduction and survival outcome in
epithelial ovarian, tubal, and peritoneal cancer after neoadjuvant
chemotherapy. Int J Gynecol Cancer. 27:420–429. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Markman M, Federico M, Liu PY, Hannigan E
and Alberts D: Significance of early changes in the serum CA-125
antigen level on overall survival in advanced ovarian cancer.
Gynecol Oncol. 103:195–198. 2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Henderson JT, Webber EM and Sawaya GF:
Screening for ovarian cancer: Updated evidence report and
systematic review for the US preventive services task force. JAMA.
319:595–606. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Thériault C, Pinard M, Comamala M,
Migneault M, Beaudin J, Matte I, Boivin M, Piché A and Rancourt C:
MUC16 (CA125) regulates epithelial ovarian cancer cell growth,
tumorigenesis and metastasis. Gynecol Oncol. 121:434–443.
2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Coelho R, Marcos-Silva L, Ricardo S, Ponte
F, Costa A, Lopes JM and David L: Peritoneal dissemination of
ovarian cancer: Role of MUC16-mesothelin interaction and
implications for treatment. Expert Rev Anticancer Ther. 18:177–186.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Comamala M, Pinard M, Thériault C, Matte
I, Albert A, Boivin M, Beaudin J, Piché A and Rancourt C:
Downregulation of cell surface CA125/MUC16 induces
epithelial-to-mesenchymal transition and restores EGFR signalling
in NIH: OVCAR3 ovarian carcinoma cells. Br J Cancer. 104:989–999.
2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gubbels JA, Belisle J, Onda M, Rancourt C,
Migneault M, Ho M, Bera TK, Connor J, Sathyanarayana BK, Lee B, et
al: Mesothelin-MUC16 binding is a high affinity, N-glycan dependent
interaction that facilitates peritoneal metastasis of ovarian
tumors. Mol Cancer. 5(50)2006.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shah JS, Gard GB, Yang J, Maidens J,
Valmadre S, Soon PS and Marsh DJ: Combining serum microRNA and
CA-125 as prognostic indicators of preoperative surgical outcome in
women with high-grade serous ovarian cancer. Gynecol Oncol.
148:181–188. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jacobs IJ, Fay TN, Stabile I, Bridges JE,
Oram DH and Grudzinskas JG: The distribution of CA 125 in the
reproductive tract of pregnant and non-pregnant women. Br J Obstet
Gynaecol. 95:1190–1194. 1988.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Buamah P: Benign conditions associated
with raised serum CA-125 concentration. J Surg Oncol. 75:264–265.
2000.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bast RC Jr, Klug TL, St John E, Jenison E,
Niloff JM, Lazarus H, Berkowitz RS, Leavitt T, Griffiths CT, Parker
L, et al: A radioimmunoassay using a monoclonal antibody to monitor
the course of epithelial ovarian cancer. N Engl J Med. 309:883–887.
1983.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Matte I, Garde-Granger P, Bessette P and
Piché A: Ascites from ovarian cancer patients stimulates MUC16
mucin expression and secretion in human peritoneal mesothelial
cells through an Akt-dependent pathway. BMC Cancer.
19(406)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Worzfeld T, Pogge von Strandmann E, Huber
M, Adhikary T, Wagner U, Reinartz S and Müller R: The unique
molecular and cellular microenvironment of ovarian cancer. Front
Oncol. 7(24)2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Savant SS, Sriramkumar S and O'Hagan HM:
The role of inflammation and inflammatory mediators in the
development, progression, metastasis, and chemoresistance of
epithelial ovarian cancer. Cancers (Basel). 10(251)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sanguinete MMM, Oliveira PH, Martins-Filho
A, Micheli DC, Tavares-Murta BM, Murta EFC and Nomelini RS: Serum
IL-6 and IL-8 correlate with prognostic factors in ovarian cancer.
Immunol Invest. 46:677–688. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kampan NC, Madondo MT, McNally OM,
Stephens AN, Quinn MA and Plebanski M: Interleukin 6 present in
inflammatory ascites from advanced epithelial ovarian cancer
patients promotes tumor necrosis factor receptor 2-expressing
regulatory T cells. Front Immunol. 8(1482)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Isobe A, Sawada K, Kinose Y, Ohyagi-Hara
C, Nakatsuka E, Makino H, Ogura T, Mizuno T, Suzuki N, Morii E, et
al: Interleukin 6 receptor is an independent prognostic factor and
a potential therapeutic target of ovarian cancer. PLoS One.
10(e0118080)2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhu X, Shen H, Yin X, Long L, Chen X, Feng
F, Liu Y, Zhao P, Xu Y, Li M, et al: IL-6R/STAT3/miR-204 feedback
loop contributes to cisplatin resistance of epithelial ovarian
cancer cells. Oncotarget. 8:39154–39166. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
So KA, Min KJ, Hong JH and Lee JK:
Interleukin-6 expression by interactions between gynecologic cancer
cells and human mesenchymal stem cells promotes
epithelial-mesenchymal transition. Int J Oncol. 47:1451–1459.
2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Roomi MW, Monterrey JC, Kalinovsky T, Rath
M and Niedzwiecki A: In vitro modulation of MMP-2 and MMP-9 in
human cervical and ovarian cancer cell lines by cytokines, inducers
and inhibitors. Oncol Rep. 23:605–614. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yang WL, Godwin AK and Xu XX: Tumor
necrosis factor-alpha-induced matrix proteolytic enzyme production
and basement membrane remodeling by human ovarian surface
epithelial cells: Molecular basis linking ovulation and cancer
risk. Cancer Res. 64:1534–1540. 2004.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Morgado M, Sutton MN, Simmons M, Warren
CR, Lu Z, Constantinou PE, Liu J, Francis LL, Conlan RS, Bast RC Jr
and Carson DD: Tumor necrosis factor-α and interferon-γ stimulate
MUC16 (CA125) expression in breast, endometrial and ovarian cancers
through NFκB. Oncotarget. 7:14871–14884. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Albertsmeyer AC, Kakkassery V,
Spurr-Michaud S, Beeks O and Gipson IK: Effect of pro-inflammatory
mediators on membrane-associated mucins expressed by human ocular
surface epithelial cells. Exp Eye Res. 90:444–451. 2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Johnson DE, O'Keefe RA and Grandis JR:
Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev
Clin Oncol. 15:234–248. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lang K, Niggemann B, Zanker KS and
Entschladen F: Signal processing in migrating T24 human bladder
carcinoma cells: Role of the autocrine interleukin-8 loop. Int J
Cancer. 99:673–680. 2002.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Waugh DJ and Wilson C: The interleukin-8
pathway in cancer. Clin Cancer Res. 14:6735–6741. 2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444.
2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kisielewski R, Tołwińska A, Mazurek A and
Laudański P: Inflammation and ovarian cancer-current views. Ginekol
Pol. 84:293–297. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
34
|
Browning L, Patel MR, Horvath EB, Tawara K
and Jorcyk CL: IL-6 and ovarian cancer: Inflammatory cytokines in
promotion of metastasis. Cancer Manag Res. 10:6685–6693.
2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wang Y, Yang J, Gao Y, Du Y, Bao L, Niu W
and Yao Z: Regulatory effect of e2, IL-6 and IL-8 on the growth of
epithelial ovarian cancer cells. Cell Mol Immunol. 2:365–372.
2005.PubMed/NCBI
|
|
36
|
Kulbe H, Hagemann T, Szlosarek PW,
Balkwill FR and Wilson JL: The inflammatory cytokine tumor necrosis
factor-alpha regulates chemokine receptor expression on ovarian
cancer cells. Cancer Res. 65:10355–10362. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang F, Wang H, Jin D and Zhang Y: Serum
miR-17, IL-4, and IL-6 levels for diagnosis of endometriosis.
Medicine (Baltimore). 97(e10853)2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Jiang J, Jiang Z and Xue M: Serum and
peritoneal fluid levels of interleukin-6 and 37 as biomarkers for
endometriosis. Gynecol Endocrinol. 35:571–575. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tortorella C, Piazzolla G, Matteo M, Pinto
V, Tinelli R, Sabbà C, Fanelli M and Cicinelli E: Interleukin-6,
1β, and tumor necrosis factor α in menstrual effluents as
biomarkers of chronic endometritis. Fertil Steril. 101:242–247.
2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yao X, Huang J, Zhong H, Shen N, Faggioni
R, Fung M and Yao Y: Targeting interleukin-6 in inflammatory
autoimmune diseases and cancers. Pharmacol Ther. 141:125–139.
2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Seo KY, Chung SH, Lee JH, Park MY and Kim
EK: Regulation of membrane-associated mucins in the human corneal
epithelial cells by dexamethasone. Cornea. 26:709–714.
2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Bauer J, Lengyel G, Bauer TM, Acs G and
Gerok W: Regulation of interleukin-6 receptor expression in human
monocytes and hepatocytes. FEBS Lett. 249:27–30. 1989.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Takizawa H, Ohtoshi T, Yamashita N, Oka T
and Ito K: Interleukin 6-receptor expression on human bronchial
epithelial cells: Regulation by IL-1 and 6. Am J Physiol.
270:L346–L352. 1996.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Böttger E, Grangeiro de Carvalho E, Meese
S, Kun JF and Esen M: Expression of interleukin-6 family receptors
in NK92 cells is regulated by cytokines and not through direct
interaction with Plasmodium falciparum-infected erythrocytes. J
Interferon Cytokine Res. 33:65–71. 2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Liu CC, Jewett MC, Chin JW and Voigt CA:
Toward an orthogonal central dogma. Nat Chem Biol. 14:103–106.
2018.PubMed/NCBI View Article : Google Scholar
|