Introduction
To date, more than 170 human papillomavirus (HPV)
genotypes have been reported, of which HPV16 is the most common
type in the oral cavity (1). Our
previous meta-analysis revealed that oral HPV infection was
significantly associated with current smoking and sexual behavior
in healthy individuals, but not with drinking (1). Additionally, men are more susceptible
to oral HPV infection than women (1,2),
indicating a sex-specific susceptibility to HPV infection in the
oral cavity. A higher seroconversion rate in response to genital
HPV infection among women may be associated with the low prevalence
of HPV16 in the oral cavity (2). A
bimodal age peak (i.e., 20-29 years and 60-69 years) for HPV16
infection in the oral cavity exists in Japanese people without oral
cancer or oral pre-malignant lesions (3). This finding suggests that a weakened
immune system due to aging may be related to oral HPV infection.
Thus, it is hypothesized that age-related changes in oral health
(e.g., chronic periodontitis and denture use) may be associated
with oral HPV infection in older people. However, the relationship
between HPV infection and oral health status in older Japanese
people remains unknown.
In the present investigation, we examined the HPV16
viral load in the oral cavity and its association with the oral
bacterial count (4). An increased
number of oral bacteria was significantly associated with a high
copy number of HPV16 DNA (4). This
result indicates that poor oral hygiene induces viral replication
of HPV in the oral cavity. However, the relationship of oral HPV16
infection with oral hygiene has not been fully elucidated among
Japanese people.
The oropharynx is a more common site for HPV16
infection than the oral cavity and nasopharynx (5). Viral organisms are thought to become
trapped in the tonsillar crypts and then infect basal keratinocytes
by invading the epithelium (5).
Thus, it is likely that sensitivity to HPV infection is associated
with the histological features of the local region. In the oral
cavity, periodontal pockets are thought to serve as an important
reservoir of viral organisms. Additionally, inflammatory
periodontal tissue may provide a chance for exposed basal cells to
become infected with HPV.
The objective of this study was to conduct a
preliminary investigation into the association between HPV16 DNA
prevalence and oral health status in older people. Males are
significantly associated with a higher prevalence of oral HPV, and
this sex difference may be an important factor affecting oral HPV
infection rates (2,3). Therefore, we targeted older women to
exclude the effect of sex differences on oral HPV infection in this
study.
Materials and methods
Subjects
We targeted 52 women aged ≥ 60 years who visited the
Department of Oral Health of Hiroshima University Hospital from
August 2018 to February 2019. We excluded patients with oral cancer
or potentially malignant oral disorders (e.g., leukoplakia,
erythroplakia or lichen planus) (n=1), those receiving chemotherapy
and radiotherapy for cancer (n=3), those with severe
immunodeficiency and auto-immune disease (n=2) and current smokers
(n=0) because such clinical factors are thought to be risk factors
for oral HPV infection (1).
Finally, we analyzed 46 female patients aged ≥ 60 years (mean age
74.6 years, range 60-94 years). The study design was approved by
the Ethics Committee of Hiroshima University (no. E-1022) and all
participants signed an informed consent agreement.
Oral rinse sample collection and DNA
extraction
An oral rinse sample was collected after subjects
rinsed with 10 ml of saline for 15 sec. Immediately after
collection, all samples were centrifuged at 2,580 x g for 10 min,
then the supernatant was decanted and the pellets were stored at
-80˚C until further processing. DNA was extracted from the oral
rinse sample and purified using a PureLink™ Microbiome DNA
Purification Kit (Thermo Fisher Scientific, Inc.).
Oral examination
After collection of the oral rinse sample, an oral
examination including a periodontal examination (i.e., assessment
of probing depth and bleeding on probing) was performed by an
experienced dentist from the Oral Health Department at Hiroshima
University Hospital. Probing depth and bleeding on probing was
examined at six sites (mesiobuccal, buccal, distobuccal,
mesiolingual, lingual and distolingual) on all remaining teeth. The
number of remaining teeth and denture use were also recorded. Next,
dental plaque accumulation was examined using a plaque disclosing
agent. Plaque Control Record scores were recorded by an experienced
dental hygienist (6). The
intrarater reliability of the dental hygienist was assessed using
an intraclass correlation coefficient. A dental hygienist
calculated PCR scores of 10 patients two times using intraoral
photographs of teeth after use of the plaque disclosing agent. The
calculated value of the intraclass correlation coefficient was
0.978, suggesting that the dental hygienist had excellent
reliability, when using previously reported criteria for intraclass
correlation coefficient (7).
Quantitation of human cell
numbers
Human cell numbers were quantified by our previously
reported method (3). Briefly,
serial 10-fold dilutions of the pUC57 vector inserted into the
human endogenous retrovirus group 3 member 1 (ERV3-1) genome were
made with copy numbers ranging from 100 to 109. DNA levels were
quantitated using a CFX connect real-time PCR detection system
(Bio-Rad Laboratories) with a reaction mixture containing 1.0 µl of
DNA, 9.0 µl of Thunderbird SYBR qPCR Mix (Toyobo Life Science), and
each pair of oligonucleotide primers. Amplifications were performed
with initial melting at 95˚C for 5 min, then 35 cycles of
denaturing at 95˚C for 30 sec, annealing at 56˚C for 30 sec, and
extension at 72˚C for 1 min. A standard curve indicating cycle
threshold (CT) value versus ERV3-1 copy number was used to estimate
the number of human cells in each sample.
HPV16 DNA detection
According to our previously reported method
(4), quantitative PCR assays were
performed using a CFX Connect real-time PCR detection system
(Bio-Rad Laboratories) to detect HPV16 DNA in the samples. We
prepared DNA samples extracted from approximately 1,000-5,000 human
cells from each PCR mixture. The primer sequences for HPV16
E6 were 5'-AAGGGCGTAACCGAAATCGGT-3' (sense) and
5'-GTTTGCAGCTCTGTGCATA-3' (antisense). A positive control
containing HPV16 DNA extracted from Caski cells was used for the
PCR reactions. Copy numbers above the detection limit in a standard
curve for HPV16 DNA were assessed as HPV16 positive. Sequencing for
the HPV16 E6 region was performed to confirm the HPV16 DNA sequence
of PCR products (140 bp). The BigDye Terminator v3.1 cycle
sequencing kit (Thermo Fisher Scientific, Inc.) was used for the
sequencing reaction. Amplified DNA fragments were sequenced with an
ABI PRISM 310 genetic analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc.).
Detecting periodontal disease-related
bacteria by PCR
The bacterial 16S rRNA gene was employed to detect
periodontal disease-related bacteria using PCR. One microliter of
DNA sample was used for the PCR. PCR products were amplified with
GoTaq® Green Master Mix (Promega) and previously
reported primers. Amplifications were performed with initial
melting at 95˚C for 2 min, then 35 cycles of denaturing at 95˚C for
1 min, annealing at 57˚C for 1 min, and extension at 72˚C for 1
min. PCR primer sets reported previously are as follows (8,9):
Porphyromonas gingivalis, 5'-AGGCAGCTTGCCATACT GCG-3'
(sense) and 5'-ACTGTTAGCAACTACCGATGT-3' (antisense); Tannerella
forsythia, 5'-GCGTATGTAACCTGCCC GCA-3' (sense) and
5'-TGCTTCAGTGTCAGTTATACCT-3' (antisense); Treponema
denticola, 5'-TAATACCGAATGTGCT CATTTACAT-3' (sense) and
5'-TCAAAGAAGCATTCCCTCTT CTTCTTA-3' (antisense); Prevotella
intermedia, 5'-TTTGTT GGGGAGTAAAGCGGG-3' (sense) and
5'-TCAACATCTCTG TATCCTGCGT-3' (antisense); and Fusobacterium
nucleatum, 5'-AGGGCATCCTAGAATTATG-3' (sense) and 5'-GGGACA
CTGAAACATCTCTGTCTCA-3' (antisense). Primer sequences of all
bacterial 16S rRNA were 5'-CGTTAGTAATCGTGGAT CAGAATG-3' (sense) and
5'-TGTGACGGGCGGTGTGTA-3' (antisense). After the PCR reaction, 9 µl
of PCR product was electrophoresed on 2% agarose gels with ethidium
bromide staining.
16S rRNA gene sequencing for taxonomic
classification of bacteria
According to our previously reported method
(4), bacterial flora analysis of
the 16S ribosomal RNA gene was performed using DNA obtained from
oral rinse samples. Briefly, amplicons were produced, cleaned and
sequenced according to the 16S Metagenomic Sequencing Library
Preparation (Illumina Inc., Part 15044223, Rev. B). After the first
and second PCR reaction, the final library was subjected to
paired-end sequencing using a MiSeq Reagent Kit v.3 on the Illumina
MiSeq platform (Illumina Inc.). After denoising the sequence data,
low quality sequences were eliminated and the sequences were
clustered into operational taxonomic units (OTUs) at 97% identity
using the CD-HIT-OTU pipeline (10). The Quantitative Insights into
Microbial Ecology (QIIME) pipeline was used to analyze the
taxonomic classification (11).
Statistical analysis
Statistical analysis was performed using SPSS
software, version 24.0 (SPSS Inc.). The χ2 test or
Fisher's exact test were used to evaluate significant differences
between positive rates of HPV16 DNA and clinical factors. The
Student's t-test or Mann-Whitney U test were used to evaluate
significant differences in age, remaining teeth, PCR record and
average percentage of bacterial genera between HPV16 DNA positive
and negative cases. If the Shapiro-Wilk test was statistically
significant, the Mann-Whitney U test was used as a non-parametric
test. Intrarater reliability was assessed using the intraclass
correlation coefficient (1,1). P < 0.05 was considered to indicate
statistical significance.
Results
Association between HPV16 DNA
positivity and clinical factors
HPV16 DNA positivity was determined in a total of 46
oral rinse samples using quantitative PCR. The CT value of
quantitative PCR was above the detection limit in a standard curve
in four of the 46 samples (8.7%). Additionally, PCR products of the
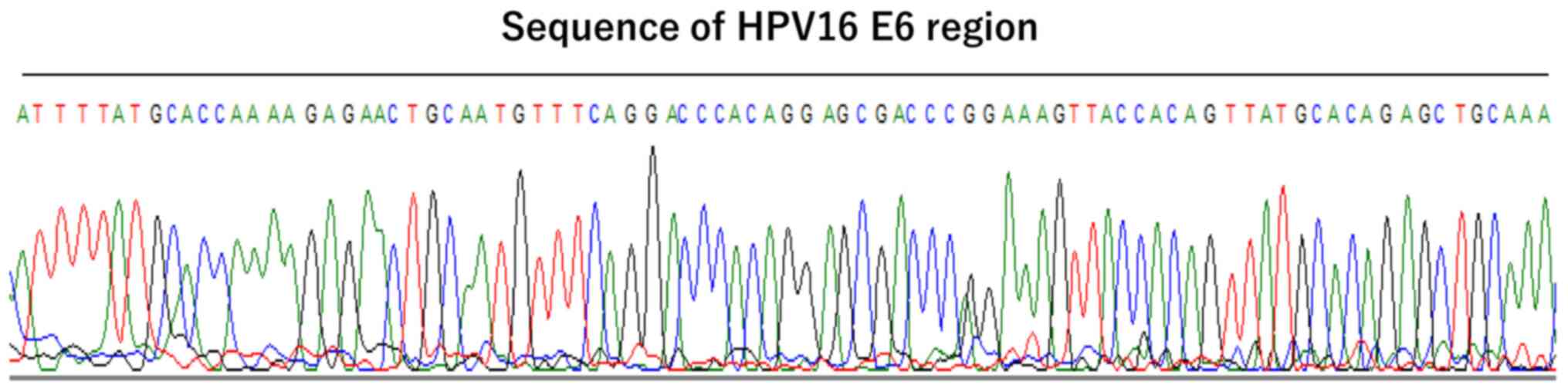
four samples were examined by sequencing for the HPV16 E6 region.
The sequence of the HPV16 E6 region was confirmed in the four
samples (Fig. 1). Thus, four cases
were determined to be HPV16 DNA positive. Table I summarizes the association between
HPV16 DNA detected by PCR and clinical factors. There was no
significant difference between the HPV16 DNA positivity rate and
clinical factors (i.e., sex, age, remaining teeth, denture use, and
medical history). Individuals with a medical history of
hyperlipidemia recorded a higher HPV16 DNA positivity rate (33.3%)
compared with those without (5.0%), but the difference was not
significant (Table I).
 | Table IAssociation between oral HPV16 DNA
and clinical parameters. |
Table I
Association between oral HPV16 DNA
and clinical parameters.
| | HPV16 DNA | |
|---|
| Clinical factor
(n) | (-) | (+) | P-value |
|---|
| Age, mean ± SD | 74.9±7.4 | 72.0±8.6 | 0.56 |
| Remaining teeth,
mean ± SD | 23.4±6.5 | 22.3±7.9 | 0.72 |
| Partial denture
user, n (%) | | | 0.35 |
|
Non-user
(32) | 30 (93.8) | 2 (6.2) | |
|
User
(14) | 12 (85.7) | 2 (14.3) | |
| Hypertension, n
(%) | | | 0.65 |
|
No (32) | 29 (90.6) | 3 (9.4) | |
|
Yes
(14) | 13 (92.9) | 1 (7.1) | |
| Diabetes, n
(%) | | | 0.50 |
|
No (39) | 36 (92.3) | 3 (7.7) | |
|
Yes (7) | 6 (85.7) | 1 (14.3) | |
| Hyperlipidemia, n
(%) | | | 0.08 |
|
No (40) | 38 (95.0) | 2 (5.0) | |
|
Yes (6) | 4 (66.7) | 2 (33.3) | |
| Stroke, n (%) | | | 0.91 |
|
No (45) | 41 (91.1) | 4 (8.9) | |
|
Yes (1) | 1 (100.0) | 0 (0.0) | |
| Heart disease,
(%) | | | 0.91 |
|
No (45) | 41 (91.1) | 4 (8.9) | |
|
Yes (1) | 1 (100.0) | 0 (0.0) | |
Association between HPV16 DNA
positivity and oral health status
Next, we examined the relationship between HPV16 DNA
positivity and dental plaque and periodontal status (Table II). HPV16 DNA positive patients
recorded higher Plaque Control Record scores than HPV16 DNA
negative patients, but a significant difference was not found.
Three of the 14 participants with periodontal pockets ≥ 6 mm were
HPV16 DNA positive (21.4%). However, no significant association was
found between probing depth and HPV16 DNA positivity. Importantly,
all HPV16 DNA positive participants exhibited bleeding on probing.
There was a significant association between HPV16 DNA positivity
and bleeding on probing (P=0.03).
 | Table IIAssociation between oral HPV16 DNA
and oral health status. |
Table II
Association between oral HPV16 DNA
and oral health status.
| | HPV16 DNA | |
|---|
| Factor (n) | (-) | (+) | P-value |
|---|
| Percentage plaque
control record scores, mean ± SD | 30.6±17.9 | 45.9±11.1 | 0.06 |
| Probing depth, n
(%) | | | 0.09 |
|
<4 mm
(20) | 20 (100.0) | 0 (0.0) | |
|
≥4 mm and
<6 mm (12) | 11 (91.7) | 1 (8.3) | |
|
≥6 mm
(14) | 11 (78.6) | 3 (21.4) | |
| Bleeding on
probing, n (%) | | | 0.03 |
|
No (26) | 26 (100.0) | 0 (0.0) | |
|
Yes
(20) | 16 (80.0) | 4 (20.0) | |
Association between HPV16 DNA
positivity and periodontal disease-related bacteria
Associations between HPV16 DNA positivity and
periodontal disease-related bacteria are summarized in Table III. T. forsythia, P.
intermedia and F. nucleatum positive participants
exhibited a higher HPV16 DNA positivity rate than T.
forsythia, P. intermedia or F. nucleatum negative
participants (12.1 vs. 0.0%, 33.3 vs. 3.8%, and 10.8 vs. 0.0%,
respectively). Importantly, there was a significant association
between HPV16 DNA positivity and P. intermedia (P=0.02).
 | Table IIIAssociation between oral HPV16 DNA
and periodontal disease-related bacteria. |
Table III
Association between oral HPV16 DNA
and periodontal disease-related bacteria.
| | HPV16 DNA | |
|---|
| Clinical factor
(n) | (-) | (+) | P-value |
|---|
| P.
gingivalis, n (%) | | | 0.47 |
|
Negative
(17) | 15 (88.2) | 2 (11.8) | |
|
Positive
(29) | 27 (93.1) | 2 (6.9) | |
| T.
forsythia, n (%) | | | 0.25 |
|
Negative
(13) | 13 (100.0) | 0 (0.0) | |
|
Positive
(33) | 29 (87.9) | 4 (12.1) | |
| T.
denticola, n (%) | | | 0.63 |
|
Negative
(25) | 23 (92.0) | 2 (8.0) | |
|
Positive
(21) | 19 (90.5) | 2 (9.5) | |
| P.
intermedia, n (%) | | | 0.02 |
|
Negative
(37) | 36 (96.2) | 1 (3.8) | |
|
Positive
(9) | 6 (66.7) | 3 (33.3) | |
| F.
nucleatum, n (%) | | | 0.41 |
|
Negative
(9) | 9 (100.0) | 0 (0.0) | |
|
Positive
(37) | 33 (89.2) | 4 (10.8 ) | |
16S rRNA gene sequence analysis in
participants with periodontitis
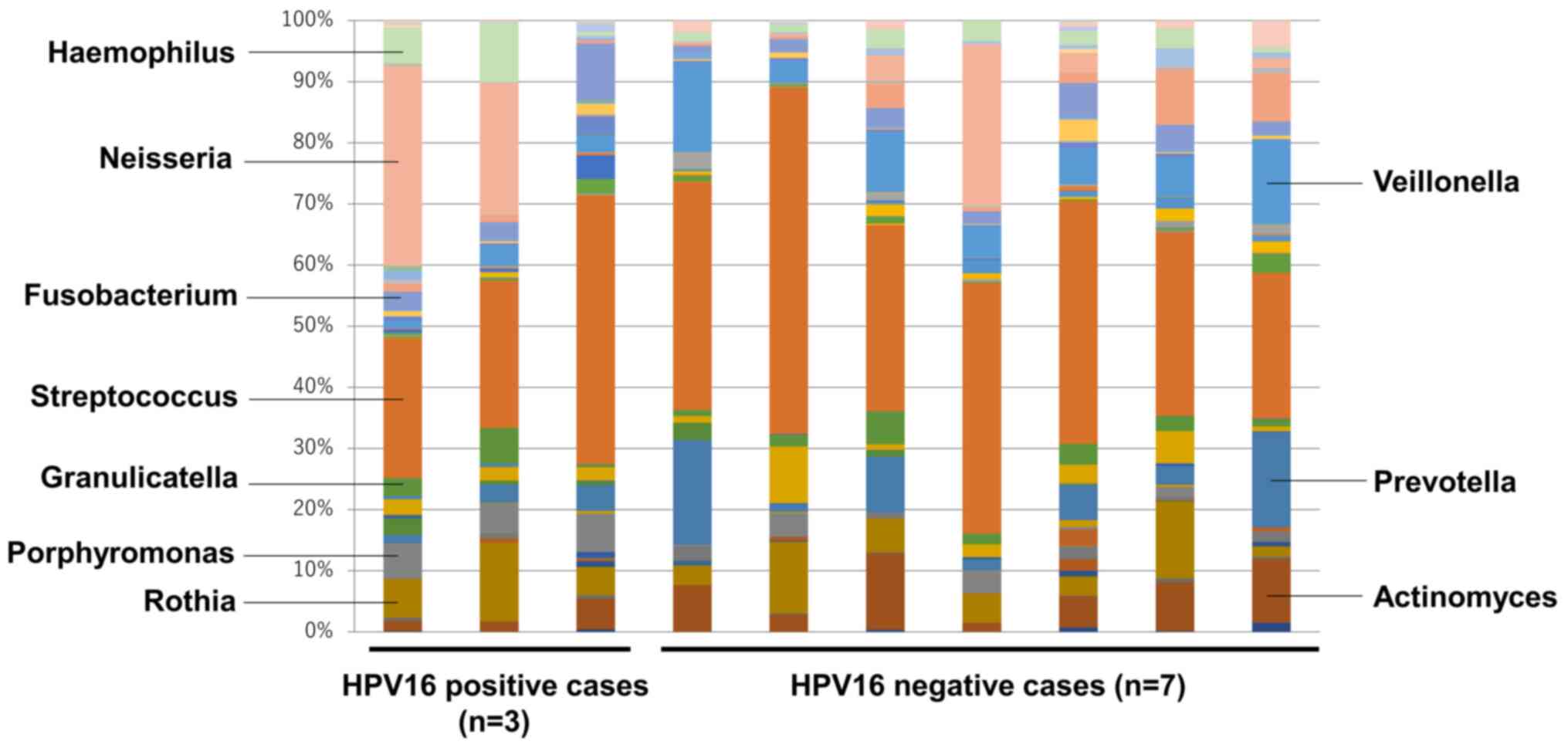
We performed analysis of 16S ribosomal RNA in
bacterial flora using oral rinse samples to examine microbiome
diversity in 10 participants with ≥ 6 mm periodontal pockets and
bleeding on probing (Fig. 2). It is
speculated that HPV16 infection may be associated with particular
bacteria in deep periodontal pockets with inflammation. Therefore,
we analyzed the association between HPV16 infection and the oral
microbiome in patients with periodontal pockets ≥ 6 mm and bleeding
on probing. We compared the percentage of each bacterial genus
between HPV16 DNA positive cases (n=3) and HPV16 DNA negative cases
(n=7). At the genus level, a total of 48 genera were found in the
two groups, with Streptococcus being most commonly detected
in both groups. Importantly, the average percentage of
Porphyromonas was significantly higher in HPV16 DNA positive
cases than in HPV16 DNA negative cases (5.57 vs. 1.44%) (Table IV). In contrast, the average
percentage of Veillonella was significantly lower in HPV16
DNA positive cases than in HPV16 DNA negative cases (2.43 vs.
8.51%) (Table IV). Additionally,
the percentage of Prevotella was lower in HPV16 DNA positive
cases than in HPV16 DNA negative cases (4.0 vs. 8.23%) (Table IV). However, there was no
significant difference between the groups. Among the bacterial
genera present in small proportions, there was a significant
difference in Oribacterium and Peptostreptococcus
between the groups (Table IV).
 | Table IVPercentage of bacteria genus in
participants with periodontal pockets ≥6 mm and bleeding on
probing. |
Table IV
Percentage of bacteria genus in
participants with periodontal pockets ≥6 mm and bleeding on
probing.
| | Average percentage
of bacteria genus, mean ± SD | |
|---|
| Genus | HPV16 DNA (-)
(n=7) | HPV16 DNA (+)
(n=3) | P-value |
|---|
|
Actinomyces | 6.83±3.95 | 2.80±1.91 | 0.15 |
|
Corynebacterium | 0.21±0.22 | 0.33±0.29 | 0.22 |
| Rothia | 6.09±4.33 | 7.93±4.38 | 0.58 |
|
Alloscardovia | 0.27±0.46 | 0.00±0.00 | 0.12 |
|
Bifidobacterium | 0.04±0.08 | 0.30±0.06 | 0.68 |
|
Scardovia | 0.24±0.36 | 0.3±0.52 | 0.52 |
|
Atopobium | 1.06±0.95 | 0.27±0.29 | 0.18 |
| Slackia | 0.01±0.04 | 0.07±0.06 | 0.27 |
|
Bacteroides | 0.00±0.00 | 0.33±0.58 | 0.52 |
|
Paludibacter | 0.50±1.00 | 0.00±0.00 | 0.38 |
|
Porphyromonas | 1.44±1.67 | 5.57±0.60 | 0.03 |
|
Tannerella | 0.23±0.40 | 0.23±0.23 | 0.52 |
|
Prevotella | 8.23±7.25 | 4.00±0.35 | 0.83 |
|
Capnocytophaga | 0.16±0.17 | 0.20±0.26 | 0.83 |
| SHD-231 | 0.01±0.04 | 0.00±0.00 | 0.83 |
|
Abiotrophia | 0.00±0.00 | 0.33±0.29 | 0.12 |
|
Granulicatella | 2.44±1.53 | 3.13±2.70 | 0.61 |
|
Lactobacillus | 0.03±0.05 | 0.00±0.00 | 0.52 |
|
Streptococcus | 36.99±10.66 | 30.27±11.74 | 0.40 |
|
Butyrivibrio | 0.04±0.08 | 0.00±0.00 | 0.52 |
|
Catonella | 0.24±0.40 | 0.07±0.06 | 0.27 |
|
Moryella | 1.10±0.80 | 0.33±0.40 | 0.21 |
|
Oribacterium | 0.93±0.78 | 0.10±0.00 | 0.03 |
|
Filifactor | 0.07±0.11 | 0.87±1.25 | 1.0 |
|
Peptostreptococcus | 0.09±0.09 | 1.67±1.93 | 0.02 |
|
Dialister | 0.14±0.26 | 0.23±0.15 | 0.52 |
|
Megasphaera | 0.84±0.97 | 0.10±0.10 | 0.27 |
|
Selenomonas | 0.06±0.05 | 0.00±0.00 | 0.38 |
|
Veillonella | 8.51±4.36 | 2.43±1.12 | 0.01 |
|
Mogibacterium | 0.03±0.05 | 0.03±0.06 | 0.67 |
|
Parvimonas | 0.80±1.23 | 0.97±0.70 | 0.27 |
|
Bulleidia | 0.36±0.40 | 0.17±0.21 | 0.38 |
| Sharpea | 0.00±0.00 | 0.07±0.12 | 0.52 |
|
Fusobacterium | 2.87±1.54 | 5.13±3.52 | 0.18 |
|
Leptotrichia | 3.46±3.66 | 0.97±0.42 | 0.67 |
|
Lautropia | 0.16±0.22 | 0.17±0.21 | 0.52 |
|
Eikenella | 0.03±0.05 | 0.13±0.15 | 0.67 |
|
Kingella | 0.24±0.40 | 0.07±0.06 | 0.83 |
|
Neisseria | 5.20±9.48 | 18.13±16.67 | 0.67 |
|
Desulfovibrio | 0.10±0.26 | 0.00±0.00 | 0.83 |
|
Campylobacter | 0.80±1.00 | 0.20±0.17 | 0.12 |
|
Cardiobacterium | 0.00±0.00 | 0.10±0.17 | 0.27 |
|
Actinobacillus | 0.27±0.46 | 0.00±0.00 | 0.83 |
|
Aggregatibacter | 0.00±0.00 | 0.07±0.06 | 0.12 |
|
Haemophilus | 0.01±0.04 | 5.37±4.57 | 0.52 |
|
Treponema | 0.24±0.40 | 0.37±0.55 | 0.18 |
| TG5 | 0.13±0.26 | 0.20±0.17 | 1.0 |
|
Mycoplasma | 0.06±0.11 | 0.13±0.15 | 0.38 |
Discussion
Several studies have investigated the localization
of HPV in periodontal tissues. Using in situ hybridization, Hormia
et al (12) detected
high-risk type HPV DNA in the junctional epithelium of periodontal
pockets of patients with periodontal disease. Other studies
reported that HPV16 DNA was detected in periodontal tissues and
gingival crevicular fluid (13,14).
These results indicate that periodontal tissues may serve as
important infection sites and reservoirs for HPV. However, HPV16
DNA was not detected in the gingival tissues obtained from
Brazilian patients with periodontal disease using quantitative PCR
(15). Such different results may
be attributed to differences in the sample type, detection method,
and clinical factors (i.e., age, sex, race and locality).
Histologic evaluation using in situ hybridization is
a reliable method to detect HPV DNA (12). Oral mucosa scraping is a useful
method for collection of a sufficient number of human cells
(16). In this study, we chose
sample collection via oral rinses due to the non-invasive nature of
this method. Oral rinse samples contain a mix of human cells,
saliva, and microorganisms. Syrjänen (16) reported that oral rinse samples
contain more bacteria and fewer human cells, compared with samples
obtained from mucosal scrapings. Thus, the large number of bacteria
may affect the quality of DNA used for PCR analysis in oral rinse
samples (16). Therefore, to
minimize the effect of bacteria on PCR analysis results, oral rinse
samples should be stored at -80˚C immediately after collection to
prevent bacteria growth.
Recent research has found a significant association
between oral health status and oral HPV infection in adults
(17,18). Both periodontal pockets ≥6 mm and
clinical attachment loss ≥7 mm were identified as significant risk
factors for oral HPV infection in Hispanic people (17). Dental plaque and gingival bleeding
were both significant risk factors for high-risk type HPV infection
in people aged 18-50 years without oral cancer and a history of HPV
vaccination (18). However, there
was no significant association between oral HPV16 DNA positivity
and oral health status (i.e., dental plaque, calculus and
periodontitis) in people aged 18-90 years without a history of
cancer or any systemic disease (19). The National Health and Nutrition
Examination Survey data in the USA found that there was no
significant relationship between oral HPV infection and periodontal
disease after adjusting for the participants' clinical parameters
(i.e., race, sex, age, smoking, drinking, sexual behavior and
education) (20). In this pilot
study, we found there was a significant relationship between
bleeding on probing (an important sign of tissue inflammation) and
HPV16 DNA positivity in older non-smoking people without oral
cancer or immunosuppressive conditions. One limitation of our study
is the small number of participants; however, our findings suggests
that deep periodontal pockets plus periodontal inflammation may be
importantly associated with HPV16 infection in periodontal tissues.
While it is still controversial whether deep inflammatory
periodontal pockets are a risk factor for HPV, it is significant
that periodontal tissue may act as a reservoir of HPV in the oral
cavity. In accordance with the previous reports by Cutress et
al (21), severe periodontitis
was defined as the presence of a pocket depth ≥6 mm. In addition,
de Souza et al (22)
reported that bleeding upon probing was significantly correlated
with local gingival tissue inflammation (i.e., number of
inflammatory cells per area). Bleeding upon probing was considered
a vital indicator of periodontal tissue inflammation due to the
presence of periodontal pathogens. Importantly, alveolar bone loss
can also be used to evaluate the severity of periodontitis.
However, we did not examine the severity of alveolar bone loss in
this study. Therefore, we presume that our method did not precisely
assess the degree of periodontitis (i.e., mild, moderate or
severe).
Increased HPV16 E6 viral copy numbers in the oral
cavity were associated with an increased number of bacteria,
suggesting that oral hygiene status may be involved in viral DNA
replication (4). Additionally, 16S
ribosomal RNA sequences analysis displayed a trend toward oral
microbiome differences between people with a high HPV16 viral load
and those with a low HPV16 viral load (4). For example, a higher percentage of an
anaerobic bacterial genus was found in people with a high HPV16
viral load. In this study, the positive rate of P.
intermedia was significantly higher in the HPV16 positive group
than in the HPV16 negative group. Thus, the presence of such
pathogenic bacteria may play a significant role in HPV infection in
the oral cavity. Salivary microbiomes with larger proportions of
Prevotella and Veillonella genera were associated
with periodontitis in Japanese adults (23). This observation suggests that
Prevotella and Veillonella play a vital role in the
development of periodontitis in Japanese people. In addition,
Neisseria and Haemophilus genera were present in much
higher proportions in the microbiota of Korean people than in those
of Japanese people (23),
indicating that there are geographical differences in the oral
microbiomes of the two countries. In the analysis of the vaginal
microbiome, diversity of vaginal microbiota was strongly associated
with HPV infection (24). There was
also a significant relationship between high-risk type HPV
infection and the Prevotella genus in the vagina of
HIV-negative participants (25).
These results indicate that changes in the composition of the
bacterial microbiome (i.e., increased anaerobic bacteria) play a
vital role in vaginal HPV infection. The Prevotella genus
may be associated with HPV infection in the vagina.
Several studies have found a significant association
between the oral herpes virus (e.g., cytomegalovirus, Epstein-Barr
virus, and herpes simplex virus type 1) and periodontal disease
(14,26,27).
Patients with advanced periodontitis showed positivity for the oral
herpes virus as well as HPV in the gingival crevicular fluid
(14). Periodontal tissues may be a
vital reservoir for viral organisms in the oral cavity.
Furthermore, oral herpes virus may impair the balance between the
host and the periodontal microbiome by increasing local
inflammatory cytokines and suppressing the host immune response
(26,27). It is likely that oral viral
infection is related to the severity of periodontitis as a result
of induction of local inflammatory cytokines in the periodontal
tissue. However, it remains unknown whether HPV plays as
significant a role as other oral viruses as an inducer of
periodontal inflammation. The characteristic oral microbiome may
play a role in favor of persistent HPV infection. The healthy
microbiome is thought to help maintain a normal immune system to
prevent viral infection of the host cells and progression of
disease (28). Since HPV16
infection is considered to be a risk factor for the development of
oral cavity cancer (29), it is
vital to clarify the relationship between HPV16 infection and the
oral microbiome. Takeshita et al (30) reported that Prevotella- and
Veillonella-dominant oral microbiomes were associated with
the active phase of periodontitis (i.e., bleeding on probing) among
Japanese people. However, Neisseria-, Haemophilus-,
Aggregatibacter- and Porphyromonas-dominant oral
microbiomes were related to a healthy periodontium (30). In this study, the Prevotella
and Veillonella genera were more prevalent in patients with
≥6 mm periodontal pockets and bleeding on probing without HPV16
infection than in those with HPV16 infection. Interestingly, the
Porphyromonas genus was significantly more prevalent in
patients with deep periodontal pockets and bleeding on probing with
HPV16 infection than in those without HPV16 infection. These
results indicate that people with both deep periodontal pocket
inflammation and oral HPV16 infection may not have a
Prevotella- or Veillonella-dominant oral microbiome
and may exhibit different features in their oral microbiomes.
However, it remains unknown which particular bacterial species or
composition of microbiome is more importantly linked to the
prevalence of oral HPV16 infection. Thus, further research is
required to clarify significant associations between oral bacteria
and oral HPV infection at the genus and species levels.
In conclusion, Oral HPV16 infection may be
associated with periodontal inflammation in older Japanese women.
However, how HPV localizes in periodontal tissues remains unknown.
Accordingly, further research will be necessary to clarify the
association between oral HPV infection in the periodontal tissue
and periodontal inflammation.
Acknowledgements
Not applicable.
Funding
The present study was supported by a Grant-in-aid
for Scientific Research (C) (grant no. 15K11290) from the Ministry
of Education, Culture, Sports and Technology of Japan.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HS designed the study, performed experiments,
analyzed and interpreted the data and wrote the paper. CYS
performed experiments, and analyzed and interpreted the data. YK,
MM, MN, MI and AS performed experiments. KO and MS discussed and
interpreted the data and aided in writing the paper. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The study design was approved by the Ethics
Committee of Hiroshima University (approval no. E-1022) and all
participants signed an informed consent agreement.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shigeishi H and Sugiyama M: Risk factors
for oral human papillomavirus infection in healthy individuals: A
systematic review and meta-analysis. J Clin Med Res. 8:721–729.
2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gillison ML, Broutian T, Pickard RK, Tong
ZY, Xiao W, Kahle L, Graubard BI and Chaturvedi AK: Prevalence of
oral HPV infection in the United States, 2009-2010. JAMA.
307:693–703. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Shigeishi H, Sugiyama M, Ohta K, Rahman MZ
and Takechi M: Higher prevalence and gene amplification of HPV16 in
oropharynx as compared to oral cavity. J Appl Oral Sci. 24:397–403.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Shigeishi H, Sugiyama M, Ohta K, Yokoyama
S, Sakuma M, Murozumi H, Kato H and Takechi M: High HPV16 E6 viral
load in the oral cavity is associated with an increased number of
bacteria: A preliminary study. Biomed Rep. 8:59–64. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Faraji F, Zaidi M, Fakhry C and Gaykalova
DA: Molecular mechanisms of human papillomavirus-related
carcinogenesis in head and neck cancer. Microbes Infect.
19:464–475. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
O'Leary TJ, Drake RB and Naylor JE: The
plaque control record. J Periodontol. 43(38)1972.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Coppieters M, Stappaerts K, Janssens K and
Jull G: Reliability of detecting ‘onset of pain’ and ‘submaximal
pain’ during neural provocation testing of the upper quadrant.
Physiother Res Int. 7:146–156. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Yasukawa T, Ohmori M and Sato S: The
relationship between physiologic halitosis and periodontopathic
bacteria of the tongue and gingival sulcus. Odontology. 98:44–51.
2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yoshida A, Suzuki N, Nakano Y, Oho T,
Kawada M and Koga T: Development of a 5' fluorogenic nuclease-based
real-time PCR assay for quantitative detection of Actinobacillus
actinomycetemcomitans and Porphyromonas gingivalis. J Clin
Microbiol. 41:863–866. 2003.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hamada R, Suehiro J, Nakano M, Kikutani T
and Konishi K: Development of rapid oral bacteria detection
apparatus based on dielectrophoretic impedance measurement method.
IET Nanobiotechnol. 5:25–31. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li W, Fu L, Niu B, Wu S and Wooley J:
Ultrafast clustering algorithms for metagenomic sequence analysis.
Brief Bioinform. 13:656–668. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hormia M, Willberg J, Ruokonen H and
Syrjänen S: Marginal periodontium as a potential reservoir of human
papillomavirus in oral mucosa. J Periodontol. 76:358–363.
2005.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Madinier I, Doglio A, Cagnon L, Lefèbvre
JC and Monteil RA: Southern blot detection of human
papillomaviruses (HPVs) DNA sequences in gingival tissues. J
Periodontol. 63:667–673. 1992.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Parra B and Slots J: Detection of human
viruses in periodontal pockets using polymerase chain reaction.
Oral Microbiol Immunol. 11:289–293. 1996.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Horewicz VV, Feres M, Rapp GE, Yasuda V
and Cury PR: Human papillomavirus-16 prevalence in gingival tissue
and its association with periodontal destruction: A case-control
study. J Periodontol. 81:562–568. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Syrjänen S: Oral manifestations of human
papillomavirus infections. Eur J Oral Sci. 126 (Suppl 1):49–66.
2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ortiz AP, González D, Vivaldi-Oliver J,
Castañeda M, Rivera V, Díaz E, Centeno H, Muñoz C, Palefsky J,
Joshipura K, et al: Periodontitis and oral human papillomavirus
infection among Hispanic adults. Papillomavirus Res. 5:128–133.
2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Dalla Torre D, Burtscher D, Sölder E,
Rasse M and Puelacher W: The correlation between the quality of
oral hygiene and oral HPV infection in adults: A prospective
cross-sectional study. Clin Oral Investig. 23:179–185.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sun CX, Bennett N, Tran P, Tang KD, Lim Y,
Frazer I, Samaranayake L and Punyadeera C: A pilot study into the
association between oral health status and human papillomavirus-16
infection. Diagnostics (Basel). 7(E11)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wiener RC, Sambamoorthi U and Jurevic RJ:
Association of periodontitis and human papillomavirus in oral rinse
specimens: Results from the National Health and Nutrition Survey
2009-2012. J Am Dent Assoc. 146:382–389. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Cutress TW, Ainamo J and Sardo-Infirri J:
The community periodontal index of treatment needs (CPITN)
procedure for population groups and individuals. Int Dent J.
37:222–233. 1987.PubMed/NCBI
|
|
22
|
de Souza PH, de Toledo BE, Rapp GE, Zuza
EP, Neto CB and Mendes AJ: Reliability of bleeding and non-bleeding
on probing to gingival histological features. J Int Acad
Periodontol. 5:71–76. 2003.PubMed/NCBI
|
|
23
|
Takeshita T, Matsuo K, Furuta M, Shibata
Y, Fukami K, Shimazaki Y, Akifusa S, Han DH, Kim HD, Yokoyama T, et
al: Distinct composition of the oral indigenous microbiota in South
Korean and Japanese adults. Sci Rep. 4(6990)2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lee JE, Lee S, Lee H, Song YM, Lee K, Han
MJ, Sung J and Ko G: Association of the vaginal microbiota with
human papillomavirus infection in a Korean twin cohort. PLoS One.
8(e63514)2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Dareng EO, Ma B, Famooto AO, Adebamowo SN,
Offiong RA, Olaniyan O, Dakum PS, Wheeler CM, Fadrosh D, Yang H, et
al: Prevalent high-risk HPV infection and vaginal microbiota in
Nigerian women. Epidemiol Infect. 144:123–137. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gao Z, Lv J and Wang M: Epstein-Barr virus
is associated with periodontal diseases: A meta-analysis based on
21 case-control studies. Medicine (Baltimore).
96(e5980)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Rodrigues PM, Teixeira AL, Kustner EC and
Medeiros R: Are herpes virus associated to aggressive
periodontitis? A review of literature. J Oral Maxillofac Pathol.
19:348–355. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Vyshenska D, Lam KC, Shulzhenko N and
Morgun A: Interplay between viruses and bacterial microbiota in
cancer development. Semin Immunol. 32:14–24. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Sugiyama M, Bhawal UK, Dohmen T, Ono S,
Miyauchi M and Ishikawa T: Detection of human papillomavirus-16 and
HPV-18 DNA in normal, dysplastic, and malignant oral epithelium.
Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 95:594–600.
2003.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Takeshita T, Nakano Y, Kumagai T, Yasui M,
Kamio N, Shibata Y, Shiota S and Yamashita Y: The ecological
proportion of indigenous bacterial populations in saliva is
correlated with oral health status. ISME J. 3:65–78.
2009.PubMed/NCBI View Article : Google Scholar
|