1. Introduction
The occurrence and development of cancer may involve
genetic as well as epigenetic changes (1). The complex processes of carcinogenesis
cannot be fully explained by genetic mutations alone, as they also
involve epigenetic alterations. Epigenetics, including DNA
methylation, RNA editing, genomic imprinting and post-translational
histone modifications, is a branch of genetics that investigates
changes in gene expression without alterations in the primary DNA
sequence. Abnormal epigenetic processes regulate gene expression,
alter gene function and promote tumorigenesis. Epigenetics is also
widely reported to have an important role in the early stages of
neoplastic development and cancer progression (2). While the early focus was on the DNA
sequence as a critical epigenetic marker in the progression of
cancer, an increasing number of subsequent studies have been
focusing on the function of histone modifications in tumorigenesis
(3,4). Histone modifications include
acetylation, phosphorylation, methylation, ADP-ribosylation,
ubiquitylation and sumoylation, among which acetylation of histones
has already been confirmed to be involved in the regulation of
various types of cancer. As more mono-ADP-ribosyltransferases have
been identified in recent years, the functions of
mono-ADP-ribosylation of histones in human disease development,
including cancer, have been further elucidated.
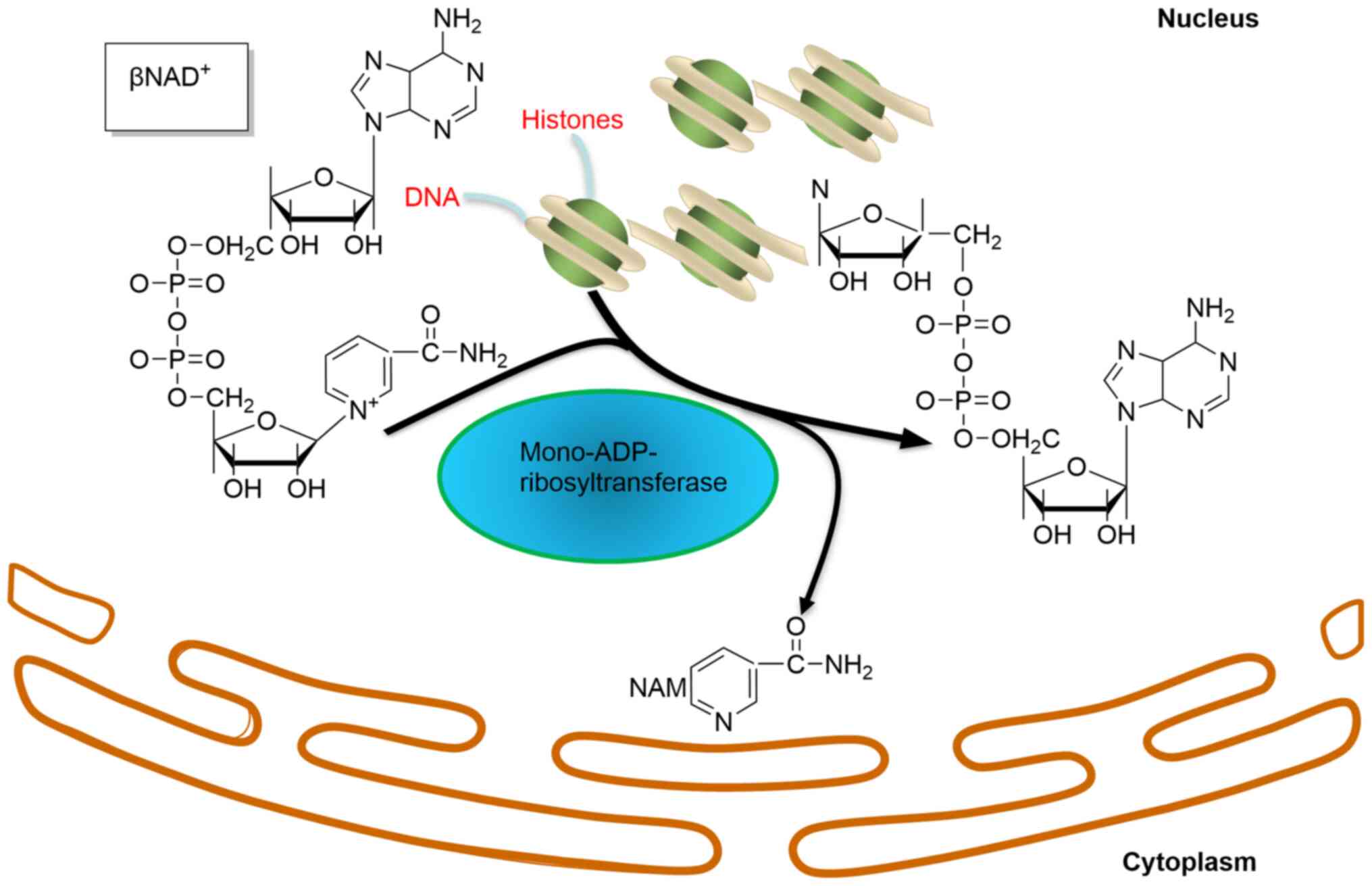
Mono-ADP-ribosyltransferase has been indicated to
transfer one ADP-ribose from the co-factor NAD+ to
target proteins (Fig. 1), and it
has been hypothesized that there are ~1,000 mono-ADP-ribosylated
proteins in cells (5). Several
mono-ADP-ribosylated proteins, such as NF-κB essential modulator
(6), inositol-requiring enzyme 1α,
proline extensin-like receptor kinase1(7), histones, RhoA and human α-defensin
1(8) have been identified to be
involved in regulating immunity, inflammation and the stress
response. However, there is still a lack of understanding of the
functions of most of these mono-ADP-ribosylated proteins due to
limitations in the methods or the tools used.
Histones, a type of basic proteins that combine with
DNA in the chromosome, include five types in eukaryotes, namely
histone (H)1, H2A, H2B, H3 and H4 (4 types of core histones). A
number of studies have indicated that poly-ADP-ribosylation of
histones have important roles in DNA repair, replication,
transcription (9,10), cell proliferation (11) and cancer, and may be associated with
histone acetylation (12),
methylation (13) and
phosphorylation (14). However,
there is a lack of research into the function of
mono-ADP-ribosylation in physiological or pathological processes.
The number of mono-ADP-ribosyltransferase types that have been
detected in mammals has increased and these enzymes have been
indicated to catalyze mono-ADP-ribosylation of histones.
Mono-ADP-ribosylation of histones has also been associated with
other modifications. The functions of mono-ADP-ribosylation of
histones may comprise roles in important physiological pathways,
which may include the development of malignant tumors (15).
2. Enzymes of mono-ADP-ribosylation in
mammals
Approximately 22 members of the
ADP-ribosyltransferase (ART) superfamily have been identified and
have been indicated to have diverse roles. Certain ARTs modify
proteins with chains of poly ADP-ribose or with mono ADP-ribose
(mADPr) (Table I). According to the
different structures of the catalytic domains, ARTs are divided
into bacterial diphtheria toxin-like ARTs (ARTDs) (16) and clostridial C2 and C3 toxin-like
ARTs (ARTCs) (17). ARTDs, which
were previously termed the poly(ADP-ribose) polymerase (PARP)
family and include 17 members (18,19),
are widely distributed in cells and are mostly concentrated in the
nucleus. Compared with ARTD1-6 possessing, ARTD7-17 (except ARTD13,
which is catalytically inactive) only catalyzes
mono-ADP-ribosylation (16,20), due to the absence of conserved
glutamate. In addition, ARTD3 has been detected as a
mono-ADP-ribosyltransferase in previous studies (21). ARTCs, as ectocellular ARTs, transfer
mADPr from NAD+ to target proteins in the cytoplasm,
cytomembrane and extracellular regions (22,23).
Therefore, ARTCs are not able to mediate mono-ADP-ribosylation of
histones, which are distributed in the nucleus. Apart from the
aforementioned, members of the sirtuin (SIRT) family (SIRT1-7
always act as histone deacetylases) possess mono-ADP-ribosylation
properties. SIRT4, as well as SIRT6 and -7, were indicated to have
endogenous mono-ADP-ribosyltransferase activity in the mitochondria
and the nucleus, respectively (24-27).
However, only a small number of these mono-ADP-ribosyltransferases,
such as ARTD3, -10, and -14 and SIRT4 and -6, have been reported to
mono-ADP-ribosylate histones in vertebrates to date (28-32).
 | Table IEnzymes of ADP-ribosylation. |
Table I
Enzymes of ADP-ribosylation.
| A, ARTDs |
|---|
| Enzymes | Subcellular
localization | Enzymatic
activities | (Refs.) |
|---|
| ARTD1 | Nucleus |
Poly-ADP-ribosyltransferase | (16-19) |
| ARTD2 | Nucleus |
Poly-ADP-ribosyltransferase | (16-19) |
| ARTD3 | Nucleus |
Mono/Poly-ADP-ribosyltransferase | (22,31) |
| ARTD4 |
Nucleus/cytoplasm |
Poly-ADP-ribosyltransferase | (16-19) |
| ARTD5 |
Nucleus/cytoplasm |
Poly-ADP-ribosyltransferase | (16-19) |
| ARTD6 |
Nucleus/cytoplasm |
Poly-ADP-ribosyltransferase | (16-19) |
| ARTD7 | Nucleus |
Mono/Poly-ADP-ribosyltransferase | (20,21) |
| ARTD8 |
Nucleus/cytoplasm |
Mono/Poly-ADP-ribosyltransferase | (20,21) |
| ARTD9 |
Nucleus/cytoplasm |
Mono/Poly-ADP-ribosyltransferase | (19) |
| ARTD10 |
Nucleus/cytoplasm |
Mono/Poly-ADP-ribosyltransferase | (20,21,30,42) |
| ARTD11 | - |
Mono/Poly-ADP-ribosyltransferase | (20,21) |
| ARTD12 | Nucleus |
Mono/Poly-ADP-ribosyltransferase | (19-21) |
| ARTD13 | Nucleus | Catalytically
inactive | (20) |
| ARTD14 | Nucleus |
Mono/Poly-ADP-ribosyltransferase | (19,29) |
| ARTD15 |
Nucleus/cytoplasm |
Mono/Poly-ADP-ribosyltransferase | (20,21) |
| ARTD16 | - |
Mono/Poly-ADP-ribosyltransferase | (20,21) |
| ARTD17 | - |
Mono/Poly-ADP-ribosyltransferase | (20,21) |
| B, ARTCs |
| Enzymes | Subcellular
localization | Enzymatic
activities | (Refs.) |
| ARTC1-5 | Ecto-cellular | - | (23,24) |
| C, special
ARTs |
| Enzymes | Subcellular
localization | Enzymatic
activities | (Refs.) |
| ADPRT1a | - |
Mono-ADP-ribosyltransferase | (38) |
| ADPRT2 | - |
Mono-ADP-ribosyltransferase | (38) |
| D, Sirtuins |
| Enzymes | Subcellular
localization | Enzymatic
activities | (Refs.) |
| SIRT1 | Nucleus | - | (27,28) |
| SIRT2 |
Nucleus/Cytoplasm | - | (27,28) |
| SIRT3 | Mitochondria | - | (27,28) |
| SIRT4 | Mitochondria |
Mono-ADP-ribosyltransferase | (25,32) |
| SIRT5 | Mitochondria | - | (27) |
| SIRT6 | Nucleus |
Mono-ADP-ribosyltransferase | (26,33) |
| SIRT7 | Nucleus
(nucleoli) |
Mono-ADP-ribosyltransferase | (25) |
3. Histones are substrates of
mono-ADP-ribosylation
All core histones and the linker histone H1 have
been reported to undergo mono-ADP-ribosylated modification
(Table II) (33). Histone H1 was indicated to be
mono-ADP-ribosylated on glutamic acid E2, E14 and E116(34), the arginine residue R34(35) and at the COOH-terminal lysine
residue K213(36). In the rat
liver, histone H2B was indicated to be mono-ADP-ribosylated on
glutamate residue 2 of the γ-COOH group (37). Both in the chromatin and in the
reconstituted recombinant nucleosomes of chicken, histone H2B E2
was a specific target of PARP3 modification with mADPr (30). In histone H2B, the residues E18 and
E19 were also indicated to be the principal sites modified by the
ARTs in response to DNA double-strand breaks (38). After hepatoma cells are alkylated,
H1 and H2B may be mono-ADP-ribosylated at the N-terminal fragment.
Mono-ADP-ribosylation also modified the Arg residues of H2A, H3 and
H4 and the glutamic residues of H2A and H2B (39). Previous studies identified K13 of
H2A, K30 of H2B, K27 and K37 of H3 and K16 of H4 as ADP-ribose
acceptor sites in poly-ADP-ribosylation (40); however, whether these are also
ADP-ribose acceptor sites in mono-ADP-ribosylation requires further
research. Therefore, the specific amino acid sites of
mono-ADP-ribosylated modifications on H2A and H4 have remained
elusive. It has remained to be determined which enzymes mediate
mono-ADP-ribosylation of histones. With ongoing research, an
increasing number of studies have identified specific enzymes
involved in regulating the mono-ADP-ribosylation of histones. Human
H2B was reported to be modified on E2 in vitro by using
recombinant ARTD10(41) and DNA
damage was able to induce mono-ADP-ribosylation of H2B E18 and E19
in vivo by specific ARTs: Protein (ADP-ribosyl) transferase
(ADPRT)1a and ADPRT2(38). In
addition, a silent information regulator 2 of yeast (SIR2)-related
protein from the protozoan parasite Trypanosoma brucei (TBSIR2RP1),
was indicated to catalyze mono-ADP-ribosylation of H2A, H2B and H4
in Trypanosoma brucei (42).
 | Table IIMono-ADP- ribosylation of
histones. |
Table II
Mono-ADP- ribosylation of
histones.
| A, H1
substrate |
|---|
| Modified amino
acid | Enzymes | Effect of the
reaction | (Refs.) |
|---|
| E2, E14E116 and
K213 | - | Regulation of H1-H1
interactions | (35,37) |
| R34 | - | Blocks the
cAMP-dependent phosphorylation of histone H1 | (36) |
| Q/N | ARTD3 | DNA repair | (22,63) |
| B, H2A
substrate |
| Modified amino
acid | Enzymes | Effect of the
reaction | (Refs.) |
| Unknown | Sir2 | Response to
oxidative stress/DNA damage | (43) |
| Unknown | Sir2 | Inhibition of
histone acetylation/silencing chromatin domains | (43,68) |
| R/E | - | Unknown | (40) |
| C, H2B
substrate |
| Modified amino
acid | Enzymes | Effect of the
reaction | (Refs.) |
| E2 | ARTD3/ARTD10 | Unknown | (31,38,42) |
| E18/E19 | ARTs,
Adprt1a/Adprt2 | Response to
oxidative stress/DNA damage | (38,39) |
| Unknown | Sir2 | Inhibition of
histone acetylation/silencing of chromatin domains | (43,68) |
| D, H3
substrate |
| Modified amino
acid | Enzymes | Effect of the
reaction | (Refs.) |
| Unknown | SIRT6, Sir2 | Inhibition histone
acetylation/silencing chromatin domains | (33,43) |
| R | - | Cell
proliferation | (11,40) |
| E, H4
substrate |
| Modified amino
acid | Enzymes | Effect of the
reaction | (Refs.) |
| R | Sir2 | Post-synthetic
modification with acetylation of core histones | (27,28,40) |
| Unknown | Sir2 | Response to
oxidative stress/DNA damage | (43) |
| Unknown | ARTD10 | Unknown | (42,66) |
| Unknown |
Sir2-relatedprotein | Inhibition histone
acetylation | (43) |
4. Methods for detecting
mono-ADP-ribosylated histones
The detection of the enzymes and specific substrates
of ADP ribosylation is an important step for identifying
mono-ADP-ribosylated histones. Radiolabeled or chemically modified
NAD+ has been widely used for detecting
mono-ADP-ribosylated proteins in vitro. In the process of
ADP ribosylation, radioactive-labeled ADP ribose of NAD+
combines with the target proteins and SDS-PAGE autoradiography may
detect these radiolabeled molecules (43). Certain studies have used self-made
ADP-ribosylated antibodies, as commercially available specific
antibodies are limited; however, the sensitivity of the antibodies
has not been satisfactory and they are not generally applicable for
wider use (44,45). Osago et al (46) have already investigated more
efficient antibodies, which were able to even detect specific
arginine mono-ADP-ribosylated peptides. Apart from the methods
mentioned above, certain other chemical tools may be used to detect
ADP-ribosylated proteins, even mono-ADP-ribosylated histones, such
as the use of ADPr (ADP-ribose)-peptides, analogues (ADPr-synthon)
and ADPr-chains (47). There are
several chemical synthesis peptides carrying mADPr, which may be
used to detect the affinity between ADP-ribose and substrates
(48). In addition, the
incorporation of benzophenone photo-cross-linkers into synthetic
peptides has been demonstrated to provide a way to probe for and
enrich ADP-ribose binding proteins (49,50).
As the methods for ADP-ribose peptide synthesis have improved, the
development of corresponding antibodies may be possible (51). Thus, the specific antibody for
histone mono-ADP-ribosylation may be improved to further
investigate the function of histones in different fields.
Mass spectrometry (MS) and selective reader domains
have also been used for detecting mono-ADP-ribosylated proteins by
identifying macrodomains, which may selectively bind ADP-ribose.
H2A, one of a number of types of macrodomain-containing proteins,
contains macro H2A, which is subdivided into macro H2A1.1, macro
H2A1.2 and macro H2A2 (52,53). Only H2A1.1 has been indicated not to
bind mono-ADP-ribose (54), while
it has remained to be determined whether mono-ADP-ribose has
connections with macro H2A1.2 and macro H2A2(52). Therefore, MS may be an important
method for identifying mono-ADP-ribosylated histones. For detecting
the specific ADP-ribosylated residues, quadruple tandem MS was
indicated to detect ADP-ribosyl-Arg and Arg-ADP-ribosylated
peptides to identify the specific arginine site of
mono-ADP-ribosylation in the target protein (55,56).
In recent years, numerous MS-based proteomics have been developed,
such as macrodomain-linked immunosorbent assay to identify
mono-ARTs (57) and liquid
chromatography-high-resolution tandem MS to identify
mono-ADP-ribose acceptor sites (58,59).
Furthermore, there are other methods allowing for the
identification of mono-ADP-ribosylation, such as a
phosphoproteomics approach via the enzymatic product of
phosphodiesterase-treated ADP-ribose (60) or an aminooxy alkyne probe for
detecting mono-ADP-ribosylated proteins (61). A mutagenesis approach has also been
employed to detect the ADP-ribose acceptor site (5), which may be a novel way to study
mono-ADP-ribosylation of histones.
5. Function of mono-ADP-ribosylated histones
in DNA damage and repair
Mono-ADP-ribosylation of histones may be involved in
DNA damage and repair (62) and the
nucleosomal surface is the main target (63). In the 1980s, Adamietz and Rudolph
(64) reported that, when AH7974
hepatoma cells were damaged by the alkylating agent dimethyl
sulfate, mono-ADP-ribosylation of histones increased by a factor of
12. Under the same conditions, the mono-ADP-ribosylated C-terminal
extension of histone H1 and the N-terminal fragment of histone H2B
was increased compared with that in untreated cells (65,66),
which may modify DNA-histone association by adding two negative
charges. TbSIR2RP1, catalyze the mono-ADP-modification of H2A and
H2B, which may occur in response to DNA damage and be involved in
DNA repair. Rulten et al (67) suggested that mono-ADP-ribosylated
H1, catalyzed by PARP3, may accelerate DNA double-strand break
repair by binding to aprataxin and polynucleotide kinase-like
factor. Changes in the types of ADP-ribosylated histones may occur
in DNA strand breaks, as in the P815 mouse mastocytoma and K562
human chronic myelogenous leukemia cell lines, mono-ADP-ribosylated
histones appeared in the absence of DNA strand breaks due to the
decrease of poly-ADP-ribose synthetase activity, whereas
poly-ADP-ribosylated histones increased following DNA stand breaks
(68).
6. Mono-ADP-ribosylation of histones in
replication and transcription
ADP ribosylation of histone is also involved in DNA
repair and replication. It has been indicated that histones are
predominantly mono-ADP-ribosylated in lysates of non-dividing
cells, while being poly-ADP-ribosylated in rapidly proliferating
cells (62,69). However, evidence for the connection
between mono-ADP-ribosylation of histones and replication is
limited. Mono-ADP-ribosylated histones present in the nuclei under
physiological conditions are considered to function in supporting
the conversion of the chromatin loop into its transcriptional
active structure (70). ARTD14, as
a mono-ADP-ribosyltransferase of histones, was indicated to
interact with aryl hydrocarbon receptor (AHR) leading to decreased
AHR transcriptional activity (28).
7. Mono-ADP-ribosylation of histones in cell
proliferation and differentiation
Mono-ADP-ribosylation of histones may promote or
inhibit cell proliferation. It has been indicated that the
mono-ADP-ribosylation of histone 3 at R117 may accelerate the
proliferation of colon carcinoma cells by regulating P300 to
increase the expression level of cyclin D1 and c-myc (11). By contrast, other studies have
indicated that after P815 mouse mastocytoma and K562 human chronic
myelogenous leukemia cell lines were treated with 5 mM butyrate or
with serum-free media for blocking cell proliferation, the level of
mono-ADP-ribosylated histones was higher compared with that in
rapidly dividing cells. Of note, there were also no
poly-ADP-ribosylated histones in the treated cells, while an
increase in poly-ADP-ribosylated histones was observed in the
rapidly dividing cells (69). The
cycle of the conversion of poly-ADP-ribosylated histones to
mono-ADP-ribosylated histones may be an important regulatory factor
in cell proliferation. In addition, a study by our group indicated
that arginine 117 of histone H3 in LoVo colon carcinoma cells with
low differentiation were modified by mono-ADP-ribosylation, while
SW480 cells with high differentiation were not (71), which suggested that
mono-ADP-ribosylated histones may vary across different colorectal
cancer cell lines with different degrees of malignancy, confirming
the hypothesis that histone mono-ADP-ribosylation may have an
important role in the development of tumors.
8. Association between mono-ADP-ribosylation
of histones and other histone modifications
A wide range of histone modifications has been
identified, which regulate signaling pathways in cell physiology
and pathology. These modifications do not exist independently and
there are connections among them. H4 is more likely to be
mono-ADP-ribosylated while it is hyper-acetylated (40,72).
Mono-ADP-ribosylation of H4 by ARTD10 may occur when K5, K8 and/or
K16 are modified by acetylation; however, these interrelations were
indicated to be relatively weak. In addition, mono-ADP-ribosylation
of H3 R117 affected the transcription and expression level of
demethylase ten-eleven translocation 1, thus regulating the
methylation of tissue factor pathway inhibitor 2 in colorectal
cancer (73). Furthermore, the
decrease of mono-ADP-ribosylation of histones in colorectal
carcinoma cells resulted in an increase of histone H3 trimethylated
at lysine 4 and phosphatase and tensin homolog, thus reducing the
phosphorylation of the PI3K/Akt signaling pathway.
Mono-ADP-ribosylation of JHDM1A/KDM2A by SIRT6 led to an increase
of histone H3 lysine 36 dimethylation levels to promote DNA repair
(32). Methylation or acetylation
of K20 in H4 may inhibit mono-ADP-ribosylation (29). When Arg34 of histone H1 is modified
by mono-ADP-ribosylation, cyclic (c)AMP-dependent phosphorylation
of histone H1 on Ser 38 may be inhibited (35). Arginine, which is located in the
NH2-terminal of the phosphate-accepting serine residue,
was indicated to be important for phosphorylation by cAMP-dependent
protein kinase. Hence, the change in the function of the arginine
residue by mono-ADP-ribosylation may affect the phosphorylation of
histones. In yeast, histone acetylation may be inhibited by
mono-ADP-ribosylation of histones, which is catalyzed by SIR2 and
may be responsible for inhibiting growth or silencing genes
(74). It was also reported that
reducing the ability of the mono-ADP-ribosyltranferase of SIR2 with
a G270A mutation may not control gene silencing and it was
hypothesized that histone acetylation may serve a bigger role
rather than just in histone mono-ADP-ribosylation (75). Therefore, the efficiency of
mono-ADP-ribosylation of histones by SIR2 requires further
investigation.
9. Hydrolytic enzymes of histone
mono-ADP-ribosylation
Mono-ADP-ribosylation is a reversible reaction,
which may be hydrolyzed by Arg-specific mono-ADP-hydrolase,
macroD1, macroD2, C6ORF130/TARG1 (76-78)
and by serine mono ADP-ribosylhydrolase-3(79). The content of macrodomain proteins
primarily originates from viruses, such as α-virus, hepatitis E
virus, severe acute respiratory syndrome coronavirus (SARS-CoV),
feline infectious peritonitis virus and hCoV-229E macrodomains
(80-82).
In a recent study, SARS-CoV-2 was reported to be
able to remove mono-ADP-ribose (MAR) from a protein substrate
(83). Whether the functions of
histone modification were regulated by these hydrolytic enzymes
requires further investigation. As the specific amino acid residues
hydrolyzed by these hydrolases are different, the acceptor sites of
histone mono-ADP-ribosylation may be confirmed by the type of
hydrolases able to hydrolyze the mono-ADP-ribosylation.
10. Conclusion
Mono-ADP-ribosylation has become a focus in the
fields of immunity, inflammation, stress response, DNA damage
response and cancer (5,84,85).
Different target proteins and even different amino acid residues
may determine the functions of mono-ADP-ribosylation. However, the
number of identified target proteins of mono-ADP-ribosylation
remains low at present, owing to the limited and simplistic
methods, not to mention the exact number and location of the
acceptor sites.
Histones are major target proteins; however,
knowledge regarding their function in pathophysiological processes
is currently limited. Histone modifications, similar to
acetylation, methylation and phosphorylation, have already been
reported to participate in multiple processes, particularly in
tumorigenesis. Studies have indicated that mono-ADP-ribosylation of
histones is able to regulate the DNA damage response, transcription
and cell proliferation, which are also important factors in
tumorigenesis. The connections between histone
mono-ADP-ribosylation and other well-known histone modifications
indicate that the combined effects of these modifications may
regulate pathophysiological processes.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by National High-Tech Research
and Development Projects (863; grant no. 2012AA02A201), the
Scientific Research Foundation of Chongqing Medical University
(grant no. 201413), the Science and Technology Research Foundation
of Chongqing Municipal Education Commission (grant no.
KJQN201800435) and the National Nature Science Foundation of China
(grant no. 30870946).
Availability of data and materials
The datasets used/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JJZ and YT performed the literature search for
relevant publications on the topic. YLW participated in drafting
the manuscript and provided critical insight. JJZ and YT confirm
the authenticity of all the raw data. All authors read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Verma M, Maruvada P and Srivastava S:
Epigenetics and cancer. Genes Dev. 18:2315–2335. 2016.
|
|
2
|
Shanmugam MK, Arfuso F, Arumugam S,
Chinnathambi A, Jinsong B, Warrier S, Wang LZ, Kumar AP, Ahn KS,
Sethi G and Lakshmanan M: Role of novel histone modifications in
cancer. Oncotarget. 9:11414–11426. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang R, Xin M, Li Y, Zhang P and Zhang M:
The functions of histone modification enzymes in cancer. Curr
Protein Pept Sci. 17:438–45. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Füllgrabe J, Kavanagh E and Joseph B:
Histone onco-modifications. Oncogene. 30:3391–403. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Feijs KL, Verheugd P and Lüscher B:
Expanding functions of intracellular resident mono-ADP-ribosylation
in cell physiology. FEBS J. 280:3519–3529. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Verheugd P, Forst AH, Milke L, Herzog N,
Feijs KL, Kremmer E, Kleine H and Lüscher B: Regulation of
NF-kappaB signalling by the mono-ADP-ribosyltransferase ARTD10. Nat
Commun. 4(1683)2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jwa M and Chang PE: PARP16 is a
tail-anchored endoplasmic reticulum protein required for the PERK-
and IRE1α-mediated unfolded protein response. Nat Cell Biol.
14:1223–1230. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Kistemaker HAV, Nardozza AP, Overkleeft
HS, van der Marel GA, Ladurner AG and Filippov DV: Synthesis and
macrodomain binding of Mono-ADP-Ribosylated peptides. Angew Chem
Int Ed Engl. 55:10634–10638. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hottiger MO: ADP-ribosylation of histones
by ARTD1: An additional module of the histone code? FEBS Lett.
585:1595–1599. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Posavec Marjanović M, Crawford K and Ahel
I: PARP, transcription and chromatin modeling. Semin Cell Dev Biol.
63:102–113. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ling F, Tang Y, Li M, Li QS, Li X, Yang L,
Zhao W, Jin CC, Zeng Z, Liu C, et al: Mono-ADP-ribosylation of
histone 3 at arginine-117 promotes proliferation through its
interaction with P300. Oncotarget. 8:72773–72787. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Verdone L, La Fortezza M, Ciccarone F,
Caiafa P, Zampieri M and Caserta M: Poly(ADP-Ribosyl)ation affects
histone acetylation and transcription. PLoS One.
10(e0144287)2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kassner I, Andersson A, Fey M, Tomas M,
Ferrando-May E and Hottiger MO: SET7/9-dependent methylation of
ARTD1 at K508 stimulates poly-ADP-ribose formation after oxidative
stress. Open Biol. 3(120173)2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tikoo K, Lau SS and Monks TJ: Histone H3
phosphorylation is coupled to poly-(ADP-ribosylation) during
reactive oxygen species-induced cell death in renal proximal
tubular epithelial cells. Mol Pharmacol. 60:394–402.
2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mareike B, Laura E, Patricia V and
Bernhard L: Intracellular Mono-ADP-ribosylation in signaling and
disease. Cells. 4:569–595. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Girolamo MD and Fabrizio G: The
ADP-Ribosyl-transferases diphtheria toxin-like (ARTDs) family: An
overview. Challenges. 9(24)2018.
|
|
17
|
Sadakierska-Chudy A and Filip MG: A
comprehensive view of the epigenetic landscape. Part II: Histone
post-translational modification, nucleosome level, and chromatin
regulation by ncRNAs. Neurotox Res. 27:172–197. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Carter-O'Connell I and Cohen MS:
Identifying direct protein targets of poly-ADP-ribose polymerases
(PARPs) using engineered PARP variants-orthogonal nicotinamide
adenine dinucleotide (NAD+) analog pairs. Curr Protoc Chem Biol.
7:121–139. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang S, Xue X, Pharmacy SO and University
CP: PARP family and clinically used PARP Inhibitors. Guangdong
Chemical Industry. 46:134–136. 2019.(In Chinese).
|
|
20
|
Pinto AF and Schüler H: Comparative
structural analysis of the putative mono-ADP-ribosyltransferases of
the ARTD/PARP family. Curr Top Microbiol Immunol. 384:153–166.
2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Loseva O, Jemth AS, Bryant HE, Schüler H,
Lehtiö L, Karlberg T and Helleday T: PARP-3 Is a
Mono-ADP-ribosylase That Activates PARP-1 in the Absence of DNA. J
Biol Chem. 285:8054–8060. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Krska D, Ravulapalli R, Fieldhouse RJ,
Lugo MR and Merrill AR: C3larvin Toxin, an ADP-ribosyltransferase
from Paenibacillus larvae. J Biol Chem. 290:1639–1653.
2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Prisilla A and Chellapandi P: Structure,
function and evolution of clostridium botulinum C2 and C3 toxins:
Insight to poultry and veterinary vaccines. Curr Protn Pept.
18:412–424. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li S and Zheng W: Mammalian sirtuins SIRT4
and SIRT7. Prog Mol Biol Transl Sci. 154:147–168. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Rahnasto-Rilla M, Lahtela-Kakkonen M and
Moaddel R: Sirtuin 6 (SIRT6) activity assays. Methods Mol Biol.
1436:259–269. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xinxin QI and Li S: Sirtuin family and its
biological characteristics. Acta Med Sin, 2016.
|
|
27
|
Balaiya S, Abu-Amero KK, Kondkar AA and
Chalam KV: Sirtuins expression and their role in retinal diseases.
Oxid Med Cell Longev. 2017(3187594)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
MacPherson L, Tamblyn L, Rajendra S,
Bralha F, McPherson JP and Matthews J:
2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase
(TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of
aryl hydrocarbon receptor transactivation. Nuclc Acids Res.
41:1604–1621. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Feijs K: Characterization of the
mono-ADP-ribosylation by ARTD10: Substrates, consequences and
reversibility. Hochschulbibliothek der Rheinisch-Westfälischen
Technischen Hochschule Aachen, 2012.
|
|
30
|
Grundy GJ, Polo LM, Zeng Z, Rulten S, Hoch
NC, Paomephan P, Xu YQ, Sweet SM, Thorne AW, Oliver AW, et al:
PARP3 is a sensor of nicked nucleosomes and monoribosylates histone
H2B(Glu2). Nat Commun. 7(12404)2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ahuja N, Schwer B, Carobbio S, Waltregny
D, North BJ, Castronovo V, Maechler P and Verdin E: Regulation of
insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase.
J Biol Chem. 282:33583–33592. 2007.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Rezazadeh S, Yang D, Biashad SA, Firsanov
D and Gorbunova V: SIRT6 mono-ADP ribosylates KDM2A to locally
increase H3K36me2 at DNA damage sites to inhibit transcription and
promote repair. Aging. 12:11165–11184. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hassa PO, Haenni SS, Elser M and Hottiger
MO: Nuclear ADP-ribosylation reactions in mammalian cells: Where
are we today and where are we going? Microbiol Mol Biol Rev.
70:789–829. 2006.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ogata N, Ueda K, Kagamiyama H and Hayaishi
O: ADP-ribosylation of histone H1. Identification of glutamic acid
residues 2, 14, and the COOH-terminal lysine residue as
modification sites. J Biol Chem. 255:7616–7620. 1980.PubMed/NCBI
|
|
35
|
Ushiroyama T, Tanigawa Y, Tsuchiya M,
Matsuura R, Ueki M, Sugimoto O and Shimoyama M: Amino acid sequence
of histone H1 at the ADP-ribose-accepting site and ADP-ribose X
histone-H1 adduct as an inhibitor of cyclic-AMP-dependent
phosphorylation. Eur J Biochem. 151:173–177. 2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Riquelme PT, Burzio LO and Koide SS: ADP
ribosylation of rat liver lysine-rich histone in vitro. J Biol
Chem. 254:3018–3028. 1979.PubMed/NCBI
|
|
37
|
Ogata N, Ueda K and Hayaishi O:
ADP-ribosylation of histone H2B. Identification of glutamic acid
residue 2 as the modification site. J Biol Chem. 255:7610–7615.
1980.PubMed/NCBI
|
|
38
|
Rakhimova A, Ura S, Hsu DW, Wang HY, Pears
CJ and Lakin ND: Site-specific ADP-ribosylation of histone H2B in
response to DNA double strand breaks. Sci Rep.
7(43750)2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Golderer G and Gröbner P: ADP-ribosylation
of core histones and their acetylated subspecies. Biochem J.
277:607–610. 1991.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Dan MB: European geosciences union general
assembly. Nuclc Acids Res. 38:6350–6362. 2016.
|
|
41
|
Kleine H, Poreba E, Lesniewicz K, Hassa
PO, Hottiger MO, Litchfield DW, Shilton B and Lüscher B:
Substrate-assisted catalysis by PARP10 limits its activity to
mono-ADP-ribosylation. Mol Cell. 32:57–69. 2008.PubMed/NCBI View Article : Google Scholar
|
|
42
|
García-Salcedo JA, Gijón P, Nolan DP,
Tebabi P and Pays E: A chromosomal SIR2 homologue with both histone
NAD-dependent ADP-ribosyltransferase and deacetylase activities is
involved in DNA repair in Trypanosoma brucei. EMBO J. 22:5851–5862.
2003.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Graves DJ, Huiatt TW, Zhou H, Huang HY and
Mcmahon KK: Regulatory role of arginine-specific
mono(ADP-Ribosyl)transferase in muscle cells. Adv Exp Med Biol.
419:305–313. 1997.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Schwab CJ, Colville MJ, Fullerton AT and
Mcmahon KK: Evidence of endogenous mono-ADP-ribosylation of cardiac
proteins via anti-ADP-ribosylarginine immunoreactivity. Proc Soc
Exp Biol Med. 223:389–396. 2010.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Meyer T and Hilz H: Production of
anti-(ADP-ribose) antibodies with the aid of a
dinucleotide-pyrophosphatase-resistant hapten and their application
for the detection of mono(ADP-ribosyl)ated polypeptides. Eur J
Biochem. 155:157–165. 2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Osago H, Terashima M, Hara N, Yamada K and
Tsuchiya M: A new detection method for arginine-specific
ADP-ribosylation of protein-a combinational use of
anti-ADP-ribosylarginine antibody and ADP-ribosylarginine
hydrolase. J Biochem Biophys Methods. 70:1014–1019. 2008.PubMed/NCBI View Article : Google Scholar
|
|
47
|
van der Heden van Noort GJ: Chemical tools
to study protein ADP-ribosylation. ACS Omega. 5:1743–1751.
2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Liu Q, Marel GAVD and Filippov DV:
Chemical ADP-ribosylation: Mono-ADPr-peptides and oligo-ADP-ribose.
Organ Biomol Chem. 17:5460–5474. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Moyle PM and Muir TW: Method for the
synthesis of mono-ADP-ribose conjugated peptides. J Am Chem Soc.
132:15878–15880. 2010.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Vivelo CA and Leung AK: Proteomics
approaches to identify mono-(ADP-ribosyl)ated and
poly(ADP-ribosyl)ated proteins. Proteomics. 15:203–217.
2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Lu AZ, Abo R, Ren Y, Gui B, Mo JR,
Blackwell D, Wigle T, Keilhack H and Niepel M: Enabling drug
discovery for the PARP protein family through the detection of
mono-ADP-ribosylation. Biochem Pharmacol. 168:97–106.
2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Han W, Li X and Fu X: The macro domain
protein family: Structure, functions, and their potential
therapeutic implications. Mutat Res. 727:86–103. 2011.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Feijs KLH, Forst AH, Verheugd P and
Lüscher B: Macrodomain-containing proteins: Regulating new
intracellular functions of mono(ADP-ribosyl)ation. Nat Rev Mol Cell
Biol. 14:443–451. 2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Forst AH, Karlberg T, Herzog N, Thorsell
AG, Gross A, Feijs KL, Verheugd P, Kursula P, Nijmeijer B, Kremmer
E, et al: Recognition of mono-ADP-ribosylated ARTD10 substrates by
ARTD8 macrodomains. Structure. 21:426–475. 2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Osago H, Yamada K, Shibata T, Yoshino KI,
Hara N and Tsuchiya M: Precursor ion scanning and sequencing of
arginine-ADP-ribosylated peptide by mass spectrometry. Anal
Biochem. 393:248–254. 2009.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Perkins DN, Pappin DJC, Creasy DM and
Cottrell JS: Probability-based protein identification by searching
sequence databases using mass spectrometry data. Electrophoresis
20: 3551-3567. Electrophoresis. 20:3551–3567. 1999.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Chen J, Lam AT and Zhang Y: A
macrodomain-linked immunosorbent assay (MLISA) for
mono-ADP-ribosyltransferases. Anal Biochem. 543:132–139.
2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Leutert M, Bilan V, Gehrig P and Hottiger
MO: Identification of ADP-ribose acceptor sites on in vitro
modified proteins by liquid chromatograph-tandem mass spectrometry.
Methods Mol Biol. 1608:137–148. 2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Larsen SC, Leutert M, Bilan V, Martello R,
Jungmichel S, Young C, Hottiger MO and Nielsen ML: Proteome-wide
identification of in vivo ADP-ribose acceptor sites by liquid
chromatography-tandem mass spectrometry. Methods Mol Biol.
1608:149–162. 2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Daniels CM, Ong SE and Leung AK:
Phosphoproteomic approach to characterize protein mono- and
poly(ADP-ribosyl)ation sites from cells. J Proteome Res.
13:3510–3522. 2014.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Morgan RK and Cohen MS: A clickable
aminooxy probe for monitoring cellular ADP-ribosylation. ACS Chem
Biol. 10:1778–1784. 2015.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Messner S and Hottiger MO: Histone
ADP-ribosylation in DNA repair, replication and transcription.
Trends Cell Biol. 21:534–542. 2011.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Karch KR, Langelier MF, Pascal JM and
Garcia BA: The nucleosomal surface is the main target of histone
ADP-ribosylation in response to DNA damage. Mol Biosys.
13:2660–2671. 2017.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Adamietz P and Rudolph A: ADP-ribosylation
of nuclear proteins in vivo. Identification of histone H2B as a
major acceptor for mono- and poly(ADP-ribose) in dimethyl
sulfate-treated hepatoma AH 7974 cells. J Biol Chem. 259:6841–6846.
1984.PubMed/NCBI
|
|
65
|
Kreimeyer A, Adamietz P and Hilz H:
Alkylation-induced mono(ADP-ribosyl)-histones H1 and H2B.
Hydroxylamine-resistant linkage in hepatoma cells. Biol Chem.
366:537–544. 1985.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Kreimeyer A, Wielckens K, Adamietz P and
Hilz H: DNA repair-associated ADP-ribosylation in vivo.
Modification of histone H1 differs from that of the principal
acceptor proteins. J Biol Chem. 259:890–896. 1984.PubMed/NCBI
|
|
67
|
Rulten SL, Fisher AEO, Robert I, Zuma MC,
Rouleau M, Ju LM, Poirier G, Reina-San-Martin B and Caldecott KW:
PARP-3 and APLF function together to accelerate nonhomologous
end-joining. Mol Cell. 41:33–45. 2011.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Boulikas T: DNA strand breaks alter
histone ADP-ribosylation. Proc Natl Acad Sci USA. 86:3499–3503.
1989.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Boulikas T: Poly(ADP-ribosylated) histones
in chromatin replication. J Biol Chem. 265:14638–14647.
1990.PubMed/NCBI
|
|
70
|
Boulikas T: Relation between
carcinogenesis, chromatin structure and poly(ADP-ribosylation)
(review). Anticancer Res. 11:489–527. 1991.PubMed/NCBI
|
|
71
|
Zhang NN, Lin T, Xiao M, Li QS, Li X, Yang
L, Wang CL and Wang YL: Transcriptome sequencing analysis of
monoADPribosylation in colorectal cancer cells. Oncol Rep.
43:1413–1428. 2020.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Böhm L, Schneeweiss FA, Sharan RN and
Feinendegen LE: Influence of histone acetylation on the
modification of cytoplasmic and nuclear proteins by
ADP-ribosylation in response to free radicals. Biochim Biophys
Acta. 1334:149–154. 1997.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Li M, Tang Y, Li Q, Xiao M, Yang Y and
Wang Y: Mono-ADP-ribosylation of H3R117 traps 5mC hydroxylase TET1
to impair demethylation of tumor suppressor gene TFPI2. Oncogene.
38:3488–3503. 2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Tanny JC, Dowd GJ, Huang J, Hilz H and
Moazed D: An enzymatic activity in the yeast Sir2 protein that is
essential for gene silencing. Cell. 99:735–45. 1999.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Michan S and Sinclair D: Sirtuins in
mammals: Insights into their biological function. Biochem J.
404:1–13. 2007.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Laing S, Unger M, Koch-Nolte F and Haag F:
ADP-ribosylation of arginine. Amino Acids. 41:257–69.
2011.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Stevens LA, Kato J, Kasamatsu A, Oda H,
Lee DY and Moss J: The ARH and macrodomain families of
α-ADP-ribose-acceptor hydrolases catalyze α-NAD+
hydrolysis. ACS Chem Biol. 14:2576–2584. 2019.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Thomas A, Deeksha M, Kerryanne C, Luca P,
Andreja M and Ivan A: MacroD1 is a promiscuous ADP-Ribosyl
hydrolase localized to mitochondria. Front Microbiol.
9(20)2018.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Abplanalp J, Leutert M, Frugier E, Nowak
K, Feurer R, Kato J, Kistemaker HVA, Filippov DV, Moss J, Caflisch
A and Hottiger MO: Proteomic analyses identify ARH3 as a serine
mono-ADP-ribosylhydrolase. Nat Commun. 8(2055)2017.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Fehr AR, Channappanavar R, Jankevicius G,
Fett C, Zhao J, Athmer J, Meyerholz DK, Ahel I and Perlman S: The
conserved coronavirus macrodomain promotes virulence and suppresses
the innate immune response during severe acute respiratory syndrome
coronavirus infection. mBio. 7:e01721–16. 2016.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Eckei L, Krieg S, Bütepage M, Lehmann A,
Gross A, Lippok BE, Grimm AR, Kümmerer BM, Rossetti G, Lüscher B
and Verheugd P: The conserved macrodomains of the non-structural
proteins of Chikungunya virus and other pathogenic positive strand
RNA viruses function as mono-ADP-ribosylhydrolases. Sci Rep.
7(41746)2017.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Li C, Debing Y, Jankevicius G, Neyts J,
Ahel I, Coutard B and Canard B: Viral macro domains reverse protein
ADP-ribosylation. J Virol. 90:8478–8486. 2016.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Alhammad YMO, Kashipathy MM, Roy A, Gagné
JP, McDonald P, Gao P, Nonfoux L, Battaile KP, Johnson DK,
Holmstrom ED, et al: The SARS-CoV-2 conserved macrodomain is a
highly efficient ADP-ribosylhydrolase enzyme. bioRxiv:
2020.05.11.089375, 2020.
|
|
84
|
Munnur D and Ahel I: Reversible
mono-ADP-ribosylation of DNA breaks. FEBS J. 284:4002–4016.
2017.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Scarpa ES, Fabrizio G and Di Girolamo M: A
role of intracellular mono-ADP-ribosylation in cancer biology. FEBS
J. 280:3551–3562. 2013.PubMed/NCBI View Article : Google Scholar
|