Introduction
Chronic silica exposure may trigger macrophage
alveolitis, induce myofibroblast transition and eventually progress
to silicosis (1). Silicosis is an
irreversible pulmonary disease and the fibrosis remains progressive
even when patients are separated from the exposure to silica
(2). In developing countries,
silica-induced silicosis remains a major concern (3). There is no effective therapy and the
molecular mechanism of silicotic fibrosis remains unclear (4).
With the development of gene chip and RNA sequencing
(RNA-seq) technologies, bioinformatic analysis has served a
significant role in screening candidate biomarkers and
investigating the mechanisms of silicosis. Comparative RNA-seq has
demonstrated that genes associated with immune cell interactions,
immune cell responses and inflammation are significantly enriched
in mice exposed to silica (5).
Genes that regulate fibrosis, redox enzymes and metalloproteinases
are upregulated in acute and chronic silicosis models (6). In vitro, macrophages (7) and lung epithelial cells (8) exposed to silica particles have
indicated the roles of certain genes in immune responses and
inflammatory pathways. These findings provide the basis to
elucidate the molecular mechanisms of silica-induced pulmonary
inflammation and fibrosis, and support research for the prevention
and treatment of silicosis.
While these studies have provided insight into the
mechanisms underlying chronic inhalation of silica, the majority of
early studies were performed using silicotic models established by
bronchial instillation and reported a limited number of genes,
pathways and functions altered by silicosis (5,6). The
present study investigated the full spectrum of mRNA expression in
changes of silicotic rats induced by chronic inhalation of silica,
which closely approximates the development of silicosis in humans
(9,10). A large number of mRNAs were
identified that were differentially expressed following chronic
exposure to silica particles, including 912 genes that were
upregulated and 426 genes that were downregulated. Furthermore,
increased levels of secreted phosphoprotein-1 (SPP1) were
identified in vivo, including the serum of silicotic
patients and lungs of silicotic rats, and in silica-treated
macrophages. Taken together, the results of the present study
indicated that the expression of mRNAs was significantly altered in
silicotic rats and suggested that certain genes are novel targets
for the diagnosis and treatment of silicosis.
Materials and methods
Rat silicosis model
The rats were housed in a temperature-control
facility at 22-24˚C and 70-75% humidity with a 12/12-h light/dark
cycle and were given free access to water and food. All
experimental and surgical procedures were approved by the Ethics
Committee for Animal Experimentation of North China University of
Science and Technology (2013-038). The silicotic model used in the
present study has been well described and documented in our
previous studies (10,11). In brief, specific pathogen-free male
Wistar rats (weight, 180±10 g; age, 3 weeks; n=20) were placed in a
HOPE-MED 8050 exposure control apparatus (HOPE Industry and Trade
Co., Ltd.) and inhaled pure air (control group) or SiO2
dust particles (80% diameter between 1 and 5 µm; s5631;
Sigma-Aldrich; Merck KGaA) at a concentration of 50±10
µg/m3. SiO2 was ground and heated at 180˚C
for 6 h for molding. The mass concentration of SiO2 was
monitored by an integrated atmospheric sampler using the
gravimetric method twice a week (10). Ten rats were randomly and equally
divided into control and silicosis model groups. Silicotic rats
were placed in the chamber for 3 h per day for 24 consecutive
weeks. The rats were anesthetized by intraperitoneal injection of
1% pentobarbital sodium (60 mg/kg) and euthanized by blood
collection from the abdominal aorta. Next, rat lung tissue was
harvested and immediately frozen in liquid nitrogen for
analyses.
Bioinformatics of mRNA sequencing in
rat lungs
Total RNA was extracted from lung tissue using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
rRNA was removed using an Epicentre Ribo-Zero™ rRNA Removal kit
(Illumina, Inc.). Subsequently, short fragments of RNA (~200 nt)
were prepared in fragmentation buffer and used as templates to
synthesize cDNAs. Following purification, double-stranded cDNA was
subjected to terminal repair. Specifically, a tail was added and
the sequencing linker was ligated. Finally, a cDNA library of total
RNA was obtained by PCR enrichment. Following the library being
qualified, its insert size was determined using an Agilent 2200
Tape Station (Agilent Technologies, Inc.), which was then sequenced
by a HiSeq 3000/HiSeq 2500 sequencing system (Illumina, Inc.).
Raw reads were filtered and aligned to the reference
genome using alignment software HISAT (version 2.0.1) (12). Next, the ratio of the genome on the
alignment and the distribution of reads on the chromosome were
compared. Finally, high quality data (clean data) were obtained.
BAM files were obtained by comparing clean data with the reference
genome using Tophat2 software (version 2.1.1) (13). Genome location information
corresponding to all reads was then counted and the coverage depth
of sequencing data was evaluated. The sequenced BAM file was mapped
to the reference rat gtf file to determine gene expression levels
and reads per kilobase of transcript per million mapped reads
values were used to calculate transcript expression levels
(14). Differential expression
analysis of mRNA was conducted by edgeR software (version 3.12)
(15) using thresholds of log
FC>1 and Q-value <0.05.
Gene ontology and pathway enrichment
analysis
The biological functions of mRNA-associated target
genes were analyzed by Gene Ontology (GO) enrichment analysis
(http://www.geneontology.org). The
pathway enrichment of mRNA-associated target genes was analyzed by
the Kyoto Encyclopedia of Genes and Genomes (KEGG) database
(version 12.0) (http://www.genome.jp/kegg/pathway.html).
Cell culture
The monocyte/macrophage NR8383 cell line was
obtained from the Cell Bank of the Chinese Academy of Sciences
(16). Cells were cultured in Ham's
F-12K medium (L450KJ; Gibco; Thermo Fisher Scientific, Inc.),
containing 20% (v/v) fetal bovine serum (SFBS; Bovogen Biologicals
Pty., Ltd.), 100 IU/ml penicillin and 100 mg/ml streptomycin at
37˚C in a humidified 5% CO2 incubator. Cells at 70-80%
confluence were treated with or without silica (50
µg/cm2).
ELISA
The samples were obtained from patients diagnosed at
different stages at the Beidaihe sanatorium (Qinhuangdao, China).
All the subjects had no serious diseases in the heart, brain, liver
or kidneys, and those >65 years old were excluded. All the
participants were male. The study period was between June 2017 and
May 2018. The human experiments were approved by the Medical Ethics
Committee of North China University of Science and Technology
(2015-046) (17). Written informed
consent was obtained from each subject to confirm their voluntary
participation in the study. The study included 18 patients who were
diagnosed with silicosis (mean age, 46.7±5.2 years) by the
Occupational Diseases Committee using diagnostic criteria for
occupational pneumoconiosis of China (GBZ 70-2015). Nine control
subjects (mean age, 48.0±7.0 years) from the same hospital were
included (Table SI). The plasma
concentration of human SPP1 was determined using a human ELISA kit
(ARG81268; Arigo Biolaboratories Corp.). The assay minimum
detectable concentration of SPP1 was 62.5 pg/ml.
Immunohistochemistry (IHC)
The samples were sequentially dehydrated, embedded
in paraffin and cut into 4 µm sections and then fixed in 4%
paraformaldehyde solution for 48 h at room temperature.
Paraffin-embedded sections of lung tissue and cells underwent
antigen retrieval (110˚C; 50 kPa; 10 mmol/l citric acid-sodium
buffer) for 80 sec, followed by quenching endogenous peroxidases
with 3% H2O2 for 15 min at room temperature.
Sections were incubated with primary antibodies against SPP1
(1:100; cat. no. AF0227; Affinity Biosciences) overnight at 4˚C.
The following day, they were incubated with goat anti-rabbit or
anti-mouse secondary antibodies (cat. no. PV-6000; Beijing
Zhongshan Jinqiao Biotechnology Co. Ltd.) at 37˚C for 30 min. Color
development was performed by 3,3'-diaminobenzidine (ZLI-9018;
OriGene Technologies, Inc.) and brown staining was considered as
positive. Sections were mounted with neutral balsam and observed by
microscopy (magnification, x400; BX53; Olympus Corporation).
Western blotting
Whole cell and lung tissue lysates were prepared
using 100-200 µl RIPA buffer (BB-3201-1; BestBio; http://bestbio.qianyan.biz/) containing a protease
inhibitor cocktail (P2714-1BTL; Sigma-Aldrich; Merck KGaA). The
sample was incubated on ice for 30 min and the supernatant was
collected following centrifugation (14,000 x g, 20 min, 4˚C). Total
protein was estimated using a BCA assay kit (PQ0012; MultiSciences
Multisciences (Lianke) Biotech Co., Ltd.). Proteins (20 µg/lane)
were separated by 10% SDS-PAGE and electrotransferred onto PVDF
membranes. The membranes were blocked with 5% dry skimmed milk for
1 h at room temperature and then incubated overnight at 4˚C with
primary antibodies against SPP1 (1:1,000; cat. no. AF0227; Affinity
Biosciences), collagen type I (Col I; 1:1000, cat. no. ab34710;
Abcam) or transforming growth factor-β1 (TGF-β1; 1:1,000; cat. no.
ARG56894; Arigo Biolaboratories Corp.). The membranes were washed
sequentially with TBST containing 0.2% Tween-20 and then incubated
with goat anti-rabbit (cat. no. 074-1506) or anti-mouse (cat. no.
074-1806) secondary antibodies (dilution, 1:5,000; KPL, Inc.) at
37˚C for 1 h. Bands were detected using the ECL™ Prime Western blot
system and average optical densities were measured using ImageJ
v6.0 software (National Institutes of Health). The results were
normalized against the Tub α expression level and corresponding
control.
Quantitative polymerase chain reaction
(qPCR)
Total RNA was isolated from rat tissues using a
RNeasy Mini-kit (Qiagen, Inc.) and quantified using a
Nano-Drop-2000 spectrophotometer (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocols. First-strand cDNA was
generated by a reverse transcription system (cat. no. ZR102;
Beijing Zoman Biotechnology Co., Ltd.), according to the
manufacturer's protocols. qPCR was performed using a 2X SYBR qPCR
mix kit (ZF102-1; Beijing Zoman Biotechnology Co., Ltd.), under the
following conditions: Initial denaturation at 94˚C for 2 min,
followed by 40 cycles of 94˚C for 15 sec and 60˚C for 30 sec in
accordance with the standard protocol of the QuantStudio™ 6 Flex
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Primer sequences were as follows. GAPDH forward,
5'-GGTGAAGGTCGGTGTGAACG-3' and reverse, 5'-CTCGCTCCTGGAAGATGGTG-3';
Dlk1 forward, 5'-CAACAATGGGACTTGCGTGG-3' and reverse,
5'-TCTCGCATGGGTTAGGGGTA-3'; Lcn2 forward,
5'-CACTTCCATCCTCGTCAGGG-3' and reverse, 5'-TTCAGTTCATCGGACAGCCC-3';
Slc26a4 forward, 5'-GTGACAATTATCGCCACCGC-3' and reverse,
5'-TGCAATAGCGTAAGCCACCA-3'; Mmp12 forward,
5'-GGAGTCCAGCCACCAACATT-3' and reverse, 5'-TTACAGATGCAGGGAAGCCC-3';
Spp1 forward, 5'-CTTTTGCCTGTTCGGCCTTG-3' and reverse,
5'-AGCCAAGTGGCTACAGCATC-3'; Fabp4 forward,
5'-CGTAGAAGGGGACTTGGTCG-3' and reverse, 5'-CCCCACCATCCAGGGTTATG-3';
Itln1, forward, 5'-TGTTGGACTGACAATGGCCC-3' and reverse,
5'-TCCAGTGACCTTCATGCCAG-3'; Defb5 forward,
5'-GCTGTCGCCCCTTTCTGTCT-3' and reverse,
5'-CAATCTGTCGAAAACTGCCAGG-3'; Lpo forward,
5'-GAAGGTGGGCTGTAATCCCC-3' and reverse, 5'-GAGGGAGAGTCCATCCTCGT-3';
Ccl7 forward, 5'-AGGGGTAGGAACGGTCTGTA-3' and reverse,
5'-TGAGGTCTCCAGGGCTTTAC-3'. The housekeeping gene, GAPDH,
was used as the endogenous control. The results were calculated
usign the 2-ΔΔCq method (18).
Statistical analysis
Statistical analysis was performed using SPSS 20.0
software (IBM Corp.). Data are presented as the mean ± standard
deviation. Two group comparisons were analyzed by the unpaired
Student's t-test. The correlation between the factors was analyzed
by Pearson correlation. P<0.05 was considered to indicate a
statistically significant difference.
Results
Differential expression profiles of
mRNAs in silicotic rat lungs
All samples in the present study were verified by
histological and biochemical evidence (9-11).
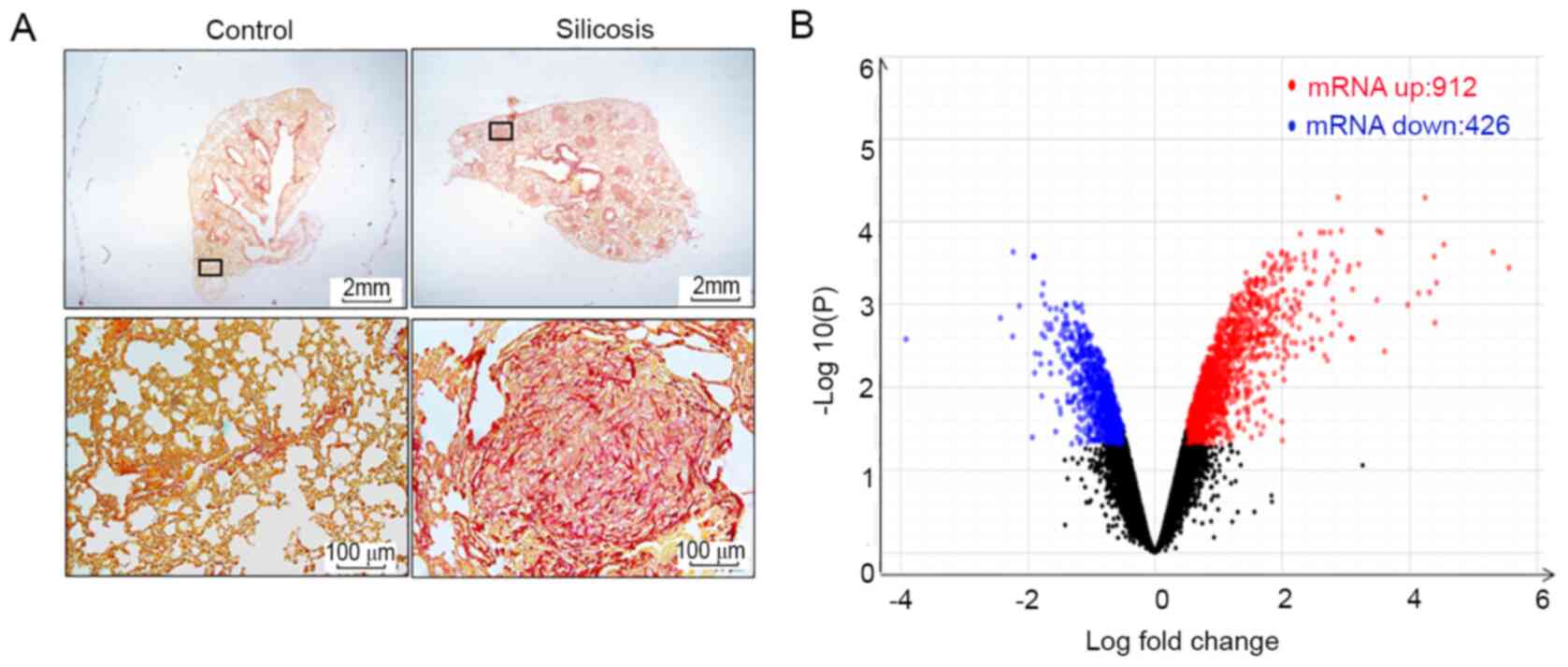
Fig. 1A demonstrates the formation
of silicotic lesions in rats exposed to silica. In RNA-sequencing
analysis, it was found that the expression of 1,338 mRNAs was
changed in silicotic rats compared with that in the control group,
including 912 upregulated and 426 downregulated mRNAs (Fig. 1B and Table SII). KEGG pathway analysis
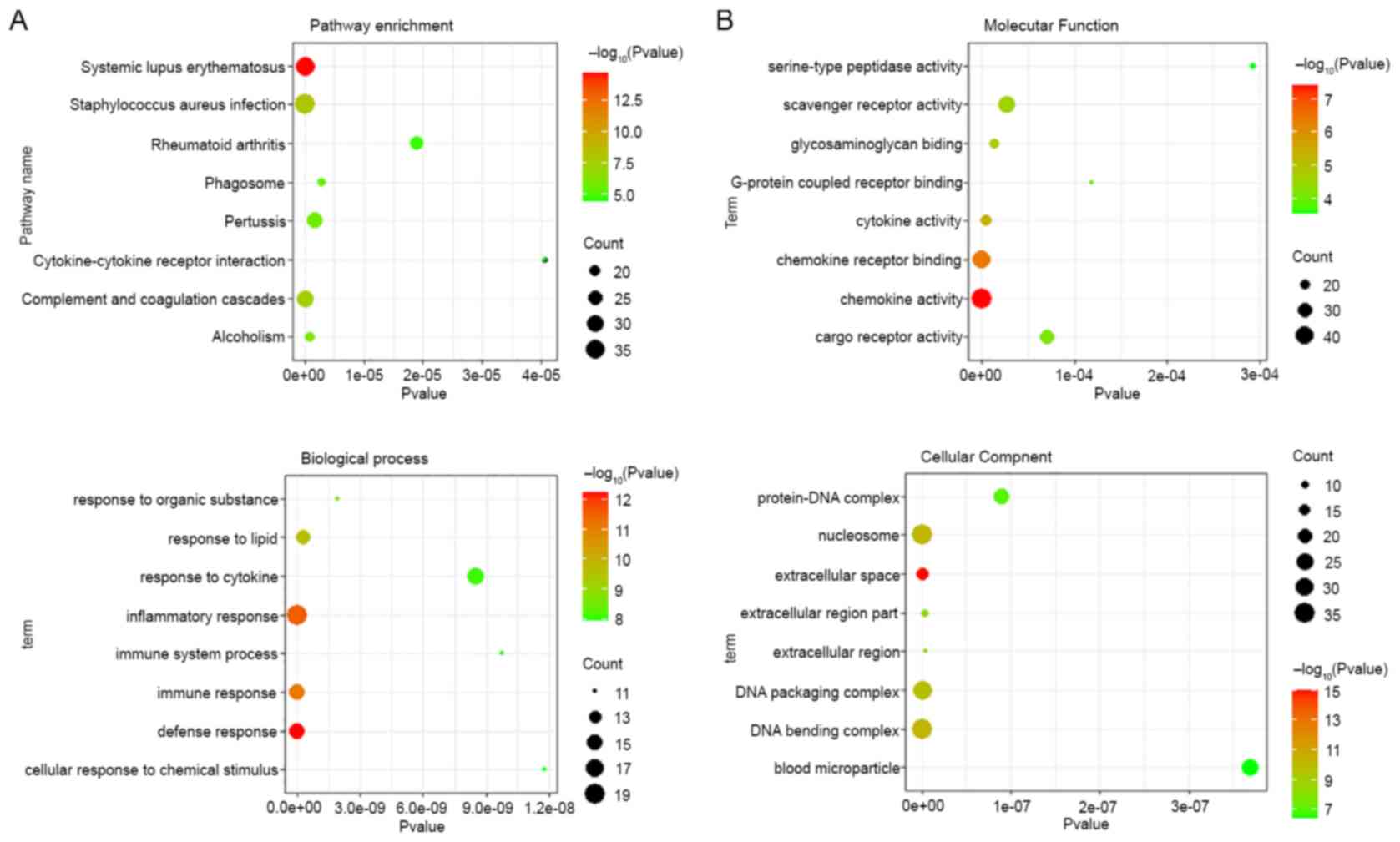
demonstrated that 30 pathways were enriched for the differentially
expressed mRNAs, and systemic lupus erythematosus, staphylococcus
aureus infection, complement and coagulation cascades, alcoholism
and pertussis were the most significantly enriched KEGG pathways
(Fig. 2A and Table SIII). As shown in Fig. 2B and Tables
SIV-SVI, GO analysis revealed that the most significantly
enriched functional terms of differentially expressed mRNAs were
the defense response, extracellular space and chemokine activity
for classifications of biological process, and cellular component
and molecular function.
Verification of differential mRNA
expression in silicotic rat lungs
On the basis of the fold-change and high abundance,
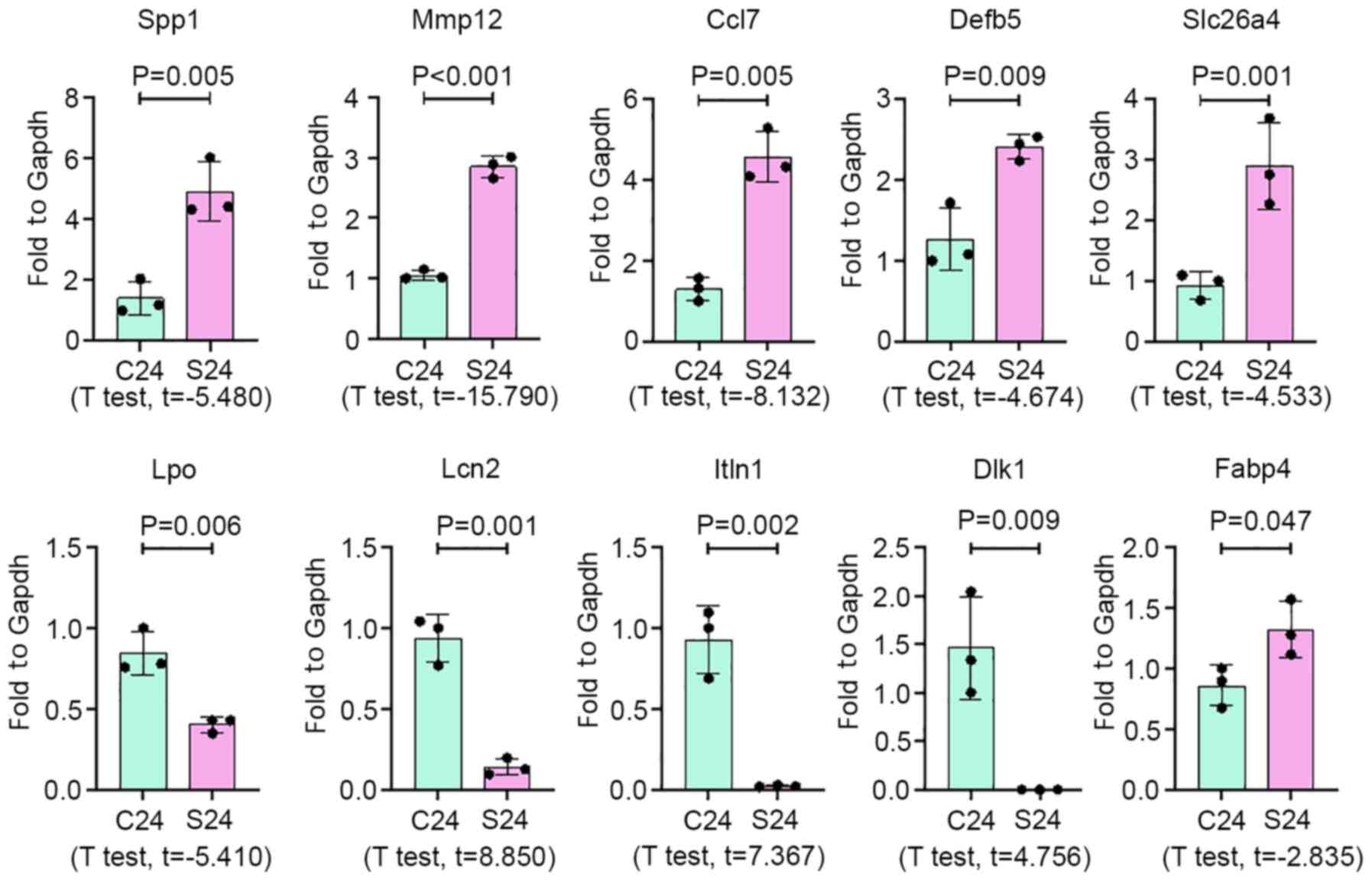
10 mRNAs were selected to validate the results of RNA-seq analysis
in silicotic rat lungs by qPCR. As a result, expression of
Spp1, Mmp12, Ccl7, Defb5, Fabp4
and Slc26a4 was increased in silicosis samples, while
Lpo, Itln1, Lcn2 and Dlk1 expression
was significantly decreased in silicotic lung tissues, compared
with the control group (Fig.
3).
SPP1 expression increases in serum
from silicosis patients, lungs of silicotic rats and silica-treated
NR8383 cells
To determine whether the mRNA findings were
representative of changes in vivo and in vitro,
ELISA, IHC and Western blot analyses of SPP1 in control and
silicosis groups were performed, the latter of which was changed
most significantly. Plasma was collected from patients with
silicosis and the expression of SPP1 was analyzed by an ELISA. SPP1
expression in patients with silicosis was increased, compared with
the control group (Fig. 4A and
Table SVII). Pearson correlation
analysis demonstrated that the expression level of SPP1 was
associated with lung function (Fig.
4B). Furthermore, as shown in Fig.
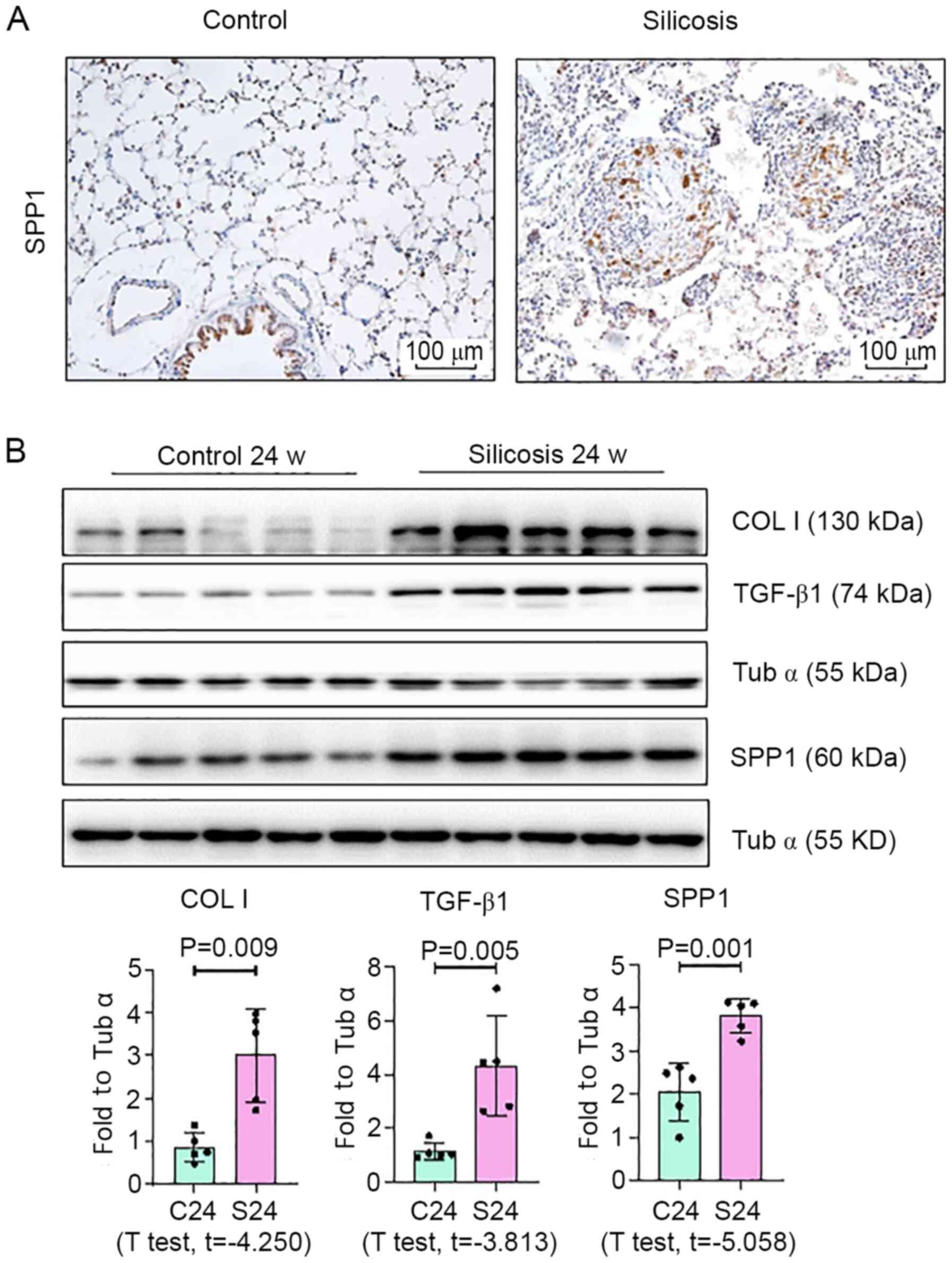
5, Western blotting revealed a marked increase in the
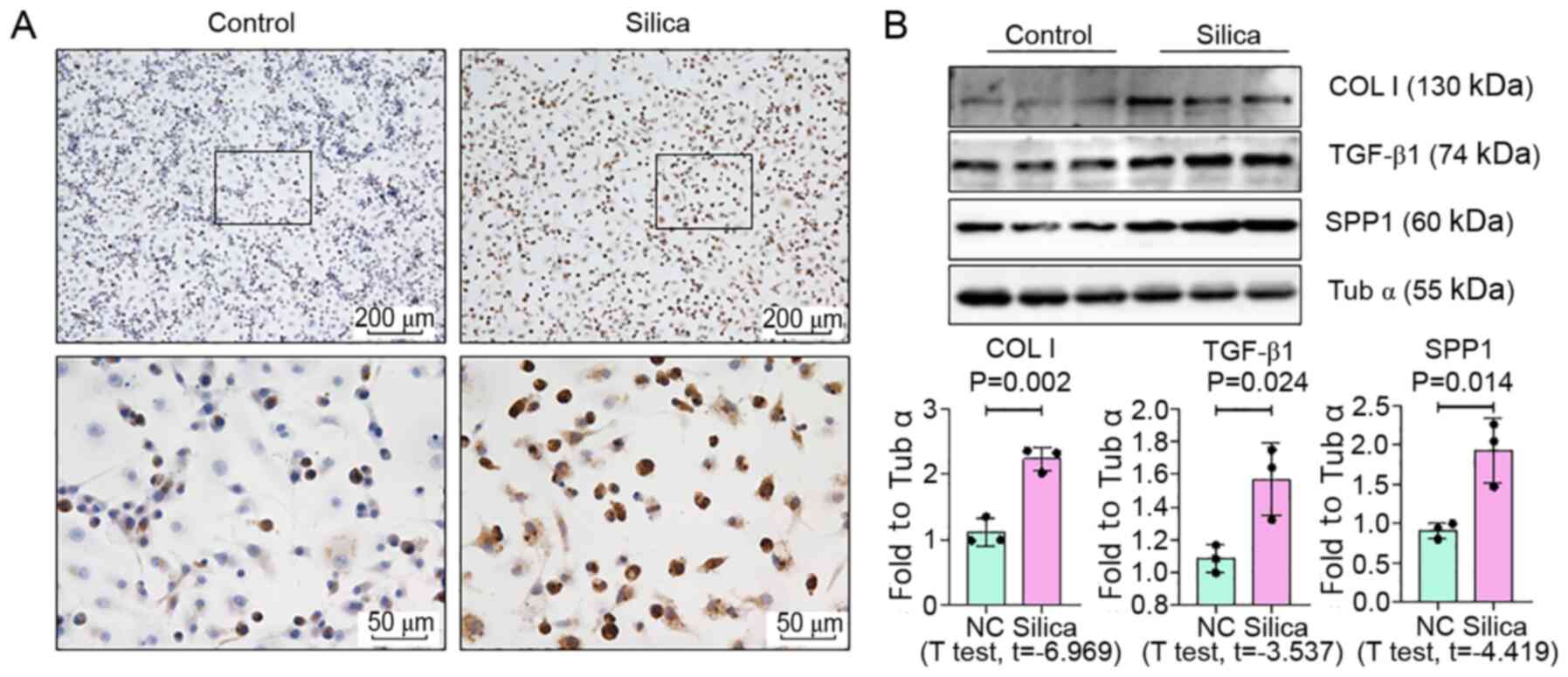
expression of SPP1 in lungs of silica-exposed rats. Furthermore,
IHC staining of lungs demonstrated that the increased level of SPP1
was mostly confined to silicotic lesions of the lung, particularly
in macrophages. In vitro, an increased level of SPP1 was
found in silica-treated NR8383 macrophages (Fig. 6).
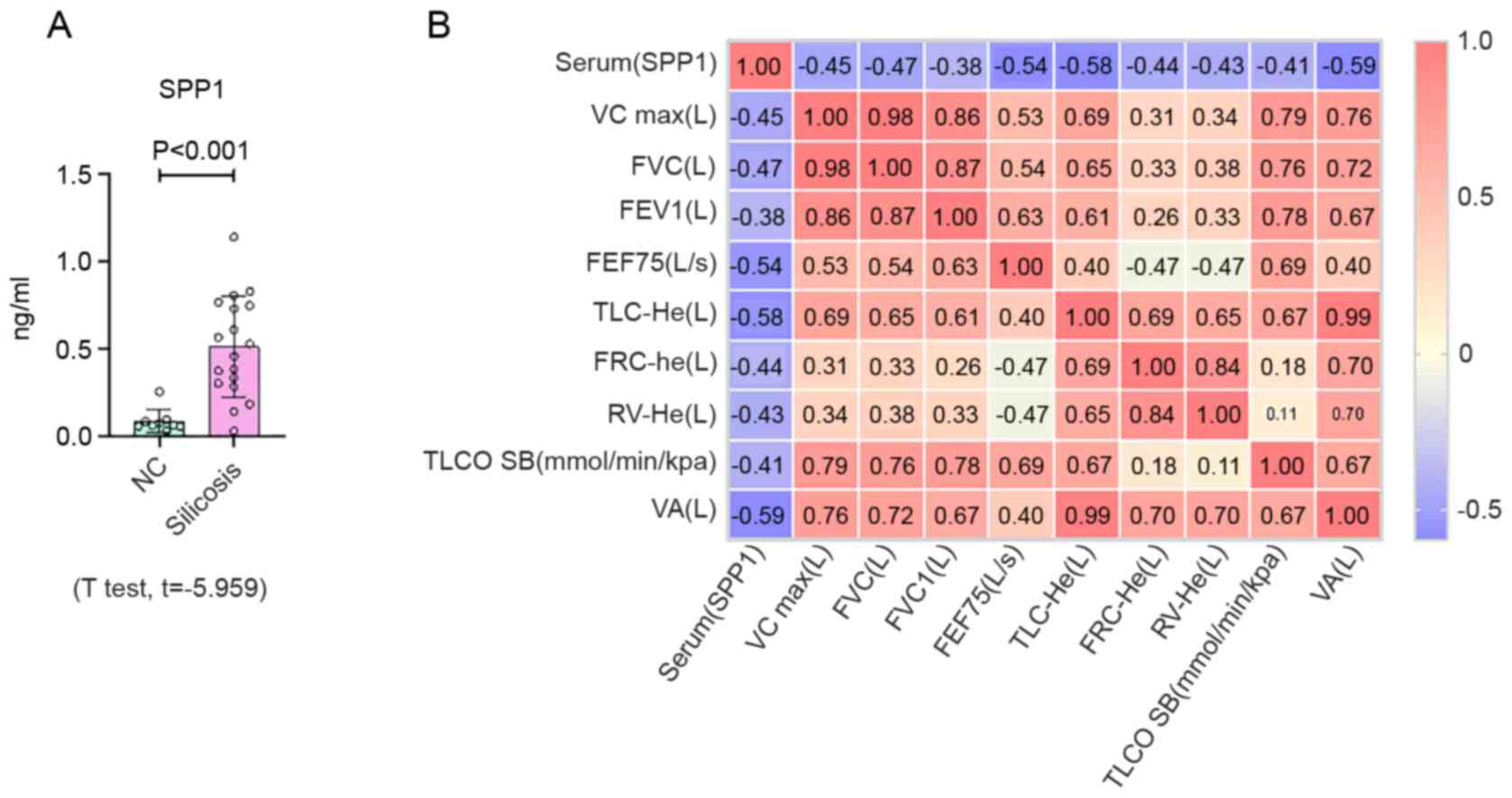 | Figure 4Expression of SPP1 in serum from
silicosis patients. (A) Protein expression levels of SPP1
determined by ELISA in plasma samples from controls (n=9) and
patients with silicosis (n=18). (B) Pearson correlation analysis of
SPP1 and lung function. Data are presented as the mean ± standard
deviation. SPP1, secreted phosphoprotein 1. SPP1, secreted
phosphoprotein 1; VC, vital capacity; FVC, forced vital capacity;
FEV1, forced expiratory volume in the first second; FEF75, forced
expiratory flow 75; TLC, total lung capacity; FRC, functional
residual capacity; RV, residual volume; TLCO SB, carbon monoxide
transfer factor determined in single breath; VA, alveolar
ventilation. |
Discussion
Molecular understanding of silicosis has been
generated from silicotic rodents with acute silicosis induced by
instillation of silica. Compared with acute silicotic models,
chronic inhalation of silica is a better method to investigate the
complicated mechanisms of silicosis, which is relevant to human
silicosis (6). In the present
study, a silicotic rat model was established by chronic silica
particle inhalation, which has been well documented by our group
(9,10), and 1,338 mRNAs were found to be
altered in the silicotic group. With the progress of RNA-seq
technologies, more differentially expressed mRNAs were identified
than in previous reports (19,20),
which provides improved understanding of the development of
silicosis.
To confirm the results obtained by RNA-seq analysis,
10 differentially expressed mRNAs were selected for verification in
silicotic lung tissues by qPCR. The results indicated that
expression of Spp1, Mmp12, Ccl7, Defb5,
Fabp4 and Slc26a4 was increased in silicosis samples,
while expression of Lpo, Itln1, Lcn2 and
Dlk1 was significantly decreased in silicotic lung tissues,
compared with the control group.
An increased level of MMP-12 is a critical player in
chronic pulmonary pathologies, including asthma, chronic
obstructive pulmonary disease and pulmonary fibrosis (21). Overexpression of SLC26A4 was
found in lungs of patients with silicosis and correlated with the
onset and prognosis of silicosis (22). Although few studies have measured
the expression levels of CCL7 in patients with pulmonary fibrosis,
lung CCL7 expression is augmented in murine bleomycin- and
radiation-induced pulmonary fibroses (23,24).
Additionally, CCL2 is expressed in various cell types, including
macrophages, fibroblasts, endothelial cells and epithelial cells,
which exhibits increased expression during silicosis (25-27).
FABP4 deteriorates renal interstitial fibrosis by promoting
inflammation and lipid metabolism disorders (28). Dlk1 has been reported to accelerate
fibroblast-to-myofibroblast differentiation and induces myocardial
fibrosis (29).
Among the upregulated mRNAs, SPP1 has been
demonstrated to be a marker of IPF (30). Gene or protein overexpression of
SPP1 (also named osteopontin) is induced in the lungs,
bronchoalveolar lavage fluid and serum of humans and rodents
following exposure to various drugs and pathological agents
(31). It contains an Arg-Gly-Asp
motif that binds to the integrin family of adhesion molecules and
is highly upregulated in mice with bleomycin-induced lung fibrosis
and idiopathic pulmonary fibrosis (IPF) patients (30,32,33).
The present study found that Spp1 had the highest false
discovery rate among dysregulated mRNAs in silicotic rats. Notably,
clinical correlation analyses have demonstrated that high SPP1
plasma levels in primary myelofibrosis patients correlate with a
more severe fibrosis degree and shorter overall survival time
(34). In line with these data, it
was found that the expression level of SPP1 was increased in serum
from patients with silicosis. Pearson correlation analysis
demonstrated that the expression level of SPP1 was associated with
lung function. Furthermore, IHC staining demonstrated positive
expression of SPP1 in macrophages of silicotic lesions. It has been
reported that macrophages with high SPP1 expression represent a
profibrotic macrophage population in IPF lungs, which promote
activation of myofibroblasts (35).
Furthermore, global SPP1 ablation correlates with decreased
fibrosis and decreased TGF-β in dystrophic muscle (36). Additionally, SPP1 promotes
fibroblast differentiation and induces upregulation of collagen
type I expression in pathological fibrosis associated with liver,
skin and lung tissues (37-39).
Deletion of SPP1 in bleomycin-induced lung fibrosis decreases
upregulated expression of collagen type 1 and MMP2 (40,41).
High expression of SPP1 was also found in silica-treated
macrophages. These results indicated high SPP1 expression during
silicosis, which may strongly contribute toward lung fibrosis in
occupational exposure to silica.
In conclusion, it was found that the expression of
mRNAs was significantly altered in silicotic rats, which suggests
that certain genes may be novel targets for the diagnosis and
treatment of silicosis.
Supplementary Material
Occupational history of patients
exposed to silica.
Different genes in silica-treated rat
lungs
KEGG pathway
GO-Biological Process
GO-Cellular Component
GO-Molecular Function
Correlation analysis between lung
function index and SPP1.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81972988), Natural Science
Foundation of Hebei Province (grant no. H2020209052), Science and
Technology Research Project of Hebei Province universities (grant
no. ZD2019077), The Science and Technology Plan Project of Tangshan
City (grant no. 20130206b), and Open Fund of Key Laboratory of
Functional and Clinical Translational Medicine of Xiamen Medical
Collage (grant no. XMMC-FCTM201902).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FY and HX designed the study. WC performed all the
experiments. BZ, TL, FJ and YL collected experimental samples. WC
and HX interpreted the data and drafted the manuscript. FY, WC and
HX confirmed the authenticity of the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
All experimental and surgical procedures involving
animals were approved by the Ethics Committee for Animal
Experimentation of North China University of Science and Technology
(2013-038). The human experiments were approved by the Medical
Ethics Committee of North China University of Science and
Technology (2015-046). Written informed consent was obtained from
each subject to confirm their voluntary participation in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li J, Yao W, Hou JY, Zhang L, Bao L, Chen
HT, Wang D, Yue ZZ, Li YP, Zhang M and Hao CF: Crystalline silica
promotes rat fibrocyte differentiation in vitro, and fibrocytes
participate in silicosis in vivo. Biomed Environ Sci. 30:649–660.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siribaddana AD, Wickramasekera K, Palipana
WM, Peiris MD, Upul BK, Senevirathna KP and Dassanayake DL: A study
on silicosis among employees of a silica processing factory in the
Central Province of Sri Lanka. Ceylon Med J. 61:6–10.
2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cainelli F, Tanko MN and Vento S: . Silica
exposure and silicosis: Action is needed now. South Med J.
103(1078)2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kulkarni GK: Prevention and control of
silicosis: A national challenge. Indian J Occup Environ Med.
11:95–96. 2007.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chen J, Yao Y, Su X, Shi Y, Song X, Xie L,
You J, Tian L, Yang L, Fang A and Xiong J: Comparative RNA-Seq
transcriptome analysis on silica induced pulmonary inflammation and
fibrosis in mice silicosis model. J Appl Toxicol. 38:773–782.
2018.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Langley RJ, Mishra NC, Peña-Philippides
JC, Rice BJ, Seagrave JC, Singh SP and Sopori ML: Fibrogenic and
redox-related but not proinflammatory genes are upregulated in
Lewis rat model of chronic silicosis. J Toxicol Environ Health A.
74:1261–1279. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chan JYW, Tsui JCC, Law PTW, So WKW, Leung
DYP, Sham MMK, Tsui SKW and Chan CWH: RNA-Seq revealed
ATF3-regulated inflammation induced by silica. Toxicology.
393:34–41. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chan JYW, Tsui JCC, Law PTW, So WKW, Leung
DYP, Sham MMK, Tsui SKW and Chan CWH: Profiling of the
silica-induced molecular events in lung epithelial cells using the
RNA-Seq approach. J Appl Toxicol. 37:1162–1173. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Hui Z, Dingjie X, Yuan Y, Zhongqiu W, Na
M, Mingjian B, Yu G, Guangyuan L, Xuemin G, Shifeng L, et al:
Silicosis decreases bone mineral density in rats. Toxicol Appl
Pharmacol. 348:117–122. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shifeng L, Hong X, Xue Y, Siyu N, Qiaodan
Z, Dingjie X, Lijuan Z, Zhongqiu W, Xuemin G, Wenchen C, et al:
Ac-SDKP increases alpha-TAT 1 and promotes the apoptosis in lung
fibroblasts and epithelial cells double-stimulated with TGF-beta1
and silica. Toxicol Appl Pharmacol. 369:17–29. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cai W, Xu H, Zhang B, Gao X, Li S, Wei Z,
Li S, Mao N, Jin F, Li Y, et al: Differential expression of lncRNAs
during silicosis and the role of LOC103691771 in myofibroblast
differentiation induced by TGF-β1. Biomed Pharmacother.
125(109980)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14(R36)2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chen Y, Xu D, Yao J, Wei Z, Li S, Gao X,
Cai W, Mao N, Jin F, Li Y, et al: Inhibition of miR-155-5p exerts
anti-fibrotic effects in silicotic mice by regulating meprin α. Mol
Ther Nucleic Acids. 19:350–360. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Gao X, Xu H, Zhang B, Tao T, Liu Y, Xu D,
Cai W, Wei Z, Li S, Zhang H, et al: Interaction of
N-acetyl-seryl-aspartyl-lysyl-proline with the
angiotensin-converting enzyme 2-angiotensin-(1-7)-Mas axis
attenuates pulmonary fibrosis in silicotic rats. Exp Physiol.
104:1562–1574. 2019.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sellamuthu R, Umbright C, Roberts JR,
Young SH, Richardson D, McKinney W, Chen BT, Li S, Kashon M and
Joseph P: Molecular mechanisms of pulmonary response progression in
crystalline silica exposed rats. Inhal Toxicol. 29:53–64.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sellamuthu R, Umbright C, Roberts JR,
Cumpston A, McKinney W, Chen BT, Frazer D, Li S, Kashon M and
Joseph P: Molecular insights into the progression of crystalline
silica-induced pulmonary toxicity in rats. J Appl Toxicol.
33:301–312. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Garbacki N, Di Valentin E, Piette J,
Cataldo D, Crahay C and Colige A: Matrix metalloproteinase 12
silencing: A therapeutic approach to treat pathological lung tissue
remodeling? Pulm Pharmacol Ther. 22:267–278. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jiang QT, Han L, Liu X and Zhu BL:
Upregulation of Slc26a4 in the Early Development of Silicosis via
GEO Database Analysis in vivo and in vitro. Biomed Environ Sci.
32:938–943. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kaminski N, Allard JD, Pittet JF, Zuo F,
Griffiths MJ, Morris D, Huang X, Sheppard D and Heller RA: Global
analysis of gene expression in pulmonary fibrosis reveals distinct
programs regulating lung inflammation and fibrosis. Proc Natl Acad
Sci USA. 97:1778–1783. 2000.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Johnston CJ, Williams JP, Okunieff P and
Finkelstein JN: Radiation-induced pulmonary fibrosis: Examination
of chemokine and chemokine receptor families. Radiat Res.
157:256–265. 2002.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Paine R III, Rolfe MW, Standiford TJ,
Burdick MD, Rollins BJ and Strieter RM: MCP-1 expression by rat
type II alveolar epithelial cells in primary culture. J Immuno.
150:4561–4570. 1993.PubMed/NCBI
|
|
26
|
Brieland JK, Jones ML, Clarke SJ, Baker
JB, Warren JS and Fantone JC: Effect of acute inflammatory lung
injury on the expression of monocyte chemoattractant protein-1
(MCP-1) in rat pulmonary alveolar macrophages. Am J Respir Cell Mol
Biol. 7:134–139. 1992.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Barrett EG, Johnston C, Oberdorster G and
Finkelstein JN: Antioxidant treatment attenuates cytokine and
chemokine levels in murine macrophages following silica exposure.
Toxicol Appl Pharmacol. 158:211–220. 1999.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Qiao Y, Liu L, Yin L, Xu L, Tang Z, Qi Y,
Mao Z, Zhao Y, Ma X and Peng J: FABP4 contributes to renal
interstitial fibrosis via mediating inflammation and lipid
metabolism. Cell Death Dis. 10(382)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yang XQ, Zhao FF and Wang DX: Metabolism
reprogramming: New insights of Dlk1 into cardiac fibrosis. Eur
Heart J. 40(3574)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Pardo A, Gibson K, Cisneros J, Richards
TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M and
Kaminski N: Up-regulation and profibrotic role of osteopontin in
human idiopathic pulmonary fibrosis. PLoS Med.
2(e251)2005.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Sabo-Attwood T, Ramos-Nino ME,
Eugenia-Ariza M, Macpherson MB, Butnor KJ, Vacek PC, McGee SP,
Clark JC, Steele C and Mossman BT: Osteopontin modulates
inflammation, mucin production, and gene expression signatures
after inhalation of asbestos in a murine model of fibrosis. Am J
Pathol. 178:1975–1985. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
El Deeb S, Abdelnaby R, Khachab A, Blasius
K, Tingart M and Rath B: Osteopontin as a biochemical marker and
severity indicator for idiopathic hip osteoarthritis. Hip Int.
26:397–403. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lorenzen JM, Nickel N, Krämer R, Golpon H,
Westerkamp V, Olsson KM, Haller H and Hoeper MM: Osteopontin in
patients with idiopathic pulmonary hypertension. Chest.
139:1010–1017. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ruberti S, Bianchi E, Guglielmelli P,
Rontauroli S, Barbieri G, Tavernari L, Fanelli T, Norfo R, Pennucci
V, Fattori GC, et al: Involvement of MAF/SPP1 axis in the
development of bone marrow fibrosis in PMF patients. Leukemia.
32:438–449. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Morse C, Tabib T, Sembrat J, Buschur KL,
Bittar HT, Valenzi E, Jiang Y, Kass DJ, Gibson K, Chen W, et al:
Proliferating SPP1/MERTK-expressing macrophages in idiopathic
pulmonary fibrosis. Eur Respir J. 54(1802441)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Vetrone SA, Montecino-Rodriguez E,
Kudryashova E, Kramerova I, Hoffman EP, Liu SD, Miceli MC and
Spencer MJ: Osteopontin promotes fibrosis in dystrophic mouse
muscle by modulating immune cell subsets and intramuscular
TGF-beta. J Clin Invest. 119:1583–1594. 2009.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Dong J and Ma Q: Osteopontin enhances
multi-walled carbon nanotube-triggered lung fibrosis by promoting
TGF-beta1 activation and myofibroblast differentiation. Part Fibre
Toxicol. 14(18)2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hunter C, Bond J, Kuo PC, Selim MA and
Levinson H: The role of osteopontin and osteopontin aptamer
(OPN-R3) in fibroblast activity. J Surg Res. 176:348–358.
2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Morimoto Y, Hirahara K, Kiuchi M, Wada T,
Ichikawa T, Kanno T, Okano M, Kokubo K, Onodera A, Sakurai D, et
al: Amphiregulin-producing pathogenic memory T helper 2 cells
instruct eosinophils to secrete osteopontin and facilitate airway
fibrosis. Immunity. 49:134–150.e6. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Berman JS, Serlin D, Li X, Whitley G,
Hayes J, Rishikof DC, Ricupero DA, Liaw L, Goetschkes M and O'Regan
AW: Altered bleomycin-induced lung fibrosis in
osteopontin-deficient mice. Am J Physiol Lung Cell Mol Physiol.
286:L1311–L1318. 2004.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Takahashi F, Takahashi K, Okazaki T, Maeda
K, Ienaga H, Maeda M, Kon S, Uede T and Fukuchi Y: Role of
osteopontin in the pathogenesis of bleomycin-induced pulmonary
fibrosis. Am J Respir Cell Mol Biol. 24:264–271. 2001.PubMed/NCBI View Article : Google Scholar
|