Introduction
The proangiogenic effects of leptin, an essential
adipokine secreted from fat tissue, have an important role in the
development of cancer (1). It has
been shown that the inhibition of leptin-induced angiogenesis
results in decreased levels of vascular endothelial growth factor
(VEGF)/VEGFR2, hypoxia inducible factor (HIF) 1α, NF-κB, IL-1 and
Notch and reduced tumor growth in breast cancer (2,3).
Notch, as one of these target genes, affects various cell programs
responsible for proliferation and apoptosis, thereby is essential
for vasculogenesis and angiogenesis in cancer progression (3). It has been shown that leptin is one of
the regulators of Notch expression in breast cancer. In addition,
leptin induces the expression levels of IL-1 and VEGF/VEGFR2, as
well as the expression levels of Notch1-4/Jagged-1/delta-like
canonical Notch ligand (Dll) 4, which are important for the Notch
signaling pathway. All these effects of leptin are related to the
JAK-2/STAT-3 signaling pathway, the NF-κB transcription factor, and
HIF-1α, which is a key regulator of hypoxia response (4).
The interaction of IL-1, as another target gene,
with leptin, has an important role in tumor inflammation,
proliferation, and angiogenesis (5). IL-1 is a potent proinflammatory
cytokine that influences tumor invasion and IL-1R expression levels
are increased in tumor cells (6).
Co-expression of IL-1R Type I, leptin/leptin receptor (ObR) and
VEGF has been demonstrated in breast cancer (2).
Notch, IL-1 and leptin crosstalk outcome (NILCO)
signaling, a novel mechanism that interacts with proinflammatory
and proangiogenic signals, is critical for cell proliferation and
angiogenesis in cancer (4,7). Leptin mediates proangiogenic actions
by upregulating VEGF/VEGFR2 through the NILCO pathway (3,8). The
expression profiles of the mediators of the NILCO pathway have not
been studied yet in detail in human colorectal cancer. The present
study aimed to assess the NILCO pathway and the related gene and
protein expression levels in human colorectal cancer tissues
compared with normal colon tissue. In addition, leptin, IL-6 and
VEGFA plasma concentrations were determined in cancer patients and
in healthy individuals.
Materials and methods
Study subjects
The present study was approved by the Ethics
Committee of Eskisehir Osmangazi University for Clinical Research
(approval no. 80558721/90) and was performed following the ethical
standards of The Helsinki Declaration. Tissue specimens from tumor
and adjacent normal colon tissue (n=40) were collected from the
Department of General Surgery, Hospital of Eskisehir Osmangazi
University, Eskisehir, Turkey. Healthy individuals were 25-50 years
old without any disease. Written consent was obtained for the use
of tissue and blood samples and for the publication of the derived
data in the present study.
Tissue samples
In the present study, tumor and adjacent normal
colon tissues from patients with colorectal cancer were collected
between May 2017 and January 2018, and examined. All specimens were
obtained during routine surgery performed in patients with colon
cancer. Forty consecutive patients, aged 50 years or older, who
underwent colectomy due to colon cancer were enrolled in the study
after obtaining informed consent. Patients who had received
neoadjuvant therapy and patients with diabetes, body mass index
>30 kg/m², coexisting or previous cancer history, and family
history of colorectal cancer were excluded.
Besides the colorectal cancer tissues,
macroscopically normal colonic tissues were obtained from the
distal edge of the resection, which were confirmed as normal by
pathologists at the Hospital of Eskisehir Osmangazi University.
Samples approved by two pathologists were included in the study.
Samples for which pathologists did not agree were excluded.
Pathologists classified samples according to pathological type,
tumor stage and metastasis status. Immediately after excision,
tissue samples were fixed in RNAlater solution (Qiagen GmbH) and
stored at -80˚C.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from tumor and adjacent
normal colon tissues using the GeneJet RNA Purification kit (Thermo
Fisher Scientific, Inc.). The concentration and purity of the RNA
were measured using a NanoDrop 1000 (Thermo Fisher Scientific,
Inc.). The 260/280 values were 2.1, thus RNA purity was termed as
good (9). Isolated RNA samples were
converted to cDNA using the RevertAid First Strand cDNA Synthesis
kit (Thermo Fisher Scientific, Inc.) at 42˚C for 60 min and 70˚C
for 5 min. cDNA samples were stored at -80˚C until further
analysis. The mRNA expression levels of leptin, ObRb, Notch-1,
Notch-4, IL-1α, IL-1β, IL-1R, IL-6, JAK-2, STAT-1, STAT-3, VEGFA,
VEGFR1, VEGFR2, TNF-α, NF-κB, Jagged-1, HIF-1α and TNF receptor 1
(TNFR1) were measured using the SYBR Green qPCR kit (Thermo Fisher
Scientific, Inc.). After 10 min at the 95˚C activation period, the
cycling conditions were 20 sec at 95˚C, 30 sec at 55˚C and 20 sec
at 72˚C, for 45 cycles. The β-actin gene was used as an internal
control. Primer sequences are listed in Table I. Relative fold changes in mRNA
expression were calculated using the formula
2-ΔΔCq (10).
 | Table ISequences of primers used in reverse
transcription-quantitative PCR. |
Table I
Sequences of primers used in reverse
transcription-quantitative PCR.
| Gene | Forward primer
(5'-3') | Reverse primer
(5'-3') |
|---|
| Leptin |
ACAGAAAGTCACCGGTTTGG |
GCTCTTAGAGAAGGCCAGCA |
| ObRb |
AGGACGAAAGCCAGAGACAACC |
GCCTGGGCCTCTATCTCCCA |
| IL-1α |
CTTCTGGGAAACTCACGGCA |
AGCACACCCAGTAGTCTTGC |
| IL-1β |
CCTGAGCTCGCCAGTGAAAT |
GTCGGAGATTCGTAGCTGGA |
| IL-1R |
TGTGGTCCCTGTGTAAAGTCC |
TGCCTGAGGTCTTGGAAAAAC |
| IL-6 |
CTTCTCCACAAACATGTAACAAGAG |
TTTCACCAGGCAAGTCTCCTC |
| STAT-1 |
TGCTGAGGTTTAGCTGTCAGT |
AAGTCTCTTGGCTAGTGCAG |
| HIF-1a |
ACTTGGCAACCTTGGATTGGA |
GTGCTGAGTAACCACCACTTA |
| STAT-3 |
GGCATTCGGGAAGTATTGTCG |
GGTAGGCGCCTCAGTCGTATC |
| β-actin |
GGACTTCGAGCAAGAGATGG |
AGCACTGTGTTGGCGTACAG |
| TNF-α |
CACAGTGAAGTGCTGGCAAC |
GATCAAAGCTGTAGGCCCCA |
| TNFR1 |
ACCAAGTGCCACAAAGGAAC |
CTGCAATTGAAGCACTGGAA |
| NF-κB |
AGGACGTGGAGTCAGGCTAT |
TCTTGAGAAGGCTCAGCAGC |
| VEGFA |
TGTCTAATGCCCTGGAGCCT |
TTAACTCAAGCTGCCTCGCC |
| Notch-1 |
TCCTAGTTTGGGAGGAGCAGA |
CACTGGCATGACACACAACAG |
| Notch-4 |
GGATCCCCCAAAATGAAGGG |
TCTGCTCTGGTGGGCATACA |
| Jagged-1 |
GCACGCGTCATTGTGTTACC |
GCGCAGCCTTTTATTCCCTT |
| JAK-2 |
TGAGTTCGAAGCTAGCAGGG |
AAGCCCGTCACAGTTGTCTC |
| VEGFR1 |
GCTGTTTTCTCTCGGATCTCCA |
TCCGAGCCTGAAAGTTAGCA |
| VEGFR2 |
CGGTCAACAAAGTCGGGAGA |
CAGTGCACCACAAAGACACG |
Determination of leptin, VEGF, IL-1
and IL-6 plasma concentrations by ELISA
Plasma samples from healthy individuals and patients
with colorectal cancer were stored at -20˚C until further analysis.
Plasma leptin, VEGF, IL-1 and IL-6 concentrations were determined
using commercially available human-specific ELISA kits (VEGF, cat.
no. BMS277/2; Invitrogen; Thermo Fisher Scientific, Inc.; Leptin,
IL-1 and IL-6, cat. nos. KAP2281, KAP1211 and KAP1261,
respectively; DIAsource ImmunoAssays SA), as recommended by the
manufacturers. Optical density measurements were obtained using a
plate reader system (Awareness Technology, Inc.).
Protein isolation and western
blotting
Total protein was extracted from ~100 mg of tissue
using M-PER Mammalian Protein Extraction Reagent, according to the
manufacturer's instructions (Thermo Fisher Scientific, Inc.). The
protein concentration was determined using a BCA Protein Assay kit
(Thermo Fisher Scientific, Inc.). A total of 50 µg per sample was
separated by electrophoresis using Bolt™ 4-12% Bis-Tris Plus gels
(Thermo Fisher Scientific, Inc.). The IBlot 2 Dry Blotting system
(Thermo Fisher Scientific, Inc.) was used for the transfer to
nitrocellulose membrane, according to the manufacturer's
instructions. The blocking solution was prepared from the iBind™
Flex Solution kit (Thermo Fisher Scientific, Inc.) and the membrane
was incubated in the blocking solution for 5 min. After blocking,
membranes were incubated with primary and secondary antibodies for
2 h on the iBind™ Flex Western device for 2.5 h, according to the
manufacturer's instructions (Thermo Fisher Scientific, Inc.). The
β-actin antibody (1:1,000; cat. no. ab8227) was purchased from
Abcam. Primary antibodies targeting Jagged-1 (1:500; cat. no.
AP0531), STAT-1 (1:500; cat. no. A12075), STAT-3 (1:500; cat. no.
A1192), JAK-2 (1:500; cat. no. A11497), HIF-1α (1:500; cat. no.
A11945), phosphorylated (p) STAT-1 (1:500; cat. no. AP0135),
pSTAT-3 (1:500; cat. no. AP0070), pJAK-2 (1:500; cat. no. AP0531),
IκB (a1:500; cat. no. A16929) and pIκB (1:500; cat. no. AP0614)
were purchased from ABclonal Biotech Co., Ltd.. Anti-rabbit
HRP-conjugated secondary antibodies used were purchased from Abcam
(1:2,000; cat. no. ab205718). Immunodetection was performed using
chemiluminescence (West-Dura; Thermo Fisher Scientific, Inc.). The
signal intensity was visualized on a ChemiDoc-It Imaging System
(Bio-Rad Laboratories, Inc.), quantified by densitometry using
ImageJ 1.49v (National Institutes of Health) and the means and SDs
were calculated for each tissue type.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism version 6 (GraphPad Software, Inc.). To determine
the tumor characteristics, one-sample χ2 tests were
used. Descriptive statistics are reported as n (sample size), mean
and SD for continuous variables, and as n (sample size), median and
25 and 75th percentiles for categorical variables. Continuous
normally distributed measurements were compared across two groups
by Student's t-test. Continuous variables that did not show normal
distribution were compared by the Mann-Whitney U-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Patient characteristics
Patient demographic data and tumor characteristics
are listed in Table II.
 | Table IIPatient demographic data and tumor
characteristics. |
Table II
Patient demographic data and tumor
characteristics.
| Clinicopathological
feature | Number of
patients |
|---|
| Sex | |
|
Female | 22 |
|
Male | 18 |
| Average weight
(kg) | 73 |
| Average height
(cm) | 165 |
| Tumor
localization | |
|
Right
colon | 5 |
|
Left
colon | 6 |
|
Sigmoid
colon | 13 |
|
Cecum | 5 |
|
Rectum | 11 |
| Pathologic
type | |
|
Tubular
adenocarcinoma | 35 |
|
Mucinous
carcinoma | 5 |
| TNM
classification | |
|
T2 | 2 |
|
T3 | 33 |
|
T4 | 5 |
| Lymph node
metastasis | |
|
N1 | 7 |
|
N2 | 7 |
| Distant
metastasis | |
|
M0 | 35 |
|
M1 | 5 |
Tumor staging findings
Tissues from patients with T2, T3 and T4 stage
cancer were included in the present study. Individuals with T1
stage cancer are not taken into operation. One-sample χ2
tests were used to determine the difference between T2, T3, and T4
distributions of tumor stages in patients. In this analysis, the
expected frequency for each stage would be 40/3, therefore 13.33
samples for each stage. The P-value obtained as a result of the
chi-square test analysis was P<0.001. This finding indicated
that the distribution of tumor stages in patients was not equal and
that the patients were mostly at the T3 stage (Fig. S1A).
Tumor pathological findings
To detect the difference in the distribution between
the tubular adenocarcinoma and mucinous carcinoma type in the
patients, a one-sample Chi-square test was applied. In this
analysis, the expected frequency for each type would be 40/2,
therefore 20 samples per cancer type. The P-value obtained as a
result of the analysis was P<0.001. According to this result,
the distribution of pathological types was not equal and most
patients were presented with tubular adenocarcinoma (Fig. S1B).
Tumor localization findings
To determine the difference in the distribution
between the right colon, left colon, sigmoid colon, cecum and
rectum tumor localization in the patients, a one-sample chi-square
test was used. In this analysis, the expected frequency for each
site could be assumed to be equal. The P-value obtained as a result
of the analysis was P=0.199. According to this result, the tumor
localization distribution in patients was found to be equal
(Fig. S1C).
Gene expression levels
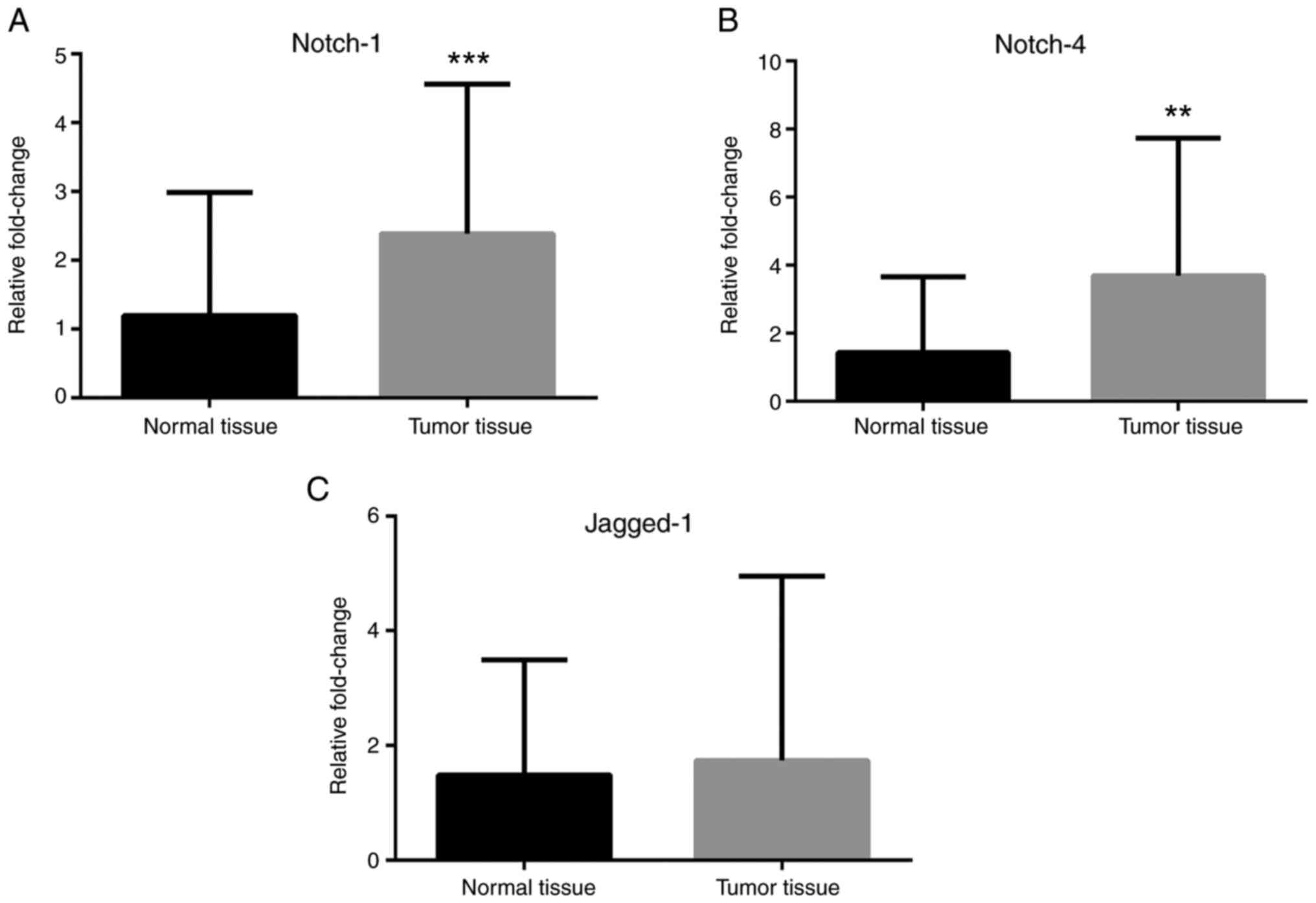
RT-qPCR analyses revealed that Notch-1 and Notch-4
mRNA expression levels were greater in cancer tissues compared with
normal tissues (Fig. 1A and
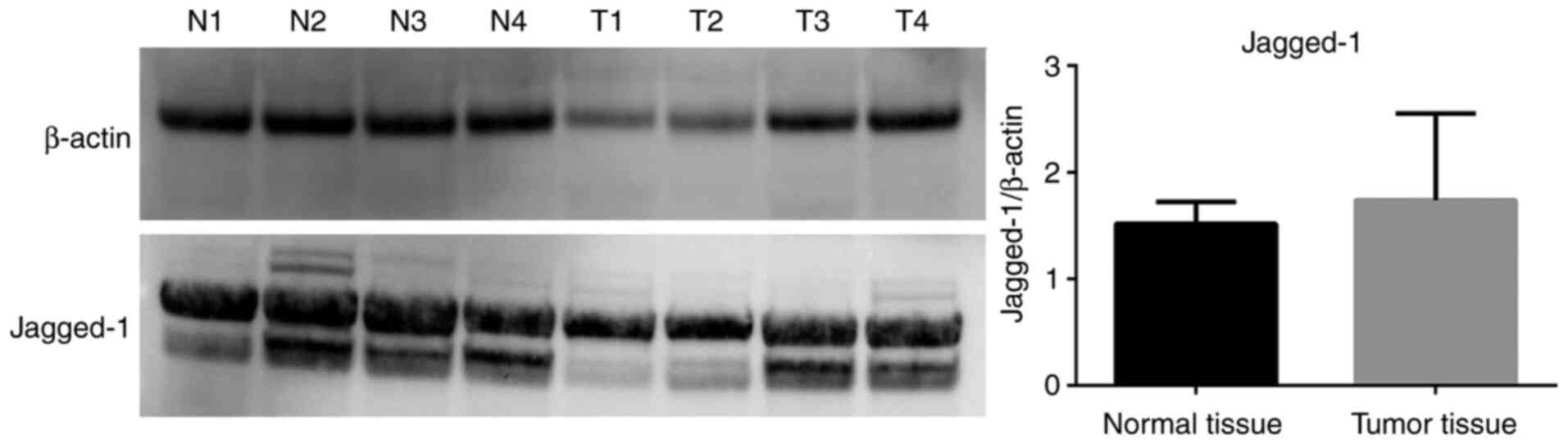
B), while no difference in Jagged-1
mRNA levels was observed (Fig. 1C).
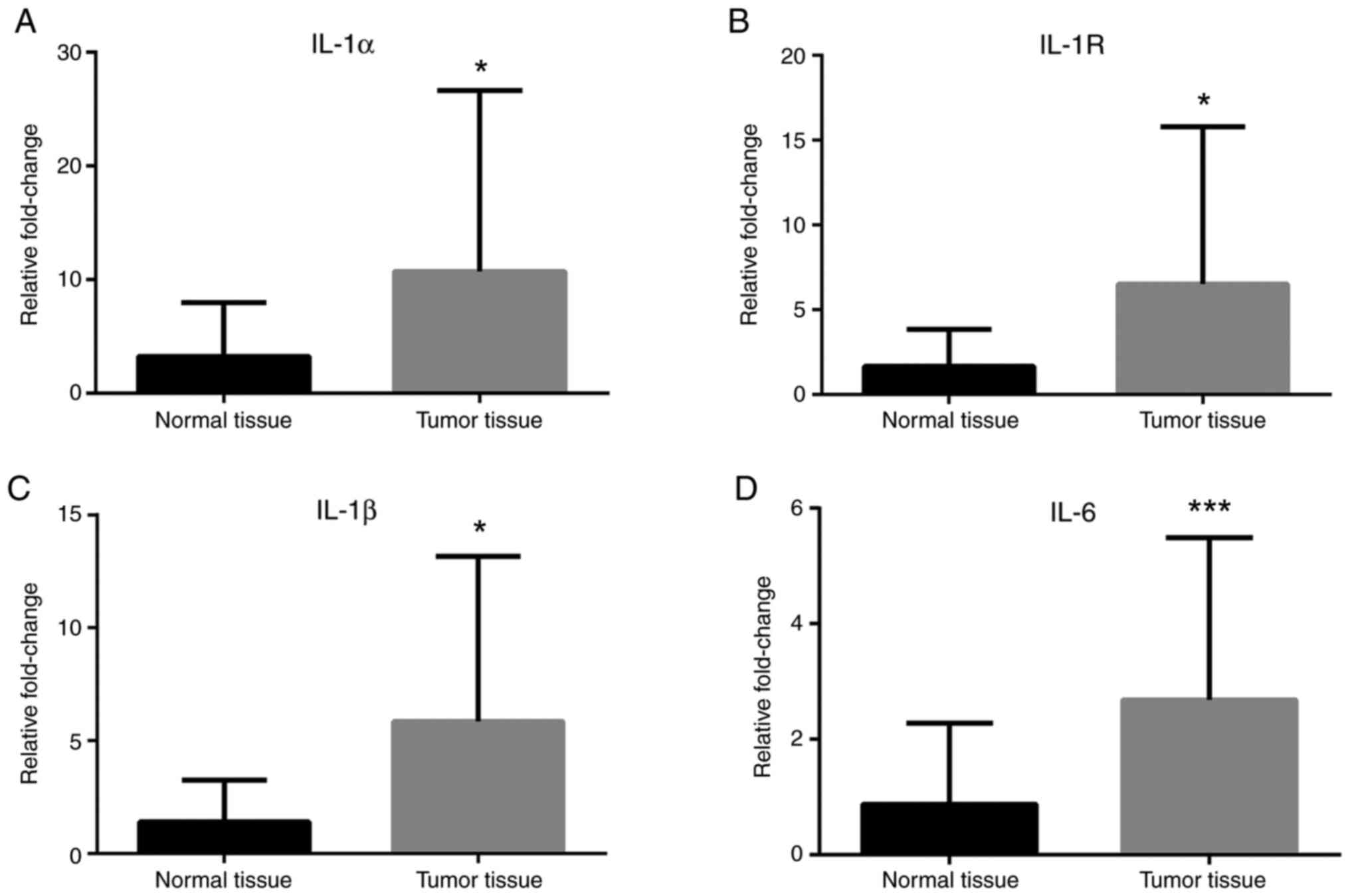
IL-1α, IL-1R, IL-1β and IL-6 mRNA expression levels were greater in
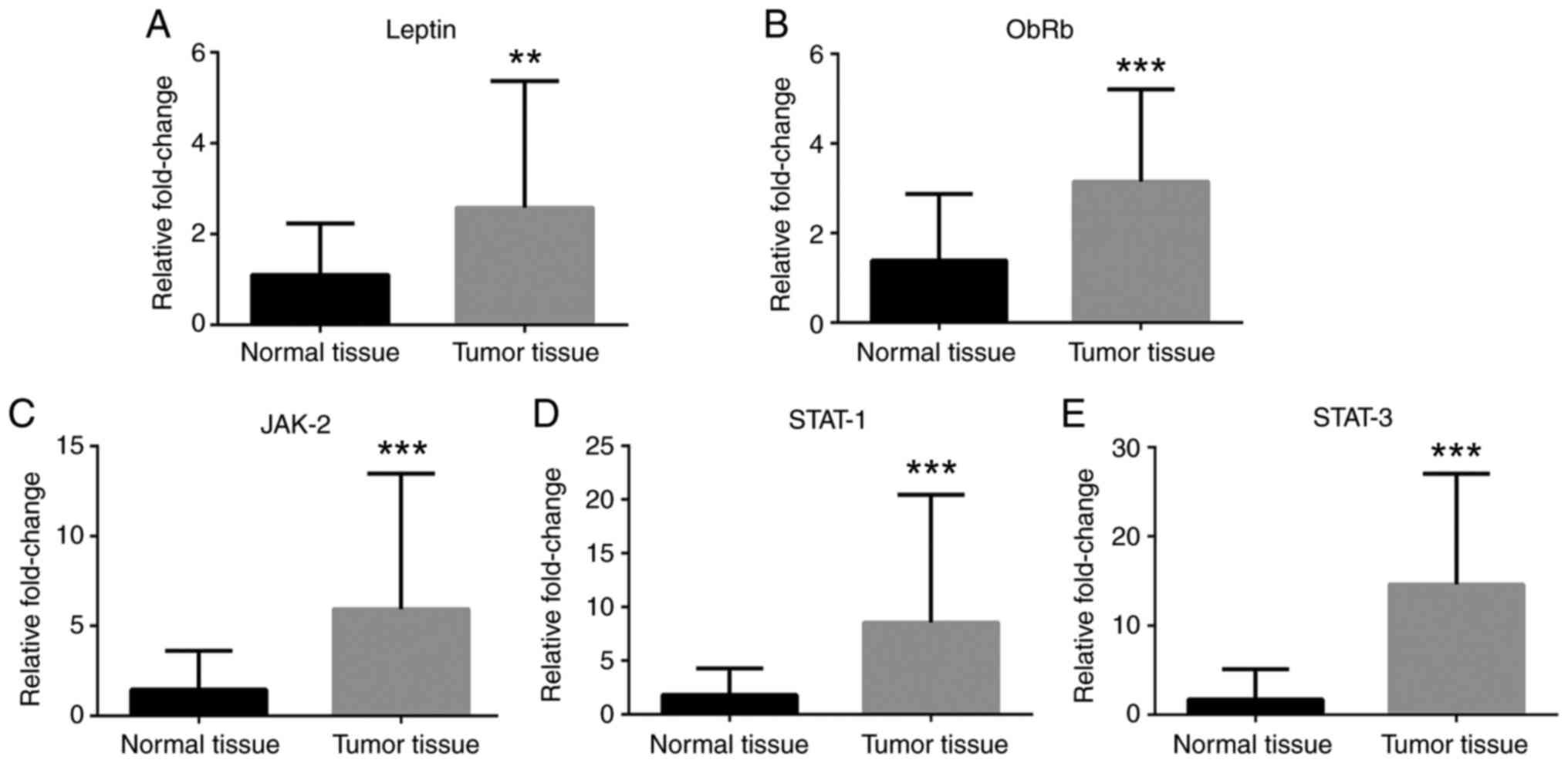
cancer tissues compared with normal tissues (Fig. 2). Leptin, ObRb, JAK-2, STAT-1 and
STAT-3 mRNA expression levels were greater in cancer tissues
compared with normal tissues (Fig.
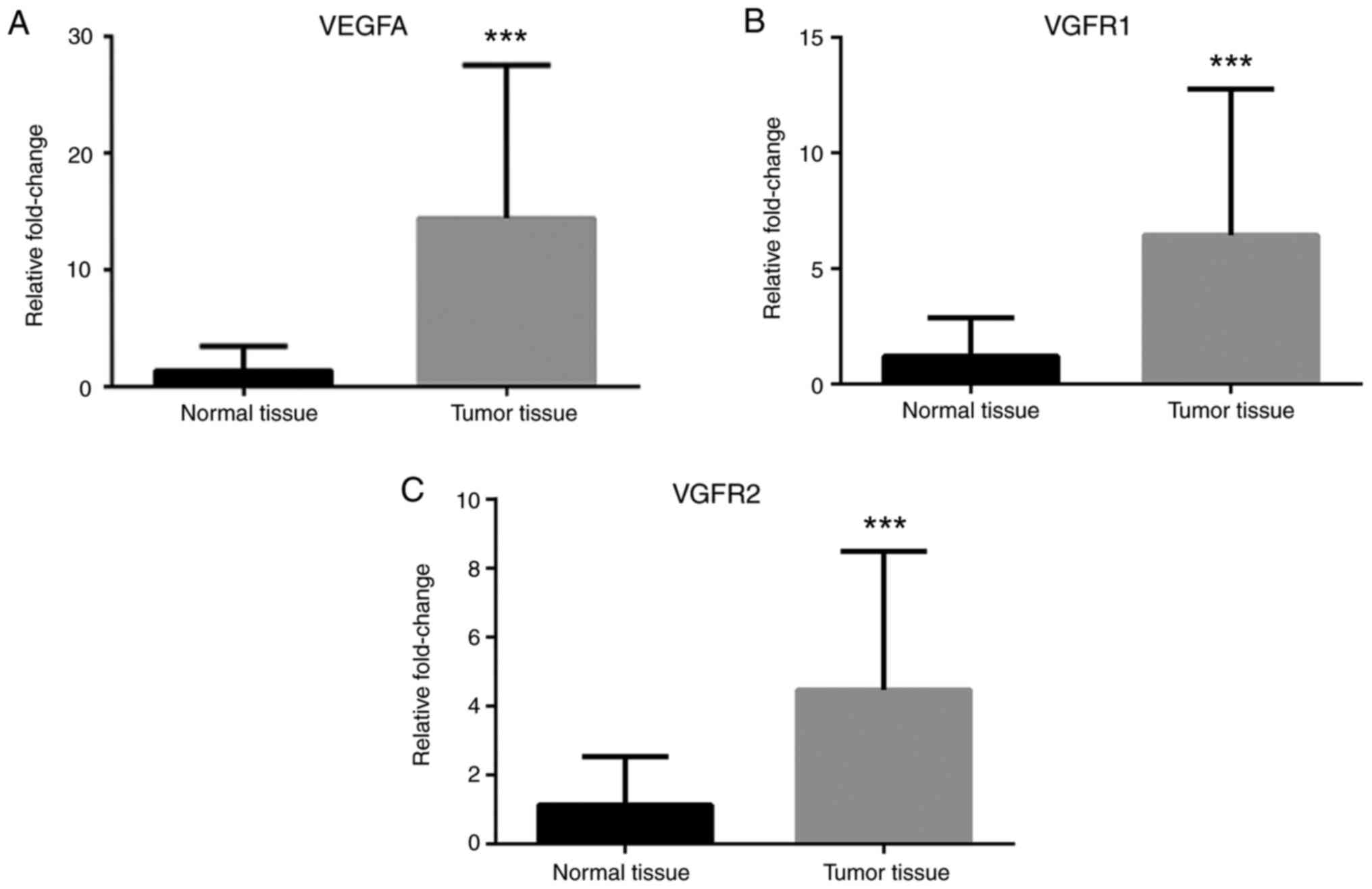
3). Additionally, VEGFA, VEGFR1 and VEGFR2 mRNA expression
levels were greater in cancer tissues compared with normal tissues
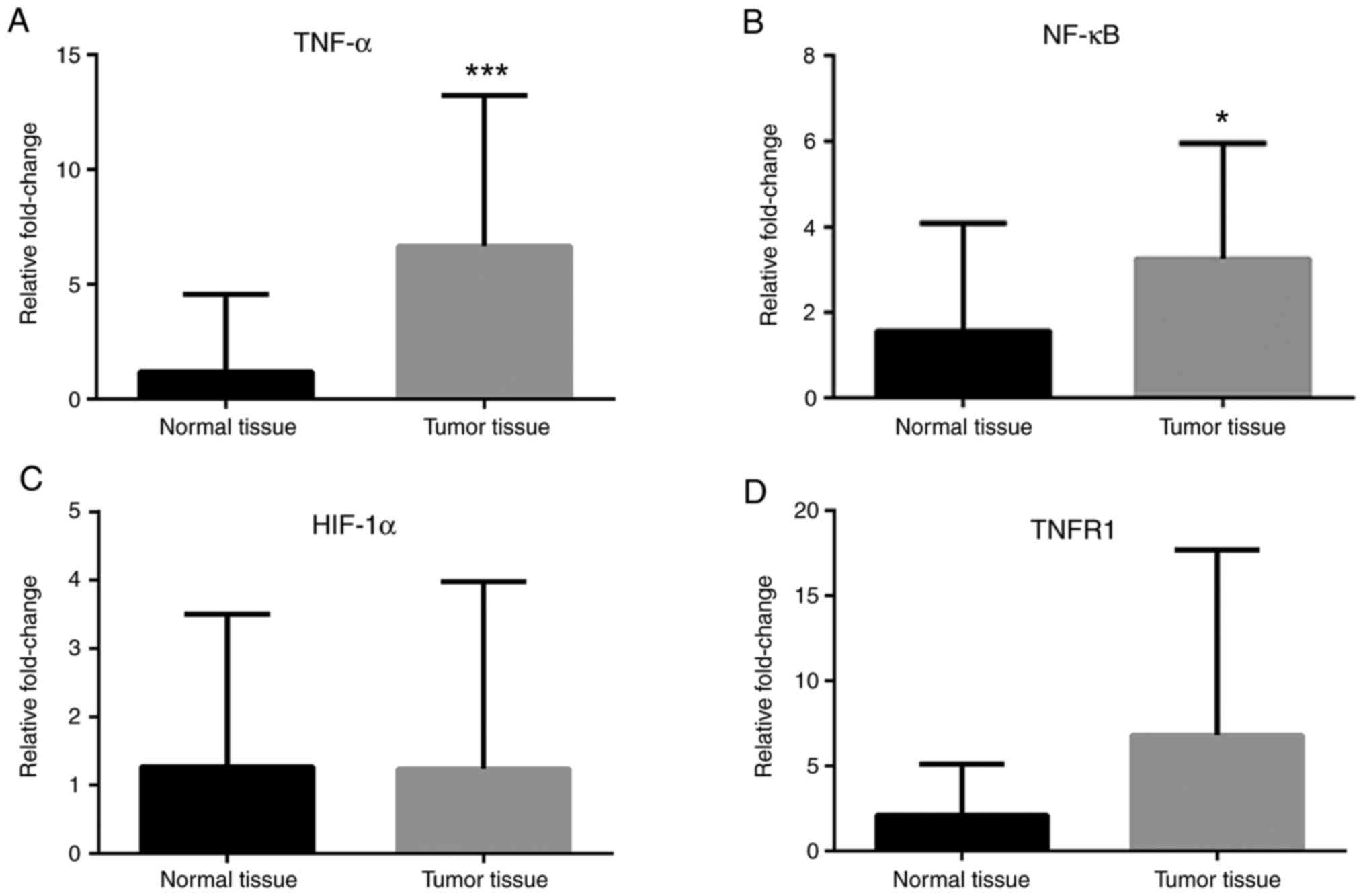
(Fig. 4). Finally, TNF-α and NF-κB
mRNA expression levels were greater in cancer tissues compared with
normal tissues (Fig. 5A and
B), while HIF-1α and TNFR1 mRNA
expression levels were unchanged (Fig.
5C and D).
Protein expression levels
Western blot analyses revealed that Jagged-1 protein
expression levels were not significantly different in tumor tissues
compared with normal tissues (Fig.
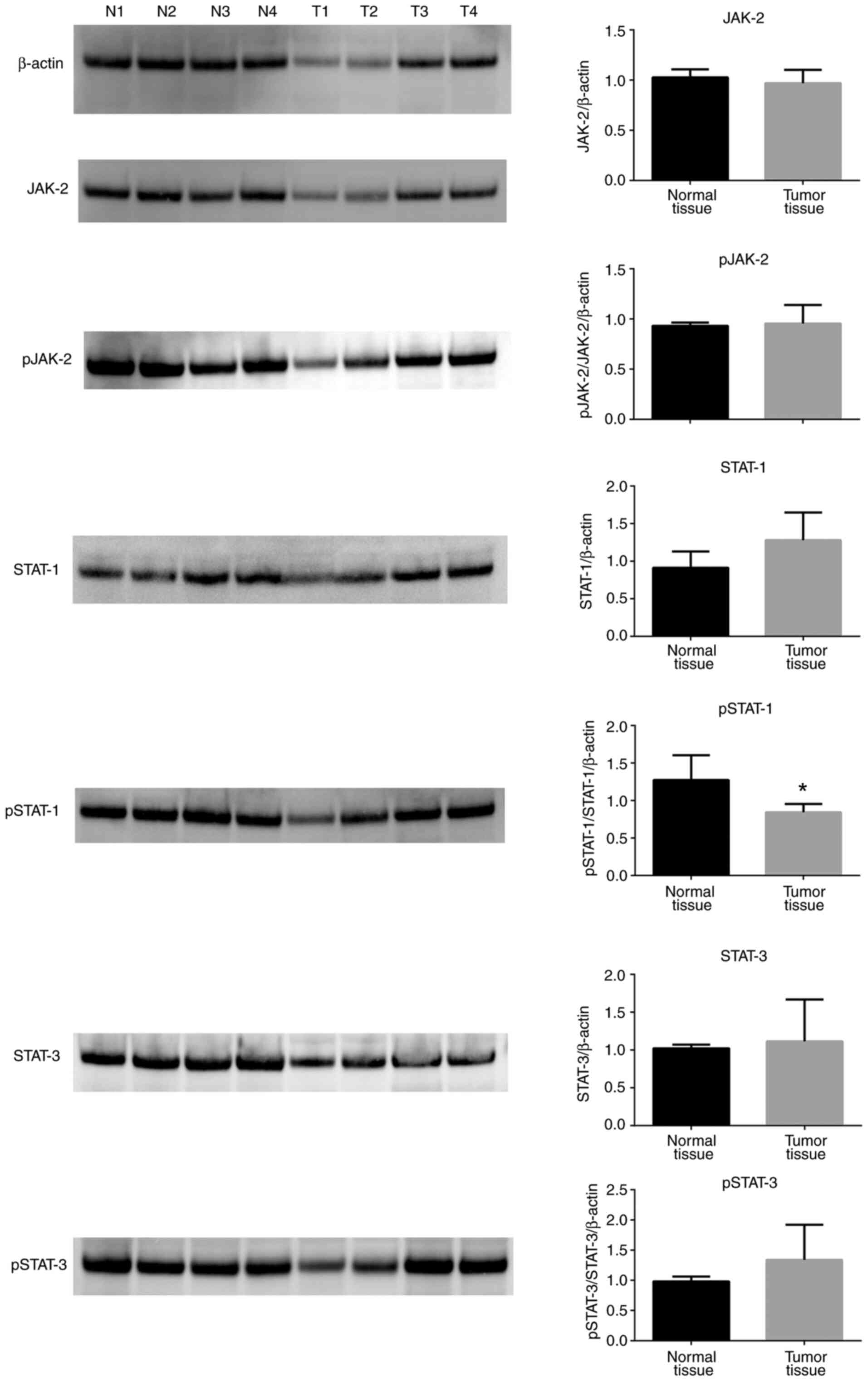
6). JAK-2, pJAK-2, STAT-1, STAT-3 and pSTAT-3 protein
expression levels were unchanged between tumor tissues and normal
tissues (Fig. 7), and only pSTAT-1
protein levels were significantly decreased in tumor tissues
compared with normal controls (Fig.
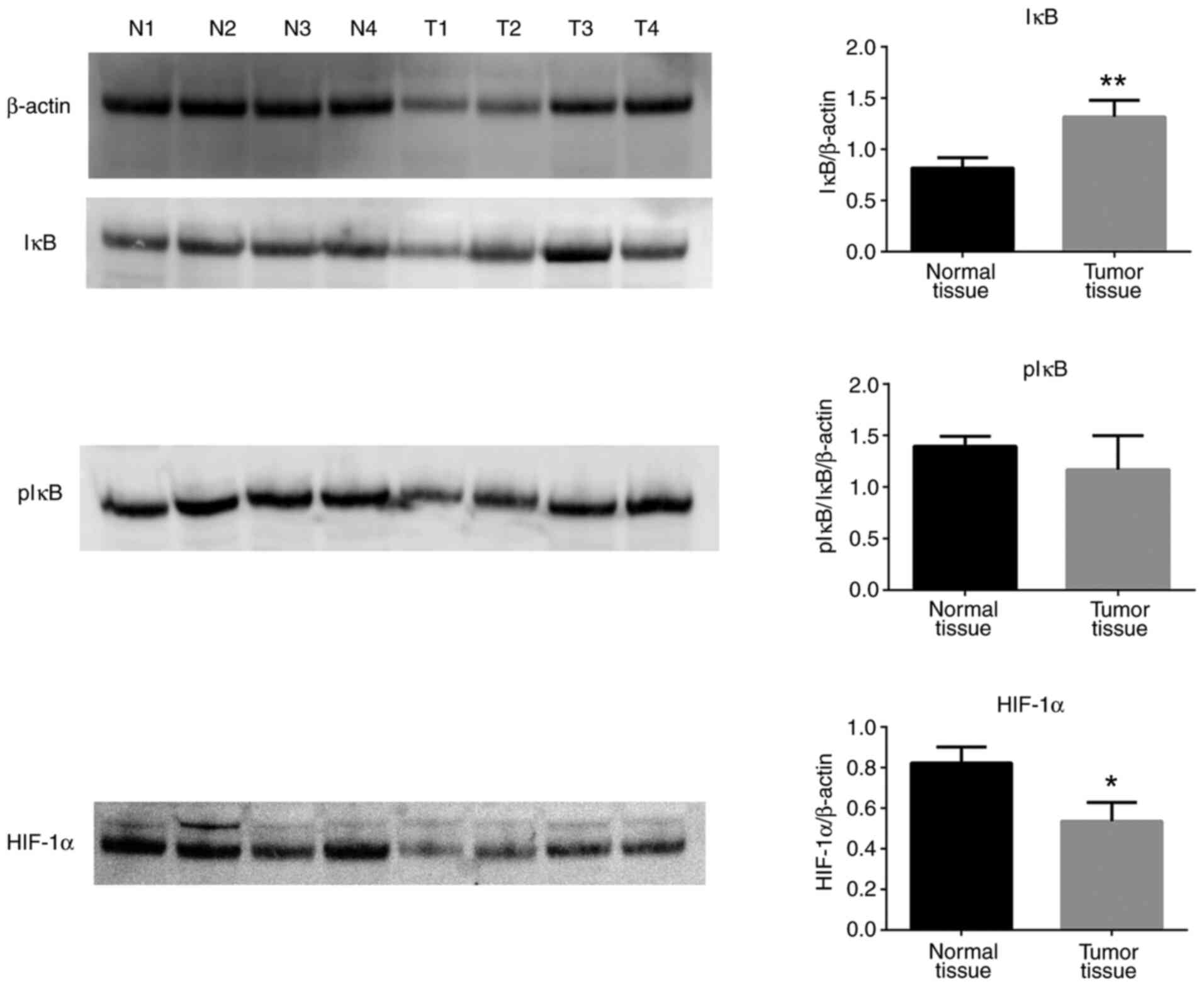
7). Of note, total IκB protein expression levels were increased
in tumor tissues compared with normal tissues, while pIκB protein
levels were unchanged (Fig. 8).
Finally, HIF-1α protein expression levels were significantly
decreased in tumor tissues compared with normal controls (Fig. 8).
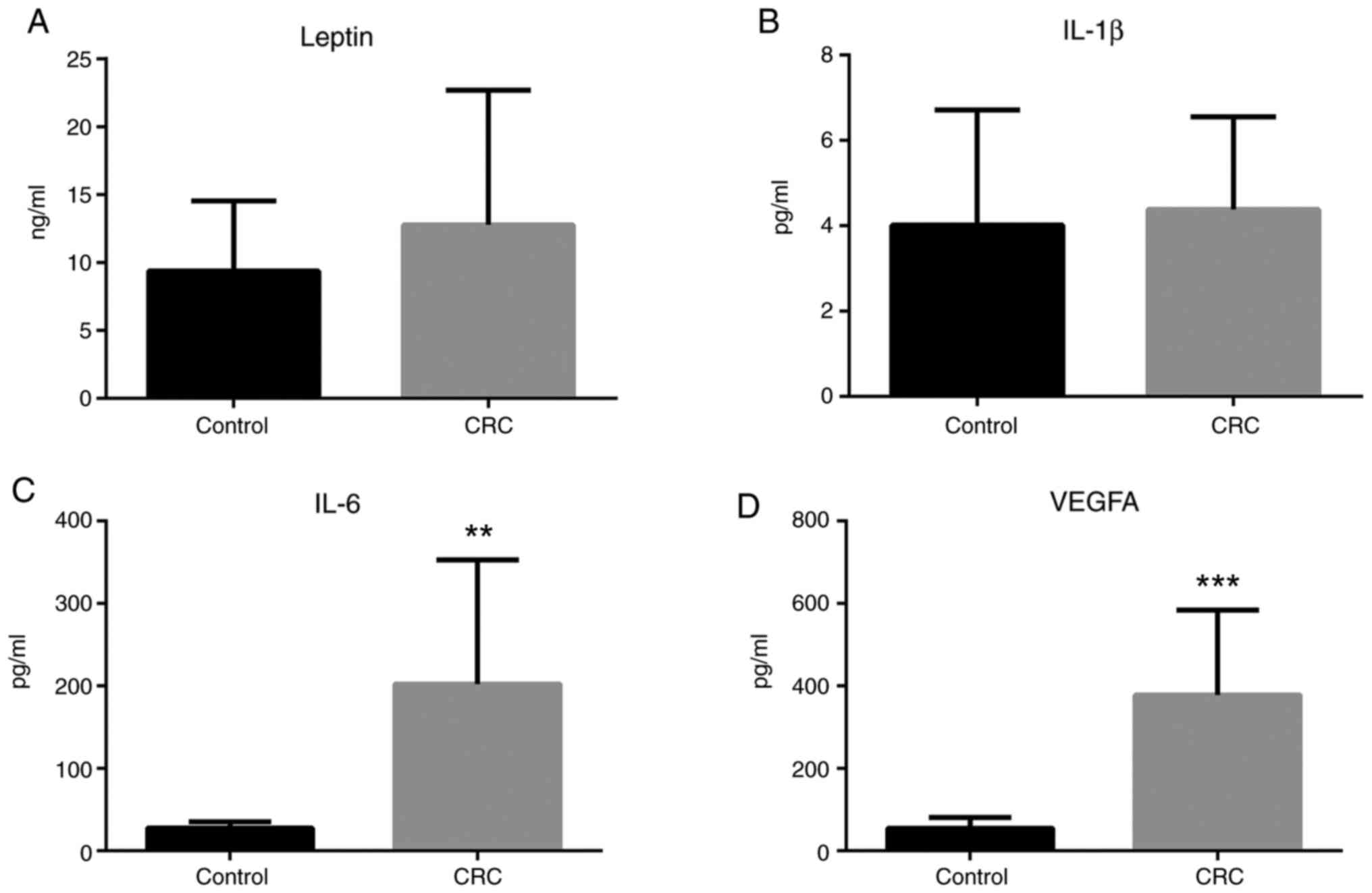
Leptin, IL-1β, IL-6, and VEGFA plasma
concentrations
ELISA analyses revealed that leptin plasma
concentration was somewhat increased in patients with colorectal
cancer, albeit the result was not statistically significant
(Fig. 9A). IL-1β plasma
concentration was unchanged in patients with colorectal cancer
compared to healthy individuals (Fig.
9B). Notably, IL-6 and VEGFA plasma concentrations were
significantly increased in patients with colorectal cancer compared
to healthy individuals (Fig. 9C and
D).
Discussion
Analysis of the patient cohort used in the present
study revealed that the histologic types and tumor grades of the
adenocarcinomas were not evenly distributed. The patients had
mostly T3 tubular adenocarcinomas, thus the present results were
predominantly attributable to this particular group of patients. It
has been shown that leptin levels are increased in multiple types
of cancer, including colorectal carcinomas (11-13).
Effects of leptin are mediated by multiple signaling pathways,
including the JAK-2/STAT-3 pathway. STAT-3 contributes to
oncogenesis by stimulating cell cycling and inhibiting apoptosis.
Endo et al (14) suggested
that leptin may promote the progression of colorectal cancer
through the ObRb/STAT-3 pathway. This previous report found a
marked increase of ObR expression levels in colon tumors compared
with normal epithelium (14). In
the present study, leptin, ObR, JAK-2, STAT-1, and STAT-3 mRNA
expression levels in colorectal tumor tissues were greater compared
with normal colon tissues. Of note, the leptin protein levels in
the blood samples of the patients with colorectal cancer were not
changed. The correlations between mRNA and protein concentrations
are typically poor. Furthermore, it is unclear to what extent the
relative concentration differences on the mRNA level are
transferred to the protein level (15).
The present results, which are mainly consistent
with previously published literature, demonstrated that adipokines
are closely related to tumorigenesis. Additionally, leptin and
IL-1-induced signals, which collaborate in many pathologic states,
have important roles in inflammation, proliferation, and
angiogenesis (7).
The IL-1 family of cytokines have proinflammatory
and angiogenic properties. The expression of IL-1 is associated
with an aggressive phenotype of breast cancer, while in
vitro studies have revealed that IL-1 may influence progression
and metastasis of lung cancer, colorectal cancer, and malignant
melanoma (5). The current study has
shown that the mRNA expression levels of IL-1α and IL-1β were
increased in the cancer tissues compared to the normal tissues. In
cancer patients, IL-6 plasma levels were significantly elevated
compared with the control group.
IL-1 is a potent leptin inducer and IL-1 receptor
type 1 (IL-1R TypeI) partially mediates the effects of leptin
(16). Immunohistochemical analyses
have shown that IL-1R TypeI, leptin/ObR, and VEGF are coexpressed
in cancer cells (2), which
indicates that the interaction of leptin and IL-1 may have an
important role in angiogenesis and tumor progression. Consistent
with these findings from previous studies, the current study has
demonstrated that IL-1R TypeI mRNA expression levels were greater
in the cancer tissues compared with the normal tissues.
The Notch signaling pathway is crucial in colorectal
cancer progression, as well as in self-renewal and homeostasis of
the normal intestinal epithelium (17). Notch signaling regulates the balance
between cell proliferation, differentiation, and apoptosis
(18,19). This pathway has a key role in tumor
angiogenesis and is associated with worse prognosis and survival in
cancer patients. Ligands and receptors of Notch are membrane-bound
proteins (3). The expression of
both the Notch-1 and Notch-2 receptors and of the Jagged-1 ligands
have been demonstrated to be increased in colorectal cancer cell
lines (20). In the present study,
Notch-1 and Notch-4 mRNA expression levels in the cancer tissues
were increased compared with the normal tissues, but the expression
levels of Jagged-1 were unchanged.
Previous studies have reported that leptin is a
regulator of Notch expression in breast cancer. In addition, leptin
induces the expression of IL-1 and of VEGF/VEGFR2, as well as the
expression of Notch1-4/Jagged-1/Dll-4(4). This information indicates that NILCO
may serve an essential role in the progression of cancer. NILCO
emerges as a mechanism that induces cell proliferation and
activates proangiogenic and proinflammatory pathways, thus serving
a crucial role in the progression of different types of cancer
(4,7).
Leptin mediates proangiogenic actions either by
transactivating VEGFR2 independent of VEGF or by upregulating
VEGF/VEGFR2 through the NILCO pathway (3). In the present study, the results
demonstrated that the mRNA expression levels of VEGFA, VEGFR1, and
VEGFR2 and the blood VEGFA protein levels were increased, which
suggests that leptin in cancer tissues may have proangiogenic
properties.
The increase in the levels of angiogenic factors,
such as TNF-α, VEGF, IL-1, IL-6 and leptin, indicated an increase
in the extent of angiogenesis. In the present study, the increase
in VEGF, IL-1, IL-6, leptin and ObRb mRNA expression levels is
consistent with the previous literature and confirms that these
genes contribute to the progression of cancer (3).
It has been found that ObRb is expressed in human
lung cancer A549 and H157 cells and leptin release increases the
production of immuno-inflammatory cytokines, such as IL-6, VEGF and
prostaglandin (21). Angiogenesis
is an extremely important process contributing to the progression
of cancer. The growth factor VEGF released from endothelial cells
has an important role in angiogenesis. Since VEGF plays a role in
tumor angiogenesis as well as proliferation, invasion and
metastasis of tumor cells, it is one of the main targets in cancer
treatment (3). According to these
two studies, the relationship between leptin, IL-6 and VEGF plays
an important role in cancer development and prognosis. This
information is in line with the results of the present study, where
leptin, IL-6 and VEGF levels were significantly increased in cancer
tissues. In addition, hypoxia induces angiogenesis, through the
transcriptional activity of HIF-1α. It has been reported that
hypoxia-induced HIF-1α is associated with an increase in the levels
of leptin and VEGF expression levels in breast cancer. (3). By contrast, there was no significant
change in the mRNA expression levels of HIF-1α in the present
study.
TNF-α and IL-6 synergistically promote colorectal
cancer cell growth and cytokine production (22). Migration, invasion, and infiltration
of colorectal cancer cells are potentiated by TNF-α (23). Popivanova et al (24) have shown that blocking TNF-α reduces
colorectal carcinogenesis. In the present study, the results
demonstrated that TNF-α mRNA expression levels were increased in
tumor tissues, while TNFR1 mRNA expression levels were unchanged.
TNF-α-induced NF-κB regulates the transcription of genes associated
with cell growth and proliferation. In resting cells, NF-κB is
inactive and sequestered in the cytoplasm by inhibitory proteins,
such as the inhibitor of κB (IκB) (25). NF-κB is activated through either the
classical or alternative pathways. The classical pathway is
activated by inflammatory cytokines, IL-1, and TNFα. IκB allows
NFkB nuclear translocation and activates the transcription of the
target genes in the nucleus, which leads to development of
inflammation and tumor progression by increasing cell proliferation
and migration (26). NF-κB
expression in colorectal cancer is associated with proliferation
and angiogenesis and shortened survival (27,28).
In the present study, the mRNA expression levels of NF-κB and the
protein levels of IκB were increased in tumor tissues, whereas
there was no significant change in the protein levels of pIκB.
These findings indicated that the levels of NF-κB in the tumor
tissues were increased but activation and translocation of NF-κB to
the nucleus were inconclusive.
The significance of the present findings is
highlighted by the fact that the present study was conducted in
human colorectal cancer tissues, and that the results were
compatible with the findings of previous studies. The limitations
of the present study were that the patients were not classified
according to their metastasis status and that there was no detailed
immunohistochemical analysis for the NILCO interaction. Further
studies will be required in the future with patients classified
according to their metastasis status and with detailed functional
and protein interaction experimental analyses.
In conclusion, the present study demonstrated that
increased levels of leptin, inflammatory cytokines, and the
interaction between these molecules may have an important role in
colorectal cancer progression. The current findings give valuable
information about the intracellular mechanisms related to NILCO,
which are essential for tumor biology. Future studies on the
inhibition of NILCO may yield novel treatment options for
colorectal cancer.
Supplementary Material
(A) Tumor staging distribution.
One-sample chi-square test was used to determine the difference
between T2, T3, and T4 distributions of tumor stages in patients.
In this analysis, the expected frequency for each grade would be
40/3, therefore 13.33 samples in each stage. The P-value obtained
as a result of the analysis was P<0.001. (B) Tumor pathological
type distribution. To detect the difference between the
distribution of tubular adenocarcinoma and mucinous carcinoma in
the patients, a one-sample chi-square test was applied. In this
analysis, the expected frequency for each type would be 40/2,
therefore 20 samples per cancer type. The P-value obtained as a
result of the analysis was P<0.001. (C) Tumor localization
distribution. To determine the difference between the right colon,
left colon, sigmoid colon, cecum, and rectum tumor localization
distributions in the patients, a one-sample chi-square test was
applied. The P-value obtained as a result of the analysis was
P=0.199. Therefore, the tumor localization appeared to be random in
the patients included in the present study.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by The Scientific and
Technological Research Council of Turkey (TUBITAK; grant no.
116S628).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
NE, RO and MO confirm the authenticity of all the
raw data. NE performed the analysis and publication of the results,
specifically, RT-qPCR, western blotting experiments and ELISA
experiments. RO performed the analysis and publication of the
results, specifically, RT-qPCR, western blotting and ELISA
experiments. MO performed the analysis and publication of the
results, specifically, RT-qPCR, western blotting and ELISA
experiments. SE performed the diagnosis of CRC, detection of the
sample groups and isolation of the CRC tissue. FY performed the
diagnosis of CRC, detection of the sample groups and isolation of
the CRC tissue. EI performed the diagnosis of CRC, detection of the
sample groups and isolation of the CRC tissue. EvC performed
pathological evaluations and diagnoses. FC performed pathological
evaluations and diagnoses. ErC performed the analysis and
publication of the results. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Eskisehir Osmangazi University for Clinical Research
(approval no. 80558721/90) and was performed following the ethical
standards of The Helsinki Declaration. Tissue specimens from tumor
and adjacent normal colon tissues were collected from the
Department of General Surgery, Hospital of Eskisehir Osmangazi
University, Eskisehir, Turkey. Written consent was obtained from
all the subjects participating in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
van Kruijsdijk RC, van der Wall E and
Visseren FL: Obesity and cancer: The role of dysfunctional adipose
tissue. Cancer Epidemiol Biomarkers Prev. 18:2569–2578.
2009.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhou W, Guo S and Gonzalez-Perez RR:
Leptin pro-angiogenic signature in breast cancer is linked to IL-1
signalling. Br J Cancer. 104:128–137. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Gonzalez-Perez RR, Lanier V and Newman G:
Leptin's pro-angiogenic signature in breast cancer. Cancers
(Basel). 5:1140–1162. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Guo S and Gonzalez-Perez RR: Notch, IL-1
and leptin crosstalk outcome (NILCO) is critical for leptin-induced
proliferation, migration and VEGF/VEGFR-2 expression in breast
cancer. PLoS One. 6(e21467)2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang B, Wood IS and Trayhurn P: Hypoxia
induces leptin gene expression and secretion in human
preadipocytes: Differential effects of hypoxia on adipokine
expression by preadipocytes. J Endocrinol. 198:127–134.
2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Elaraj DM, Weinreich DM, Varghese S,
Puhlmann M, Hewitt SM, Carroll NM, Feldman ED, Turner EM and
Alexander HR: The role of interleukin 1 in growth and metastasis of
human cancer xenografts. Clin Cancer Res. 12:1088–1096.
2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lipsey CC, Harbuzariu A, Daley-Brown D and
Gonzalez-Perez RR: Oncogenic role of leptin and Notch interleukin-1
leptin crosstalk outcome in cancer. World J Methodol. 6:43–55.
2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Daley-Brown D, Harbuzariu A, Kurian AA,
Oprea-Ilies G and Gonzalez-Perez RR: Leptin-induced Notch and IL-1
signaling crosstalk in endometrial adenocarcinoma is associated
with invasiveness and chemoresistance. World J Clin Oncol.
10:222–233. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lee S, Trivedi U, Johnson C, Farquharson C
and Bergkvist GT: Optimised isolation method for RNA extraction
suitable for RNA sequencing from feline teeth collected in a
clinical setting and at post mortem. Vet Res Commun. 43:17–27.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rao X, Huang X, Zhou Z and Lin X: An
improvement of the 2ˆ(-delta delta CT) method for quantitative
real-time polymerase chain reaction data analysis. Biostat
Bioinforma Biomath. 3:71–85. 2013.PubMed/NCBI
|
|
11
|
Cust AE, Stocks T, Lukanova A, Lundin E,
Hallmans G, Kaaks R, Jonsson H and Stattin P: The influence of
overweight and insulin resistance on breast cancer risk and tumour
stage at diagnosis: A prospective study. Breast Cancer Res Treat.
113:567–576. 2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wei EK, Giovannucci E, Fuchs CS, Willett
WC and Mantzoros CS: Low plasma adiponectin levels and risk of
colorectal cancer in men: A prospective study. J Natl Cancer Inst.
97:1688–1694. 2005.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Erkasap N, Ozkurt M, Erkasap S, Yasar F,
Uzuner K, Ihtiyar E, Uslu S, Kara M and Bolluk O: Leptin receptor
(Ob-R) mRNA expression and serum leptin concentration in patients
with colorectal and metastatic colorectal cancer. Braz J Med Biol
Res. 46:306–310. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Endo H, Hosono K, Uchiyama T, Sakai E,
Sugiyama M, Takahashi H, Nakajima N, Wada K, Takeda K, Nakagama H
and Nakajima A: Leptin acts as a growth factor for colorectal
tumours at stages subsequent to tumour initiation in murine colon
carcinogenesis. Gut. 60:1363–1371. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wegler C, Ölander M, Wiśniewski JR,
Lundquist P, Zettl K, Åsberg A, Hjelmesæth J, Andersson TB and
Artursson P: Global variability analysis of mRNA and protein
concentrations across and within human tissues. NAR Genom
Bioinform. 2(lqz010)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rene Gonzalez R, Watters A, Xu Y, Singh
UP, Mann DR, Rueda BR and Penichet ML: Leptin-signaling inhibition
results in efficient anti-tumor activity in estrogen receptor
positive or negative breast cancer. Breast Cancer Res.
11(R36)2009.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Noah TK and Shroyer NF: Notch in the
intestine: Regulation of homeostasis and pathogenesis. Annu Rev
Physiol. 75:263–288. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Baron M: An overview of the Notch
signalling pathway. Semin Cell Dev Biol. 14:113–119.
2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Guilmeau S, Flandez M, Mariadason JM and
Augenlicht LH: Heterogeneity of Jagged1 expression in human and
mouse intestinal tumors: Implications for targeting Notch
signaling. Oncogene. 29:992–1002. 2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shen Y, Wang Q, Zhao Q and Zhou J: Leptin
promotes the immune escape of lung cancer by inducing
proinflammatory cytokines and resistance to apoptosis. Mol Med Rep.
2:295–299. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
De Simone V, Pallone F, Monteleone G and
Stolfi C: Role of T H 17 cytokines in the control of
colorectal cancer. Oncoimmunology. 2(e26617)2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mueller L, von Seggern L, Schumacher J,
Goumas F, Wilms C, Braun F and Broering DC: TNF-alpha similarly
induces IL-6 and MCP-1 in fibroblasts from colorectal liver
metastases and normal liver fibroblasts. Biochem Biophys Res
Commun. 397:586–5891. 2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Popivanova BK, Kitamura K, Wu Y, Kondo T,
Kagaya T, Kaneko S, Oshima M, Fujii C and Mukaida N: Blocking
TNF-alpha in mice reduces colorectal carcinogenesis associated with
chronic colitis. J Clin Invest. 118:560–570. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
Pereira SG and Oakley F: Nuclear
factor-kappaB1: Regulation and function. Int J Biochem Cell Biol.
40:1425–1430. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hayden MS and Ghosh S: Signaling to
NF-kappaB. Genes Dev. 18:2195–2224. 2004.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Puvvada SD, Funkhouser WK, Greene K, Deal
A, Chu H, Baldwin AS, Tepper JE and O'Neil BH: NF-kB and Bcl-3
activation are prognostic in metastatic colorectal cancer.
Oncology. 78:181–188. 2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kwon HC, Kim SH, Oh SY, Lee S, Kwon KA,
Lee JH, Choi HJ, Park KJ, Lee HS, Roh MS and Kim HJ:
Clinicopathological significance of nuclear factor-kappa B, HIF-1
alpha, and vascular endothelial growth factor expression in stage
III colorectal cancer. Cancer Sci. 101:1557–1561. 2010.PubMed/NCBI View Article : Google Scholar
|