Introduction
Lung cancer is a major cancer type that threatens
human life, with limited effective treatment approaches (1). Non-small cell lung cancer (NSCLC)
accounts for >80% of lung cancer cases and is relatively
insensitive to chemotherapy or radiotherapy when compared with
small cell lung cancer (2).
Although much research has focused on the carcinogenesis behind and
the development of anti-cancer drugs for NSCLC, the prognosis of
patients with NSCLC remains poor due to drug resistance, metastasis
and recurrence (3,4). The current poor prognosis for patients
with NSCLC emphasizes the urgent need for the discovery of new
biomarkers and targets for the detection and treatment of the
metastatic and NSCLC recurrence.
MicroRNAs (miRNAs/miRs) are 18-25-nucleotide long,
non-coding RNAs that negatively regulate target gene expression by
binding to the 3' untranslated region (UTR) of target mRNAs
(5). Deregulation of miRNAs is
involved in both the initiation and development of various cancer
types (6,7). A specific miRNA may function as an
oncogene or a tumor suppressor depending on the target mRNAs in
different cellular contexts (8). A
number of miRNAs have been discovered to drive or inhibit the
progression of NSCLC by targeting critical genes in cancer-related
pathways (9-11).
Among them, the expression of several miRNAs is considered to be a
diagnostic or prognostic biomarker for patients with NSCLC
(12,13). For example, miRNA-7 serves as a
tumor suppressor in NSCLC through inhibiting the expression of
paired box 6 and protein tyrosine kinase 2 (14,15).
Comparison of miR-7 expression levels in patients with NSCLC
indicates that the miR-7 expression levels are a marker of the
therapeutic effects of gefitinib (16). Bioinformatics analysis of NSCLC
microarray data has revealed that miR-491-5p is a potential
regulator of the development of NSCLC (17). However, the molecular mechanism of
action behind how miR-491-5p regulates NSCLC is unknown.
Forkhead box (FOX)P4 is a member of the FOX
transcription factor family (18).
Overexpression of FOXP4 is observed in hepatocellular carcinoma and
osteosarcoma, when compared with normal tissues (19,20).
It has previously been shown that FOXP4 is a target of miR-138 and
that silencing FOXP4 reduces cell proliferation and cell invasion
capacity in NSCLC cells (21).
The present study aimed to investigate the effects
of miR-491-5p on proliferation and migration of A549 cells, and to
determine whether miR-491-5p directly interacted with FOXP4. It was
found that miR-491-5p was significantly downregulated in NSCLC
tissue. miR-491-5p overexpression inhibited the cell
proliferation/migration of NSCLC cells. Collectively, the present
findings suggested a tumor suppressor role of miR-491-5p in NSCLC
and may provide a potential target for the treatment of NSCLC.
Materials and methods
Clinical samples
Between January 2015 and June 2016, 40 patients (25
males, 15 females; age, 45-70 years) with NSCLC from Yantai Laiyang
Central Hospital (Laiyang, China) were enrolled in the present
study. Patients who had received preoperative anti-tumor therapy
were excluded from the present study. Written consent was received
from all participants. Tumor tissues and paired adjacent normal
tissues were collected and immediately stored at -80˚C for
subsequent RNA extraction. All experiments were approved and
conducted under the supervision of the ethics committee of Yantai
Laiyang Central Hospital.
Cell culture
The human lung epithelial cell line BEAS-2B, and
human NSCLC cell lines including A549, H157 and H1650, were
purchased from the American Type Culture Collection. All cell lines
were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (HyClone; GE Healthcare Life Sciences) in
a 37˚C incubator with 5% CO2.
Transfection of miRNA mimics and miRNA
inhibitor
miR-491-5p mimics (5'-AGUGGGGAACCCUUCCCAUGAGG-3'),
miR-NC mimics (5'-AGAAGCUGUUCCAAGGUGGGCC-3'), miR-491-5p inhibitor
(5'-CCUCAUGGAAGGGUUCCCCACU-3') and miR-NC inhibitor
(5'-GAACAUCCAGGGUCCCGUUCCU-3') were purchased from Shanghai
GenePharma Co., Ltd. For cell transfection, miRNA mimics and miRNA
inhibitor were transfected into A549 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Briefly, the cells (8x105 cells/well) were seeded into
6-well plates. 50 nM miRNA mimics or miRNA inhibitor was mixed with
Lipofectamine 2000 in Opti-MEM (Invitrogen; Thermo Fisher
Scientific, Inc.) and placed at room temperature for 10 min.
Subsequently, the mixture was added into each well. Following 6 h
of transfection at 37˚C, the culture medium was replaced with fresh
medium and cells were cultured for another 24 h at 37˚C before
being subjected to the following experiments.
Cell proliferation assay
The cell proliferation of A549 cells was detected
using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies,
Inc.) according to manufacturer's protocol. In brief, transfected
cells were seeded in 96-well plates at a density of
2x104 cells/well. At the various time points (0, 24, 48
and 72 h), 10 µl CCK-8 solution was added into each well and
cultured at 37˚C for another 2 h. Subsequently, the medium
containing CCK-8 was transferred into another 96-well plate and the
absorbance at 450 nm was detected using a microplate reader
(Bio-Rad Laboratories, Inc.).
Cell migration assay
The migration of A549 cells was measured using wound
healing assays. Cells (~5x105 cells/well) were grown in
6-well plates and cultured to 100% confluence. A wound area was
made by scratching the central area of each well using a 10 µl
pipette. The cells were cultured with serum-free DMEM to avoid cell
proliferation. IncuCyte ZOOM (Essen BioScience) was used to capture
the images of cell migration with a confocal microscope (x200
magnification). The wound areas were analyzed using Image Pro Plus
(version 6.0; Media Cybernetics, Inc.) and the wound closure
percentage was calculated as a reflection of migratory ability. The
following equation was used: Migration
rate=(Wound30/Wound0) x100%.
Western blot analysis
Protein lysates of A549 cells were collected using
RIPA lysis buffer (Sigma-Aldrich; Merck KGaA) according to the
manufacturer's protocol. Antibodies against TGF-β (cat. no. 3711;
1:1,000), MMP-2 (cat. no. 40994; 1:1,000) and MMP-9 (cat. no.
13667; 1:1,000) were purchased from Cell Signaling Technology, Inc.
FOXP4 antibodies (cat. no. ab119404; 1:1,000) were purchased from
Abcam. The GAPDH (cat. no. G8795; 1:10,000) antibody was purchased
from Sigma-Aldrich; Merck KGaA. HRP-conjugated secondary antibodies
against rabbit (cat. no. SA00001-2; 1:10,000) and mouse (cat. no.
SA00001-1; 1:10,000) were obtained from Proteintech Group, Inc.
Proteins were quantified using a BCA assay kit (Pierce; Thermo
Fisher Scientific, Inc.). Lysates containing 20 µg protein were
loaded and separated on an 8% SDS-PAGE gel. The proteins were
transferred onto a PVDF membrane and then blocked in 5% non-fat
milk at room temperature for 2 h. Subsequently, the membrane was
incubated in the primary antibodies overnight at 4˚C, followed by
incubation in the secondary antibody at room temperature for
another 1 h. The membrane was developed using ECL Western Blot
Substrate (Pierce; Thermo Fisher Scientific, Inc.). The intensity
of the bands was analyzed using ImageJ version 1.8.0 (National
Institutes of Health).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). For the detection of the expression levels of miR-491-5p,
RNA was reverse transcribed using a Mir-X miRNA First-Strand
Synthesis kit (Takara Bio, Inc.) according to the manufacturer's
protocol. miR-491-5p expression was conducted using SYBR Premix Ex
Taq II (Takara Bio, Inc.). U6 served as an internal control for
miR-491-5p. For the detection of mRNA levels of FOXP4, MMP-2 and
MMP-9, RNA was reverse transcribed into cDNA using PrimeScript RT
Master Mix (Takara Bio, Inc.). Gene expression was conducted using
SYBR Premix Ex Taq II (Takara Bio, Inc.). GAPDH served as an
internal control for the mRNA of FOXP4, MMP-2 and MMP-9. The
thermocycling parameters were as follows: Initial denaturation at
95˚C for 2 min, 40 cycles of 95˚C for 15 sec and 64˚C for 30 sec.
The primer sequences were as follows: miR-491-5p forward,
5'-GGAGTGGGGAACCCTTCC-3' and reverse, 5'-GTGCAGGGTCCGAGGT-3'; U6
forward, 5'-GTGCTCGCTTCGGCAGCACAT-3' and reverse,
5'-AATATGGAACGCTTCACGAAT-3'; FOXP4 forward,
5'-GACAGCCTACTGTGCTCACAT-3' and reverse,
5'-TTGCACTCTCCGTGTCCGTA-3'; MMP-2 forward,
5'-TACAGGATCATTGGCTACACACC-3' and reverse,
5'-GGTCACATCGCTCCAGACT-3'; MMP-9 forward,
5'-TGTACCGCTATGGTTACACTCG-3' and reverse,
5'-GGCAGGGACAGTTGCTTCT-3'; and GAPDH forward,
5'-GGAGCGAGATCCCTCCAAAA-3' and reverse,
5'-GGCTGTTGTCATACTTCTCATGG-3'. The relative expression levels were
calculated using the 2-ΔΔCq method
(22).
Lentivirus system
The FOXP4 knockdown A549 cell line (A549-FOXP4
shRNA) and control A549 cell line (A549-control shRNA) were
established using a lentivirus system. Briefly, shRNA sequences
targeting FOXP4 or non-specific sequences were synthesized and
cloned into the pLKO.1 plasmid (Shaanxi YouBio Technology Co.,
Ltd.). The pLKO.1-FOXP4 shRNA or pLKO.1-control shRNA was
co-transfected with packaging plasmids into 293 cells to obtain the
virus. The virus was collected 48 h later and transfected into
target cells using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37˚C for 48 h. The cells that
were successfully infected with virus were screened by exposure to
puromycin (10 µM; Sigma-Aldrich; Thermo Fisher Scientific, Inc.)
for 72 h.
Dual luciferase reporter assay
FOXP4 was predicted to be targeted by miR-491-5p
using TargetScan (www.targetscan.org). The 3'UTR of FOXP4 mRNA was
amplified from cDNA of BEAS-2B cells and ligated into the pGL3
plasmid. The amplification of the FOXP4 3'UTR was achieved using
PrimeSTAR Max DNA polymerase (Takara Bio, Inc.) with two primers.
The primer sequences were: FOXP4 3'UTR forward,
5'-GGGCCTGTAGTGACCGGCAG-3' and FOXP4 3'UTR reverse,
5'-AATTGTTTTTATTGCATTGCATTGT-3'. Two site mutations were introduced
into pGL3-FOXP4 3'UTR-wild-type (WT) to construct the pGL3-FOXP4
3'UTR-mutant (Mut) using a QuickChange Site-Directed Mutagenesis
kit (Stratagene; Agilent Technologies, Inc.) following the
manufacturer's protocol. pGL3-FOXP4 3'UTR-WT and pGL3-FOXP4
3'UTR-Mut were co-transfected into A549 cells with miR-NC mimics or
miR-491-5p mimics using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). After 48 h, the luciferase
activity of each well was calculated using a Dual Luciferase
Reporter System (Promega Corporation). The firefly luciferase
activity was normalized to Renilla luciferase activity.
Overexpression of FOXP4
Full-length FOXP4 was amplified from A549 cDNA using
PrimeSTAR Max DNA polymerase (Takara Bio, Inc.) and inserted into
the pcDNA3.1 plasmid (YouBio). The primer sequences were: FOXP4
forward, 5'-GCTTGGTACCGAATGATGGTGGAATCTGCCTCG-3' and FOXP4 reverse,
5'-CCGCTCGAGGGACAGTTCTTCTCCCGGCA-3'. For overexpression of FOXP4, 2
µg pcDNA3.1-FOXP4 was mixed with Lipofectamine® 2000 in
serum-free DMEM for 15 min and then added into cultured cells
(2x106 A549 cells/well in 6-well plates) at room
temperature. The cells were transfected for 48 h at 37˚C before
being subjected to further experiments.
Statistical analysis
All data were analyzed using GraphPad Prism 6
(GraphPad Software, Inc.) and are presented as the mean ± SD
deviation of 3 independent repeats. The results in Figs. 1A and B, and 2B
and E were analyzed using paired
Student's t-test, while the results in Fig. 1C were analyzed using Pearson
correlation analysis. The data from Fig. 2A were analyzed using a one-way ANOVA
and post-hoc Tukey's tests. The comparisons between two groups in
the remaining figures were conducted using unpaired Student's
t-tests. Differences among more than two groups were calculated
using one-way ANOVAs followed by post hoc Student-Newman-Keul's or
Tukey's tests. P<0.05 was considered to indicate a statistically
significant difference.
Results
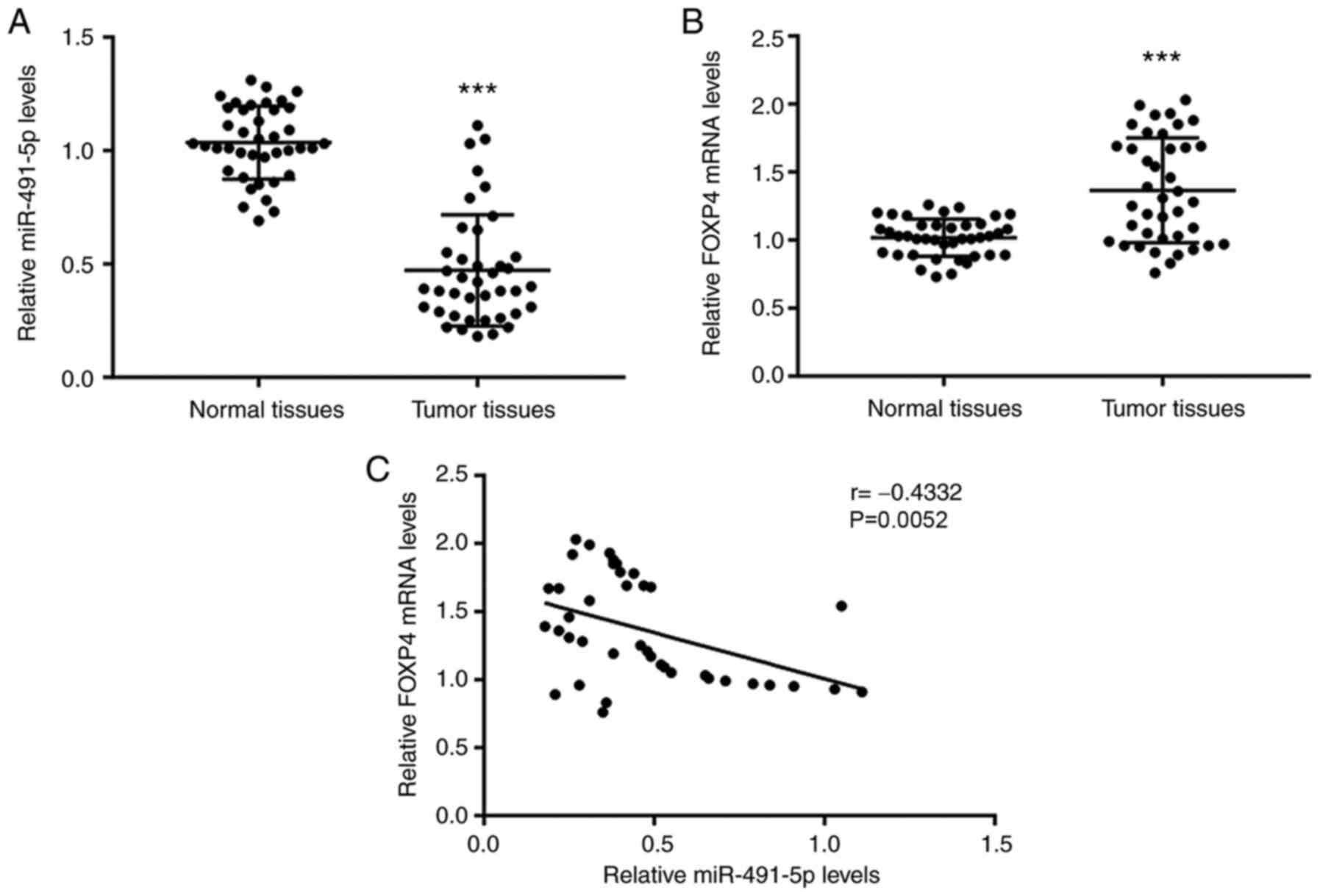
Lower miR-491-5p and higher FOXP4
expression is found in NSCLC tissues
RT-qPCR was first used to detect the miR-491-5p
expression levels and FOXP4 mRNA expression levels in tumor tissues
and matched normal tissues from 40 patients with NSCLC. Compared
with the normal tissues, there was significantly lower miR-491-5p
levels and higher FOXP4 mRNA levels in the tumor tissues (Fig. 1A and B). Pearson correlation analysis confirmed
that miR-491-5p levels were negatively associated with FOXP4 mRNA
levels in NSCLC tumor tissues (Fig.
1C).
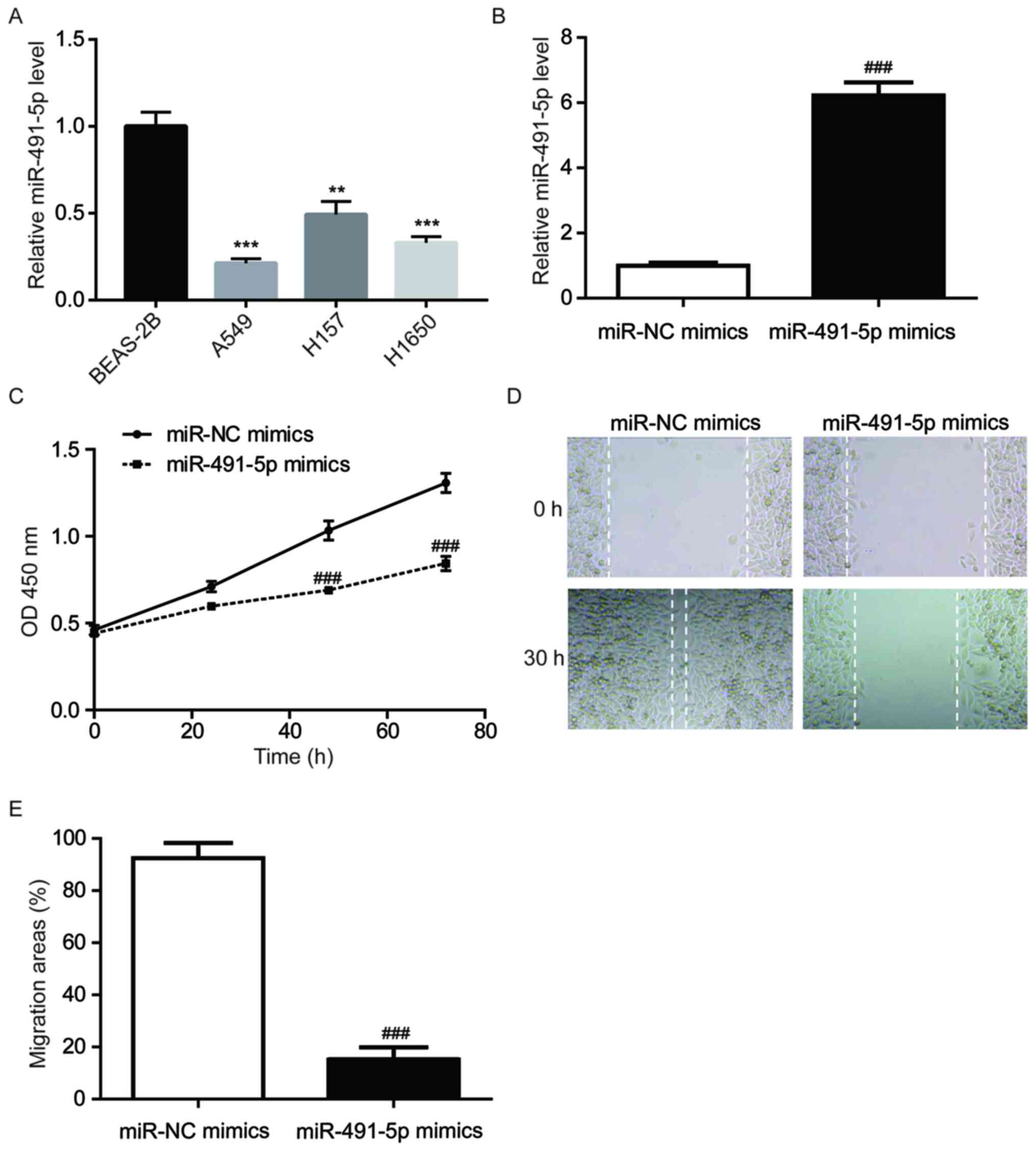
miR-491-5p mimics inhibits A549 cell
proliferation/migration
RT-qPCR was performed to detect the miR-491-5p
expression levels in a normal lung cell line (BEAS-2B) and NSCLC
cell lines (A549, H157 and H1650). Compared with BEAS-2B cells,
there was significantly lower miR-491-5p expression in all NSCLC
cell lines (Fig. 2A). A549 cells
were used for the subsequent experiments as they had the lowest
miR-491-5p expression levels among the three cell lines. To
investigate the function of miR-491-5p in NSCLC, miR-491-5p mimics
were transfected into A549 cells. Significantly increased
miR-491-5p levels were observed in the miR-491-5p mimics group
compared to the miR-NC mimics group (Fig. 2B). In comparison with the miR-NC
mimics group, miR-491-5p mimics significantly decreased A549 cell
proliferation (Fig. 2C). In
addition, compared with the miR-NC mimics group, miR-491-5p mimics
reduced the wound closure areas in A549 cells, indicating that
miR-491-5p inhibited the cell migratory ability of A549 cells
(Fig. 2D and E).
miR-491-5p targets and decreases FOXP4
expression levels in A549 cells
To validate the regulatory effect of miR-491-5p on
FOXP4 expression, miR-491-5p was overexpressed in A549 cells and
the FOXP4 expression levels were analyzed. Consistent with their
association in NSCLC tumor tissues, compared with the miR-NC mimics
group, miR-491-5p mimics significantly decreased FOXP4 at the mRNA
and protein expression levels in A549 cells (Fig. 3A-C). The expression levels of
miR-491-5p significantly decreased in cells transfected with
miR-491-5p inhibitor compared with the control transfection
(Fig. S1). Downregulation of
miR-491-5p increased FOXP4 mRNA and protein levels in A549 cells
(Fig. 3D-F). A putative binding
site between miR-491-5p and the FOXP4 3'UTR sequence was predicted
using TargetScan software (Fig.
3G). To determine whether FOXP4 was a direct target of
miR-491-5p in NSCLC cells, a dual luciferase assay was used to
investigate the binding sites between miR-491-5p and FOXP4 mRNA.
Compared with the miR-NC mimics group, miR-491-5p mimics
significantly decreased the luciferase activity of A549 cells
transfected with FOXP4 3'UTR-WT but not FOXP4 3'UTR-Mut, indicating
a regulatory relationship between miR-491-5p and FOXP4 in A549
cells (Fig. 3H).
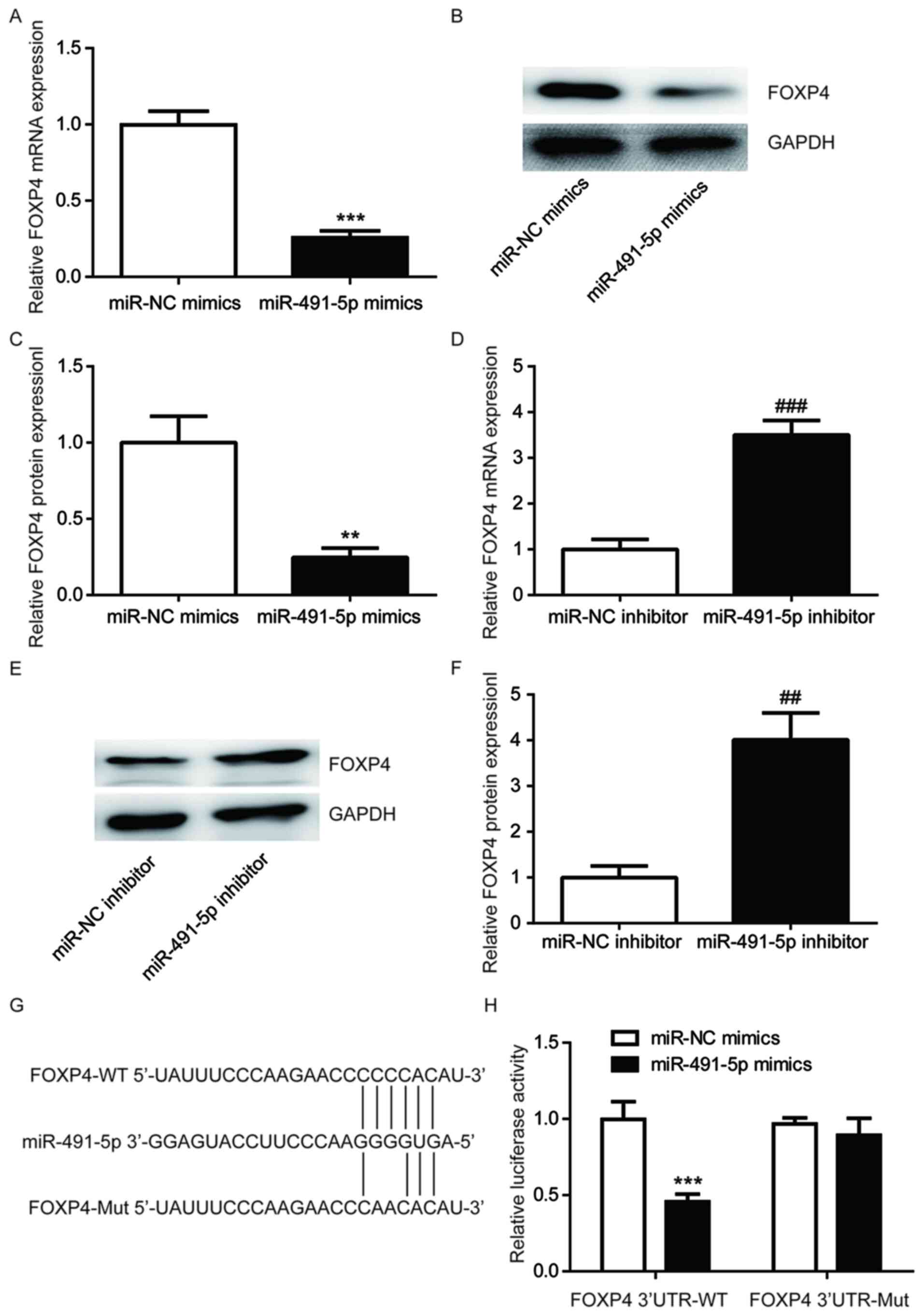 | Figure 3miR-491-5p binds to the 3'UTR of FOXP4
in A549 cells. Compared with miR-NC mimics group, miR-491-5p mimics
reduces FOXP4 (A) mRNA and (B and C) protein expression levels in
A549 cells. (D) Compared with the miR-NC inhibitor group,
miR-491-5p inhibitor increases FOXP4 (D) mRNA and (E and F) protein
expression levels in A549 cells. (G) The putative binding site
between miR-491-5p and the FOXP4 3'UTR, as well as the constructed
FOXP4 3'UTR-Mut are shown. (H) Compared with the miR-NC mimics
group, miR-491-5p mimics led to a reduction in luciferase activity
of A549 cells transfected with pGL3-FOXP4 3'UTR-WT (H).
**P<0.01, ***P<0.001 vs. miR-NC mimics;
##P<0.01 and ###P<0.001 vs. miR-NC
inhibitor. FOXP4, forkhead box P4; miR, microRNA; Mut, mutant; NC,
negative control; UTR, untranslated region; WT, wild-type. |
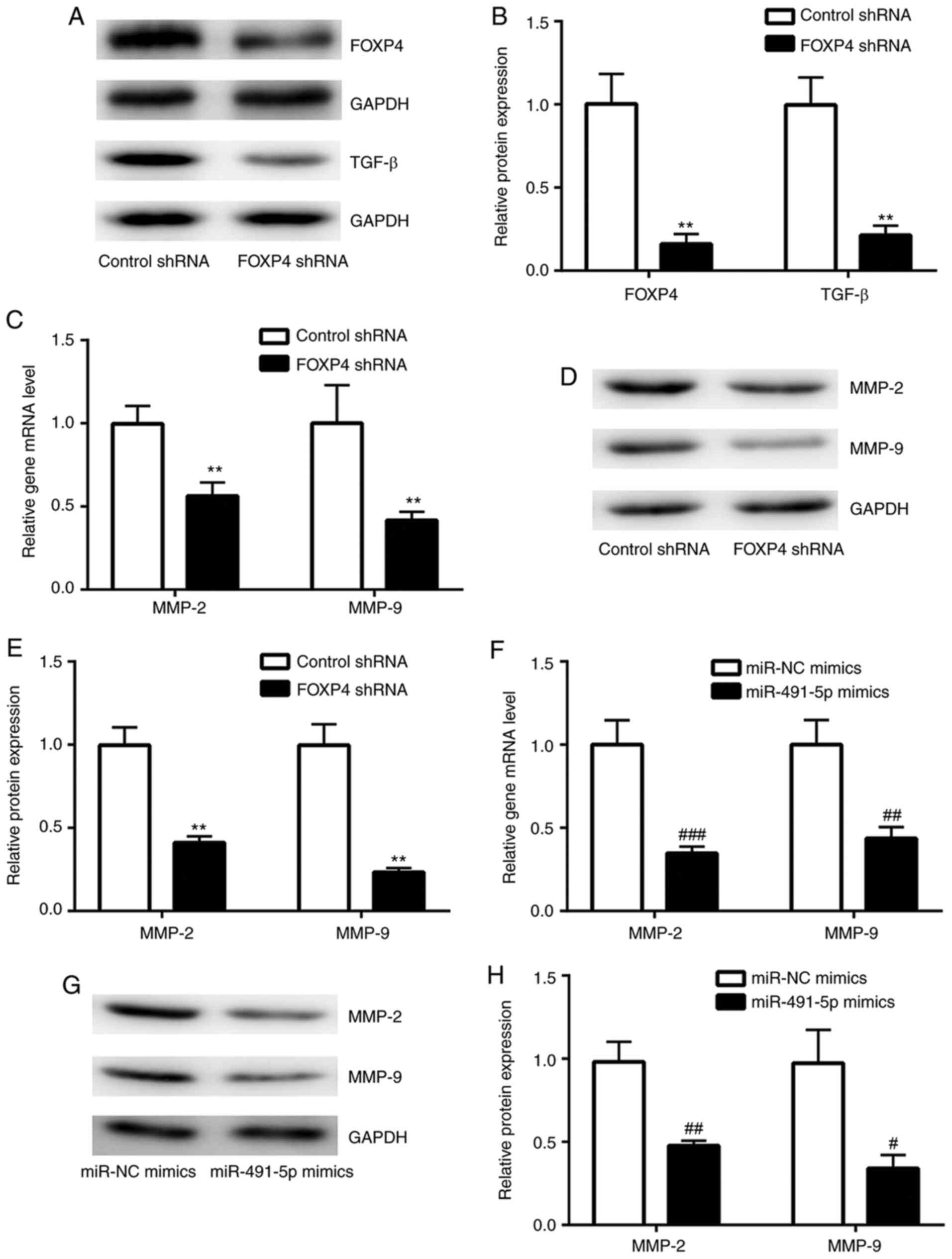
FOXP4 knockdown suppresses TGF-β
signaling in A549 cells
FOXP4 regulates target gene expression through its
interaction with FOXP1/FOXP2(23).
FOXP2 stimulates cancer cell migration by promoting TGF-β
expression (24). A FOXP4-knockdown
model was constructed in A549 cells using a lentivirus system to
study the function of miR-491-5p. As shown in Fig. 4A and B, FOXP4 expression was greatly reduced in
FOXP4-knockdown A549 cells compared with control shRNA A549 cells.
In FOXP4-knockdown A549 cells, TGF-β protein expression levels were
significantly reduced compared with the control shRNA A549 cells
(Fig. 4A and B). MMP-2 and MMP-9 are downstream target
genes of TGF-β (25). It was found
that in the FOXP4-knockdown group, there were significantly reduced
mRNA and protein expression levels of MMP-2 and MMP-9 in comparison
with the control shRNA group (Fig.
4C-E). Notably, overexpression of miR-491-5p also significantly
decreased MMP-2 and MMP-9 expression levels in A549 cells, in
comparison with the miR-NC mimics group (Fig. 4F-H), suggesting the existence of an
miR-491-5p/FOXP4/TGF-β axis in NSCLC cells.
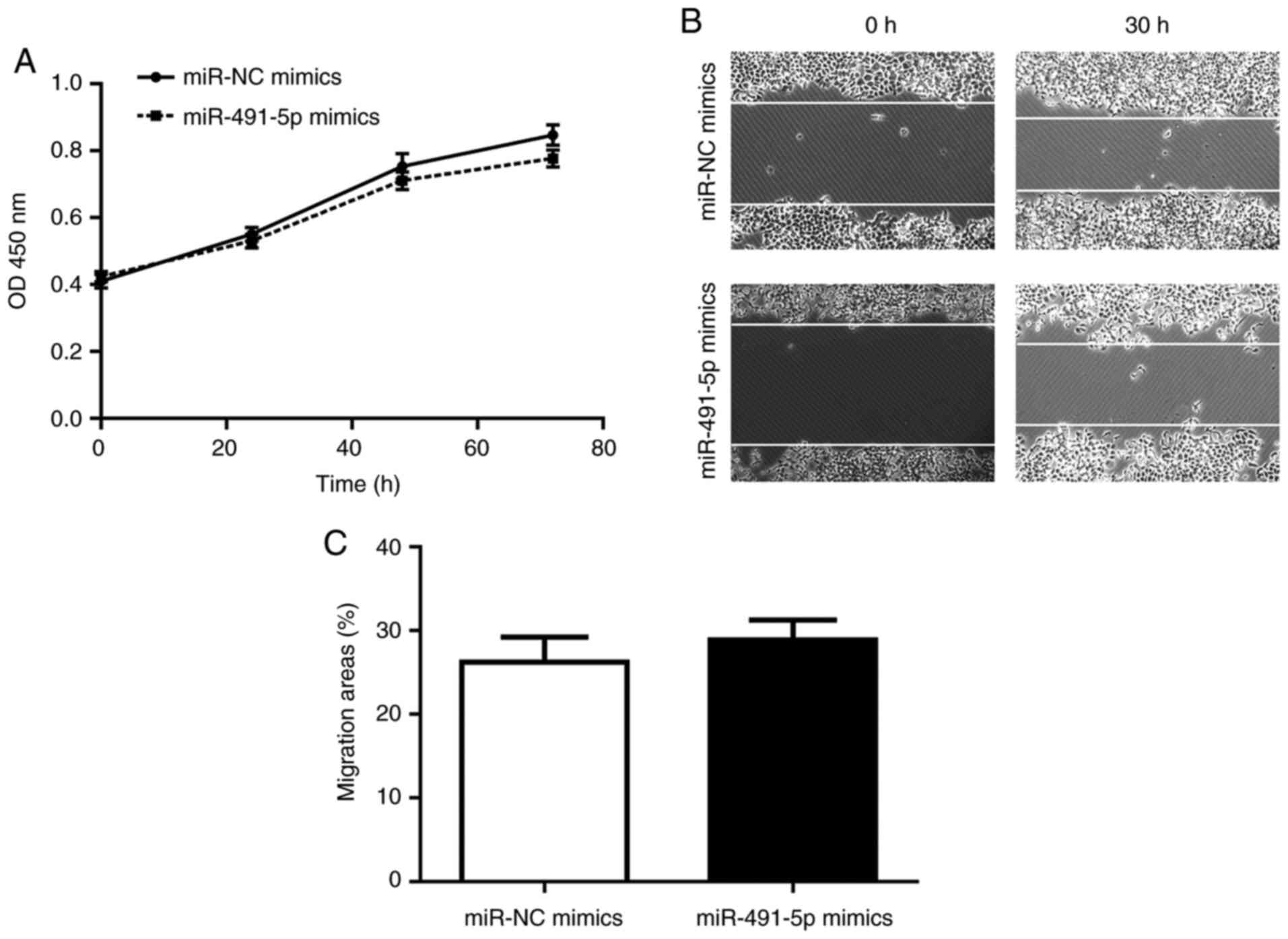
miR-491-5p mimics do not affect cell
proliferation/migration in FOXP4-knockdown A549 cells
Subsequently, the present study sought to
investigate whether FOXP4 was indispensable to miR-491-5p on its
regulation of cell proliferation/migration. In FOXP4-knockdown A549
cells, miR-491-5p mimics exerted a slight, but non-significant
effect on cell proliferation/migration compared to the miR-NC
mimics group (Fig. 5A-C).
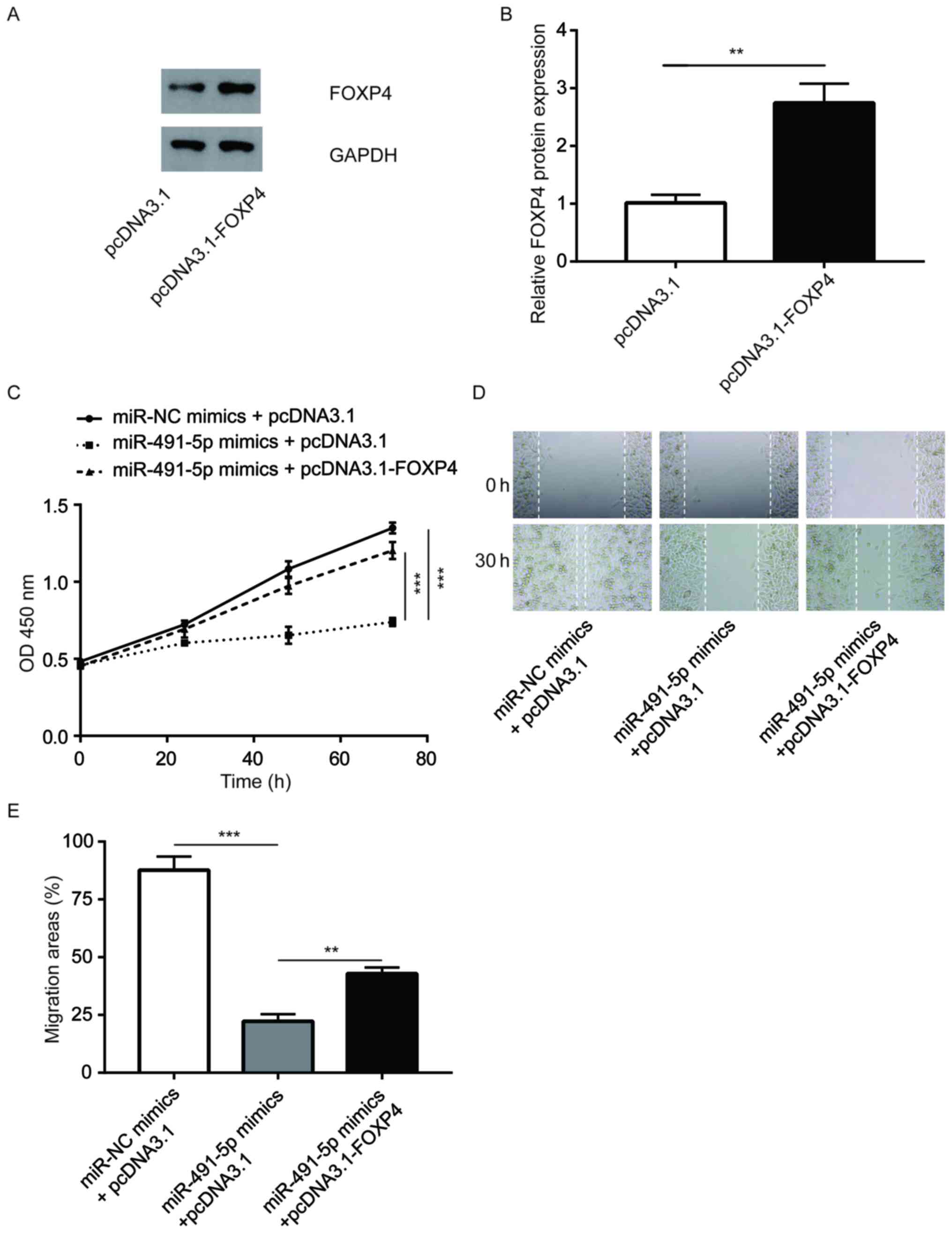
FOXP4 is pivotal for the function of
miR-491-5p in NSCLC cells
The pcDNA3.1-FOXP4 vector was constructed to
overexpress FOXP4 in A549 cells, which was found to effectively
increase FOXP4 protein expression levels (Fig. 6A and B). It was observed that the overexpression
of FOXP4 reversed the inhibition of cell proliferation induced by
miR-491-5p mimics in A549 cells (Fig.
6C). In addition, FOXP4 elevation partially reversed the
inhibition of cell migration induced by miR-491-5p mimics in A549
cells (Fig. 6D and E). The data suggested that miR-491-5p
partially relied on the regulation of FOXP4 to control NSCLC cell
proliferation and migration.
Discussion
Currently, the prognosis for patients with NSCLC is
not satisfactory and attempts to improve the clinical outcome have
largely relied on the discovery of novel targets and biomarkers.
miRNAs have captured the attention of researchers in terms of their
function during the initiation and development of NSCLC (2,6). The
present study showed that miR-491-5p inhibited NSCLC cell
proliferation/migration by targeting FOXP4.
Accumulating evidence suggests that miR-491-5p is a
tumor suppressor in various types of cancer, including breast
cancer, prostate cancer and gastric cancer (26-28).
The present study found that miR-491-5p levels were decreased in
tumor tissues compared with matched normal tissues from patients
with NSCLC. In addition, enhanced expression levels of miR-491-5p
led to a reduction of cell proliferation/migration in A549 cells.
These data are consistent with a recent study which showed that
miR-491-5p is a tumor suppressor in NSCLC (29). miR-491-5p suppresses cancer
progression by targeting various oncogenes according to the context
within the type of cell (30,31).
FOXP4 promotes NSCLC progression by accelerating
cell proliferation and enhancing the invasive ability of NSCLC
cells (21). In the present study,
miR-491-5p mimics led to a decrease in FOXP4 expression. miR-491-5p
inhibitor, on the contrary, increased FOXP4 expression in A549
cells. FOXP4 has been validated to be a target gene of miR-491-5p
in human osteosarcoma (19). Using
a dual luciferase reporter assay, the present study showed that
miR-491-5p could directly bind to the 3'UTR of FOXP4 mRNA in A549
cells.
Previous studies have indicated that miR-491-5p
targets MMP-9 and insulin-like growth factor 2 mRNA-binding protein
1 to suppress cell proliferation and cell migration in non-small
cell lung cancer (29,32). MMP-2 and MMP-9 are key regulators of
metastasis in cancer cells (33).
The TGF-β signaling pathway plays a critical role in mediating
metastasis, partly by promoting the expression of MMP-2 and
MMP-9(34). In the current study,
it was discovered that knockdown of FOXP4 reduced the TGF-β protein
expression levels and decreased the expression of MMP-2 and MMP-9
in A549 cells. Meanwhile, in FOXP4-knockdown A549 cells,
transfection of miR-491-5p mimics did not inhibit cell
proliferation or cell migration. Additionally, overexpression of
FOXP4 was able to partially reverse the miR-491-5p mimics-induced
inhibition of cell proliferation/migration in A549 cells. Thus,
these data suggested that the inhibitory effect of miR-491-5p on
cell proliferation and migration mainly relies on its regulation of
FOXP4 in NSCLC cells.
In conclusion, the present study revealed a tumor
suppressor role for miR-491-5p in NSCLC. Mechanistically,
miR-491-5p may have inhibited the cell proliferation/migration of
A549 cells, as well as the activation of the TGF-β signaling
pathway by directly binding to the 3'UTR of FOXP4 mRNA. The present
findings enhance the current understanding of the role of
miR-491-5p in NSCLC, indicating miR-491-5p as a promising target
for the treatment of patients with NSCLC.
Supplementary Material
The miR-491-5p expression levels of
A549 cells transfected with miR-491-5p inhibitors and NC
inhibitors. ***P<0.001. miR, microRNA; NC, negative
control.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PL conceived and designed the study. FW performed
the majority of the experiments and wrote the manuscript. AJ
assisted with the experiments. ZZ and JL participated in the
analysis and data interpretation. PL and FW confirmed the
authenticity of all the raw data. All authors read and approved the
final manuscript and agreed to be accountable for all aspects of
the research.
Ethics approval and consent to
participate
This study was performed in accordance with standard
guidelines and was approved by the Ethics Committee of Yantai
Laiyang Central Hospital. Written informed consent was obtained
from all the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Sauer AM, Chen MS Jr,
Kagawa-Singer M, Jemal A and Siegel RL: Cancer statistics for Asian
Americans, Native Hawaiians, and Pacific Islanders, 2016:
Converging incidence in males and females. CA Cancer J Clin.
66:182–202. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Wood SL, Pernemalm M, Crosbie PA and
Whetton AD: The role of the tumor-microenvironment in lung
cancer-metastasis and its relationship to potential therapeutic
targets. Cancer Treat Rev. 40:558–566. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Laskin JJ and Sandler AB: State of the art
in therapy for non-small cell lung cancer. Cancer Invest.
23:427–442. 2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Paliouras AR, Monteverde T and Garofalo M:
Oncogene-induced regulation of microRNA expression: Implications
for cancer initiation, progression and therapy. Cancer Lett.
421:152–160. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ye MF, Zhang JG, Guo TX and Pan XJ:
MiR-504 inhibits cell proliferation and invasion by targeting LOXL2
in non small cell lung cancer. Biomed Pharmacother. 97:1289–1295.
2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hao J, Zhao S, Zhang Y, Zhao Z, Ye R, Wen
J and Li J: Emerging role of microRNAs in cancer and cancer stem
cells. J Cell Biochem. 115:605–610. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhou Q, Huang SX, Zhang F, Li SJ, Liu C,
Xi YY, Wang L, Wang X, He QQ, Sun CC and Li DJ: MicroRNAs: A novel
potential biomarker for diagnosis and therapy in patients with
non-small cell lung cancer. Cell Prolif. 50(e12394)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Xiong K, Shao LH, Zhang HQ, Jin L, Wei W,
Dong Z, Zhu YQ, Wu N, Jin SZ and Xue LX: MicroRNA-9 functions as a
tumor suppressor and enhances radio-sensitivity in radio-resistant
A549 cells by targeting neuropilin 1. Oncol Lett. 15:2863–2870.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang Y, Wang Y and Wang J: MicroRNA-584
inhibits cell proliferation and invasion in non-small cell lung
cancer by directly targeting MTDH. Exp Ther Med. 15:2203–2211.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kulda V, Svaton M, Mukensnabl P, Hrda K,
Dvorak P, Houdek Z, Houfkova K, Vrzakova R, Babuska V, Pesek M and
Pesta M: Predictive relevance of miR-34a, miR-224 and miR-342 in
patients with advanced squamous cell carcinoma of the lung
undergoing palliative chemotherapy. Oncol Lett. 15:592–599.
2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang J, Wang T, Zhang Y, Wang H, Wu Y,
Liu K and Pei C: Upregulation of serum miR-494 predicts poor
prognosis in non-small cell lung cancer patients. Cancer Biomark.
21:763–768. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cao Q, Mao ZD, Shi YJ, Chen Y, Sun Y,
Zhang Q, Song L and Peng LP: MicroRNA-7 inhibits cell
proliferation, migration and invasion in human non-small cell lung
cancer cells by targeting FAK through ERK/MAPK signaling pathway.
Oncotarget. 7:77468–77481. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Luo J, Li H and Zhang C: MicroRNA-7
inhibits the malignant phenotypes of non-small cell lung cancer in
vitro by targeting Pax6. Mol Med Rep. 12:5443–5448. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mou K, Gu W, Gu C, Zhang J, Qwang W, Ren G
and Tian J: Relationship between miR-7 expression and treatment
outcomes with gefitinib in non-small cell lung cancer. Oncol Lett.
12:4613–4617. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Teufel A, Wong EA, Mukhopadhyay M, Malik N
and Westphal H: FoxP4, a novel forkhead transcription factor.
Biochim Biophys Acta. 1627:147–152. 2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tian W, Liu J, Pei B, Wang X, Guo Y and
Yuan L: Identification of miRNAs and differentially expressed genes
in early phase non-small cell lung cancer. Oncol Rep. 35:2171–2176.
2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yin Z, Ding H, He E, Chen J and Li M:
Up-regulation of microRNA-491-5p suppresses cell proliferation and
promotes apoptosis by targeting FOXP4 in human osteosarcoma. Cell
Prolif. 50(e12308)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang G and Sun Y, He Y, Ji C, Hu B and Sun
Y: MicroRNA-338-3p inhibits cell proliferation in hepatocellular
carcinoma by target forkhead box P4 (FOXP4). Int J Clin Exp Pathol.
8:337–344. 2015.PubMed/NCBI
|
|
21
|
Yang T, Li H, Thakur A, Chen T, Xue J, Li
D and Chen M: FOXP4 modulates tumor growth and independently
associates with miR-138 in non-small cell lung cancer cells. Tumour
Biol. 36:8185–8191. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sin C, Li H and Crawford DA:
Transcriptional regulation by FOXP1, FOXP2, and FOXP4 dimerization.
J Mol Neurosci. 55:437–448. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Song XL, Tang Y, Lei XH, Zhao SC and Wu
ZQ: miR-618 inhibits prostate cancer migration and invasion by
targeting FOXP2. J Cancer. 8:2501–2510. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Da C, Liu Y, Zhan Y, Liu K and Wang R:
Nobiletin inhibits epithelial-mesenchymal transition of human
non-small cell lung cancer cells by antagonizing the TGF-β1/Smad3
signaling pathway. Oncol Rep. 35:2767–2774. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hui Z, Yiling C, Wenting Y, XuQun H,
ChuanYi Z and Hui L: miR-491-5p functions as a tumor suppressor by
targeting JMJD2B in ERα-positive breast cancer. FEBS Lett.
589:812–821. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Xu Y, Hou R, Lu Q, Zhang Y, Chen L, Zheng
Y and Hu B: MiR-491-5p negatively regulates cell proliferation and
motility by targeting PDGFRA in prostate cancer. Am J Cancer Res.
7:2545–2553. 2017.PubMed/NCBI
|
|
28
|
Sun R, Liu Z, Tong D, Yang Y, Guo B, Wang
X, Zhao L and Huang C: miR-491-5p, mediated by Foxi1, functions as
a tumor suppressor by targeting Wnt3a/β-catenin signaling in the
development of gastric cancer. Cell Death Dis.
8(e2714)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gong F, Ren P, Zhang Y, Jiang J and Zhang
H: MicroRNAs-491-5p suppresses cell proliferation and invasion by
inhibiting IGF2BP1 in non-small cell lung cancer. Am J Transl Res.
8:485–495. 2016.PubMed/NCBI
|
|
30
|
Sun D, Han S, Liu C, Zhou R, Sun W, Zhang
Z and Qu J: Microrna-199a-5p functions as a tumor suppressor via
suppressing connective tissue growth factor (CTGF) in follicular
thyroid carcinoma. Med Sci Monit. 22:1210–1217. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang Q, Li Q, Xu T, Jiang H and Xu LG:
miR-491-5p suppresses cell growth and invasion by targeting Notch3
in nasopharyngeal carcinoma. Oncol Rep. 35:3541–3547.
2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Pirooz HJ, Jafari N, Rastegari M,
Fathi-Roudsari M, Tasharrofi N, Shokri G, Tamadon M, Sazegar H and
Kouhkan F: Functional SNP in microRNA-491-5p binding site of MMP9
3'-UTR affects cancer susceptibility. J Cell Biochem.
119:5126–5134. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Stetler-Stevenson WG: The role of matrix
metalloproteinases in tumor invasion, metastasis, and angiogenesis.
Surg Oncol Clin N Am. 10:383–392. 2001.PubMed/NCBI
|
|
34
|
Kim S, Lee J, You D, Jeong Y, Jeon M, Yu
J, Kim SW, Nam SJ and Lee JE: Berberine suppresses cell motility
through downregulation of TGF-β1 in triple negative breast cancer
cells. Cell Physiol Biochem. 45:795–807. 2018.PubMed/NCBI View Article : Google Scholar
|