Introduction
T cell acute lymphoblastic leukemia (T-ALL) is a
malignant hematological disease involving the infinite expansion of
defective naive T cells (1,2). Children diagnosed with T-ALL account
for ~15% of all cases of ALL (3).
Currently, chemoradiotherapy and hematopoietic stem cell
transplantation (HSCT) are the main therapeutic regimens used for
ALL (1,4). With regards to the use of
chemoradiotherapy, drug resistance and the tolerance of patients to
the drugs used in the later stages of treatment are the main
drawbacks for its use (5). On the
other hand, HSCT, is associated with high costs and severe
complications, such as graft-versus-host disease, which greatly
limits its use and effectiveness (6-8).
Even in cases in which initial treatment may seem effective,
relapses can often occur unexpectedly (9). As such, effective treatment regimens
for T-ALL are urgently required, particularly for children who are
diagnosed with early T cell progenitor ALL (10). Currently, the development of novel
strategies which can overcome the current obstacles is a major
challenge to effectively treat the disease (11).
MicroRNAs (miRNAs/miRs), as non-coding RNAs, play an
important role in regulating mRNA expression (12,13).
Researchers have found that miRNAs are involved in various cellular
processes, such as cell cycle progression and determining cell
fate, through affecting proliferation, differentiation, metabolism
and apoptosis (14). Consequently,
miRNAs are regarded as targets for cancer therapeutic intervention,
rendering them promising candidates (15,16).
The focus of research on miRNAs is increasing. Previous studies
have demonstrated that miR-325 can potently inhibit cell growth,
such as that of hepatocellular carcinoma and non-small cell lung
cancer (NSCLC), by targeting high mobility group box1 and aquaporin
5 (17-19).
Bcl-2-associated athanogene (BAG)2 is an
anti-apoptotic oncogene, which plays a pivotal role in various
diseases, such as numerous types of cancer, Alzheimer's disease,
Parkinson's disease and spinocerebellar ataxia type-3(20). Previous studies have demonstrated
that BAG2 is highly expressed in a number of tumor types, such as
multiple myeloma, colorectal cancer, ovarian cancer and lung cancer
(21-24).
However, the roles of miR-325 and BAG2 in T-ALL, as well as the
interaction between the two, remain to be determined. As such, the
present study aimed to investigate the roles of miR-325 and BAG2 in
a T-ALL cell line (Jurkat cells) and to further explore the
underlying mechanisms of action.
Materials and methods
Cell lines and clinical samples
In the present study, human T-ALL cell lines,
including TALL-1, KOPTK1, Jurkat, CCRF-CEM and Molt16, were
purchased from the American Type Culture Collection (ATCC). The
cells mentioned above were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin, (NanJing SunShine
Biotechnology Co., Ltd.) and 100 µg/ml streptomycin (Sunshine
Biotechnology, Nanjing, China). The cells were maintained in a
humidified atmosphere with 5% CO2 at 37˚C.
Clinical samples were obtained from Zibo Central
Hospital (Zibo, China) from February 2018 to April 2019. Blood
samples were obtained from 20 pediatric patients (age range, 3.6-14
years; 10 males, 10 females) who were diagnosed with T-ALL and 20
healthy donors (age range, 3-15.2 years; 10 males, 10 females). All
fresh blood samples were immediately separated into several
portions, snap-frozen in liquid nitrogen and stored at -80˚C prior
to protein and RNA extraction. The present study was approved by
the Ethical Review Committee of Zibo Central Hospital. All
participants and their legal guardians agreed to the use of their
samples in the present study and written informed consents were
obtained from all of the legal guardians of all participants.
Transient transfection
0.2 µM BAG2-specific small interfering RNA
(BAG2-siRNA; 5'-GGGAAGAACUCUCACCGUUTT-3'; Santa Cruz Biotechnology,
Inc.), 0.2 µM control-siRNA (5'-UUCUCCGAACGUGUCACGUTT-3'; Santa
Cruz Biotechnology, Inc.), 1 µg control-plasmid (cat. no.
sc-437275; Santa Cruz Biotechnology, Inc.), 1 µg BAG2-plasmid (cat.
no. sc-404540-ACT; Santa Cruz Biotechnology, Inc.), 100 nM mimic
control (Shanghai GenePharma Co., Ltd.) and 100 nM miR-325 mimic
(Shanghai GenePharma Co., Ltd.) were transfected into the Jurkat
cell line, separately, using Lipofectamine® 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol at 37˚C. Immediately following 48 h of
transfection, the cells were collected for protein and RNA
extraction.
Bioinformatics analysis
Bioinformatics analysis software TargetScan version
7.2 (http://www.targetscan.org/vert_72/) was used to
predict the potential targets of miR-325.
Luciferase reporter assay
The pGL-3 plasmid vector (Promega Corporation) was
employed in the luciferase reporter assay, which contained the
wild-type (BAG2-WT) and mutant-type BAG2-mutant, containing
mutations in the binding region of miR-325 with the BAG2 gene. The
BAG2 gene was designed to contain the predicted miR-325 binding
site. For the luciferase assay, 293T cells were cultured in a
24-well plate at a density of 5x104 cells/well overnight
at 37˚C prior to transfection. The cells were co-cultured with the
pGL-3 plasmid vectors and miR-325 mimic using a
Lipofectamine® 3000 reagent kit at 37˚C for 48 h. After
48 h of transfection, the luciferase activity was measured using
the Dual-luciferase Reporter Assay system (Promega Corporation)
according to the manufacturer's protocol, and the results were
normalized to Renilla luciferase activity.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
To further confirm the expression levels of miR-325
and BAG2, RT-qPCR was performed. Total RNA was extracted from the
cell lines using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and RNA was extracted from clinical
samples using the miRNeasy Mini kit (Qiagen, Inc.), according to
the manufacturer's instructions. A NanoDrop-1000 spectrophotometer
(Thermo Fisher Scientific, Inc.) was applied to quantify the
extracted RNA. RNA was reverse transcribed into cDNA using
Superscript III Reverse Transcriptase (Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. The reaction
conditions for RT were as follows: 70˚C for 5 min, 37˚C for 5 min
and 42˚C for 60 min. The quantitative expression of each gene was
determined using SYBR-Green I (Thermo Fisher Scientific, Inc.).
Thermocycling conditions used for the qPCR were as follows: Initial
denaturation at 95˚C for 5 min; followed by 38 cycles of 15 sec at
95˚C, 1 min at 60˚C and 30 sec at 72˚C; and a final extension for
10 min at 72˚C. The primers were synthesized and purified by Sangon
Biotech (Shanghai) Co., Ltd. Primer sequences were listed as
following: miR-325 forward,
5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACACUUAC-3' and reverse,
5'-ACACTCCAGCTGGGCCUAGUAGGUGUCCAGU-3'; BAG2 forward,
5'-CTTTGAGAGAAGCAGCAACTG-3' and reverse,
5'-TGACACTTCAACGGTGAGAG-3'; U6 forward, 5'-ATACAGAGAAAGTTAGCACGG-3'
and reverse, 5'-GGAATGCTTCAAAGAGTTGTG-3'; GAPDH forward,
5'-TTTGGTATCGTGGAAGGACTC-3' and reverse,
5'-GTAGAGGCAGGGATGATGTTCT-3'. GAPDH (for mRNA) and U6 (for miRNA)
were used as the internal controls. Gene expression was calculated
using the 2-ΔΔCq method (25).
Western blot analysis
Following transfection, the Jurkat cells were
harvested and lysed in RIPA lysis buffer (Beyotime Institute of
Biotechnology) containing protease inhibitor cocktail
(Sigma-Aldrich; Merck KGaA). The clinical samples were lysed using
a Total Protein Extraction kit (Beijing Solarbio Science &
Technology Co., Ltd.). The protein concentration was then
determined using a BCA kit (Beyotime Institute of Biotechnology).
Individual samples (15 µg/lane) were separated by 12% SDS-PAGE and
transferred electrophoretically onto PVDF membranes (EMD
Millipore). The membranes were blocked using 5% non-fat milk
contained in TBST (0.1% Tween 20) for 1 h at room temperature,
followed by incubation with primary antibodies against BAG2 (cat.
no. ab79406; Abcam; working dilution, 1:1,000), Bcl-2 (cat. no.
ab182858; Abcam; working dilution, 1:1,000), Bax (cat. no. ab32503;
Abcam; working dilution, 1:1,000) and GAPDH (cat. no. ab9485;
Abcam; working dilution, 1:1,000) overnight at 4˚C. The following
day, the membranes were incubated with a horseradish
peroxidase-conjugated anti-rabbit IgG secondary antibody (cat. no.
7074; Cell Signaling Technology, Inc.; working dilution, 1:2,000)
at room temperature for 1 h. To observe the protein bands, an ECL
kit (Amersham Pharmacia Biotech) was used for staining and the
membranes were then photographed immediately. Band densities were
quantified using Gel-Pro Analyzer densitometry software (version
6.3; Media Cybernetics, Inc.).
In vitro proliferation assay
To assess the effects of BAG2-siRNA and miR-325 on
proliferation in vitro, a standard MTT assay was performed.
Briefly, the Jurkat cells were seeded in 96-well plates at a
density of 4,000 cells/well in complete medium, as described above,
and cultured overnight. The following day, the cells were
transfected with control-siRNA, BAG2-specific siRNA,
control-plasmid, mimic control, miR-325 mimic, miR-325 mimic +
control-plasmid, or miR-325 mimic + BAG2-plasmid in fresh medium at
37˚C for 48 h following the same conditions as aforementioned. The
Jurkat cells were separately centrifuged and examined at 0, 12, 24
and 48 h immediately following the end of the transfection. A total
of 150 µl DMSO (Sigma-Aldrich; Thermo Fisher Scientific, Inc.) was
added to dissolve the purple formazan. Untreated cells were used as
controls and regarded to have 100% viability. The data were
measured using a BioTek microplate reader (BioTek Instruments,
Inc.) at the absorbance of 570 nm.
Flow cytometry (FCM) for
apoptosis
To verify the effects on apoptosis in vitro,
Jurkat cells were harvested following transfection at the
concentration of 1x106 cells/tube. The cells were then
washed, pelleted and stained with Annexin V-FITC (Beyotime
Institute of Biotechnology) and PI on ice in the dark for 15 min.
The samples were then analyzed using a flow cytometer (FACSCalibur;
BD Biosciences) and quadrants 2 and 3 (Q2 + Q3) were used for
calculating the extent of apoptosis. FlowJo software (version
7.6.1; FlowJo LLC) was applied to analyze the data.
Statistical analysis
Unless otherwise stated, the data in the present
study are expressed as the means ± SD, with n=3 or more replicates.
GraphPad Prism 6.0 (GraphPad Software) was used for statistical
analysis. The statistical significance of differences between
groups was determined using unpaired Student's t-tests or one-way
ANOVAs followed by Tukey's post hoc tests. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of miR-325 in clinical
samples and human T-ALL cell lines
Previous studies have demonstrated that miR-325 can
suppress cancer cell proliferation and accelerate apoptosis
(17-19);
however, to the best of our knowledge, there are no previous
studies investigating the role of miR-325 in T-AL. In the present
study, miR-325 expression levels in clinical samples from healthy
donors and pediatric patients diagnosed with T-ALL, as well as in
T-ALL cell lines (Jurkat, CCRF-CEM, TALL-1 and KOPTK1) were
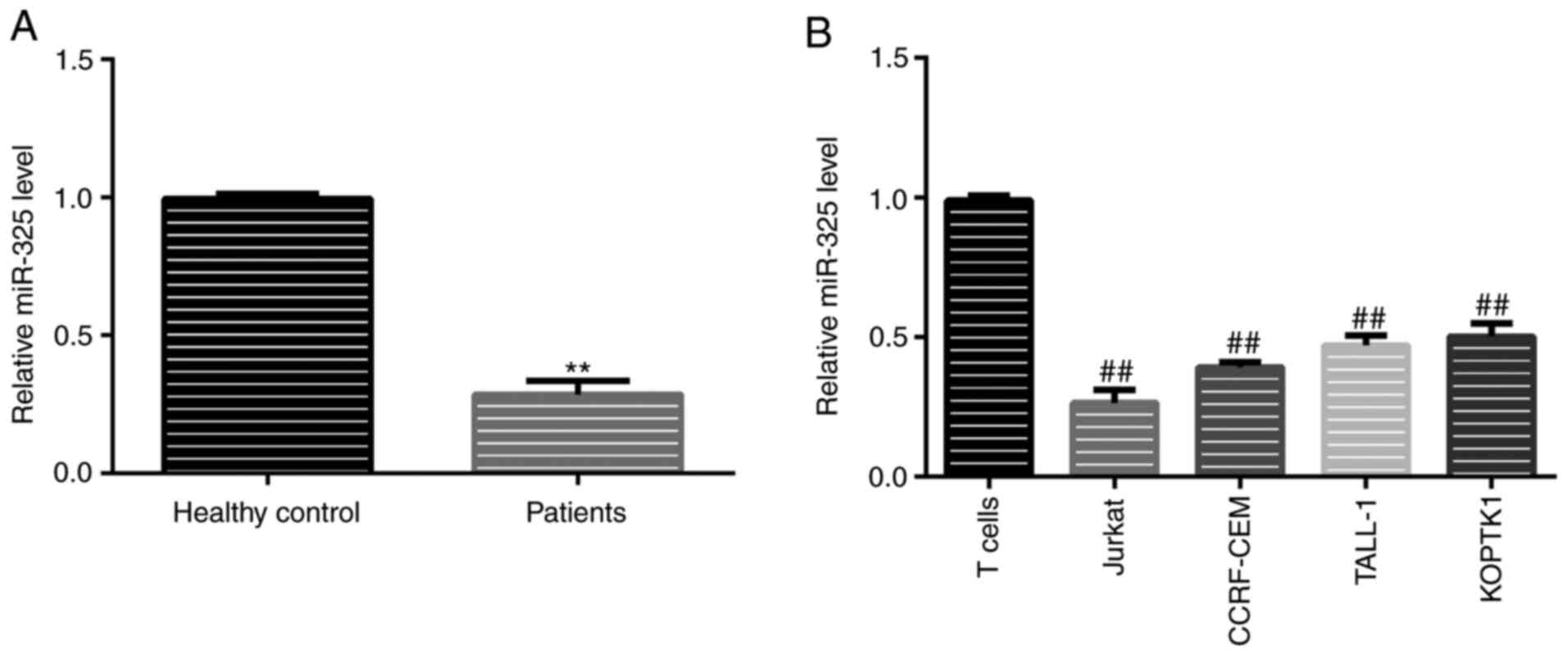
detected using RT-qPCR. The results revealed that, compared with
the healthy samples, the levels of miR-325 in the patient samples
were significantly lower (Fig. 1A).
A similar trend was observed in the cell lines, with the Jurkat
cells exhibiting the lowest expression levels of miR-325 (Fig. 1B). For this reason, Jurkat cells
were selected for use in further in vitro experiments.
Verification of the interaction
between miR-325 and BAG2
To date, the experimental approach available for the
verification of miRNAs and their target gene include the
dual-luciferase report assay, which can detect the interaction
between miRNAs and their corresponding target genes (26). In the present study, TargetScan,
which is a popular miRNA target predication algorithm, was used to
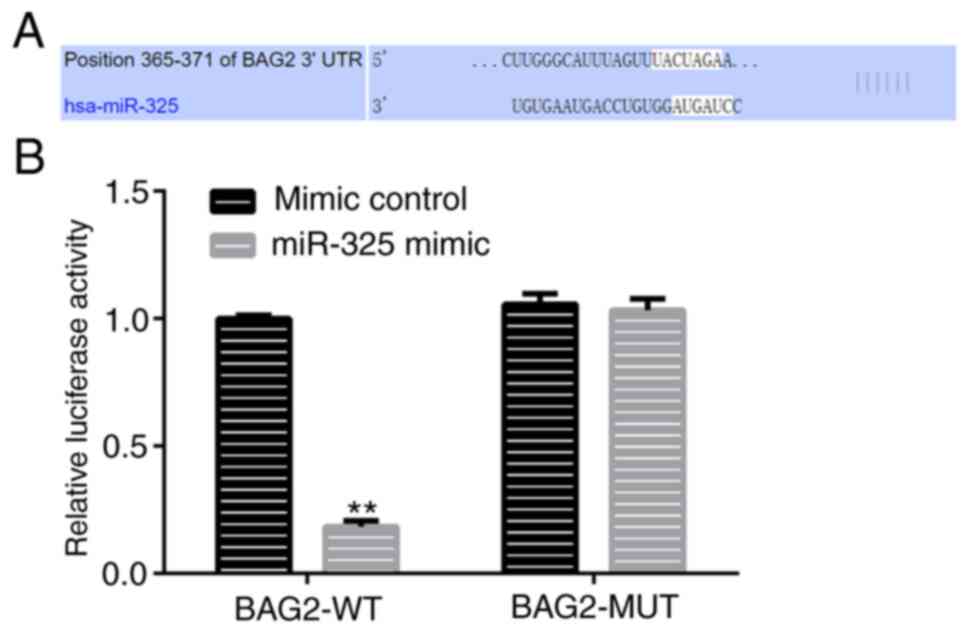
identify the target of miR-325. The results revealed that miR-325
shared a binding site with BAG2 (Fig.
2A). In addition, a dual-luciferase report assay was performed
and the results further verified the interaction between miR-325
and BAG2 (Fig. 2B). In view of this
hypothesis, it was confirmed that BAG2 was a direct target gene of
miR-325.
Expression of BAG2 in clinical samples
and human T-ALL cell lines
As it was found that BAG2 was a target of miR-325,
as mentioned above, it was hypothesized that the expression levels
of BAG2 in patient samples and T-ALL cell lines would exhibit an
opposite trend to that of miR-325. To verify this hypothesis, the
mRNA expression levels of BAG2 in patient samples and T-ALL cell
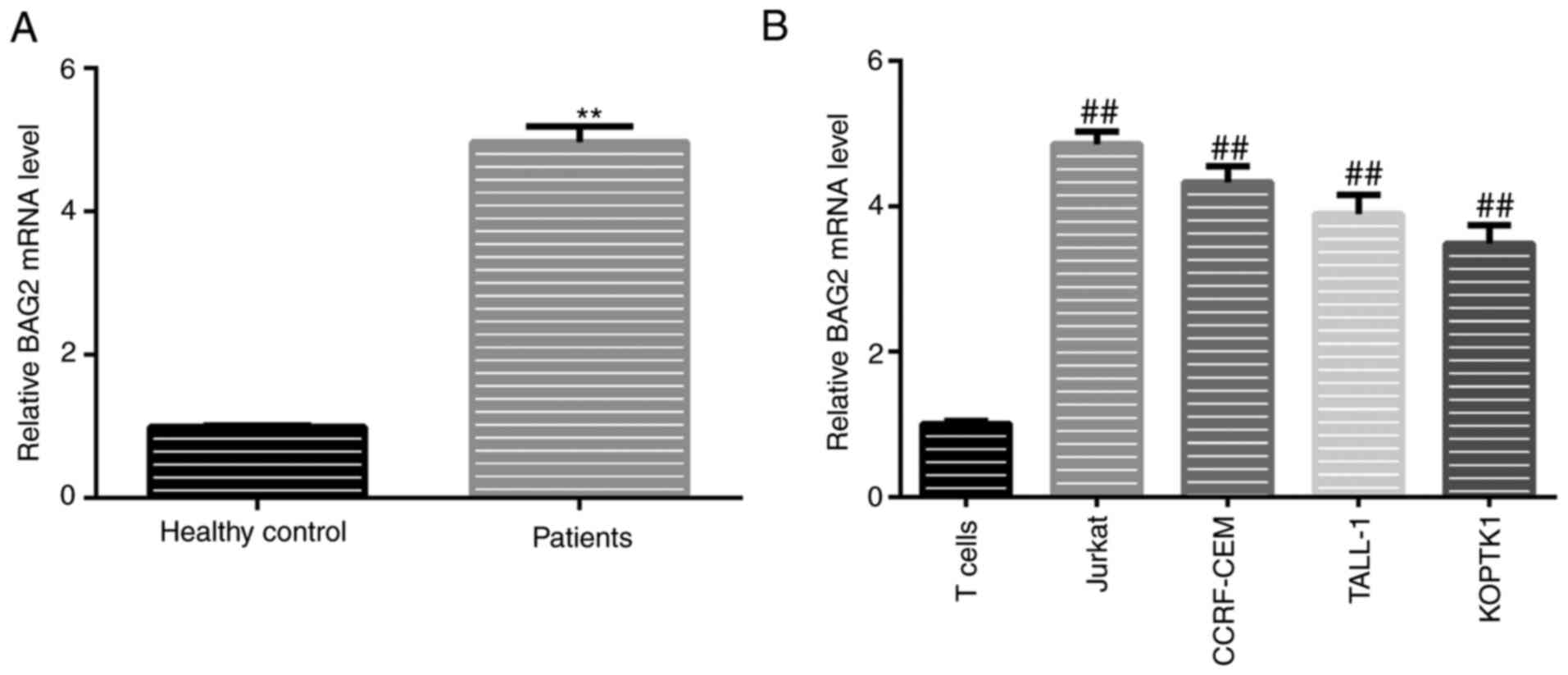
lines was detected using RT-qPCR. The results revealed that the
expression of BAG2 in patients diagnosed with T-ALL was
significantly higher compared with the healthy donors (Fig. 3A). It was also found the trend for
the expression levels of miR-325 in T-ALL cell lines was in
accordance with that in patient samples, with significantly higher
levels in all T-ALL cell lines (Fig.
3B). Moreover, the BAG2 expression levels were highest in the
Jurkat cells compared with all other T-ALL cell lines (Fig. 3B).
BAG2 knockdown inhibits the
proliferation and promotes the apoptosis of Jurkat cells
To explore the role of BAG2 in cell proliferation
and apoptosis, siRNA technology was applied in the present study.
BAG2-specific siRNA and control-siRNA were generated and
co-cultured with Jurkat cells for 48 h, respectively. The RT-qPCR
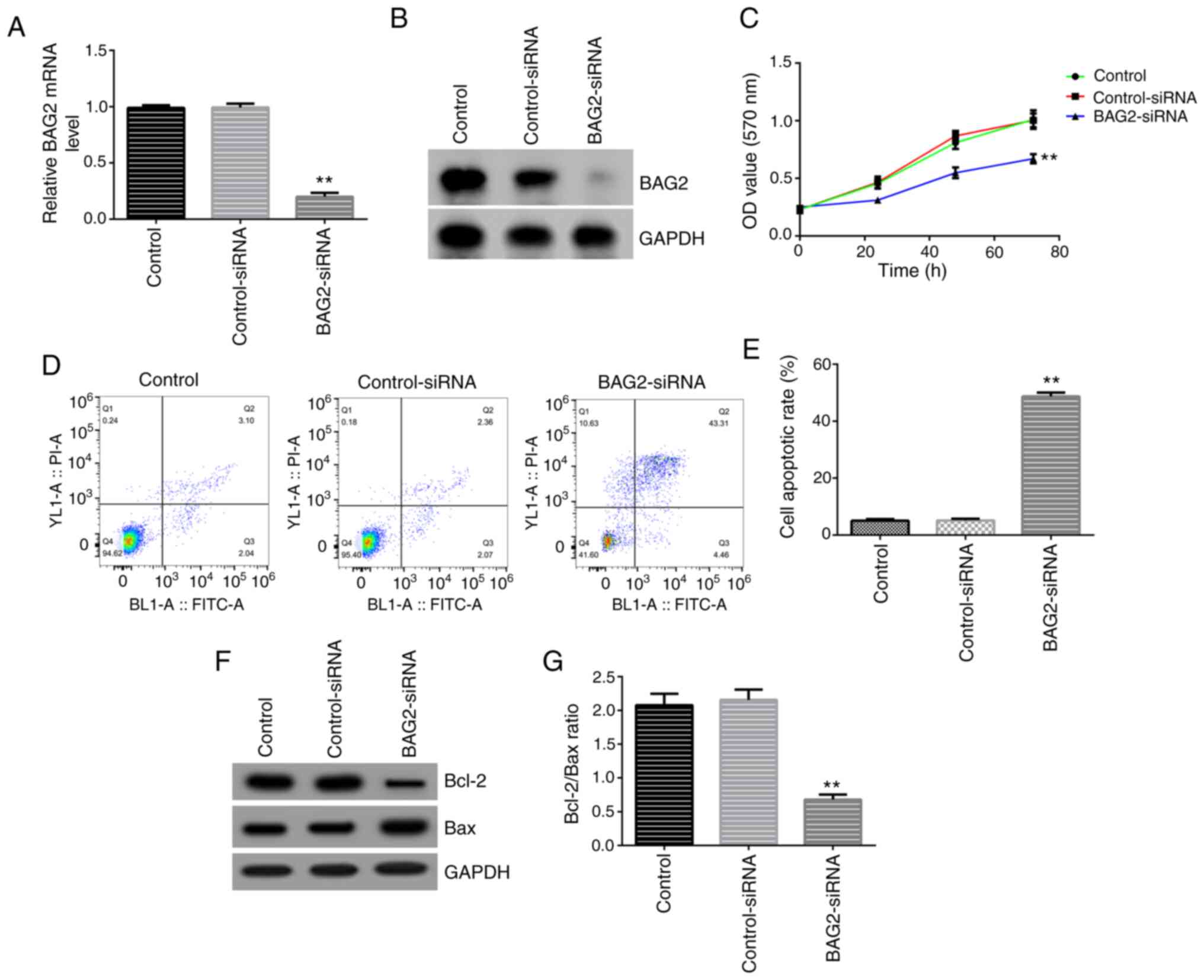
and western blotting data revealed that the expression of BAG2 in
Jurkat cells was notably decreased following transfection with
BAG2-siRNA, compared with the control-siRNA transfected group,
demonstrating a successful transfection (Fig. 4A and B). Subsequently, the biological behaviors,
namely the proliferation and apoptosis of Jurkat cells, following
BAG2 knockdown were assessed. Firstly, an MTT assay was performed
at 0, 24, 48 and 72 h. The analysis revealed that the knockdown of
BAG2 markedly inhibited the proliferation of Jurkat cells (Fig. 4C). Secondly, using Annexin V-FITC/PI
double staining, the cells were analyzed using FCM. It was found
that the percentage of apoptotic cells was markedly increased in
the group transfected with BAG2-siRNA (Fig. 4D and E). Finally, the expression levels of the
apoptosis-related proteins, Bcl-2 and Bax, were examined using
western blot analysis. The data demonstrated that the protein
expression levels of Bcl-2 were decreased and the Bax protein
expression levels were increased (Fig.
4F), with the ratio of Bcl-2/Bax being significantly decreased
(Fig. 4G) compared with the
control-siRNA group. Taken together, these data showed that BAG2 is
a key target which can directly influence the cell proliferation
and apoptosis of Jurkat cells.
Overexpression of miR-325 can
partially inhibit cell proliferation and induce apoptosis by
downregulating the expression of BAG2
Based on the previous results, it was hypothesized
that miR-325 might protect Jurkat cells from proliferation and
accelerate apoptosis in a BAG2-dependent manner. Then, to determine
whether the overexpression of miR-325 can protect Jurkat cells from
proliferation and accelerate apoptosis in a BAG2-dependent manner,
Jurkat cells were first transfected with various plasmids, miRNA
mimics or a combination of these for 48 h. RT-qPCR and western blot
analyses were carried out to identify the transfection efficiency.
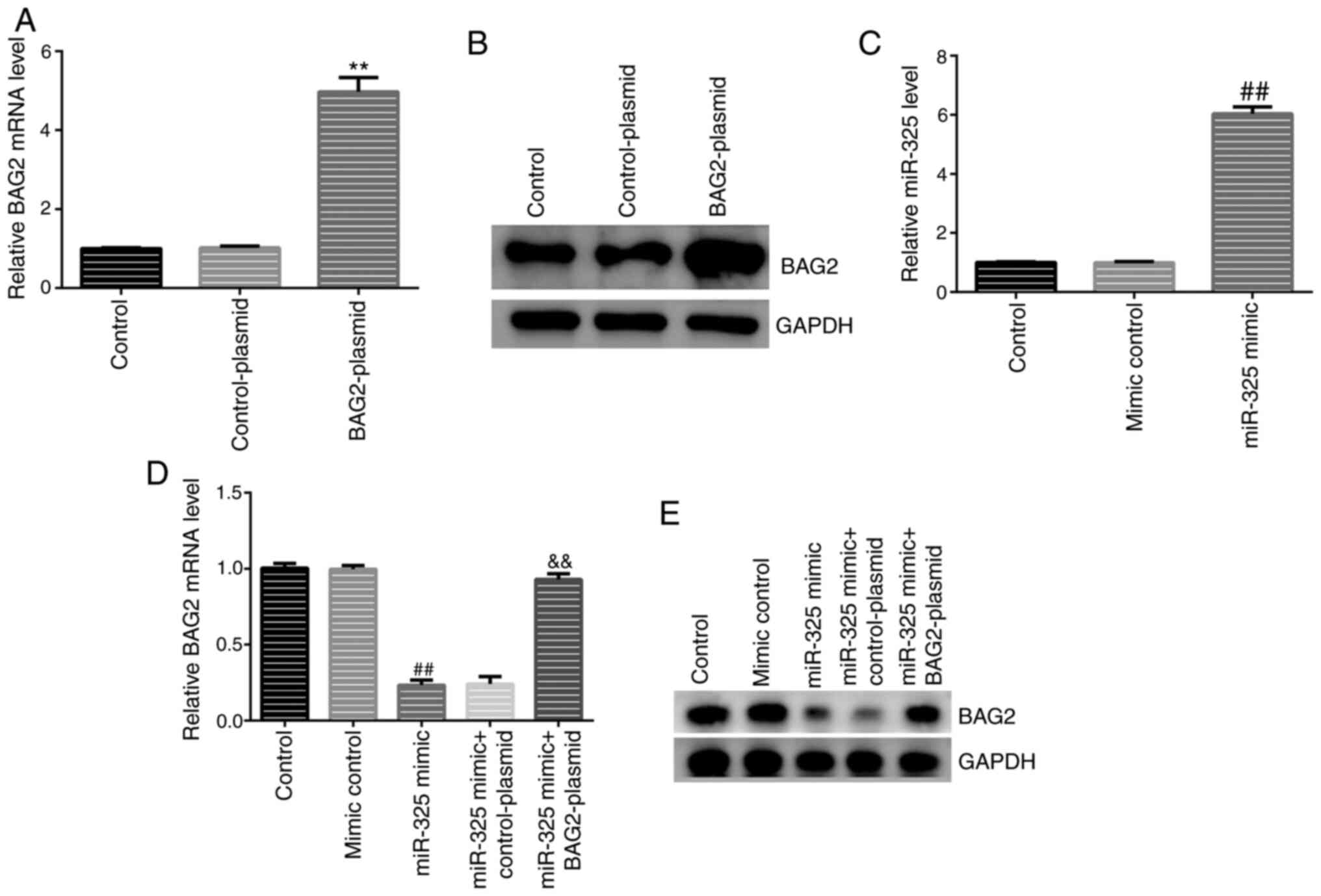
It was found that, compared with the control group, the expression
levels of BAG2 and miR-325 were significantly increased following
transfection with BAG2-plasmid and miR-325 mimic alone (Fig. 5A-C), demonstrating the successful
transfections. It was also found that the expression levels of BAG2
were notably decreased in the group transfected with miR-325 mimic
(Fig. 5D and E), as hypothesized.
The analysis revealed that the overexpression of
miR-325 reduced the expression levels of BAG2 in Jurkat cells and
the downregulatory effect was reversed by the introduction of the
BAG2-plasmid (Fig. 5D and E). To better understand the biological
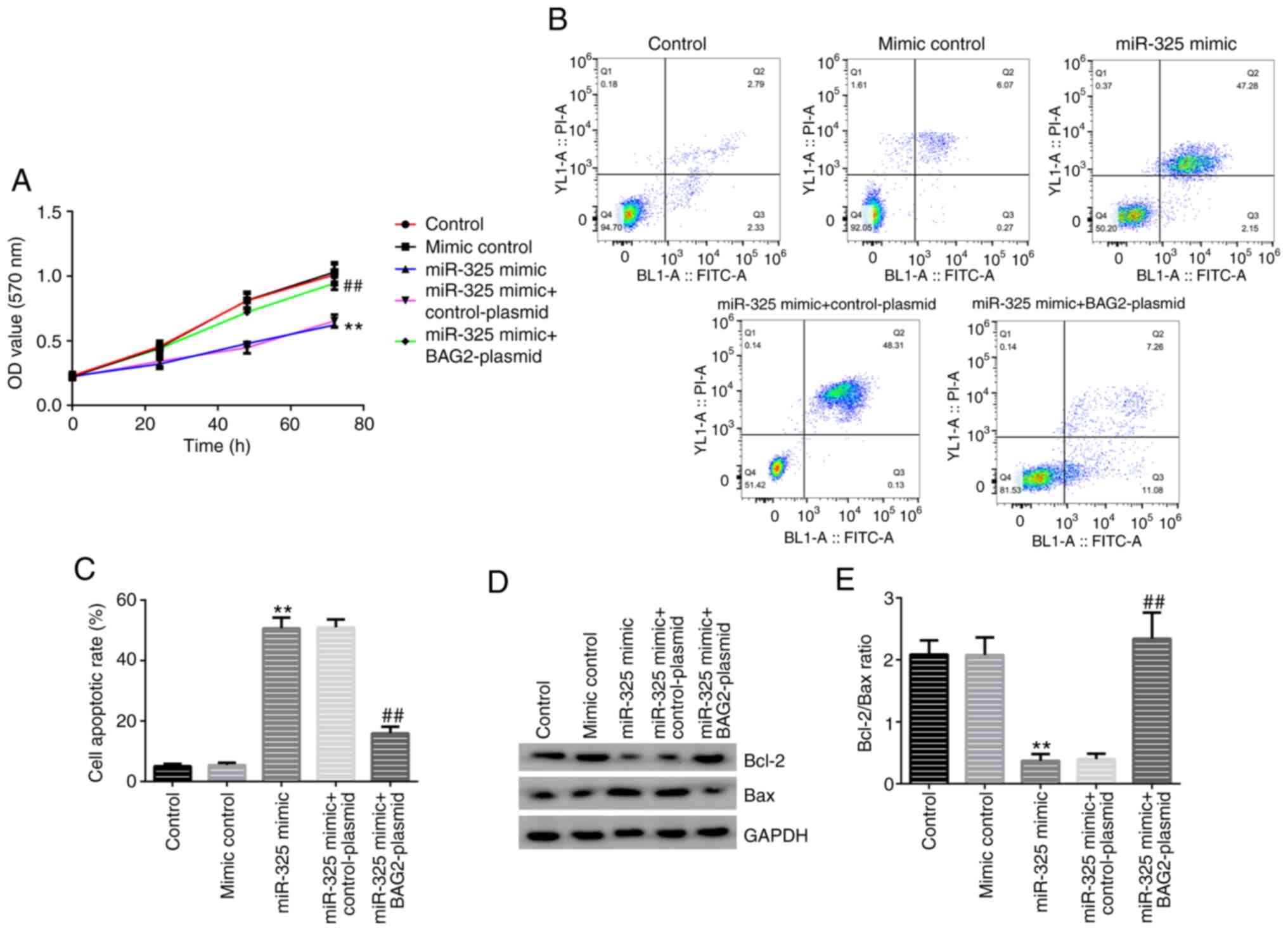
effects of miR-325 mimic on cell proliferation and apoptosis
following knocking down of the expression levels of BAG2, an MTT
assay, western blot analysis and FCM were performed. In the group
transfected with the miR-325 mimic alone, the cell proliferation of
the Jurkat cells was inhibited (Fig.
6A) and the percentage of apoptotic cells was significantly
increased (Fig. 6B and C). In addition, the effect on the protein
expression levels of Bcl-2 and Bax, was investigated. A decreased
Bcl-2 expression and an increased Bax expression were observed
(Fig. 6D and E). Moreover, the data also demonstrated
that the overexpression of BAG2 reversed the biological behaviors
caused by miR-325 mimic transfection. These results demonstrated
that expression of BAG2 was necessary for the induction of
apoptosis of Jurkat cells by miR-325.
Discussion
It has previously been demonstrated that BAG2 plays
a crucial role in the progression of tumor cell growth (27). BAG2, as an anti-apoptotic gene, can
promote cell proliferation, inhibit cell apoptosis and arrest the
cell cycle, and a raised BAG2 expression is found in a number of
tumor types, such as thyroid cancer (28) and breast cancer (22). Ge et al (29) found that upregulation of BAG2 might
be associated with underlying TNF-related apoptosis-inducing ligand
(TRAIL)-resistance mechanisms in NSCLC. However, its biological
function in T-ALL is not yet well understood.
In the present study, preliminary analysis with
RT-qPCR and western blot analysis suggested that the mRNA and
protein expression levels of BAG2 were higher in the blood samples
of patients diagnosed with T-ALL and in T-ALL cell lines compared
with those in healthy donor samples. To discover the role of BAG2
on proliferation and apoptosis, BAG2 expression was silenced in
Jurkat cells using siRNA technology. The data displayed a
significant decrease in cell expansion and an increased portion of
apoptotic cells in the group transfected with BAG2-siRNA. This
suggested that BAG2 indeed affects cell growth and survival.
miRNAs, as non-coding small RNAs, are regulators of
gene expression at the mRNA level (28,30).
An increasing number of studies have focused on oncogenic miRNAs,
which determine the tumor cell fate and oncogenic miRNAs have been
shown to be promising targets for therapy (31,32).
T-ALL is a refractory and relapsing malignant cancer which has been
the focus of numerous clinicians and studies in recent years
(33,34). In recent years, the research of
miRNA in T-ALL has attracted more and more attention (1). In 2004, Chen et al (35) identified three miRNAs that are
specifically expressed in hematopoietic cells, which were
dynamically regulated in the period of early hematopoiesis and
lineage commitment. miR-142-3p (36), miR-155(37), miR-146a (38) and miR-150(39) have been reported to play roles in
T-ALL. The present study found that miR-325 shared a binding site
with BAG2 using TargetScan and the result was in accordance with
the results of the dual-luciferase reporter assay. As previously
reported, miRNAs are small RNAs which play a significant role in
regulating gene expression by interfering with mRNA translation or
promoting mRNA degradation (40).
The results described above demonstrated the following 3 points: i)
Compared with healthy donor samples, the expression of miR-325 was
markedly lower and the level of BAG2 was markedly higher in
patients with T-ALL and in T-ALL cell lines; ii) BAG2 knockdown can
influence cell proliferation and apoptosis; and iii) miR-325 shares
a binding site with BAG2. Taken together, it was hypothesized that
BAG2 may be targeted by miR-325 in T-ALL. To verify this
hypothesis, Jurkat cells were transfected with miR-325 mimics in
vitro and cell proliferation was significantly inhibited
compared with the control group. This was consistent with previous
studies (17-19),
demonstrating that miR-325 has an inhibitory effect on the
proliferation of cancer cells. Additionally, the biological effects
of miR-325 on the proliferation and apoptosis of Jurkat cells were
reversed by the introduction of BAG2.
The BAG family was first identified as a group of
proteins that prevent cell death through their interaction with
Bcl-2(20). The Bcl-2 family genes
(Bcl-2 and Bax) play critical roles in the apoptosis of T-ALL cells
(41,42). In the present study, the results
also demonstrated that the protein expression levels of Bcl-2 were
decreased and the levels of Bax were increased simultaneously
following the downregulation of the levels of BAG2. This indicated
that Bcl-2/Bax may be the downstream genes of BAG2 regulated by
miR-325. The antitumor biological effects may be dependent on the
miR-325/BAG2/Bcl-2/Bax pathway; however, this requires verification
in future studies.
In conclusion, the findings of the present study
demonstrated that the miR-325/BAG2 axis may be a promising
therapeutic target for the treatment of T-ALL. miR-325 inhibited
the proliferation and promoted the apoptosis of T-ALL cells in a
BAG2-dependent manner.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FYW contributed to study design, data collection,
statistical analysis, data interpretation and manuscript
preparation. FLW and SZ contributed to data collection and
statistical analysis. XX contributed to data collection,
statistical analysis and manuscript preparation. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical Review
Committee of Zibo Central Hospital. All participants and their
legal guardians agreed to the use of their samples in the present
study and written informed consents were obtained from all of the
legal guardians of all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Correia NC and Barata JT: MicroRNAs and
their involvement in T-ALL: A brief overview. Adv Biol Regul.
74(100650)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Litzow MR and Ferrando AA: How i treat
T-cell acute lymphoblastic leukemia in adults? Blood. 126:833–841.
2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hounjet J, Habets R, Schaaf MB, Hendrickx
TC, Barbeau LM, Yahyanejad S, Rouschop KM, Groot AJ and Vooijs M:
The anti-malarial drug chloroquine sensitizes oncogenic NOTCH1
driven human T-ALL to γ-secretase inhibition. Oncogene.
38:5457–5468. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lange A, Dlubek D, Zdziarski R,
Chodorowska A, Mordak-Domagala M, Klimczak A, Lange J and Jaskula
E: Donor lymphocyte infusions to leukemic bone lesions are
therapeutically effective in a ph+ all patient with
post-HSCT relapse. J Immunotoxico. 11:347–352. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Follini E, Marchesini M and Roti G:
Strategies to overcome resistance mechanisms in T-cell acute
lymphoblastic leukemia. Int J Mol Sci. 20(3021)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ali N, Flutter B, Rodriguez RS, Paghaleh
ES, Barber LD, Lombardi G and Nestle FO: Xenogeneic
graft-versus-host-disease in NOD-scid IL-2rγnull mice display a
T-effector memory phenotype. PLoS One. 7(e44219)2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gill S, Tasian SK, Ruella M, Shestova O,
Li Y, Porter DL, Carroll M, Desnoyers GD, Scholler J, Grupp SA, et
al: Preclinical targeting of human acute myeloid leukemia and
myeloablation using chimeric antigen receptor-modified T cells.
Blood. 123:2343–2354. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Riegel C, Boeld TJ, Doser K, Huber E,
Hoffmann P and Edinger M: Efficient treatment of murine acute GvHD
by in vitro expanded donor regulatory T cells. Leukemia.
34:895–908. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kuhlen M, Willasch AM, Dalle JH, Wachowiak
J, Yaniv I, Ifversen M, Sedlacek P, Guengoer T, Lang P, Bader P, et
al: Outcome of relapse after allogeneic HSCT in children with ALL
enrolled in the ALL-SCT 2003/2007 trial. Br J Haematol. 180:82–89.
2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pui CH, Pei DQ, Cheng C, Tomchuck SL,
Evans SN, Inaba H, Jeha S, Raimondi SC, Choi JK, Thomas Paul G and
Dallas MH: Treatment response and outcome of children with T-cell
acute lymphoblastic leukemia expressing the gamma-delta T-cell
receptor. Oncoimmunology. 8(1599637)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hefazi M and Litzow MR: Recent advances in
the biology and treatment of T cell acute lymphoblastic leukemia.
Curr Hematol Malig Rep. 13:265–274. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Drury RE, O'Connor D and Pollard AJ: The
clinical application of microRNAs in infectious disease. Front
Immunol. 8(1182)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lin T, Zhou SM, Gao H, Li YQ and Sun LJ:
MicroRNA-325 is a potential biomarker and tumor regulator in human
bladder cancer. Technol Cancer Res Treat.
17(1533033818790536)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yooa B, Ghosha SK, Kumar M, Moore A, Yigit
MV and Medarova Z: Design of nanodrugs for miRNA targeting in tumor
cells. J Biomed Nanotechnol. 10:1114–1122. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ingenito F, Roscigno G, Affinito A, Nuzzo
S, Scognamiglio I, Quintavalle C and Condorelli G: The Role of
Exo-miRNAs in cancer: A focus on therapeutic and diagnostic
applications. Int J Mol Sci. 20(4687)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li H, Huang W and Luo R: The microRNA-325
inhibits hepatocellular carcinoma progression by targeting high
mobility group box 1. Diagn Pathol. 10(117)2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang Z, Han Y, Sun G, Liu X, Jia X and Yu
X: MicroRNA-325-3p inhibits cell proliferation and induces
apoptosis in hepatitis B virus-related hepatocellular carcinoma by
down-regulation of aquaporin 5. Cell Mol Biol Lett.
24(13)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yao SH, Zhao TJ and Jin H: Expression of
microRNA-325-3p and its potential functions by targeting HMGB1 in
non-small cell lung cancer. Biomed Pharmacother. 70:72–79.
2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Qin L, Guo J, Zheng Q and Zhang H: BAG2
structure, function and involvement in disease. Cell Mol Biol Lett.
21(18)2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ge F, Zhang L, Tao SC, Kitazato K, Zhang
ZP, Zhang XE and Bi LJ: Quantitative proteomic analysis of tumor
reversion in multiple myeloma cells. J Proteome Res. 10:845–855.
2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yang KM, Bae E, Ahn SG, Pang K, Park Y,
Park J, Lee J, Ooshima A, Park B, Kim J, et al: Co-chaperone BAG2
determines the pro-oncogenic role of cathepsin B in triple-negative
breast cancer cells. Cell Rep. 21:2952–2964. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang XY, Hong SS, Zhang M, Cai QQ, Zhang
MX and Xu CJ: Proteomic alterations of fibroblasts induced by
ovarian cancer cells reveal potential cancer targets. Neoplasma.
65:104–112. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yue X, Zhao Y, Liu J, Zhang C, Yu H, Wang
J, Zheng T, Liu L, Li J, Feng Z and Hu W: BAG2 promotes
tumorigenesis through enhancing mutant p53 protein levels and
function. Elife. 4(e08401)2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xia W, Cao G and Shao N: Progress in miRNA
target prediction and identification. Sci China C Life Sc.
52:1123–1130. 2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sun L, Chen G, Sun AQ, Wang Z, Huang H,
Gao Z, Liang W, Liu C and Li K: BAG2 promotes proliferation and
metastasis of gastric cancer via ERK1/2 signaling and partially
regulated by miR186. Front Onco. 10(31)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Selmansberger M, Feuchtinger A, Zurnadzhy
L, Michna A, Kaiser JC, Abend M, Brenner A, Bogdanova T, Walch A,
Unger K, et al: CLIP2 as radiation biomarker in papillary thyroid
carcinoma. Oncogene. 34:3917–3925. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ge Y, Yan D, Deng H, Chen W and An G:
Novel molecular regulators of tumor necrosis factor-related
apoptosis-inducing ligand (TRAIL)-induced apoptosis in NSCLC cells.
Clin Lab. 61:1855–1863. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jamali L, Tofigh R, Tutunchi S, Panahi G,
Borhani F, Akhavan S, Nourmohammadi P, Ghaderian SMH, Rasouli M and
Mirzaei H: Circulating microRNAs as diagnostic and therapeutic
biomarkers in gastric and esophageal cancers. J Cell Physiol.
233:8538–8550. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kwan JY, Psarianos P, Bruce JP, Yip KW and
Liu FF: The complexity of microRNAs in human cancer. J Radiat Res.
57 (Suppl):106–111. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
D'Angelo B, Benedetti E, Cimini A and
Giordano A: MicroRNAs: A puzzling tool in cancer diagnostics and
therapy. Anticancer Res. 36:5571–5575. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Pehlivan KC, Duncan BB and Lee DW: CAR-T
cell therapy for acute lymphoblastic leukemia: Transforming the
treatment of relapsed and refractory disease. Curr Hematol Malig
Rep. 13:396–406. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Vairy S, Garcia JL, Teira P and
Bittencourt H: CTL019 (tisagenlecleucel): CAR-T therapy for
relapsed and refractory B-cell acute lymphoblastic leukemia. Drug
Des Devel Ther. 12:3885–3898. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lv M, Zhang X, Jia H, Li D, Zhang B, Zhang
H, Hong M, Jiang T, Jiang Q, Lu J, et al: An oncogenic role of
miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by
targeting glucocorticoid receptor-a and cAMP/PKA pathways.
Leukemia. 26:769–777. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhang H, Ji W, Huang R, Li L, Wang X, Li
L, Fu X, Sun Z, Li Z, Chen Q and Zhang M: MicroRNA-155 is a
potential molecular marker of natural killer/T-cell lymphoma.
Oncotarget. 7:53808–53819. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Saki N, Abroun S, Soleimani M, Mortazavi
Y, Kaviani S and Arefian E: The roles of miR-146a in the
differentiation of Jurkat T-lymphoblasts. Hematology. 19:141–147.
2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Abe F, Kitadate A, Ikeda S, Yamashita J,
Nakanishi H, Takahashi N, Asaka C, Teshima K, Miyagaki T, Sugaya M
and Tagawa H: Histone deacetylase inhibitors inhibit metastasis by
restoring a tumor suppressive microRNA-150 in advanced cutaneous
T-cell lymphoma. Oncotarget. 8:7572–7585. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Takahashi RU, Prieto-Vila M, Hironaka A
and Ochiya T: The role of extracellular vesicle microRNAs in cancer
biology. Clin Chem Lab Med. 55:648–656. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Senkevitch E, Li W, Hixon JA, Andrews C,
Cramer SD, Pauly GT, Back T, Czarra K and Durum SK: Inhibiting
Janus Kinase 1 and BCL-2 to treat T cell acute lymphoblastic
leukemia with IL7-Rα mutations. Oncotarget. 9:22605–22617.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zadi Heydarabad M, Vatanmakanian M,
Abdolalizadeh J, Mohammadi H, Azimi A, Mousavi Ardehaie R,
Movasaghpour A and Farshdousti Hagh M: Apoptotic effect of
resveratrol on human T-ALL cell line CCRF-CEM is unlikely exerted
through alteration of BAX and BCL2 promoter methylation. J Cell
Biochem. 119:10033–10040. 2018.PubMed/NCBI View Article : Google Scholar
|