Introduction
Infantile hemangioma (IH) is the most common type of
soft tissue tumor in infants, with an incidence of 4-10% (1,2). The
growth of IH typically increases rapidly during the first three
months, particularly between the first five and eight weeks of
life, and ~80% of its maximum growth is completed by the age of
three months (3,4). Although most IHs resolve naturally
without treatment, 10-15% of cases have complications, such as
obstruction, ulceration or disfigurement, and require intervention.
IH results from angiogenesis and angiogenesis-related disorders.
However, the factors that trigger IH development have remained to
be elucidated. Therefore, studies investigating the molecular
mechanisms of the tumorigenesis and progression of IH are
required.
Circular RNA (circRNA) is a type of noncoding RNA
(ncRNA) composed of transcripts from exons. It was first discovered
by Sanger et al (5) in a
virus by using electron microscopy in 1976. Over the last few
years, circRNAs have been indicated to have multiple functions,
including acting as microRNA (miRNA/miR) sponges, influencing gene
expression by regulating splicing or transcription and interacting
with RNA-binding proteins (6-8).
An increasing number of studies have demonstrated the functions of
circRNAs in various diseases. Furthermore, circRNAs may serve as
prognostic and diagnostic biomarkers for various diseases (9). A previous study by our group, in which
the profiles of circRNAs in IH and adjacent normal tissues were
compared, revealed that circRNAs participate in angiogenesis and
vascular development-related biological processes (10). However, to the best of our
knowledge, plasma circRNAs have so far remained to be
systematically evaluated in IH. As circRNAs are excreted from cells
in exosomes, their recovery from the blood is a viable option.
The present study aimed to determine the circRNA
profiles of three pairs of plasma samples from patients with
proliferative IH and healthy control individuals using
high-throughput microarray and explore the roles of these circRNAs
in IH using bioinformatic methods. The results enhance the current
understanding of the roles of circRNAs in the pathogenesis of IH
and may aid in the discovery of blood-based biomarkers for
diagnosing IH.
Materials and methods
Patient samples
In the present study, the inclusion criteria were as
follows: Proliferative IH diagnosed by two independent doctors
according to clinical and pathological features (red soft lump that
appears within 3 months of birth and rapidly increases in size,
with an elevated skin temperature) (3) and Doppler ultrasound (frequently
subcutaneous, with variable echogenicity; intralesional vessels not
visible on gray-scale imaging; high vascular density on color
Doppler; increased echogenicity and decreased vascularity with
involution) (11). Furthermore,
only patients aged 3-6 months were considered to ensure that the
IHs were still in the proliferative phase. The exclusion criteria
were as follows: i) Administration of any treatment for IH prior to
participating in the study and ii) the presence of another disease.
Blood samples from a total of 17 patients with IH were collected at
the Department of Plastic Surgery of Shandong Provincial Hospital
(Jinan, China) between May 2018 and April 2019. In addition, blood
samples were collected from 17 normal age-matched healthy
volunteers. The volunteers came from the children in the physical
examination center of Shandong Provincial Hospital (Jinan, China)
and blood was collected with the consent of their
parents/guardians. Plasma was obtained from the blood samples via
centrifugation and stored at -80˚C. A total of three randomly
selected sample pairs were used for microarray analysis. The other
14 sample pairs were used for further validation of the microarray
results. The demographic characteristics of the cohort and the
clinicopathological features of the patients with IH are provided
in Table I.
 | Table IBaseline characteristics of patients
with IH and control subjects. |
Table I
Baseline characteristics of patients
with IH and control subjects.
| Characteristic | IH (n=17) | Control (n=17) |
|---|
| Sex | | |
|
Male | 6 (35.3) | 6 (35.3) |
|
Female | 11 (64.7) | 11 (64.7) |
| Age (months) | | |
|
<3 | 12 (70.6) | 11 (64.7) |
|
≥3 | 5 (29.4) | 6 (35.3) |
| History of
treatment | | |
|
Yes | 0 (0) | |
|
No | 17 (100.0) | |
| Location of IH | | |
|
Head, face,
neck | 6 (35.3) | |
|
Extremity | 3 (17.6) | |
|
Trunk | 6 (35.3) | |
|
Multiple
sites | 2 (11.8) | |
RNA isolation and quality control
RNA from each sample was isolated using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The purity and concentration of the
total RNA samples were determined using a NanoDrop ND-1000
spectrometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.). RNA integrity was assessed through electrophoresis on a
denaturing agarose gel.
Microarray
Sample labeling and array hybridization were
performed according to the manufacturer's protocol (Arraystar human
circular RNA v2; design ID 074301). In brief, total RNAs were
digested with RNase R (Epicentre; Illumina, Inc.) to remove linear
RNAs and enrich circRNAs. The enriched circRNAs were amplified and
transcribed into fluorescent complementary (c)RNAs using a random
priming method according to the Arraystar's protocol (Super RNA
Labeling Kit; Arraystar, Inc.). The labeled cRNAs were purified
using an RNeasy Mini Kit (Qiagen GmbH). The concentration and
specific activity of the labeled cRNAs (pmol Cy3/µg cRNA) were
measured using a NanoDrop ND-1000. Thereafter, 1 µg of each labeled
cRNA was fragmented by adding 5 µl of 10X blocking agent and 1 µl
of 25X fragmentation buffer, followed by heating at 60˚C for 30
min. Subsequently, 25 µl of 2X hybridization buffer was added to
dilute the labeled cRNA. Next, 50 µl of hybridization solution was
dispensed into the gasket slide and assembled on the circRNA
expression microarray slide. The slides were incubated for 17 h at
65˚C in an Agilent Hybridization Oven (Agilent Technologies, Inc.).
The hybridized arrays were washed, fixed and scanned using an
Agilent Scanner (G2505C; Agilent Technologies, Inc.).
Data analysis
Raw data were extracted using the Agilent Feature
Extraction software (version 11.0.1.1; Agilent Technologies, Inc.).
A series of data-processing steps, including quantile normalization
and low-intensity filtering, were performed using the limma package
of the R software (https://rstudio.com/). The fold-change (FC; i.e.,
ratio of group averages) in the plasma circRNA levels between
patients with IH and normal subjects was determined for each
circRNA. The statistical significance of the differences was
estimated using the Student's t-test. circRNAs with FCs≥1.5 and
P≤0.05 were considered to be significantly differentially
expressed.
Validation of microarray data using
reverse transcription-quantitative PCR (RT-qPCR)
The total RNA from the three paired plasma samples
of IH cases and healthy children used for microarray analysis was
isolated using the TRIzol reagent and reverse-transcribed (2X PCR
master mix; Arraystar, Inc.; 95˚C for 10 min, followed by 40 cycles
of 95˚C for 10 sec and 60˚C for 60 sec) to synthesize cDNA using a
GeneAmp PCR System 9700 (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the kit's protocols. GAPDH was
selected as the internal reference. All cDNAs were assembled in the
ViiA 7 real-time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) (2X Master Mix, 5 µl; 10 µM PCR specific primer
F, 0.5 µl; 10 µM PCR specific primer R, 0.5 µl; water was added to
create the total volume of 8 µl). The reaction was performed as
follows: 95˚C for 10 min, followed by 40 cycles of 95˚C for 10 sec
and 60˚C for 60 sec. All targets and references were evaluated in
triplicate wells. The relative expression levels of each target
circRNA were calculated using the 2-ΔΔCq method
(12). Primer sequences are
presented in Table II. The
identified circRNAs, which were consistent with the regulatory
direction of the microarray results and significantly
differentially expressed, were further verified in the remaining 14
sample pairs.
 | Table IISequences of primers used for
PCR. |
Table II
Sequences of primers used for
PCR.
| Gene name | Bidirectional
primer sequence | Annealing
temperature (˚C) | Product length
(bp) |
|---|
| GAPDH (human) | F:
5'GGGAAACTGTGGCGTGAT3' | | |
| | R:
5'GAGTGGGTGTCGCTGTTGA3' | 60 | 299 |
|
hsa_circRNA_001490 | F:
5'AACTCATACTATGGGTGGTGACTT3' | | |
| | R:
5'CACATATCCTATGTTCATCAATCTG3' | 60 | 93 |
|
hsa_circRNA_103372 | F:
5'AGAAGGCAGCCAACCAGAT3' | | |
| | R:
5'TTCAGATTTCTTTAGCCACTCAA3' | 60 | 120 |
|
hsa_circRNA_103361 | F:
5'TTTAACTAGCACTGCTTGTCGGA3' | | |
| | R:
5'ATCACCTTTGGCAGGATCTTCA3' | 60 | 84 |
|
hsa_circRNA_104310 | F:
5'GGACACGGTCTTTCTTATTCAGG3 ' | | |
| | R:
5'ACAGTGATGGTCGAAACGGTG3' | 60 | 63 |
|
hsa_circRNA_023016 | F:
5'TGTTGGATTTGGGAATGACCT3' | | |
| | R:
5'CCACACACATCCCACTACCCT3' | 60 | 147 |
|
hsa_circRNA_102101 | F:
5'TTGTTGGGACATGTATATTGTACAC3' | | |
| | R:
5'CTATATGCTTTATATGCCTTTCCTG3' | 60 | 93 |
|
hsa_circRNA_103546 | F:
5'GAGTGCAGCTCCCCCTATAAG3' | | |
| | R:
5'CCCTGTACTTTGAAAGCCTCTT3' | 60 | 151 |
|
hsa_circRNA_062683 | F:
5'GCTGTCCTCCACCATGAAGA 3' | | |
| | R:
5'CCAGGCTCACATCTCCCTT3' | 60 | 140 |
|
hsa_circRNA_103573 | F:
5'CGGTGTTGTTCTTTGCCATTT3' | | |
| | R:
5'TGCAGGGGAATGATGAATACAG3 ' | 60 | 96 |
|
hsa_circRNA_101566 | F:
5'TTATACCCAGTAAAGGAGAAAGC3' | | |
| | R:
5'CCTCAGAGTTTGAGAAATAACATAC3' | 60 | 91 |
Construction of circRNA-miRNA-mRNA
networks
The circRNA-targeted miRNAs were predicted using
Arraystar's homemade miRNA target prediction software (Arraystar,
Inc.) based on the data obtained from TargetScan, miRanda and
statistical analysis (methods listed in Data S1). The network was constructed
using Cytoscape software (version 2.8.3) (https://cytoscape.org/). Gene ontology (GO) term
analysis was performed using the Database for Annotation,
Visualization and Integrated Discovery (DAVID) tool (http://david.abcc.ncifcrf.gov/) to explore the
potential functions of the target genes in the categories
biological process, cellular component and molecular function. The
Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database
resource for understanding the high-level functions and effects of
a biological system. KEGG pathway enrichment analysis was performed
using the KEGG Orthology-Based Annotation System (KOBAS; http://kobas.cbi.pku.edu.cn/).
Statistical analysis
All data were expressed as the mean ± standard error
of the mean or n (%) and analyzed using the Student's t-test with
SPSS 22.0 software (IBM Corp.). GraphPad Prism 8 (GraphPad
Software, Inc.) and Microsoft Excel 2019 (Microsoft Corp.) were
used for other statistical analyses. P<0.05 was
considered to indicate statistical significance.
Results
Demographic characteristics of
patients and volunteers
In the present study, 17 patients with IH (6 males
and 11 females) and 17 age-matched healthy volunteers (6 males and
11 females) were collected. Among the patients with IH, 12 cases
were <3 months old and 5 cases were ≥3 months old. Among the
volunteers, 11 cases were <3 months old and 6 were ≥3 months
old. The sex and age distribution of patients and volunteers were
not statistically different.
Differentially expressed circRNAs
detected using microarray analysis
A microarray analysis of 13,617 circRNAs in the
plasma of patients with IH and healthy controls was performed.
Relevant data were uploaded to the Gene Expression Omnibus (GEO)
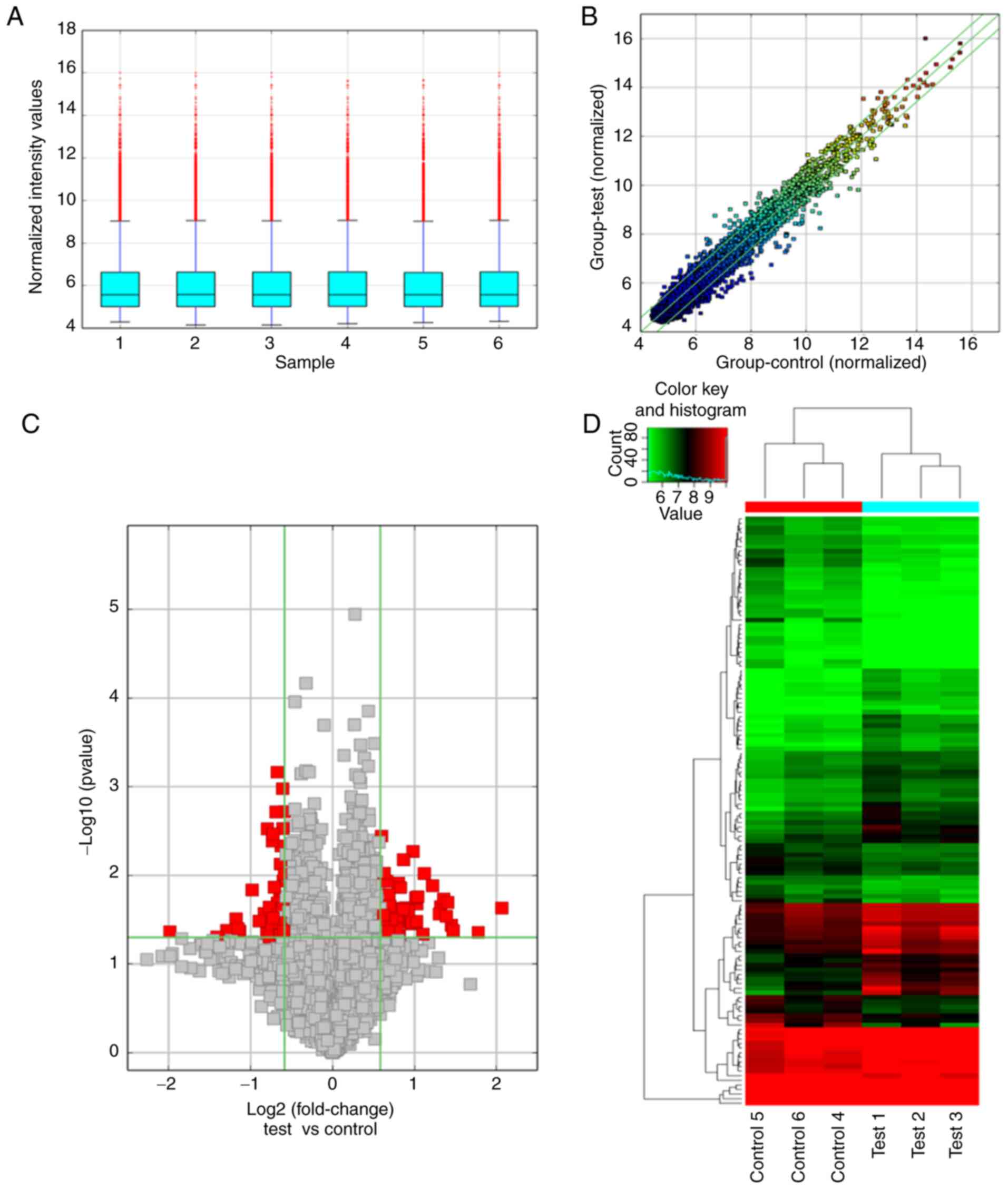
database (accession no. GSE162905). A box plot was used to indicate
the intensity values of the six samples after normalization
(Fig. 1A). The scatter plot
(Fig. 1B) and volcano plot
(Fig. 1C) revealed differences in
circRNA expression between the test and control groups. Finally,
data analysis and screening based on FC>1.5 and P<0.05
revealed 72 upregulated and 56 downregulated circRNAs. Among the
upregulated circRNAs, 48 were exonic, 15 were intronic, 5 were
sense overlapping, 3 were intergenic and 1 was antisense. The
downregulated circRNAs included 51 exonic, 2 intronic and 3 sense
overlapping circRNAs. In addition, the distribution of these
circRNAs on chromosomes was summarized, indicating that they were
distributed on each chromosome. The distinct differential
expression of these 128 circRNAs was visualized by hierarchical
clustering in the heat map (Fig.
1D). These results indicated that the present differential
expression profile was reliable.
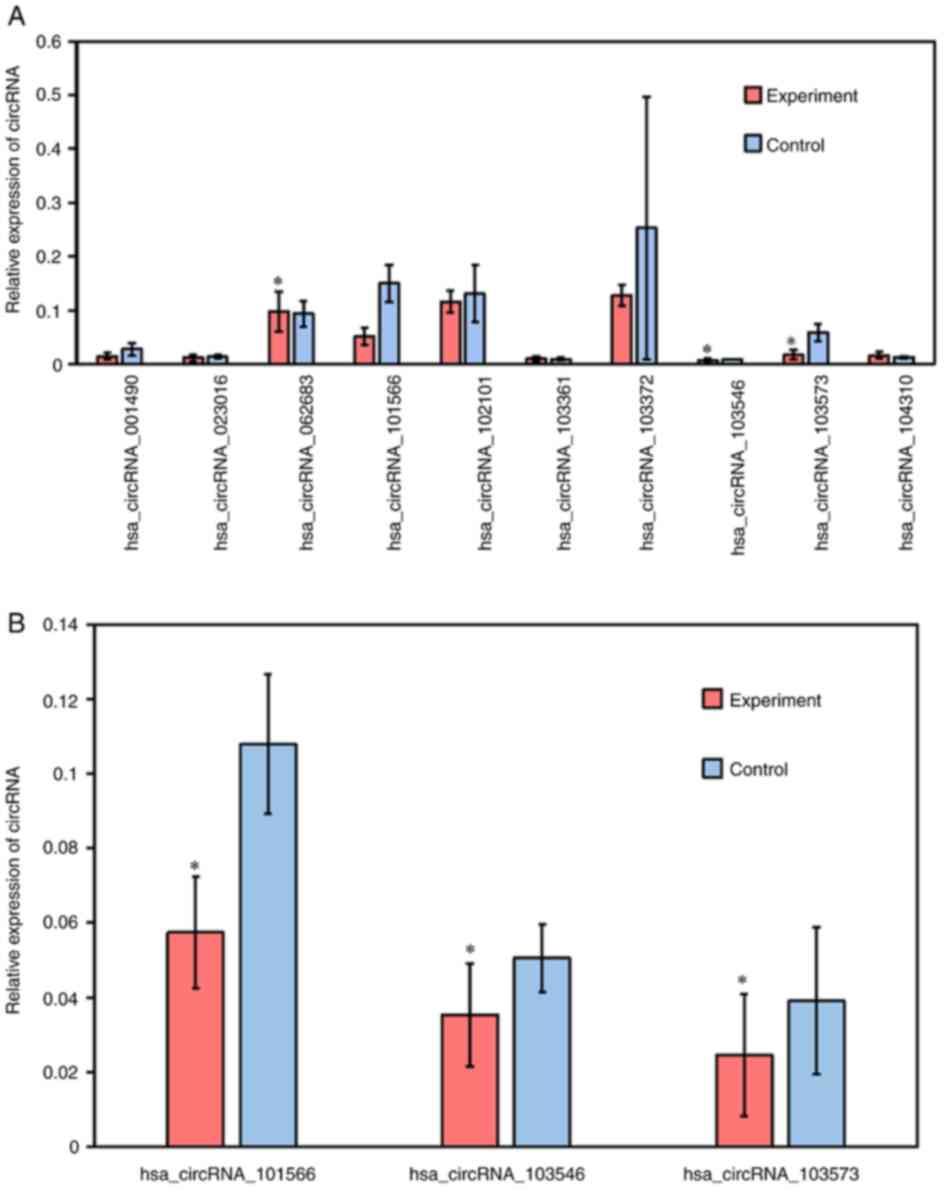
RT-qPCR verification of the selected
differentially expressed circRNAs
Based on known data of microarray analysis, 10
circRNAs we selected for RT-qPCR validation, including 6
upregulated and 4 downregulated circRNAs that were exonic
(FC>1.5, P<0.05, raw intensity of the samples were >100
and the number of bases was 200-3,000; Table III). The original signal intensity
was high and the number of bases was 200-3,000. The results
suggested that the three downregulated circRNAs [Homo
sapiens (hsa)_circRNA_101566, hsa_circRNA_103546 and
hsa_circRNA_103573] were significantly differentially expressed
(Fig. 2A), consistent with the
microarray results. However, no upregulated circRNAs were detected.
Analysis of another seven samples revealed similar expression
levels or no significant differences between patients with IH and
healthy children. The three circRNAs were further verified using
the remaining 14 sample pairs and significantly different
expression levels were detected (Fig.
2B).
 | Table IIISelected differentially expressed
circRNAs. |
Table III
Selected differentially expressed
circRNAs.
| circRNA | P-value | Absolute FC | Direction of
regulation | Chromosome | circRNA type |
|---|
|
hsa_circRNA_001490 | 0.023331583 | 4.1779908 | Up | 5 | Exonic |
|
hsa_circRNA_103372 | 0.044010096 | 3.4234124 | Up | 3 | Exonic |
|
hsa_circRNA_103361 | 0.036601471 | 2.7282583 | Up | 3 | Exonic |
|
hsa_circRNA_104310 | 0.020127023 | 2.6489823 | Up | 7 | Exonic |
|
hsa_circRNA_062683 | 0.045776034 | 2.1446458 | Up | 22 | Exonic |
|
hsa_circRNA_023016 | 0.017327291 | 2.0521739 | Up | 11 | Exonic |
|
hsa_circRNA_102101 | 0.043224524 | 3.961216 | Down | 17 | Exonic |
|
hsa_circRNA_103546 | 0.039832007 | 2.2214431 | Down | 3 | Exonic |
|
hsa_circRNA_103573 | 0.032726148 | 1.8586376 | Down | 3 | Exonic |
|
hsa_circRNA_101566 | 0.022844096 | 1.703774 | Down | 15 | Exonic |
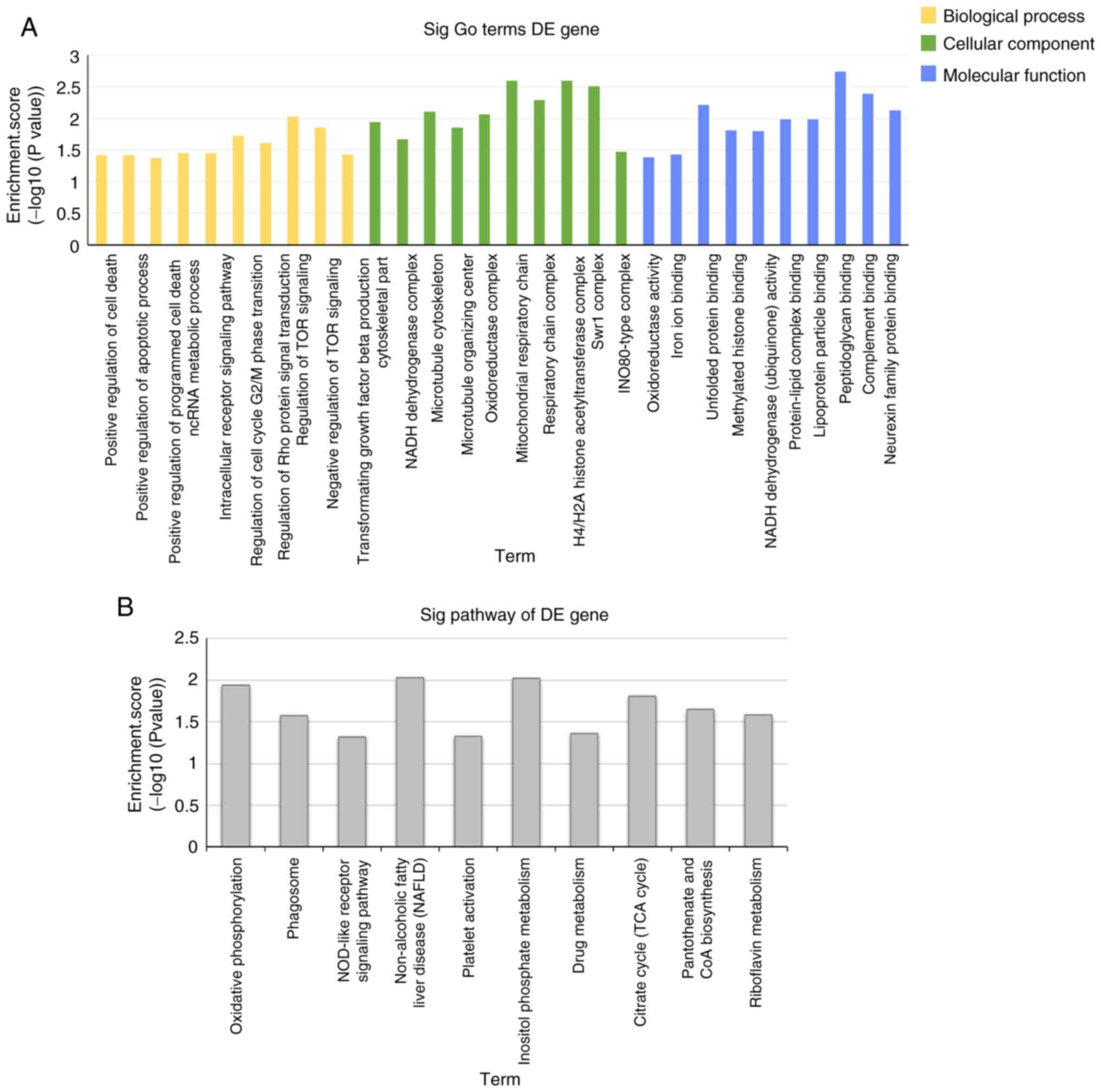
MiRNA prediction and bioinformatics
analysis of circRNAs
Based on the results of RT-qPCR, the miRNA response
elements of the circRNAs were predicted using the Arraystar miRNA
target prediction software. To further investigate the functions of
these target genes, GO and KEGG analyses of the differentially
expressed circRNAs were performed using DAVID and KOBAS. Based on
the RT-qPCR results, three downregulated circRNAs
(hsa_circRNA_101566, hsa_circRNA_103546 and hsa_circRNA_103573)
were selected for further analysis. The top 10 significant pathways
in the 3 categories of the GO analysis (biological process,
molecular function and cellular component), including positive
regulation of cell death, regulation of TOR signaling and ncRNA
metabolic process, are presented in Fig. 3A. The KEGG analysis revealed the top
10 pathways associated with the downregulated circRNAs (Fig. 3B), including oxidative
phosphorylation, citrate cycle, CoA biosynthesis pathway and
NOD-like receptor signaling pathway. These pathways have a vital
role in the development of angiogenesis and vascular diseases.
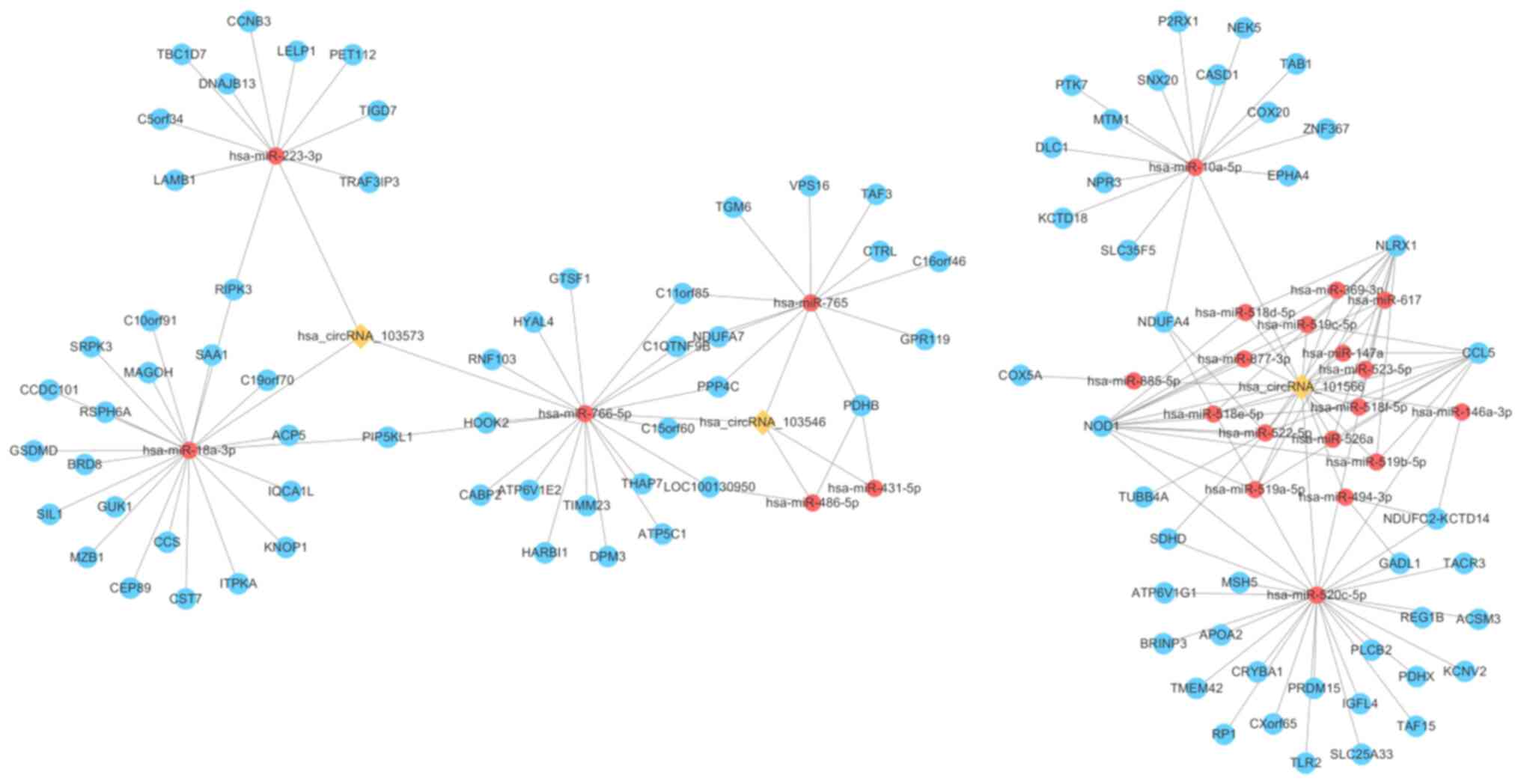
Construction of the circRNA-miRNA-mRNA
network
To further evaluate the relationship between the
confirmed differentially expressed circRNAs and the corresponding
miRNAs and to investigate the mechanisms underlying IH development,
Cytoscape was used to illustrate the
hsa_circRNA_101566/hsa_circRNA_103546/hsa_circRNA_103573-miRNA-mRNA
network (Fig. 4). The number of
genes targeted by hsa_circRNA_101566 was the highest among all
circRNAs evaluated (Fig. 4). As the
amount of raw data was large, miRNAs related to angiogenesis and
partially related genes were selected (referring to the increase of
other tumors' angiogenesis while promoting the growth of other
tumors, so its influence on blood vessels may be indirectly
regulated), including hsa_miR_765, hsa_miR_486, hsa_miR_18a,
hsa_miR_10a, hsa_miR_223 and hsa_miR_522 (13-20).
The remaining targets are listed in Table SI. It was indicated that
hsa_circRNA_101566, hsa_circRNA_103546 and hsa_circRNA_103573
contain 18, 4 and 3 potential miRNA response elements, respectively
(Fig. 4).
Discussion
To the best of our knowledge, the present study was
the first study to detect and profile circRNAs in the plasma of
patients with IH. The results of the present study revealed that
the downregulated circRNAs, namely hsa_circRNA_101566,
hsa_circRNA_103546 and hsa_circRNA_103573, targeted at least 25
different miRNAs. In recent years, the role of circRNAs in vascular
diseases has been reported (21,22)
and certain miRNAs have been confirmed to be involved in the
pathogenesis of IH (23-25).
In the present study, three pairs of plasma specimens were first
analyzed using a microarray. A total of 72 upregulated and 56
downregulated circRNAs (FC≥1.5, P<0.05) were identified.
Previous studies have analyzed circRNA expression in IH tumor
tissues using microarray (10) and
RNA-seq (26); however, circRNA
expression in plasma samples from IH patients has not been
previously examined. Li et al (27), by comparing a large cohort,
determined that microarray is more sensitive and efficient than RNA
sequencing (RNA-seq). In addition, certain studies have suggested
that plasma samples from cancer patients produce even more accurate
results compared with those obtained using tissue samples to study
circRNAs for diagnostic applications (28,29).
In the present study, a preliminary analysis of the
128differentially expressed circRNAs screened was performed and 6
upregulated and 4 downregulated circRNAs were selected for
validation using RT-qPCR, including the 3 downregulated circRNAs
hsa_circRNA_101566, hsa_circRNA_103546 and hsa_circRNA_103573. The
results were consistent with the microarray results. Studies on
circRNAs involved in IH were first performed by our group in
2017(10), in which microarray
analysis detected 234 upregulated and 374 downregulated circRNAs
from four pairs of tumor tissue. Subsequently, Li et al
(26) identified 124 upregulated
and 125 downregulated circRNAs using RNA-seq analysis of three
pairs of tumor tissue. However, the circRNAs validated by the two
groups are not the same and exhibited less overlap in the
experimental results. Of the circRNAs screened for statistically
significant differences in the present study, only one
(hsa_circRNA_104310) was mentioned in the study by Fu et al
(10). The reasons for this were
further analyzed. First, as only three or four sample pairs were
used for the analysis in the three studies, this led to sample
errors of the microarray or RNA-seq results. Furthermore, the
circRNAs validated by Li et al (26) were randomly selected and it was not
based on any specific strategy to select the most relevant circRNAs
in their methods. Generally, it is best to choose the most distinct
circRNAs or the circRNAs that may serve an important role in the
regulatory network. Finally, while the present study was the first
to analyze circRNAs in plasma from pediatric patients with IH,
previous studies have analyzed circRNAs in tumor samples. There is
likely a difference in circRNA expression between plasma and tumor
tissue and this difference should be further explored. These
results suggest that the use of different analytical methods and
different samples may yield different results and that the
development of IH may involve different regulatory pathways in
tumor tissue and plasma; these results are consistent with the
cell- and tissue-specific expression of circRNAs (30). Therefore, for circRNA investigations
in IH, large samples and multicenter studies are required. In
addition, the lack of comparison of tumor and plasma samples from
the same patient is also a limitation of the present study. In the
future, both tumor and plasma specimens from patients will be
collected for validation to obtain more convincing results.
A previous study indicated that hsa_miR_520c
expression was significantly upregulated in IHs, suggesting that
this miRNA is important in IHs (13), whereas bioinformatics analysis
(using TargetScan and miRanda) suggested that hsa_circRNA_101566
and hsa_circRNA_103546 are ‘miRNA sponges’ of hsa_miR_520c.
Therefore, targeting of hsa_miR_520c by hsa_circRNA_101566 and
hsa_circRNA_103546 may be involved in IH development. Furthermore,
certain circRNA-targeted miRNAs (hsa_miR_765, hsa_miR_486,
hsa_miR_18a, hsa_miR_10a, hsa_miR_223 and hsa_miR_522) have been
indicated to participate in promoting angiogenesis or
inducing tumor cell proliferation (14-20)
and may be related to angiogenesis and angiogenesis-related
disorders in IHs.
GO is a bioinformatics tool originally designed to
unify the representation of genes and gene products of various
species (31). The ontology
includes three categories: Cellular component, molecular function
and biological process. GO analysis of these target genes, which
indirectly regulate the three downregulated circRNAs, revealed
their involvement in physiological function that are closely
related to the development of IH. Yang et al (32) demonstrated that ras homolog family
member A (RhoA), indirectly controlled by hsa_circRNA_101566, is
able to regulate cellular apoptosis. In the absence of RhoA, the
apoptosis of human umbilical vein endothelial cells induced by
TNF-α was inhibited. Conversely, Del Re et al (33) reported that RhoA/Rho kinase
activation induces a mitochondrial death pathway and cardiomyocyte
apoptosis, which may be correlated with the proliferation of
hemangioma-derived endothelial cells during IH proliferation. Of
note, this study confirmed that TOR has an important role in the
vital activity of cells, while it has been indicated that mammalian
target of rapamycin (mTOR) is present in humans. Medici and Olsen
(34) demonstrated that rapamycin
inhibits the proliferation of hemangioma-derived endothelial cells
by decreasing hypoxia-inducible factor-1 (HIF-1) expression in
endothelial cells and that the mTOR-related signaling pathway may
be one of the mechanisms of IH pathogenesis. Thus, rapamycin is
able to inhibit angiogenesis as an mTOR inhibitor and mTOR-related
signaling pathways may contribute to the pathogenesis of IH.
Certain target genes of hsa_circRNA_101566 are involved in the
negative regulation of mTOR signaling, suggesting that mTOR
signaling is upregulated and may contribute to proliferation in
IH.
KEGG is a knowledge database for systematic analysis
of gene function, linking genomic information with higher-order
functional information (35). KEGG
analysis suggested that pathways, such as the pantothenate and CoA
biosynthesis pathways and the NOD-like receptor and citrate cycle
pathways, are influenced by the differential expression of the
circRNAs. Succinate dehydrogenase (SDH) in the citrate cycle
consists of four subunits (36),
namely SDHA, SDHB, SDHC and SDHD. The downregulated
hsa_circRNA_103546 is able to indirectly regulate the low
expression of SDHD. Selak et al (37) reported that the accumulation of
succinate due to SDH inhibition led to activation of HIF-1α, which
may also be involved in the pathogenesis of IH. In addition,
hsa_circRNA_101566 indirectly regulates NOD-like receptor family
member X1 (NLRX1), which may be regulated by miR-520c involved in
the NOD-like receptor signaling pathway (38), whereas NLRX1 is able to inhibit
NF-κB and thus indirectly inhibit vascular endothelial growth
factor (VEGF) (39). It was
previously suggested that VEGF contributes to IH development
(40) and its downregulation
weakens the inhibitory effect on VEGF, which may also be a
promising finding in IH. The hypoxia theory, one of the most widely
accepted theories regarding the pathogenesis of IHs, states that
hypoxic stress triggers the overexpression of angiogenic factors,
such as VEGF, via the HIF-1α pathway (41,42).
In support of this, the present study hypothesized that the
hsa_circRNA_101566/hsa-miR-520c-5p/NLRX1 signaling axis has a role
in the molecular mechanisms of IH development. However, further
studies are required to determine the function of circRNAs and
circRNA-miRNA-mRNA networks in IH.
In conclusion, the present study was the first, to
the best of our knowledge, to detect and profile circRNAs in the
plasma of patients with IH. A total of 128 differentially expressed
circRNAs, among which 72 were upregulated and 56 downregulated,
were screened. Furthermore, three downregulated circRNAs
(hsa_circRNA_101566, hsa_circRNA_103546, and hsa_circRNA_103573)
were verified using RT-qPCR. The roles of differentially expressed
circRNAs were explored using GO and KEGG pathway analyses. It was
determined that hsa_circRNA_101566, which is able to regulate the
mTOR signaling pathway, may be an important regulatory molecule in
IH development and that the VEGF signaling pathway may be regulated
indirectly by targeting hsa_miR_520c. However, a limitation of the
present study was the small number of samples used; further larger
studies are required in order to determine whether these circRNAs
may be used as biomarkers. Furthermore, functional verification of
the roles of the relevant RNA molecules in vitro and in
vivo is required. These studies may provide novel ideas for
accurate diagnosis and a better understanding of the pathogenesis
of IH.
Supplementary Material
Supplementary Data
The information of the differentially
expressed circRNAs and the miRNAs interacting with the
circRNAs
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the National Natural Science
Foundation of China (grant no. 81873938).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the Gene Expression Omnibus
repository (https://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
JB was involved with conceptualization and design of
the study, study supervision and statistical analysis. ZL, YC
performed experimental operations, statistical analysis, data
analysis and manuscript writing. ZZ and XL were involved with
acquisition of data. RH helped conceptualize design and supervise
the study. JB and RH checked and approved the authenticity of the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
statement
This study was approved by the Ethics Committee of
Shandong Provincial Hospital (Jinan, China; no. 2018-051) and
performed according to the Ethical Guidelines of the Declaration of
Helsinki. Written informed consent was obtained from the guardian
of each subject.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hoornweg MJ, Smeulders MJC, Ubbink DT and
van der Horst CM: The prevalence and risk factors of infantile
haemangiomas: A case-control study in the Dutch population.
Paediatr Perinat Epidemiol. 26:156–162. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kilcline C and Frieden IJ: Infantile
hemangiomas: How common are they? A systematic review of the
medical literature. Pediatr Dermatol. 25:168–173. 2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Tollefson MM and Frieden IJ: Early growth
of infantile hemangiomas: What parents' photographs tell us.
Pediatrics. 130:e314–e320. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chang LC, Haggstrom AN, Drolet BA, Baselga
E, Chamlin SL, Garzon MC, Horii KA, Lucky AW, Mancini AJ, Metry DW,
et al: Growth characteristics of infantile hemangiomas:
Implications for management. Pediatrics. 122:360–367.
2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73(3852e3856)1976.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and characterizing circRNA-protein
interaction. Theranostics. 7:4183–4191. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Fu C, Lv R, Xu G, Zhang L, Bi J, Lin L,
Liu X and Huo R: Circular RNA profile of infantile hemangioma by
microarray analysis. PLoS One. 12(e0187581)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Johnson CM and Navarro OM: Clinical and
sonographic features of pediatric soft-tissue vascular anomalies
part 1: Classification, sonographic approach and vascular tumors.
Pediatr Radiol. 47:1184–1195. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Strub GM, Kirsh AL, Whipple ME, Kuo WP,
Keller RB, Kapur RP, Majesky MW and Perkins JA: Endothelial and
circulating C19MC microRNAs are biomarkers of infantile hemangioma.
JCI Insight. 1(e88856)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xie BH, He X, Hua RX, Zhang B, Tan GS,
Xiong SQ, Liu LS, Chen W, Yang JY, Wang XN and Li HP: Mir-765
promotes cell proliferation by downregulating INPP4B expression in
human hepatocellular carcinoma. Cancer Biomark. 16:405–413.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Shi XF, Wang H, Xiao FJ, Yin Y, Xu QQ, Ge
RL and Wang LS: MiRNA-486 regulates angiogenic activity and
survival of mesenchymal stem cells under hypoxia through modulating
Akt signal. Biochem Biophys Res Commun. 70:670–677. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yang Q, Wang X, Cui J, Wang P, Xiong M,
Jia C, Liu L, Ning B, Li L, Wang W, et al: Bidirectional regulation
of angiogenesis and miR-18a expression by PNS in the mouse model of
tumor complicated by myocardial ischemia. BMC Complement Altern
Med. 14(183)2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fei D, Zhang X, Liu J, Tan L, Xing J, Zhao
D and Zhang Y: Long noncoding RNA FER1L4 suppresses tumorigenesis
by regulating the expression of PTEN targeting miR-18a-5p in
osteosarcoma. Cell Physiol Biochem. 51:1364–1375. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang X, Ling CC, Li L, Qin Y, Qi J, Liu X,
You B, Shi Y, Zhang J, Jiang Q, et al: MicroRNA-10a/10b represses a
novel target gene mib1 to regulate angiogenesis. Cardiovasc Res.
110:140–150. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Henn D, Abu-Halima M, Wermke D, Falkner F,
Thomas B, Köpple C, Ludwig N, Schulte M, Brockmann MA, Kim YJ, et
al: MicroRNA-regulated pathways of flow-stimulated angiogenesis and
vascular remodeling in vivo. J Transl Med. 17(22)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hao Z, Chao Y, Meiyuan C, Zhu L, Se T,
Jianxin J and Sun C: MiR-522 contributes to cell proliferation of
hepatocellular carcinoma by targeting DKK1 and SFRP2. Tumor Biol.
37:11321–11329. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li CY, Ma L and Yu B: Circular RNA
hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells
proliferation and angiogenesis. Biomed Pharmacother. 95:1514–1519.
2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Boeckel JN, Jaé N, Heumüller AW, Chen W,
Boon RA, Stellos K, Zeiher AM, John D, Uchida S and Dimmeler S:
Identification and characterization of hypoxia-regulated
endothelial circular RNA. Circ Res. 117:884–890. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bertoni N, Pereira LM, Severino FE, Moura
R, Yoshida WB and Reis PP: Integrative meta-analysis identifies
microRNA-regulated networks in infantile hemangioma. BMC Med Genet.
17(4)2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li D, Li P, Guo Z, Wang H and Pan W:
Downregulation of miR-382 by propranolol inhibits the progression
of infantile hemangioma via the PTEN-mediated AKT/mTOR pathway. Int
J Mol Med. 39:757–763. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li J, Li Q, Chen L, Gao Y, Zhou B and Li
J: Competitive endogenous RNA networks: Integrated analysis of
non-coding RNA and mRNA expression profiles in infantile
hemangioma. Oncotarget. 9:11948–11963. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li J, Li Q, Chen L, Gao Y and Li J:
Expression profile of circular RNAs in infantile hemangioma
detected by RNA-Seq. Medicine (Baltimore).
97(e10882)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li S, Teng S, Xu J, Su G, Zhang Y, Zhao J,
Zhang S, Wang H, Qin W, Lu ZJ, et al: Microarray is an efficient
tool for circRNA profiling. Brief Bioinform. 20:1420–1433.
2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tan H, Gan L, Fan XM, Liu L and Liu S:
Diagnostic value of circular RNAs as effective biomarkers for
cancer: A systematic review and meta-analysis. Oncotargets Ther.
12:2623–2633. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li J, Li H, Lv XT, Yang Z, Gao M, Bi Y,
Zhang Z, Wang S, Cui Z, Zhou B and Yin Z: Diagnostic performance of
circular RNAs in human cancers: A systematic review and
meta-analysis. Mol Genet Genom Med. 7(e00749)2019.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Chen B and Huang S: Circular RNA: An
emerging non-coding RNA as a regulator and biomarker in cancer.
Cancer Lett. 418:41–50. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yuan F, Pan X, Chen L, Zhang YH, Huang T
and Cai YD: Analysis of protein-protein functional associations by
using gene ontology and KEGG pathway. Biomed Res Int.
2019(4963289)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yang L, Tang L, Dai F, Meng G, Yin R, Xu X
and Yao W: Raf-1/CK2 and RhoA/ROCK signaling promote TNF-α-mediated
endothelial apoptosis via regulating vimentin cytoskeleton.
Toxicology. 389:74–84. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Del Re DP, Miyamoto S and Brown JH:
RhoA/Rho kinase up-regulate Bax to activate a mitochondrial death
pathway and induce cardiomyocyte apoptosis. J Biol Chem.
282:8069–8078. 2007.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Medici D and Olsen BR: Rapamycin inhibits
proliferation of hemangioma endothelial cells by reducing
HIF-1-dependent expression of VEGF. PLoS One.
7(e42913)2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Moosavi B, Zhu XL, Yang WC and Yang GF:
Molecular pathogenesis of tumorigenesis caused by succinate
dehydrogenase defect. Eur J Cell Biol. 99(151057)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Selak MA, Armour SM, MacKenzie ED,
Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB
and Gottlieb E: Succinate links TCA cycle dysfunction to
oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer
Cell. 7:77–85. 2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Liu Y, Tang N, Cao K, Wang S, Tang S, Su H
and Zhou J: Negative-pressure wound therapy promotes wound healing
by enhancing angiogenesis through suppression of NLRX1 via miR-195
upregulation. Int J Low Extrem Wounds. 17:144–150. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Xu W, Li S, Yu F, Zhang Y, Yang X, An W,
Wang W and Sun C: Role of Thrombospondin-1 and Nuclear Factor-κB
signaling pathways in antiangiogenesis of infantile hemangioma.
Plast Reconstr Surg. 142:310e–321e. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Przewratil P, Sitkiewicz A and
Andrzejewska E: Local serum levels of vascular endothelial growth
factor in infantile hemangioma: Intriguing mechanism of endothelial
growth. Cytokine. 49:141–147. 2010.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Drolet BA and Frieden IJ: Characteristics
of infantile hemangiomas as clues to pathogenesis: Does hypoxia
connect the dots? Arch Dermatol. 146:1295–1299. 2010.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Leaute-Labreze C, Prey S and Ezzedine K:
Infantile haemangioma: Part I. Pathophysiology, epidemiology,
clinical features, life cycle and associated structural
abnormalities. J Eur Acad Dermatol Venereol. 25:1245–1253.
2011.PubMed/NCBI View Article : Google Scholar
|