Introduction
Osteosarcoma (OS) is the most common malignant bone
tumor in teenagers and often occurs during rapid-growth of the
metaphysis of long bones. Its clinical incidence is rather high,
accounting for 60% of malignant bone tumors in adolescents
(1,2). Previous trauma history, genetic
factors, virus infection and radioactive stimulation are all
closely related to the etiology of OS (3). Due to the high malignancy, rapid
growth and invasiveness and early metastasis of OS, most OS
patients are diagnosed at an advanced stage (4). It is therefore necessary to
investigate the molecular mechanism underlying OS pathogenesis and
explore potentially valuable biomarkers and therapeutic targets for
OS treatment.
Long non-coding RNA (lncRNA) is a form of non-coding
RNA with a length of ~200 nucleotides in eukaryotic cells. lncRNA
plays an important role in genomic imprinting, chromatin
modification, transcriptional activation and intranuclear
transport, as well as in the proliferation, apoptosis, migration
and invasion of cancer cells (5,6).
Accumulating evidence suggests that many lncRNAs participate in the
tumorigenesis and progression of multiple cancers, including OS.
lncRNA HOXA11-AS promotes the proliferation and invasion of gastric
cancer by scaffolding the chromatin modification factors polycomb
repressive complex 2, lysine-specific histone demethylase 1A and
DNA (cytosine-5)-methyltransferase 1(7). lncRNA SNHG1 acts on microRNA
(miR)-326, regulating the expression of RNA binding protein NOB1
and promoting the tumorigenesis of OS (8). Highly conserved and enriched in many
types of cells, non-coding RNA activated by DNA damage (NORAD) is
an lncRNA which plays an important role in cancer biology (9). As a novel competitive endogenous RNA
(ceRNA), NORAD can enhance hypoxia-induced epithelial-mesenchymal
transition by targeting miR-125a-3p, thereby promoting the
metastasis of pancreatic cancer (10). However, the role and mechanism of
NORAD in OS have not been fully elucidated.
miRs are a class of small non-coding RNAs consisting
of ~18-22 nucleotides. miRs can inhibit the translation or regulate
the degradation of their target mRNAs by binding to their
3'-untraslation regions (3'-UTRs) (11). miRs also participate in many
cellular processes, including the differentiation, proliferation,
metastasis and apoptosis of cancer cells (11-14).
miR-486 inhibits the progression of OS by regulating the protein
kinase C-δ pathway (15). The
down-regulation of miR-664a disrupts the migration of OS cells via
the regulation of maternally expressed 3(16) and miR-491 inhibits lung metastasis
and chemotherapy resistance of OS by targeting αB-crystallin
(17).
miR-155 plays an important role in the progression
of multiple types of cancers and other human diseases. It has been
suggested that macrophage-derived exosomes containing miR-155
inhibit the proliferation of fibroblasts and promote fibroblast
inflammation in myocardial injury (18). Down-regulation of miR-155 can
inhibit the anus kinase-signal transducer and activator of
transcription 3 pathway by up-regulating the expression of
suppressor of cytokine signaling 3, thus suppressing the
proliferation and promoting the apoptosis of lymphoma cells
(19). miR-155 can also regulate
the proliferation, invasion and apoptosis of renal cancer cells via
regulation of the GSK-3β/β-catenin pathway (20). The function of miR-155 in OS has
also been reported. Specifically, through the regulation of the
phosphatase and tensin homolog deleted on chromosome
ten/phosphoinositide 3-kinase/AKT/mechanistic target of rapamycin
pathway, miR-155 can modulate doxorubicin-induced autophagy in OS
cells (21). However, to the best
of our knowledge, no previous studies have explored the mechanism
of miR-155 dysregulation in OS.
Through the Starbase database, a potential binding
site between NORAD and miR-155-5p was identified. As NORAD has been
reported to be an oncogenic lncRNA in several cancers (9,10), it
was hypothesized that NORAD could promote OS progression by
regulating miR-155-5p. In the present study, the expression and
biological functions of NORAD and miR-155-5p were investigated in
OS. Additionally, NORAD was validated to contain a conserved target
site of miR-155-5p. Based on this discovery, it was concluded that
NORAD could regulate the proliferation, migration and invasion of
OS cells by targeting miR-155-5p.
Materials and methods
Sample collection
OS tissues and paired adjacent tissues were
surgically removed from patients (aged 7-35 years old; mean age,
25.2 years old) in Zhongnan Hospital of Wuhan University between
July 2016 and November 2018. The collection and use of samples was
approved by the Ethics Committee of Zhongnan Hospital and the
Ethics Review Board of Ezhou Central Hospital. Patients or the
parents of patients that were minors provided their written
informed consent prior to study commencement. The tissue samples
were obtained during surgery and the surgery was a part of
comprehensive treatment program that aimed to prolong the prognosis
of the patients. No patients had received radiotherapy or
chemotherapy before surgery. The detailed parameters of all
patients are shown in Tables I and
II. All samples were confirmed as
OS through clinical, imaging and histological methods (22). After collection, the samples were
immediately stored at -80˚C until use. To facilitate subsequent
analyses, the above 30 patients were divided into a low expression
group (n=15) and a high expression group (n=15) according to the
cutoff value of 2.72 for NORAD expression. in their tumor
tissues.
 | Table ICorrelation between NORAD levels and
clinical features in osteosarcoma patients. |
Table I
Correlation between NORAD levels and
clinical features in osteosarcoma patients.
| Parameter | Total number | Low NORAD
expression | High NORAD
expression | χ2 | P-value |
|---|
| Sex | | | | 0.5357 | 0.4642 |
|
Male | 16 | 7 | 9 | | |
|
Female | 14 | 8 | 6 | | |
| Age (years) | | | | 0.1587 | 0.6903 |
|
<20 | 21 | 10 | 11 | | |
|
≥20 | 9 | 5 | 4 | | |
| Histological
classification | | | | 2.4000 | 0.3012 |
|
Osteoblastic | 15 | 6 | 9 | | |
|
Chondroblastic | 10 | 5 | 5 | | |
|
Fibroblastic | 5 | 4 | 1 | | |
| Tumor grade | | | | 7.0335 | 0.0080a |
|
Low | 11 | 9 | 2 | | |
|
High | 19 | 6 | 13 | | |
| Enneking stage | | | | 6.7857 | 0.0336a |
|
I | 8 | 7 | 1 | | |
|
II | 14 | 6 | 8 | | |
|
III | 8 | 2 | 6 | | |
| Tumor size
(cm) | | | | 4.8214 | 0.0281a |
|
<8 | 16 | 11 | 5 | | |
|
≥8 | 14 | 4 | 10 | | |
| Metastasis | | | | 9.6000 | 0.0019a |
|
No | 10 | 9 | 1 | | |
|
Yes | 20 | 6 | 14 | | |
 | Table IICorrelation between miR-155-5p levels
and clinical features in osteosarcoma patients. |
Table II
Correlation between miR-155-5p levels
and clinical features in osteosarcoma patients.
| Parameter | Total number | Low miR 155-5p
expression | High miR-155-5p
expression | χ2 | P-value |
|---|
| Sex | | | | 2.3295 | 0.1269 |
|
Male | 16 | 7 | 9 | | |
|
Female | 14 | 10 | 4 | | |
| Age (years) | | | | 0.5236 | 0.4693 |
|
<20 | 21 | 11 | 10 | | |
|
≥20 | 9 | 6 | 3 | | |
| Histological
classification | | | | 1.3575 | 0.5073 |
|
Osteoblastic | 15 | 7 | 8 | | |
|
Chondroblastic | 10 | 7 | 3 | | |
|
Fibroblastic | 5 | 3 | 2 | | |
| Tumor grade | | | | 8.2935 | 0.0040a |
|
Low | 11 | 10 | 1 | | |
|
High | 19 | 7 | 12 | | |
| Enneking stage | | | | 8.8244 | 0.0121a |
|
I | 9 | 7 | 2 | | |
|
II | 13 | 9 | 4 | | |
|
III | 8 | 1 | 7 | | |
| Tumor size
(cm) | | | | 4.6930 | 0.0303a |
|
<8 | 16 | 12 | 4 | | |
|
≥8 | 14 | 5 | 9 | | |
| Metastasis | | | | 7.3692 | 0.0066a |
|
No | 10 | 9 | 1 | | |
|
Yes | 20 | 8 | 13 | | |
Cell culture and transfection
The human OS cell lines 143B, HOS, MG63, Saos-2 and
U2OS, the normal osteoblast cell line hFOB and HEK-293T were
purchased from the American Type Culture Collection. Both OS cell
lines (Saos-2 and U20S) and HEK-293T cells were cultured in a
Dulbecco's Modified Eagle's medium (DMEM; HyClone; Cytiva)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin
(Gibco; Thermo Fisher Scientific, Inc.). The culture conditions
were 5% CO2 and 37˚C. hFOB cells were cultured under the
same conditions in a DMEM/Ham's F-12 (1:1) medium supplemented with
10% FBS and 0.3 mg/ml G418 (Gibco; Thermo Fisher Scientific, Inc.).
NORAD plasmids, NORAD siRNA and its control (si-NC), miR-155-5p
mimics and its control (miR-NC), miR-155-5p inhibitors
(miR-155-5p-in) and its control (inh-NC) were designed and
constructed by GenePharma Co., Ltd. (Shanghai, China).
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) was
used to conduct transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA in tissues and Saos-2/U2OS cells was
extracted with a TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) miR was extracted using a
mirPremier® microRNA Isolation Kit (Sigma-Aldrich; Merck
KGaA). Total RNA was reverse transcribed using a First Strand cDNA
Synthesis Kit (Thermo Fisher Scientific Inc.). PCR was performed
using a LightCycler FastStart DNA Master plus a SYBR Green I kit
(Roche Diagnostics) to analyze the expression of NORAD and GAPDH
according to the manufacturer's protocol. A TaqMan microRNA assay
and TaqMan Universal Master Mix II (Applied Biosystems) were used
to analyze the contents of miR-155-5p and U6. The PCR cycles were
as follows: Initial denaturation at 95˚C for 5 min, 35 cycles of
denaturation (45 sec at 95˚C), annealing (45 sec at 60˚C) and
extension (8 min at 68˚C), and a final extension at 68˚C for 10
min. The relative expressions were calculated using the
2-ΔΔCq method (23). The
sequences of primers were as follows: NORAD forward
5'-GCCATTGGGCGAGACCTACCT-3' and reverse
5'-GTTCGGGACTTCGCTCACCTT-3'; miR-155-5p forward
5'-ACACTCCAGCTGTAAACATCCTACACTCT-3' and reverse
5'-CTCAACTGGTGTCGTGGA-3'; U6 forward 5'-GCTTCGGCAGCACATATACTA-3'
and reverse 5'-CGAATTTGCGTGTCATCCTTG-3'; GAPDH forward
5'-TGTTCGTCATGGGTGTGAAC-3' and reverse
5'-ATGGCATGGACTGTGGTCAT-3'.
Luciferase reporter assay
The NORAD sequence was amplified and inserted to a
pmirGLO plasmid (Promega Corporation) downstream of the luciferase
gene to produce pMIR-NORAD-wild type (NORAD-wt). To obtain the
mutant (mut) plasmid (NORAD-mut), a fragment containing a target
region of the mutation was designed. Then, NORAD-wt, NORAD-mut,
miR-155-5p mimics and miR-NC were transfected into HEK-293T cells
at a final concentration of 50 nM using Lipofectamine®
3000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h of
transfection, luciferase activity was analyzed using a
Dual-Luciferase Reporter Assay System (Promega Corporation). The
ratio of firefly luciferase activity to Renilla luciferase
activity was calculated and used as relative luciferase
activity.
RNA immunoprecipitation (RIP)
An EZ-Magna RIP RNA-binding protein
immunoprecipitation kit (EMD Millipore) was used according to the
manufacturer's instruction. Cells were lysed in a complete RIPA
buffer containing protease and RNase inhibitors. Cell
(1x107 cells) extractions were incubated with a RIP
buffer containing magnetic beads tagged with human anti-AGO 2
antibody (EMD Millipore) or IgG antibody. Finally, the combined RNA
was purified with TRIzol, and the miR-155-5p and NORAD expression
were evaluated using RT-qPCR.
Cell proliferation
Cell proliferation was measured using Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) according to
the manufacturer's instructions. In brief, the transfected cells
(Saos-2 and U2OS cells were transfected with si-NC, si-NORAD,
miR-NC, miR-155-5p mimics, inh-NC, miR-155-5p-in, si-NC + inh-NC,
si-NORAD + inh-NC, si-NC + miR-155-5p-in, si-NORAD + miR-155-5p-in,
respectivly) were seeded into individual wells of a 96-well plate
at a density of 1x104 per well and incubated for 0, 24,
48 and 72 h (37˚C, 5% CO2), respectively. A 10 µl volume
of CCK-8 solution was added to each well and incubated for a
further 1 h. The OD values at 450 nm were measured using a
microplate reader (Bio-Rad Laboratories, Inc.).
Transwell assay
Transfected cells (Saos-2 and U2OS cells were
transfected with si-NC, si-NORAD, miR-NC, miR-155-5p mimics,
inh-NC, miR-155-5p-in, si-NC + inh-NC, si-NORAD + inh-NC, si-NC +
miR-155-5p-in, si-NORAD + miR-155-5p-in, respectivly) were seeded
into a 24-well plate at a density of 5x104 cells/well
and cultivated at 37˚C, 5% CO2 equipped with Transwell
chambers (8 µm pore size; BD Biosciences). The cells were suspended
in the upper chamber in a serum-free medium (~1x105
cells/well), and the lower chamber was loaded with a medium
containing 10% FBS. After 24 h, the cells on the surface of the
upper chamber were gently wiped off with a cotton swab, and the
cells that migrated or invaded the lower chamber were fixed with
methanol (37˚C 15 min) and then stained with crystal violet (37˚C,
30 min). Finally, the stained cells were visualized under an
inverted microscope (magnification, x400) and counted in five
random fields.
Bioinformatics analysis
In the present study, StarBase database (version
3.0; http://starbase.sysu.edu.cn/) was used
to identify the binding sites between miR-155-5p and NORAD.
Statistical analysis
All experiments were repeated three times, with
three replicates each time. All statistical analyses were conducted
using SPSS 20.0 (SPSS, Inc.) and Graphpad Prism 7 software
(GraphPad Software, Inc.). Measurement data are presented as the
mean ± standard deviation. One-Sample Kolmogorov-Smirnov test was
used to examine whether the data are normally distributed or not.
For normally distributed data, comparisons between two groups were
performed using unpaired t-test. The comparisons among three or
more groups were performed with one-way ANOVA followed by Tukey's
post hoc test. For data with a skewed distributed, the comparison
between two groups was performed by paired sample Wilcoxon
signed-rank test. The relationship between NORAD/miR-155-5p and
clinicopathological characteristics was determined with the
χ2 test. P<0.05 indicated a statistically significant
difference.
Results
NORAD expression is increased in OS
tissues in comparison with controls and is closely associated with
the clinicopathological characteristics of OS
To investigate whether NORAD was expressed at
abnormal levels in OS, RT-qPCR was used to determine the expression
level of NORAD in tumor tissues and adjacent tissues from 30 OS
patients. The results showed that the expression level of NORAD in
tumor tissues was significantly higher than that in matched control
tissues (Fig. 1A; P<0.001).
Additionally, the expression of NORAD was examined in 143B, HOS,
MG63, Saos-2, U2OS and hFOB cells. It was found that the expression
of NORAD in all OS cell lines was higher than that in hFOB cells
(Fig. 1B, P<0.001). To
understand the clinical significance of NORAD in OS, the
relationship between the expression level of NORAD and the clinical
characteristics of OS was evaluated. The above 30 patients were
divided into a low expression group (n=15) and a high expression
group (n=15) according to the expression level of NORAD in their
tumor tissues. The results suggested that a high NORAD expression
level was related to tumor grade, Enneking stage, tumor size and
the status of metastasis (Table I;
P<0.05). However, the expression level of NORAD was not
associated with the sex, age or histological subtype of the
patients. All results suggested that the expression level of NORAD
was increased in OS vs. matched control tissues and the patients
with a high expression level of NORAD had an unfavorable
prognosis.
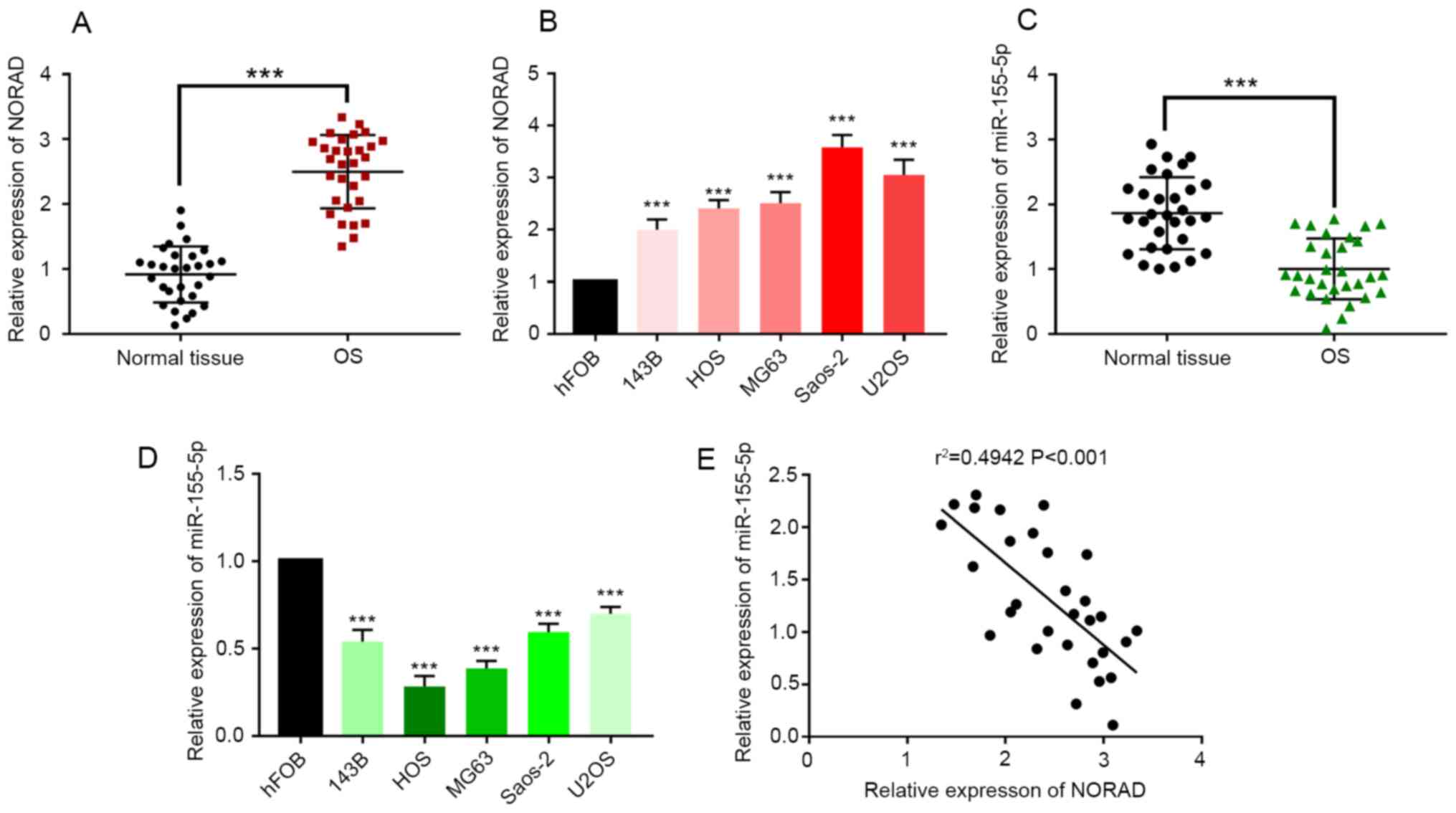 | Figure 1NORAD expression is increased and
miR-155-5p expression reduced in OS tissues and cells in comparison
with controls. (A) RT-qPCR was performed to measure the expression
of NORAD in OS tumor and adjacent normal tissues from 30 patients.
(B) RT-qPCR was performed to measure the expression of NORAD in
143B, HOS, MG63, Saos-2, U2OS and hFOB cells. (C) RT-qPCR was
performed to measure the expression of miR-155-5p in OS tumor and
adjacent normal tissues from 30 patients. (D) RT-qPCR was performed
to measure the expression of miR-155-5p in 143B, HOS, MG63, Saos-2,
U2OS and hFOB cells. (E) Correlation analysis between the
expression levels of NORAD and miR-155-5p in 30 OS tumor tissues.
***P<0.001. NORAD, non-coding RNA activated by DNA
damage; miR, microRNA; OS, osteosarcoma; RT-qPCR, reverse
transcription quantitative PCR. |
miR-155-5p expression is reduced in OS
tissues and cell lines in comparison with controls
miR-155-5p was significantly reduced in both OS
tissues and OS cell lines in comparison with controls (Fig. 1C and D; P<0.001). Furthermore, a reduced
expression level of miR-155-5p was associated with tumor grade,
Enneking stage, tumor size and the status of metastasis (Table II, P<0.05). These results
supported the hypothesis that miR-155-5p can act as a tumor
suppressor in OS. A correlation analysis was conducted between the
expression levels of NORAD and miR-155-5p in the 30 OS tumor tissue
samples. The results indicated a negative relationship between
NORAD and miR-155-5p expressions (Fig.
1E; r=-0.6248; P<0.001), suggesting a potential regulatory
relationship between NORAD and miR-155-5p.
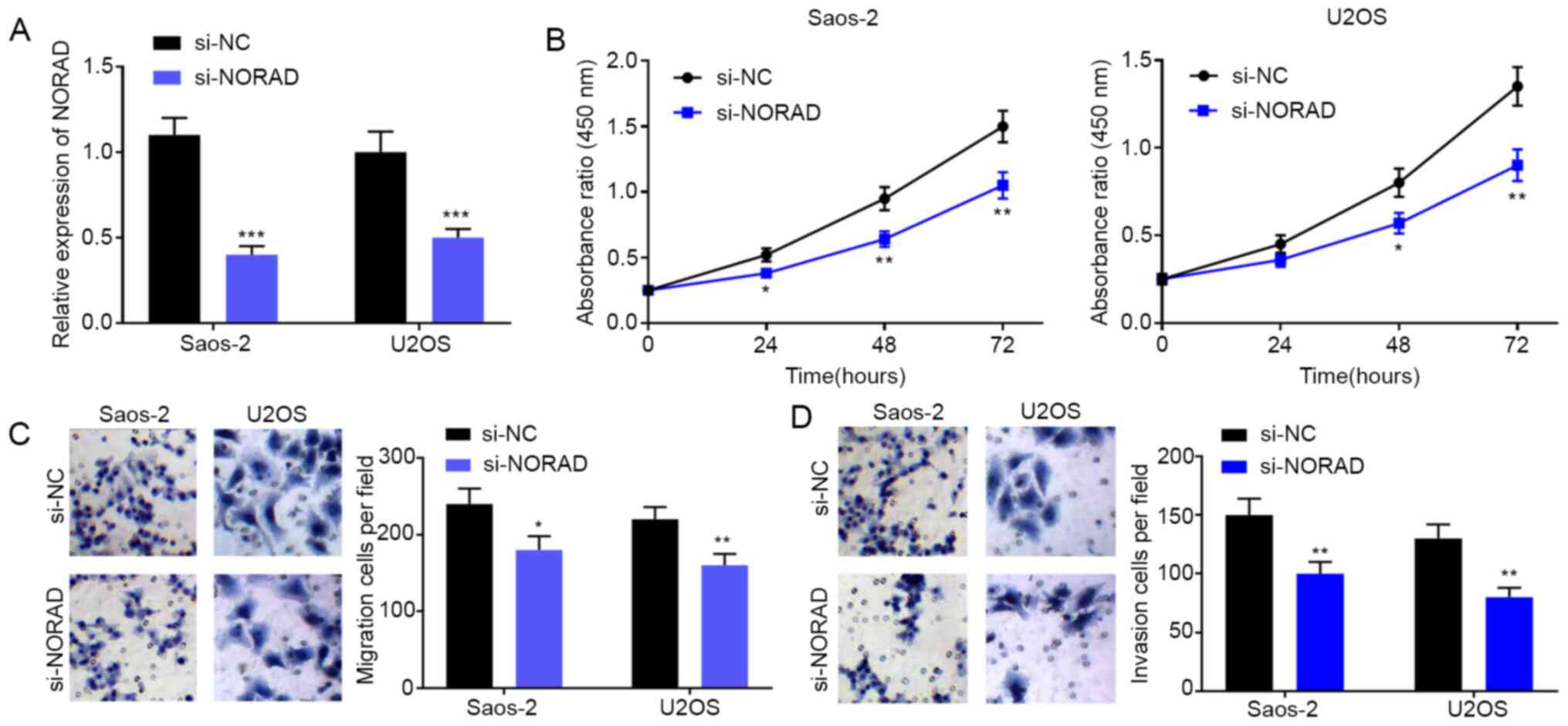
NORAD knockout inhibits the
proliferation, migration and invasion of OS cells
To explore the effect of NORAD on the malignant
phenotypes of OS, NORAD siRNA and NC siRNA were transfected into
Saos-2 and U2OS cells to observe their effects. The NORAD
expression level in the si-NORAD group was significantly lower than
that in the si-NC group (Fig. 2A;
P<0.001). A CCK-8 assay was performed to investigate the effect
of NORAD on cell proliferation, and the result showed that the
proliferation of OS cells was significantly inhibited after NORAD
knockdown (Fig. 2B). Furthermore,
the effect of NORAD knockdown on the migration and invasion of OS
cells was studied. Transwell assay showed that the count of
migrating and invading cells in the si-NORAD group was
significantly lower than that in the si-NC group (Fig. 2C; P<0.001). These results
suggested that NORAD modulated the malignant phenotypes of OS
cells.
miR-155-5p inhibits the proliferation,
invasion and migration of OS cells
The function of miR-155-5p in OS was explored in the
present study. After validating transfection efficiency (Fig. 3A; P<0.01), the CCK-8 assay was
performed to investigate the effect of miR-155-5p on cell
proliferation. The result indicated that the proliferation of OS
cells was inhibited by miR-155-5p but promoted by miR-155-5p
inhibitors in comparison with controls (Fig. 3B). Furthermore, the effects of
miR-155-5p on the invasion and migration of OS cells were studied.
The Transwell assay showed that the count of migrating and invading
OS cells was reduced with treatment with miR-155-5p mimics but
increased with treatment with miR-155-5p inhibitor in comparison
with controls (Fig. 3C and D; P<0.001). Collectively, these results
support the hypothesis that miR-155-5p acted as a tumor suppressor
in OS.
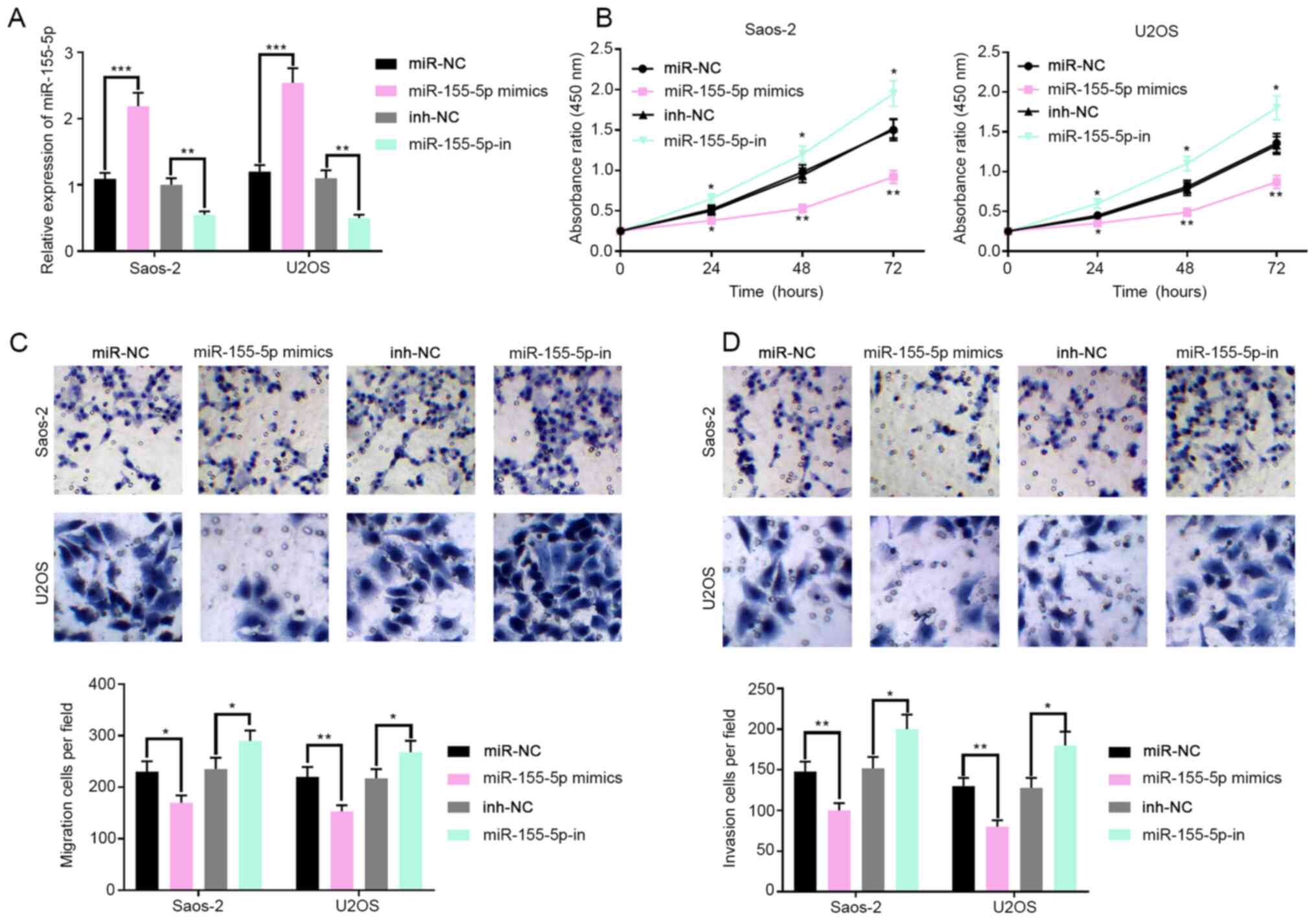 | Figure 3miR-155-5p regulates the
proliferation, invasion and migration of OS cells. (A) Transfection
efficiency of miR-155-5p mimics and inhibitors in OS cell lines was
validated by RT-PCR. (B) After the transfection of miR-155-5p
mimics or inhibitors, the proliferation of OS cells in different
groups was compared with the CCK-8 assay. (C) After the
transfection of miR-155-5p mimics or inhibitors, the migration of
OS cells in different groups was compared with the Transwell assay.
(D) After the transfection of miR-155-5p mimics or inhibitors, the
invasion of OS cells in different groups was compared with the
Transwell assay. *P<0.05, **P<0.01,
***P<0.001 vs. NC or NC-in. miR, microRNA; OS,
osteosarcoma; RT-qPCR, reverse transcription quantitative PCR; NC,
non-coding. |
NORAD acts as a sponge of miR-155-5p
in OS
Using StarBase, a miR-155-5p binding site was
identified in NORAD (Fig. 4A).
RT-qPCR suggested that the knockdown of NORAD significantly
up-regulated the expression level of miR-155-5p in both OS cell
lines, while the transfection of miR-155-5p mimics did not change
the expression level of NORAD (Fig.
4B and C). To investigate
whether NORAD could directly interact with miR-155-5p, a dual
luciferase reporter assay was performed by constructing NORAD-wt
and NORAD-mut luciferase reporter plasmids. Subsequently, the
NORAD-wt or NORAD-mut luciferase reporter plasmid was
co-transfected into HEK-293T cells with miR-155-5p mimics or
miR-NC. The results showed that the luciferase activity in the
HEK-293T cells co-transfected with NORAD-wt and miR-155-5p mimics
was significantly inhibited. However, the luciferase activity in
the HEK-293T cells co-transfected with NORAD-mut and miR-155-5p
mimics showed no statistically significant changes (Fig. 4D). A RIP experiment was also
conducted and the results implied that NORAD and miR-155-5p formed
a complex in OS cells (Fig. 4E).
Collectively, these data suggested that miR-155-5p interacted with
its binding site on NORAD to negatively regulate the expression of
NORAD. Subsequently, a CCK-8 experiment showed that the knockdown
of NORAD could inhibit the proliferation of OS cells, but this
effect could be partially neutralized by the co-transfection with
miR-155-5p inhibitors (Fig. 4F).
The Transwell assay also suggested that the knockdown of NORAD
inhibited the migration and invasion of Saos-2 cells, and this
effect could also be partially reversed by miR-155-5p inhibitors
(Fig. 4G and H). It was concluded that NORAD may
regulate the malignant phenotypes of OS cells via regulating
miR-155-5p expression.
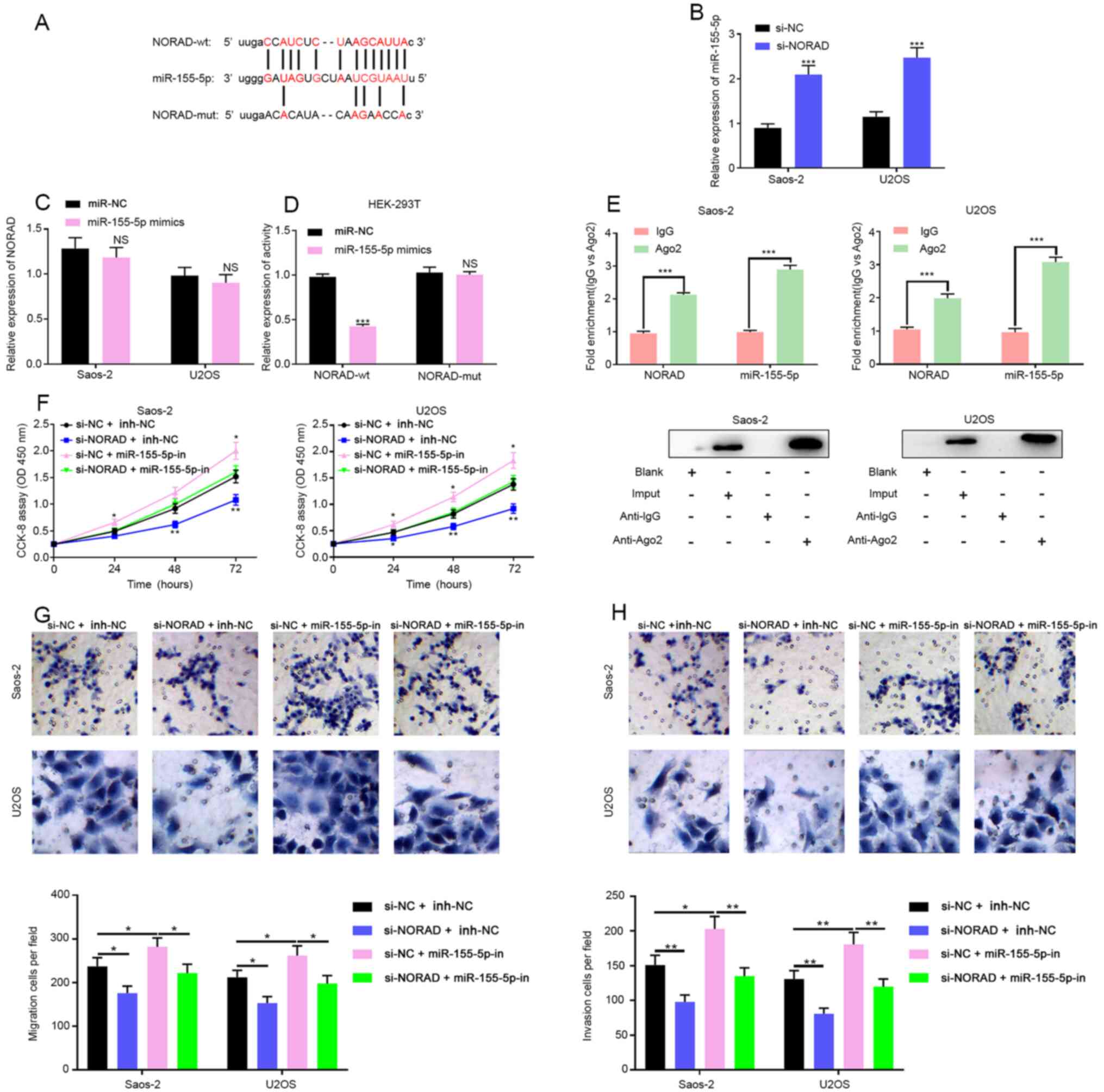 | Figure 4NORAD acts as a miR-155-5p sponge in
OS. (A) StarBase predicted a possible binding site of miR-155-5p on
NORAD. (B) The expression of miR-155-5p was examined by RT-qPCR
after NORAD knockdown in OS cells. (C) The expression of NORAD was
examined by RT-qPCR after transfection of miR-155-5p into OS cells.
(D) Luciferase reporter assay was used to assess the binding
between NORAD and miR-155-5p. (E) Direct binding between NORAD and
miR-155-5p was detected by the RNA immunoprecipitation assay. (F)
Proliferation of OS cells in si-NORAD + miR-NC, si-NORAD +
miR-155-5p-in, si-NC + miR-155-5p-in and si-NC + miR-NC groups was
measured with the Transwell assay. (G) Migration and (H) invasion
of OS cells in si-NORAD + miR-NC, si-NORAD + miR-155-5p-in, si-NC +
miR-155-5p-in and si-NC + miR-NC groups were measured with the
Transwell assay. *P<0.05, **P<0.01,
***P<0.001 as indicated. NORAD, non-coding RNA
activated by DNA damage; miR, microRNA; OS, osteosarcoma; RT-qPCR,
reverse transcription quantitative PCR; si, short interfering. |
Discussion
In recent years, studies have shown that lncRNAs
play important roles in the progression of many diseases (24-26).
According to previous reports, NORAD expression is increased in
many tumors and contributes to their progression (27-31).
One example is that the overexpression of NORAD promoted the
invasion and migration of malignant melanoma cells by regulating
the miR-205/EGLN2 pathway (31).
The present study aimed to establish whether NORAD played a similar
role in OS. The RT-qPCR experiment indicated that NORAD expression
was increased in OS tissues and cell samples. The correlation
between NORAD expression and the clinicopathological features of OS
patients was then investigated, and the results suggested that a
high expression level of NORAD was correlated with the tumor grade,
Enneking stage, tumor size, and tumor metastasis, indicating that
NORAD may play an oncogenic role in the pathological process of OS.
In the present study, NORAD siRNA was also used to knockdown the
expression level of NORAD in OS cell lines. The results suggested
that the down-regulation of NORAD could inhibit the proliferation,
migration and invasion of OS cells. From these results, it was
concluded that NORAD indeed acted as an oncogene in the progression
of OS, which was consistent with its role in other
malignancies.
miRs are important factors in tumorigenesis and
tumor progression and the up-regulation or down-regulation of miR
expression levels can affect the biological behavior of tumor
cells. For example, miR-30a-5p can inhibit the metastasis of human
colon cancer by targeting ITGB3(32); the down-regulation of miR-362-3p and
miR-329 can accelerate the progression of human breast cancer
(33). In the present study, it was
validated that miR-155-5p was down-regulated in OS tissues. The
correlation between the expression level of miR-155-5p and the
clinicopathological features of OS patients was also analyzed, and
the results showed that a low miR-155-5p expression level was
correlated with the tumor grade, Enneking stage, tumor size and
tumor metastasis, indicating that miR-155-5p may act as a tumor
suppressor in OS. In a previous study, miR-155-5p was shown to
inhibit the proliferation of OS cells and induce cell apoptosis
(34), which was consistent with
the results of the present study.
Accumulating evidence suggests that lncRNAs can bind
to miRs to act as an endogenous miR sponge, thus regulating the
expression and biological function of miRs (35). lncRNA SNHG3, lncRNA SNH5 and lncRNA
FBXL19-AS1 have been reported to interact with miRs to regulate the
progression of OS (24,36,37).
To further clarify the mechanism underlying the role of NORAD in
the carcinogenesis of OS, a bioinformatics analysis was performed
to identify miR-155-5p as a potential target of NORAD. RT-qPCR,
luciferase assay and RIP experiment were used to confirm that NORAD
could indeed bind to miR-155-5p. It was also confirmed that the
miR-155-5p expression in OS cell lines was significantly increased
after the NORAD expression was knocked down. The proliferation
activity of OS cells with inhibited expressions of both NORAD and
miR-155-5p was stronger than that of the OS cells in the si-NORAD
group but weaker than that of the OS cells in the miR-155-5p-in
group. Thereby, it was concluded that NORAD acted as an oncogene by
sponging miR-155-5p.
There are several limitations to the present study.
First of all, in vivo experiments are required to further
confirm the findings of this study. Additionally, NORAD may act
through other miRNA targets to affect the biological behaviors of
OS, and these potential miRNA targets remain to be screened and
validated. Additionally, the downstream genes and signaling
pathways of miR-155-5p involved in OS progression remain to be
validated. In addition to acting as ceRNA, lncRNA can also
participate in cancer progression via other mechanisms, including
chromatin modification, regulation of RNA processing (splicing,
editing, localization, translation, degradation),
post-translational regulation of protein activity and localization,
facilitation of ribonucleoprotein complex formation, and gene
silencing through the production of endogenous siRNA (38-40).
It is also important to find out whether NORAD promotes OS
progression via the above mechanisms.
In conclusion, NORAD expression is increased in OS
tissues and may be used as a marker for the prognosis of patients
with OS. NORAD directly targets miR-155-5p and inhibits its
expression, thus promoting the progression of OS. Therefore, NORAD
may be used as a new therapeutic target in OS management.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FX, YW and BZ conceived the study and performed the
experiments. LY, JW, SZ and ZX analyzed and interpreted the data.
FX, JW, LY, YW and SZ drafted and critically revised the manuscript
for important intellectual content. YW and BZ confirmed the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics Review
Board of Ezhou Central Hospital and patients or their guardians
provided their informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Berner K, Johannesen TB, Berner A,
Haugland HK, Bjerkehagen B, Bøhler PJ and Bruland ØS: Time-trends
on incidence and survival in a nationwide and unselected cohort of
patients with skeletal osteosarcoma. Acta Oncol. 54:25–33.
2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lee L, Fei L, Pope J and Wagner LM: Early
lymphocyte recovery and outcome in osteosarcoma. J Pediatr Hematol
Oncol. 39:179–183. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
O'Brien K and Chaman CL: An investigation
into the nutritional status and interventions taken in paediatric
and adolescent patients undergoing map chemotherapy for
osteosarcoma. Clin Nutr. 3(271)2018.
|
|
4
|
Bellini G, Di Pinto D, Tortora C, Manzo I,
Punzo F, Casale F and Rossi F: The role of mifamurtide in
chemotherapy-induced osteoporosis of children with osteosarcoma.
Curr Cancer Drug Targets. 17:650–656. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z and Wang J: lncRNA HOXA11-AS promotes
proliferation and invasion of gastric cancer by scaffolding the
chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res.
76:6299–6310. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang J, Cao L, Wu J and Wang Q: Long
non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326
and promotes tumorigenesis in osteosarcoma. Int J Oncol. 52:77–88.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lee S, Kopp F, Chang TC, Sataluri A, Chen
B, Sivakumar S, Yu H, Xie Y and Mendell JT: Noncoding RNA NORAD
regulates genomic stability by sequestering PUMILIO proteins. Cell.
164:69–80. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li H, Wang X, Wen C, Huo Z, Wang W, Zhan
Q, Cheng D, Chen H, Deng X, Peng C and Shen B: Long noncoding RNA
NORAD, a novel competing endogenous RNA, enhances the
hypoxia-induced epithelial-mesenchymal transition to promote
metastasis in pancreatic cancer. Mol Cancer. 16(169)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Du Y, Liu XH, Zhu HC, Wang L, Ning JZ and
Xiao CC: miR-543 promotes proliferation and Epithelial-Mesenchymal
transition in prostate cancer via targeting RKIP. Cell Physiol
Biochem. 41:1135–1146. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mesci A, Huang X, Taeb S, Jahangiri S, Kim
Y, Fokas E, Bruce J, Leong HS and Liu SK: Targeting of CCBE1 by
miR-330-3p in human breast cancer promotes metastasis. Br J Cancer.
116:1350–1357. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu Y, Niu Z, Lin X and Tian Y: miR-216b
increases cisplatin sensitivity in ovarian cancer cells by
targeting PARP1. Cancer Gene Ther. 24:208–214. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zehentmayr F, Hauser-Kronberger C,
Zellinger B, Hlubek F, Schuster C, Bodenhofer U, Fastner G,
Deutschmann H, Steininger P, Reitsamer R, et al: Hsa-miR-375 is a
predictor of local control in early stage breast cancer. Clin
Epigenetics. 8(28)2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
He M, Wang G, Jiang L, Qiu C, Li B, Wang J
and Fu Y: miR-486 suppresses the development of osteosarcoma by
regulating PKC-δ pathway. Int J Oncol. 50:1590–1600.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sahin Y, Altan Z, Arman K, Bozgeyik E,
Koruk OM and Arslan A: Inhibition of miR-664a interferes with the
migration of osteosarcoma cells via modulation of MEG3. Biochem
Biophys Res Commun. 490:1100–1105. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang SN, Luo S, Liu C, Piao Z, Gou W, Wang
Y, Guan W, Li Q, Zou H, Yang ZZ, et al: miR-491 inhibits
osteosarcoma lung metastasis and chemoresistance by targeting
αB-crystallin. Mol Ther. 25:2140–2149. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang C, Zhang C, Liu L, A X, Chen B, Li Y
and Du J: Macrophage-Derived mir-155-Containing exosomes suppress
fibroblast proliferation and promote fibroblast inflammation during
cardiac injury. Mol Ther. 25:192–204. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li XD, Li XM, Gu JW and Sun XC: miR-155
regulates lymphoma cell proliferation and apoptosis through
targeting SOCS3/JAK-STAT3 signaling pathway. Eur Rev Med Pharmacol
Sci. 21:5153–5159. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wei RJ, Zhang CH and Yang WZ: miR-155
affects renal carcinoma cell proliferation, invasion and apoptosis
through regulating GSK-3β/β-catenin signaling pathway. Eur Rev Med
Pharmacol Sci. 21:5034–5041. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang L, Tang B, Han H, Mao D, Chen J, Zeng
Y and Xiong M: miR-155 affects osteosarcoma MG-63 cell autophagy
induced by adriamycin through regulating PTEN-PI3K/AKT/mTOR
signaling pathway. Cancer Biother Radiopharm. 33:32–38.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen H, Liu T, Ouyang H, Lin S, Zhong H,
Zhang H and Yang Y: Upregulation of FTX promotes osteosarcoma
tumorigenesis by increasing SOX4 expression via miR-214-5p. Onco
Targets Ther. 13:7125–7136. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ruocco M, Baroncelli R, Cacciola SO, Pane
C, Monti MM, Firrao G, Vergara M, Magnano di San Lio G, Vannacci G
and Scala F: Polyketide synthases of Diaporthe helianthi and
involvement of DhPKS1 in virulence on sunflower. BMC Genomics.
19(27)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu XS, Wang F, Li HF, Hu YP, Jiang L,
Zhang F, Li ML, Wang XA, Jin YP, Zhang YJ, et al: lncRNA-PAGBC acts
as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO
Rep. 18:1837–1853. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Shi HJ, Wang MW, Sun JT, Wang H, Li YF,
Chen BR, Fan Y, Wang SB, Wang ZM, Wang QM and Wang LS: A novel long
noncoding RNA FAF inhibits apoptosis via upregulating FGF9 through
PI3K/AKT signaling pathway in ischemia-hypoxia cardiomyocytes. J
Cell Physiol. 234:21973–21987. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ruan Z, Wang S, Yu W and Deng F: lncRNA
MALAT1 aggravates inflammation response through regulating PTGS2 by
targeting miR-26b in myocardial ischemia-reperfusion injury. Int J
Cardiol. 288(122)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Lan T, Yuan K, Yan X, Xu L, Liao H, Hao X,
Wang J, Liu H, Xie K, Li J, et al: lncRNA SNHG10 facilitates
hepatocarcinogenesis and metastasis by modulating its homolog
SCARNA13 via a positive feedback loop. Cancer Res. 79:3220–3234.
2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sun Y, Wang J, Pan S, Yang T, Sun X, Wang
Y, Shi X, Zhao X, Guo J and Zhang X: LINC00657 played oncogenic
roles in esophageal squamous cell carcinoma by targeting miR-615-3p
and JunB. Biomed Pharmacother. 108:316–324. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wu X, Lim ZF, Li Z, Gu L, Ma W, Zhou Q, Su
H, Wang X, Yang X and Zhang Z: NORAD expression is associated with
adverse prognosis in esophageal squamous cell carcinoma. Oncol Res
Treat. 40:370–374. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang J, Li XY, Hu P and Ding YS: lncRNA
NORAD contributes to colorectal cancer progression by inhibition of
miR-202-5p. Oncol Res. 26:1411–1418. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yang X, Cai JB, Peng R, Wei CY, Lu JC, Gao
C, Shen ZZ, Zhang PF, Huang XY, Ke AW, et al: The long noncoding
RNA NORAD enhances the TGF-β pathway to promote hepatocellular
carcinoma progression by targeting miR-202-5p. J Cell Physiol.
234:12051–12060. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chen Y, Cao K, Li J, Wang A, Sun L, Tang
J, Xiong W, Zhou X, Chen X, Zhou J and Liu Y: Overexpression of
long non-coding RNA NORAD promotes invasion and migration in
malignant melanoma via regulating the MIR-205-EGLN2 pathway. Cancer
Med. 8:1744–1754. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wei W, Yang Y, Cai J, Cui K, Li RX, Wang
H, Shang X and Wei D: miR-30a-5p suppresses tumor metastasis of
human colorectal cancer by targeting ITGB3. Cell Physiol Biochem.
39:1165–1176. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Rice MA, Ishteiwy RA, Magani F, Udayakumar
T, Reiner T, Yates TJ, Miller P, Perez-Stable C, Rai P, Verdun R,
et al: The microRNA-23b/-27b cluster suppresses prostate cancer
metastasis via Huntingtin-interacting protein 1-related. Oncogene.
35:4752–4761. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bhattacharya S, Chalk AM, Ng AJ, Martin
TJ, Zannettino AC, Purton LE, Lu J, Baker EK and Walkley CR:
Increased miR-155-5p and reduced miR-148a-3p contribute to the
suppression of osteosarcoma cell death. Oncogene. 35:5282–5294.
2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Chen J, Wu Z and Zhang Y: lncRNA SNHG3
promotes cell growth by sponging miR-196a-5p and indicates the poor
survival in osteosarcoma. Int J Immunopathol Pharmacol.
33(2058738418820743)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang Z, Wang Z, Liu J and Yang H: Long
non-coding RNA SNHG5 sponges miR-26a to promote the tumorigenesis
of osteosarcoma by targeting ROCK1. Biomed Pharmacother.
107:598–605. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Pan R, He Z, Ruan W, Li S, Chen H, Chen Z,
Liu F, Tian X and Nie Y: lncRNA FBXL19-AS1 regulates osteosarcoma
cell proliferation, migration and invasion by sponging miR-346.
Onco Targets Ther. 11:8409–8420. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yang H, Jiang Z, Wang S, Zhao Y, Song X,
Xiao Y and Yang S: Long non-coding small nucleolar RNA host genes
in digestive cancers. Cancer Med. 8:7693–7704. 2019.PubMed/NCBI View Article : Google Scholar
|