Introduction
Ankylosing spondylitis (AS) is a chronic
immune-mediated disease from the larger group of spondyloarthritis
(SpA) characterized mainly by axial damage (1). The pathogenesis of AS includes the
presence of a particular genetic marker, human leukocyte antigen
(HLA)-B27, that interacts with environmental factors capable of
initiating development of the disease (2-4).
Many clinical studies based on animal models have attempted to
explain the complex pathogenesis of AS, but to date, the pathogenic
mechanism of the disease remains partially unknown (5,6).
AS can be considered a systemic condition,
presenting both musculoskeletal (axial and peripheral involvement)
and extraarticular manifestations, the most common being ocular,
digestive, cardiovascular and pulmonary ones. The close link
between the gut and SpA is well known; inflammatory bowel disease
(IBD) and AS having etiopathogenic similarities and being
considered distinct phenotypes of the same immuno-mediated disease
(7,8). A high percentage, up to 70%, of
patients with AS have subclinical intestinal inflammation and up to
10% of them will develop a clinically manifested IBD (9,10).
The importance of the intestinal microbiota in the
development of various systemic diseases has been highlighted over
time. It has been shown that under germ-free conditions,
HLA-B27/β2-microglobulin-transgenic rats do not present any
manifestation of the disease; by introducing commensal intestinal
bacteria in this sterile environment, the development of both
intestinal and joint inflammation was observed (11). Furthermore, in this rat model there
was evidence of an impaired mucosal immunity (12) and a specific intestinal dysbiosis
characterized by decreased species of Rikenellaceae and
increased Prevotella species (13).
A recently published article attempts to explain the
role of intestinal microbiota in the development of AS based on a
literature review (14). Thus, in
the first place, is the interaction between the antigen HLA-B27 and
the intestinal bacterial structures that can lead to a misfolding
of HLA-B27 and to an unfolded protein response of the endoplasmic
reticulum (14,15). This unfolded response is responsible
for the induction of pro-inflammatory proteins (16) as well as for the phenomenon of
autophagy (17). Moreover, there is
a molecular mimicry between bacterial peptides presented by HLA-B27
and various self-peptides which may induce cross-immune responses
(14,18).
On the other hand, changes were observed in the
intestinal mucosa characterized by an increased intestinal
permeability (14,19,20),
by an increased secretion of A immunoglobulins (IgA) (21) and proinflammatory cytokines mediated
by the activation of Th17 lymphocytes (22-25).
Taking all this into consideration, our study aimed
to analyze intestinal dysbiosis in patients with AS in terms of
composition, highlighting the correlations with different
demographic, clinical and paraclinical features.
Patients and methods
We conducted a prospective, case-control study in
Northeastern Romania which included 60 subjects. The enrolled cases
were distributed as follows: 28 cases in the AS group and 32
healthy controls. The individuals included in the study were
enrolled at the 1st Rheumatology Clinic of the Rehabilitation
Hospital Iasi from April 2016 to March 2017. All the included cases
expressed their informed consent to participate in the study.
Approval was obtained from the Ethics Committees of the Grigore T
Popa University of Medicine and Pharmacy and Rehabilitation
Hospital Iasi from which the cases were selected.
The inclusion criteria were: Age over 18 years;
signed consent by the participant to be included in the study;
definite diagnosis of AS. Patients diagnosed with AS met the 1984
modified New York Diagnostic Criteria (26). All subjects included in the analysis
completed a food questionnaire regarding the food components on
which their diet was based during the last month. The selected
group of patients with AS was as homogeneous as possible in terms
of diet, excluding cases in which probiotic medication were
prescribed.
Exclusion criteria consisted of: Patient refusal to
participate in this study, uncertain diagnosis of AS, serious
infections in the last 3 months (tuberculosis, Clostridium
difficile), colorectal cancer, antibiotic therapy during the
last 3 months.
For each case, a monitoring form was completed which
included demographic data [name, age, area of origin, ethnicity,
occupation, smoking status, body mass index (BMI)], year of
diagnosis, family and personal pathological history, and current
treatment. The group of patients with AS was divided in two
clinical subgroups representing a pure axial form and a form
associated with peripheral manifestations. For evaluating disease
activity, the BASDAI (Bath Ankylosing Spondylitis Disease Activity
Index) and BASFI (Bath Ankylosing Spondylitis Functional Index)
scores were used (1,22).
Intestinal microbiota analysis was performed by
real-time polymerase chain reaction (qPCR) in stool samples. From
each case included in the study, 20 g of feces was obtained. The
stool samples were transported (within a maximum of 4 h from
sampling) to the microbiology laboratory and were frozen at a
temperature of -80˚C for one week maximum until deoxyribonucleic
acid (DNA) extraction.
DNA extraction from fecal samples
For DNA extraction from feces, the
GenElute™ Stool DNA Isolation Kit (Sigma Aldrich; Merck
KGaA) was used. DNA extraction included the following steps: i)
From the 20 g of feces collected from patients, 200 mg was isolated
and added to a special extraction tube (Bead Tube) along with 1 ml
of Lysis Buffer L; ii) 100 µl of another special lysis solution
(Lysis Additive A) was added; iii) mixed for 3 min, then
centrifuged for 2 min at 14,000 rpm; iv) from the obtained
supernatant, 600 µl was transferred to another DNA tube
(DNAase-free microcentrifuge tube) over which 100 µl of Binding
Buffer I was added and the mixture was incubated for 10 min on ice,
then centrifuged for 2 min; v) from the newly obtained supernatant,
700 µl was separated in a 2-ml tube (DNAase-free microcentrifuge
tube) over which 700 µl of ethanol was added and centrifuged; vi)
from the ethanol clarification supernatant, 600 µl was separated,
introduced into a specific DNA binding tube and centrifuged for 1
min at 6,000 rpm; vii) the DNA-binding column was mixed with 500 µl
of wash buffer (SK buffer) and centrifuged for 1 min; viii) the
washed DNA binding column was introduced into an Elution tube and
50 µl of Elution Buffer (E) was added, centrifuged for 2 min at
2,000 rpm, and then 1 min at 14,000 rpm.
After DNA extraction from the feces, DNA quantity
and purity were checked using a NanoDrop spectrophotometer. The
purity of the samples was checked using 2 wavelengths: OD
260/280nm, respectively OD 260/230nm. The
entire phase which consisted in the extraction of pure DNA from
feces was carried out in the microbiology laboratory under the
Thermo DNA extraction hood.
qPCR
Through this study, we tried to highlight the
characteristics of different populations of the gut microbiota:
Certain genera, species and possibly a phylum. The qPCR reaction
targeted different populations of the microbiota: Total bacteria,
Bacteroides, Bifidobacterium, Clostridium coccoides
(XIVa) (C. Coccoides), Clostridium leptum
(IV) (C. Leptum), Faecalibacterium prausnitzii
(F. Prausnitzii), Lactobacillus, Escherichia coli
(E. Coli) and β-globin gene used as an internal control. The
primer structures were taken from an article published by Wang
et al (27) and verified
using OligoAnalyzer 3.1 (https://eu.idtdna.com). In addiion, the primer
annealing temperature was checked. Table I shows the primer structures and the
annealing temperatures.
 | Table IBacterial-specific 16S rRNA primers
and the annealing temperatures. |
Table I
Bacterial-specific 16S rRNA primers
and the annealing temperatures.
| Bacterial
species | Primer
direction | Sequence (5' to
3') | Annealing temp.
(˚C) |
|---|
| All bacteria | F |
ACTCCTACGGGAGGCAGCAGT | 61 |
| | R |
GTATTACCGCGGCTGCTGGCAC | |
|
Bacteroides | F |
GTCAGTTGTGAAAGTTTGC | 61.5 |
| | R |
CAATCGGGAGTTCTTCGTG | |
|
Bifidobacterium | F |
AGGGTTCGATTCTGCTCAG | 62 |
| | R |
CATCCGGCATTACCACCC | |
| C. coccoides
(XIVa) | F |
AAATGACGGTACCTGACTAA | 60.7 |
| | R |
CTTTGAGTTTCATTCTTGCGAA | |
| C. leptum
(IV) | F |
GTTGACAAAACGGAGGAAGG | 60 |
| | R |
GACGGGCGGTGTGTACAA | |
| F.
prausnitzii | F |
AGATGGCCTCGCGTCCGA | 61.5 |
| | R |
CCGAAGACCTTCTTCCTCC | |
|
Lactobacillus | F |
GCAGCAGTAGGGAATCTTCCA | 61.5 |
| | R |
GCATTYCACCGCTACACATG | |
| E. coli | F |
GTTAATACCTTTGCTCATTGA | 61 |
| | R |
ACCAGGGTATCTAATCCTGTT | |
| β-globin | F |
CAACTTCATCCACGTTCACC | - |
| | R |
GAAGAGCCAAGGACAGGTAC | |
The qPCR reaction was performed using SYBR Green
intercalary fluorochromes method that only bind to double-stranded
DNA molecules and included the following steps: i) The
amplification reaction was carried out at a final volume of 25 µl
containing: 9.8 µl SYBR Mix, 0.5 µl of each primer at a final
concentration of 0.2 µM, 0.5 µl fluorochrome ROX
(5-carboxy-X-rhodamine), 5 µl of bacterial DNA and ultra-pure water
to a volume of 20 µl (9.8 µl); ii) 1 cycle at 95˚C for 10 min; iii)
40 cycles at 95˚C for 10 sec; iv) 30 sec at normalization
temperature; and v) 30 sec at 72˚C, the annealing temperature.
In order to reduce the quantitative error of
detected bacteria and to characterize changes in bacterial copies,
abundance of 16S rRNA gene was calculated from standard curves.
Specific bacterial groups are expressed as a percentage of total
bacteria determined by universal primers. The standard curve was
constructed from decimal dilutions of the 16S rRNA amplicon using
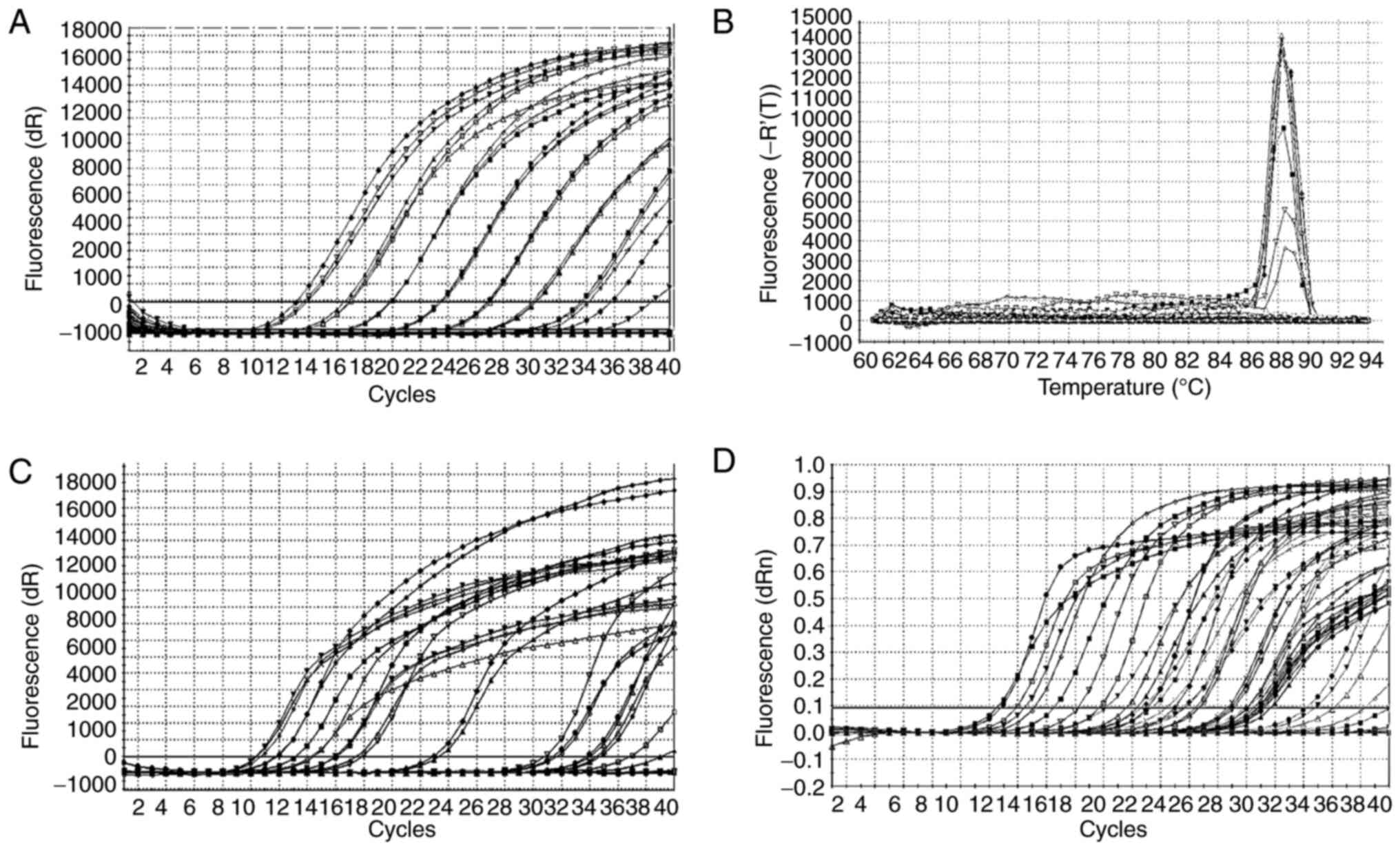
reference strains for each bacterial target. Based on this standard
curve, all sample amplification was performed (Fig. 1A). The dissociation and
amplification curves confirmed that DNA amplification occurred
specifically (Fig. 1B-D).
Following extraction, pure DNA was obtained by
spectrophotometer analysis (OD 260/280 nm, OD 260/230 nm). Data on
DNA concentration, quantity and purity are documented in Table II.
 | Table IIConcentration, quantity and purity of
the extracted DNA. |
Table II
Concentration, quantity and purity of
the extracted DNA.
| | 95% CI |
|---|
| DNA | Cases | N | Median | Standard
deviation | Min | Max |
|---|
| Concentration
(ng/µl) | AS | 28 | 48.61 | 24.81 | 38.98 | 58.23 |
| | Control | 32 | 36.86 | 26.34 | 27.36 | 46.36 |
| Quantity (µg) | AS | 28 | 2.91 | 1.48 | 2.33 | 3.49 |
| | Control | 32 | 2.21 | 1.58 | 1.64 | 2.78 |
| Purity 260/280
nm | AS | 28 | 1.97 | 0.083 | 1.94 | 2.008 |
| | Control | 32 | 2.03 | 0.17 | 1.96 | 2.09 |
| Purity 260/230
nm | AS | 28 | 0.48 | 0.35 | 0.34 | 0.62 |
| | Control | 32 | 0.44 | 0.33 | 0.32 | 0.56 |
Statistical analysis
The obtained data were centralized in the SPSS 22.0
database (IBM Corp.). Statistical analysis used both descriptive
and analytical methods at 95% significance (CI 95% CI). The
statistical tests used included ANOVA and Chi-square tests, linear
regression, and odds ratio. For comparisons between groups having a
non-linear distribution, Mann-Whitney U test and Kruskal-Wallis
method were used. A P-value less than 0.05 (P<0.05) was
considered statistically significant.
Results
Characteristics of the study
group
The demographic, clinical and paraclinical
characteristics of the cases included in this study are presented
in Table III. The compared groups
were homogeneous in terms of area of origin, smoking, social status
and BMI. We observed a higher percentage of male sex and a younger
age in the AS group. All cases were overweight.
 | Table IIIDemographic characteristics of the AS
and control groups. |
Table III
Demographic characteristics of the AS
and control groups.
| Characteristic | Subcategory | AS (n=28) | Control (n=32) |
|---|
| Sex, n (%) | Female | 11 (39.3) | 20 (62.5) |
| | Male | 17 (60.7) | 12 (37.5) |
| Age, years | Mean (SD) | 52.1 (13.6) | 61.5(10) |
| | Range | 46-57 | 57-65 |
| Area of origin, n
(%) | Urban | 20 (71.4) | 16(50) |
| | Rural | 8 (28.6) | 16(50) |
| Smoking status, n
(%) | Smokers | 10 (35.7) | 12 (37.5) |
| | Ex-smokers | 2 (7.1) | 6 (18.7) |
| | Non-smokers | 16 (57.2) | 14 (43.8) |
| BMI
(kg/m2) | Mean (SD) | 28.08 (5.2) | 26.9 (3.7) |
| | Range | 26.04-30.1 | 25.6-28.3 |
| Form of disease
(n,%) | Axial | 17 (60.7) | NA |
| | Peripheral | 11 (39.3) | NA |
| BASDAI | Mean | 4.83 | NA |
| | CI 95% | 3.87-5.79 | NA |
| BASFI | Mean | 9.11 | NA |
| | CI 95% | 4.44-13.78 | NA |
| Hemoglobin
(g/dl) | Mean (SD) | 13.01 (0.9) | 13.5 (1.6) |
| | Range | 12.6-13.3 | 12.9-14.1 |
| Iron (µg/dl) | Mean (SD) | 93.8 (31.1) | 77.7 (22.6) |
| | Range | 81.7-105.8 | 69.6-85.9 |
| Leukocytes
(/mm3x103) | Mean (SD) | 7.1 (1.8) | 6.5 (1.03) |
| | Range | 6.4-7.8 | 6.1-6.8 |
| Thrombocytes
(/mm3x103) | Mean (SD) | 293.6 (75.6) | 250.1 (35.5) |
| | Range | 264.2-322.9 | 237.2-262.9 |
| ESR (mm/h) | Mean (SD) | 23.4 (12.7) | 16.4 (10.5) |
| | Range | 18.5-28.3 | 12.6-20.2 |
| CRP (mg/dl) | Mean (SD) | 0.9 (1.3) | 0.8 (0.9) |
| | Range | 0.4-1.4 | 0.4-1.1 |
| AST (IU/l) | Mean (SD) | 28.6 (16.4) | 24.3 (15.9) |
| | Range | 22.2-35.07 | 18.5-30.05 |
| ALT (IU/l) | Mean (SD) | 24.9 (12.3) | 27.5 (15.8) |
| | Range | 20.1-29.7 | 21.8-33.2 |
| GGT (U/l) | Mean (SD) | 42.9 (23.7) | 32.8 (19.01) |
| | Range | 33.7-52.1 | 26.02-39.7 |
| Total serum
proteins (g/dl) | Mean (SD) | 7.2 (0.4) | 7.2 (0.4) |
| | Range | 7.07-7.4 | 7.06-7.4 |
| Creatinine
(mg/dl) | Mean (SD) | 0.9 (0.2) | 0.9 (0.1) |
| | Range | 0.8-1 | 0.8-0.9 |
Characteristics of the intestinal
dysbiosis in the study groups
The microbiota analysis was performed for each of
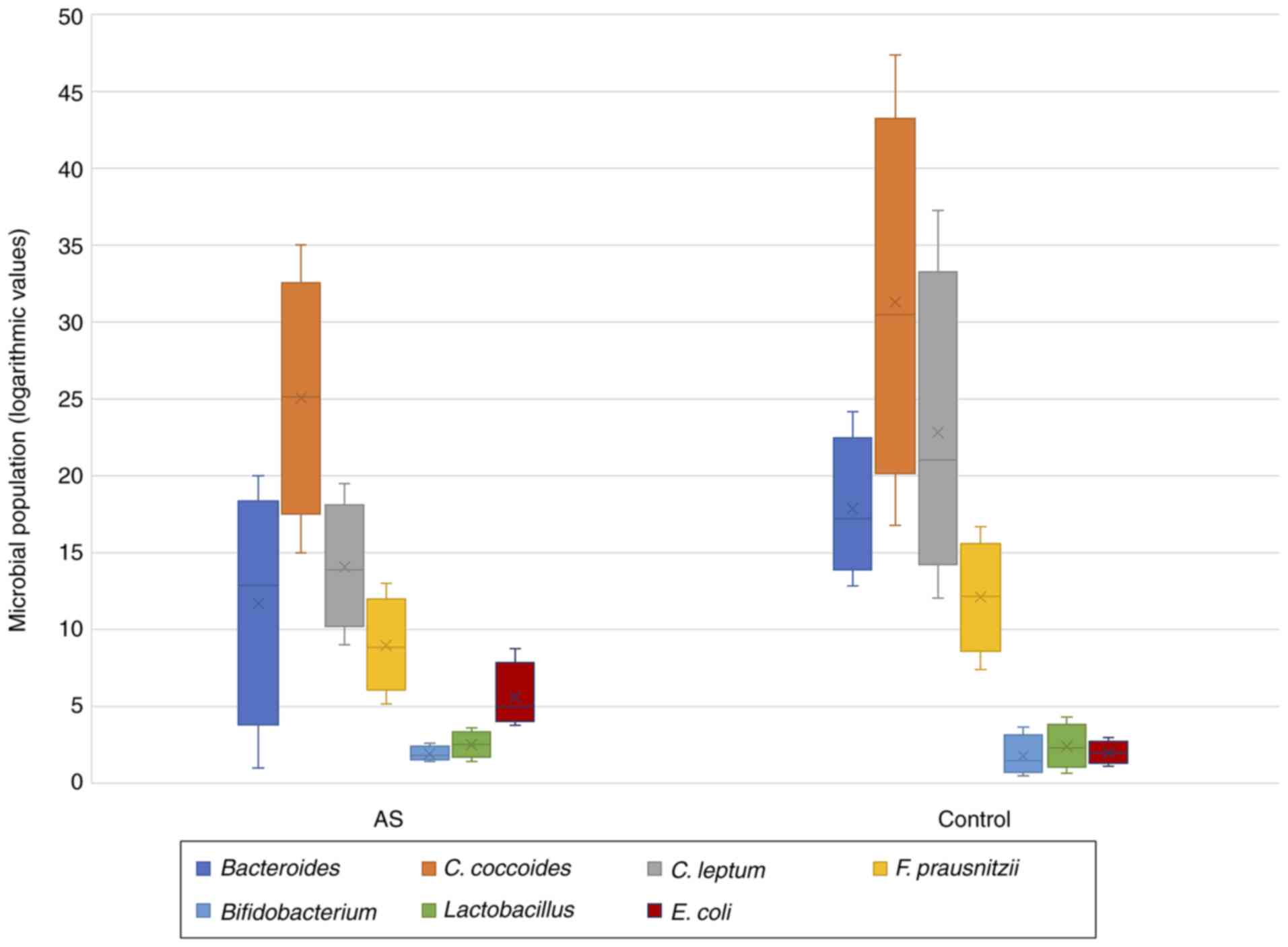
the two groups, being calculated quantitatively. The data in
Table IV show the quantitative
results expressed logarithmically.
 | Table IVQuantitative characteristics of the
microbial populations analyzed in the AS and control groups. |
Table IV
Quantitative characteristics of the
microbial populations analyzed in the AS and control groups.
| | 95% CI |
|---|
| Bacterial
population | Study group | Mean | Standard
deviation | Min | Max |
|---|
| All bacteria | AS | 2.18E+10 | 2.27E+10 | 1.3E+10 | 3.06E+10 |
| | Control | 4.13E+10 | 3.77E+10 | 2.77E+10 | 5.49E+10 |
|
Bacteroides | AS | 3.57E+09 | 4.37E+09 | 1.88E+09 | 5.26E+09 |
| | Control | 7.5E+09 | 6.96E+09 | 4.99E+09 | 1.00E+10 |
| C.
coccoides | AS | 5.13E+09 | 5.47E+09 | 3.01E+09 | 7.25E+09 |
| | Control | 1.28E+10 | 1.49E+10 | 7.44E+09 | 1.82E+10 |
| C.
leptum | AS | 2.72E+09 | 2.57E+09 | 1.72E+09 | 3.72E+09 |
| | Control | 9.64E+09 | 1.19E+10 | 5.36E+09 | 1.39E+10 |
| F.
prausnitzii | AS | 2.16E+09 | 2.73E+09 | 1.1E+09 | 3.22E+09 |
| | Control | 5.51E+09 | 5.13E+09 | 3.66E+09 | 7.36E+09 |
|
Bifidobacterium | AS | 3.72E+08 | 3.93E+08 | 2.2E+08 | 5.25E+08 |
| | Control | 4.25E+08 | 3.11E+08 | 3.13E+08 | 5.38E+08 |
|
Lactobacillus | AS | 5.21E+08 | 5.63E+08 | 3.02E+08 | 7.39E+08 |
| | Control | 8.07E+08 | 7.72E+08 | 5.29E+08 | 1.09E+09 |
| E. coli | AS | 1.06E+09 | 1.09E+09 | 6.41E+08 | 1.48E+09 |
| | Control | 8.72E+08 | 9.43E+08 | 5.32E+08 | 1.21E+09 |
In order to be able to compare the bacterial groups
according to each arm, the non-parametric Kruskal Wallis test was
used. Thus, applying the test for all combinations of 2 groups, the
following results were found. Statistically significant data were
found only for C. leptum (P=0.019) and E. coli
(P=0.013). In cases with AS, a significantly decreased level of
C. leptum was observed, associated with an increased level
of E. coli. The other analyzed microbial populations did not
show significant statistical differences with the control arm. The
group of cases with AS also showed a decreased microbial diversity
than the control group, but without any statistical value (Fig. 2).
Using the Spearman correlation coefficient,
significant correlations were found between paraclinical tests
(liver and kidney function, inflammatory syndrome) and bacterial
species (Table V). Thus, ESR and
CRP were inversely correlated with the level of Bacteroides
and directly proportional to C. coccoides and C.
leptum. Serum transaminases levels were directly proportional
to total bacteria (P=0.001). Only the ALT level was inversely
correlated with Bifidobacterium (P=0.006). Creatinine was
inversely correlated with Bacteroides (P=0.045) and E.
coli (P=0.027) and directly proportional to C. coccoides
(P<0.001) and C. leptum (P=0.005).
 | Table VCorrelations between bacterial
species and paraclinical tests. |
Table V
Correlations between bacterial
species and paraclinical tests.
| Test | Correlation | All bacteria | Bacteroides
% | C. coccoides
% | C. leptum
% | F. prausnitzii
% | Bifidobacterium
% | Lactobacillus
% | E. coli
% |
|---|
| Hb | ρ | 0.172 | 0.128 | -0.077 | -0.062 | -0.028 | 0.048 | 0.008 | -0.020 |
| | P-value | 0.056 | 0.158 | 0.397 | 0.493 | 0.759 | 0.594 | 0.928 | 0.762 |
| Leukocytes | ρ | -0.061 | 0.200 | -0.239 | -0.225 | -0.152 | 0.182 | 0.139 | 0.156 |
| | P-value | 0.498 | 0.026 | 0.008 | 0.012 | 0.091 | 0.043 | 0.123 | 0.083 |
| Thrombocytes | ρ | -0.07 | 0.063 | -0.157 | -0.101 | -0.125 | 0.129 | 0.092 | 0.132 |
| | P-value | 0.441 | 0.486 | 0.082 | 0.265 | 0.166 | 0.154 | 0.311 | 0.144 |
| Iron | Ρ | 0.133 | -0.192 | -0.039 | 0.075 | 0.168 | -0.067 | -0.018 | -0.210 |
| | P-value | 0.139 | 0.033 | 0.665 | 0.405 | 0.063 | 0.463 | 0.841 | 0.015 |
| ESR | ρ | 0.081 | -0.479 | 0.380 | 0.482 | 0.507 | -0.162 | -0.25 | -0.44 |
| | P-value | 0.369 |
<0.001 |
<0.001 |
<0.001 |
<0.001 | 0.073 | 0.005 |
<0.001 |
| CRP | ρ | -0.103 | -0.18 | 0.199 | 0.325 | 0.095 | -0.02 | -0.015 | -0.090 |
| | P-value | 0.253 | 0.045 | 0.026 |
<0.001 | 0.292 | 0.828 | 0.868 | 0.300 |
| AST | ρ | 0.329 | 0.032 | 0.026 | 0.057 | 0.120 | -0.120 | -0.169 | -0.170 |
| | P-value |
<0.001 | 0.723 | 0.776 | 0.529 | 0.183 | 0.183 | 0.061 | 0.052 |
| ALT | ρ | 0.307 | -0.009 | 0.082 | 0.101 | 0.171 | -0.247 | -0.159 | -0.210 |
| | P-value | 0.001 | 0.921 | 0.364 | 0.262 | 0.057 | 0.006 | 0.077 | 0.017 |
| GGT | ρ | 0.07 | -0.151 | 0.081 | 0.161 | 0.228 | -0.157 | -0.226 | -0.160 |
| | P-value | 0.437 | 0.094 | 0.372 | 0.074 | 0.011 | 0.081 | 0.012 | 0.076 |
| Total proteins | ρ | -0.018 | 0.275 | -0.154 | -0.227 | -0.238 | 0.071 | -0.046 | 0.219 |
| | P-value | 0.842 | 0.002 | 0.088 | 0.011 | 0.008 | 0.435 | 0.611 | 0.015 |
| Creatinine | ρ | 0.159 | -0.18 | 0.342 | 0.251 | 0.132 | -0.136 | -0.137 | -0.190 |
| | P-value | 0.077 | 0.045 |
<0.001 | 0.005 | 0.144 | 0.132 | 0.130 | 0.027 |
No correlations were found between the degree of
radiological sacroiliitis and bacterial groups (P=0.053,
Kruskal-Wallis test) or between BMI and bacterial populations
(P=0.366).
Of the 28 cases with AS, 22 patients tested positive
for the antigen HLA-B27 (human leucocyte antigen B27). Following
the statistical analysis (Mann-Whitney test), significant
correlations were highlighted between HLA-B27 and
Lactobacillus (P=0.027) and E. coli (P=0.004)
(Table VI).
 | Table VICorrelations between the antigen HLA
B27 and microbial populations in the AS group. |
Table VI
Correlations between the antigen HLA
B27 and microbial populations in the AS group.
| Test | Total bacteria | Bacteroides
% | C. coccoides
% | C. leptum
% | F. prausnitzii
% | Bifidobacterium
% | Lactobacillus
% | E. coli
% |
|---|
| Mann-Whitney U | 46.0 | 50.0 | 59.0 | 58.0 | 41.0 | 56.0 | 26.5 | 15.0 |
| Wilcoxon W | 299.0 | 71.0 | 80.0 | 79.0 | 294.0 | 309.0 | 279.5 | 268.0 |
| Z | -1.123 | -0.897 | -0.392 | -0.448 | -1.401 | -0.561 | -2.216 | -2.86 |
| P-value | 0.262 | 0.370 | 0.695 | 0.654 | 0.161 | 0.575 | 0.027 | 0.004 |
Other significant data were recorded in relation to
smoker status. Thus, correlations between C. coccoides
(P=0.033) and Bifidobacterium (P=0.006) were found in the
arm of smokers diagnosed with AS. The level of C. coccoides
was decreased, while Bifidobacterium was increased. On the
other hand, correlations with a decreased level of F.
prausnitzii (P=0.027) were found for active smokers in the
control group (Table VII).
 | Table VIICorrelations between smoking status
and microbial populations. |
Table VII
Correlations between smoking status
and microbial populations.
| Arm | Test | Total bacteria | Bacteroides
% | C. coccoides
% | C. leptum
% | F. prausnitzii
% | Bifidobacterium
% | Lactobacillus
% | E coli
% |
|---|
| Control | Z | -1.066 | -1.598 | -0.228 | -0.228 | -2.206 | -1.598 | -1.446 | -1.293 |
| | P-value | 0.286 | 0.110 | 0.819 | 0.819 | 0.027 | 0.110 | 0.148 | 0.196 |
| AS | Z | -0.768 | -1.394 | -2.138 | -1.208 | -0.232 | -2.744 | -1.465 | -1.488 |
| | P-value | 0.443 | 0.163 | 0.033 | 0.227 | 0.816 | 0.006 | 0.143 | 0.137 |
Using the Spearman correlations, we aimed to
ascertain whether there were correlations between the disease
activity quantified by BASDAI and BASFI scores and the bacterial
populations. We did not find statistically significant data between
these scores and the bacteria species (Table VIII). It should be mentioned that
the patients included in the analysis had a moderate disease
activity, with an average BASDAI of 4.83 (3.87-5.79 95% CI) and an
average BASFI of 9.11 (4.44-13.78 95% CI).
 | Table VIIICorrelations between disease activity
and microbial populations in the AS group. |
Table VIII
Correlations between disease activity
and microbial populations in the AS group.
| Score | Microbial
population | Spearman
coefficient | P-value |
|---|
| BASDAI | All bacteria | -0.336 | 0.080 |
| | Bacteroides
% | -0.371 | 0.052 |
| | C. coccoides
% | 0.111 | 0.573 |
| | C. leptum
% | 0.340 | 0.077 |
| | F. prausnitzii
% | -0.371 | 0.052 |
| | Bifidobacterium
% | 0.183 | 0.351 |
| | Lactobacillus
% | 0.331 | 0.085 |
| | E. coli
% | 0.309 | 0.110 |
| BASFI | All bacteria | -0.018 | 0.927 |
| | Bacteroides
% | -0.061 | 0.758 |
| | C. coccoides
% | 0.053 | 0.788 |
| | C. leptum
% | 0.010 | 0.960 |
| | F. prausnitzii
% | 0.104 | 0.597 |
| | Bifidobacterium
% | 0.048 | 0.809 |
| | Lactobacillus
% | 0.019 | 0.924 |
| | E. coli
% | -0.015 | 0.941 |
According to the form of the disease, patients with
AS were divided into an axial and a form with peripheral arthritis.
Using the Mann-Whitney statistical test, significant data were
found only for the Bifidobacterium bacterial group
(P=0.035). Thus, in the peripheral form of AS there was a
significant decrease of Bifidobacterium compared to the
axial form (Table IX).
 | Table IXCorrelations between microbial
populations and the form of AS. |
Table IX
Correlations between microbial
populations and the form of AS.
| Microbial
population | Form of AS | Mean rank | Sum of ranks | P-value |
|---|
| Total bacteria | Axial | 23.94 | 766.00 | 0.451 |
| | Peripheral | 20.69 | 269.00 | |
| Bacteroides
% | Axial | 22.44 | 718.00 | 0.652 |
| | Peripheral | 24.38 | 317.00 | |
| C. coccoides
% | Axial | 23.41 | 749.00 | 0.744 |
| | Peripheral | 22.00 | 286.00 | |
| C. leptum
% | Axial | 23.19 | 742.00 | 0.880 |
| | Peripheral | 22.54 | 293.00 | |
| F. prausnitzii
% | Axial | 23.09 | 739.00 | 0.940 |
| | Peripheral | 22.77 | 296.00 | |
| Bifidobacterium
% | Axial | 25.63 | 820.00 | 0.035 |
| | Peripheral | 16.54 | 215.00 | |
| Lactobacillus
% | Axial | 23.80 | 761.50 | 0.522 |
| | Peripheral | 21.04 | 273.50 | |
| E. coli
% | Axial | 23.66 | 757.00 | 0.598 |
| | Peripheral | 21.38 | 278.00 | |
Regarding the treatment followed by patients with
AS, all of them were receiving drug therapy at the time of
enrollment in the study. Thus, most cases (n=19; 67.85%) were
receiving anti-TNFα treatment, 5 cases (17.85%) were receiving
sulfasalazine (SSZ) therapy, and 4 patients (14.28%) were receiving
only symptomatic treatment with nonsteroidal anti-inflammatory
drugs (NSAIDs). Because the number of cases was relatively small
for each therapy, the statistical analysis did not have
consistency. Thus, we grouped the patients as follows: the group
that had NSAID treatment (n=4) and those on immunosuppressive
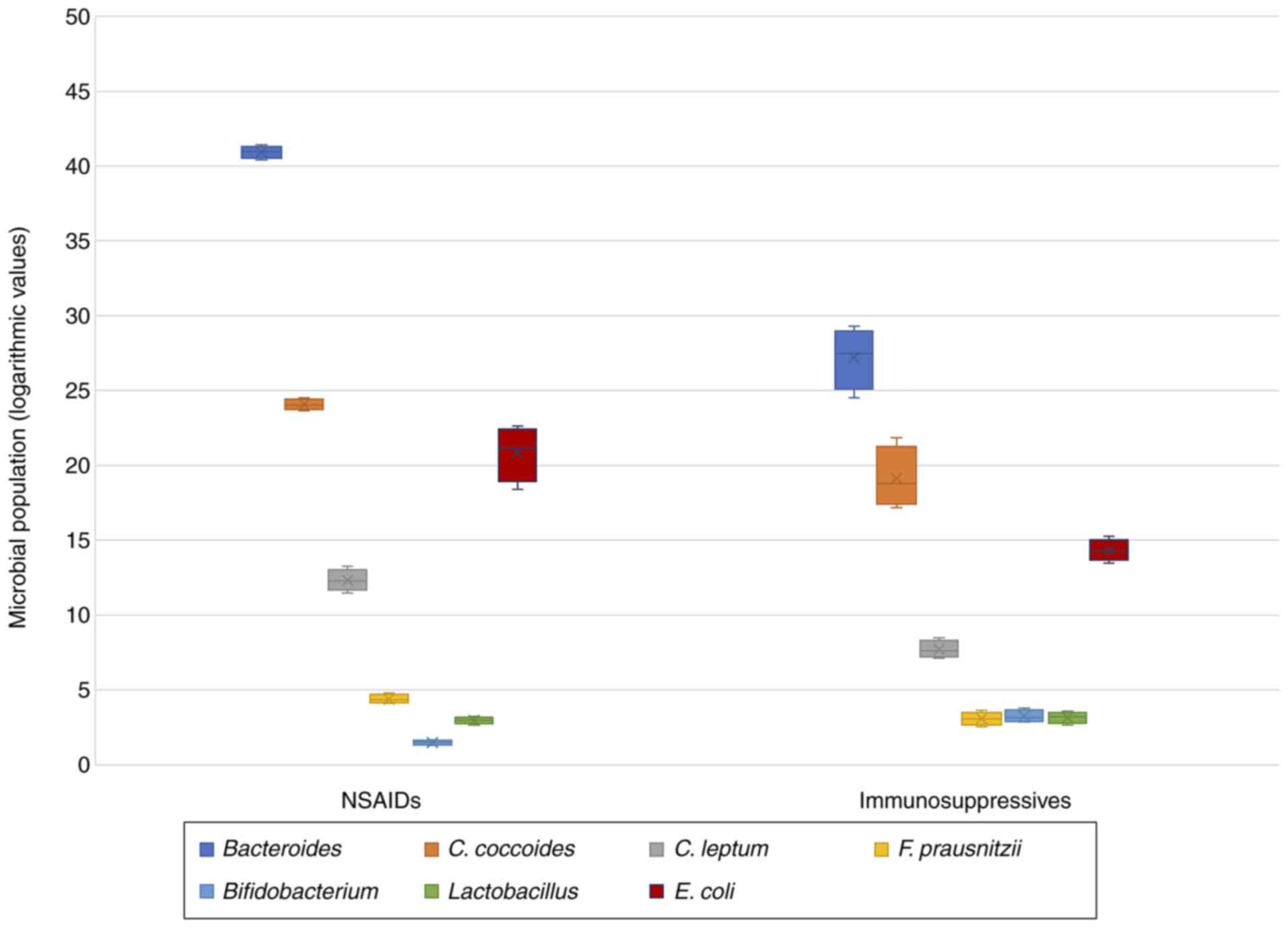
treatment (SSZ + antiTNFα; n=24). It was observed that patients who
were treated only with NSAIDs showed a decrease in bacterial
diversity (P=0.560), in Bifidobacterium (P<0.001) and
Lactobacillus (P=0.382) followed by an increase in
Bacteroides (P<0.001), C. coccoides (P=0.005),
C. leptum (P<0.001), F. prausnitzii (P=0.001) and
E. coli (P=0.001). All these data are presented in Table X and Fig. 3.
 | Table XQuantitative analysis of the
microbiome according to the treatment followed by patients with
AS. |
Table X
Quantitative analysis of the
microbiome according to the treatment followed by patients with
AS.
| | 95% CI | |
|---|
| Microbial
population | Treatment | Mean | Min | Max | Median | Standard
deviation | P-value |
|---|
| Total bacteria | IS | 2.31E+10 | 7.36E+09 | 3.88E+10 | 1.31E+10 | 2.48E+10 | 0.560 |
| | NSAIDs | 1.82E+10 | 1.44E+10 | 2.2E+10 | 1.66E+10 | 5.95E+09 | |
| Bacteroides
% | IS | 26.90 | 24.51 | 29.29 | 28.06 | 3.76 |
<0.001 |
| | NSAIDs | 40.91 | 40.41 | 41.42 | 41.00 | 0.79 | |
| C. coccoides
% | IS | 19.51 | 17.18 | 21.84 | 18.06 | 3.66 | 0.005 |
| | NSAIDs | 24.09 | 23.66 | 24.53 | 23.94 | 0.69 | |
| C. leptum
% | IS | 7.79 | 7.12 | 8.47 | 7.42 | 1.06 |
<0.001 |
| | NSAIDs | 12.37 | 11.48 | 13.26 | 12.20 | 1.40 | |
| F. prausnitzii
% | IS | 3.09 | 2.54 | 3.63 | 3.03 | 0.85 | 0.001 |
| | NSAIDs | 4.46 | 4.11 | 4.81 | 4.19 | 0.55 | |
| Bifidobacterium
% | IS | 3.33 | 2.87 | 3.79 | 2.97 | 0.72 |
<0.001 |
| | NSAIDs | 1.48 | 1.28 | 1.68 | 1.50 | 0.31 | |
| Lactobacillus
% | IS | 3.10 | 2.64 | 3.57 | 3.32 | 0.72 | 0.382 |
| | NSAIDs | 2.95 | 2.66 | 3.24 | 3.00 | 0.45 | |
| E. coli
% | IS | 14.37 | 13.48 | 15.26 | 14.20 | 1.40 | 0.001 |
| | NSAIDs | 20.52 | 18.40 | 22.64 | 21.81 | 3.33 | |
Discussion
The present study analyzed different populations of
the gut microbiome in patients with ankylosing spondylitis (AS):
Certain species, genera and possibly a phylum compared to a control
group formed by healthy individuals. The two study arms were
carefully selected to be as homogeneous as possible. Analysis of
the intestinal microbiome was performed in the feces using the qPCR
technique. We took into consideration the quantitative analysis of
the microbiome, focusing both on bacterial structures having an
anti-inflammatory role (Bifidobacterium, Lactobacillus, F.
prausnitzii), and on some with pro-inflammatory actions that
favor intestinal and systemic inflammation (Bacteroides, E.
coli).
Following the data analysis, intestinal bacterial
diversity in the AS group was decreased compared to the control.
Significant data were highlighted only for 2 bacterial species. A
significant numerical increase was observed for E. coli
associated with a decrease in C. leptum.
The data in the literature regarding gut dysbiosis
in AS are contradictory. Our results are in agreement with other
studies. Zhang et al (28)
investigated fecal microbiota in 103 cases of AS and concluded that
alpha diversity in these cases was no different from the control
group. Similar to our results, they showed a decrease in
Clostridium_XIVb, but an increase in Bacteroides.
Breban et al (29) analyzed
the intestinal microbiota in 2 cohorts: rheumatoid arthritis and SA
compared to a control group. They showed an overall decreased
microbial diversity in both groups, followed by a significant
increase in Ruminococcus gnavus in SA.
Numerous data in the literature support this
reduction in intestinal microbial diversity in patients with
chronic inflammatory autoimmune diseases (30,31).
However, there are several studies that have shown an increase in
intestinal bacteria in patients with SA, especially in those having
associated intestinal inflammation (10,32).
We demonstrated a close link between liver and
kidney function and the analyzed bacteria. Serum transaminases
levels were directly proportional to total bacteria. Only the ALT
level was inversely correlated with Bifidobacterium. Serum
creatinine was inversely correlated with Bacteroides and
E. coli and directly proportional with C. coccoides
and C. leptum.
The close connection between the intestine and the
liver is a certainty, with many authors reporting about the
presence of a liver-intestine axis (33-36).
The composition of the intestinal microbiota can modulate chronic
liver diseases by i) the production of metabolites, by altering the
integrity of the mucosal barrier, ii) by the portal system or iii)
by the liver-intestinal immune link (37).
On the other hand, important studies have
highlighted the existence of a ‘colo-renal system’ that can
influence each other. The intestinal microbiota can alter renal
function, contributing to the development of many pathologies.
There is a communication between various intestinal bacterial
groups and the cells of the renal parenchyma, which causes a
disturbance of the normal renal molecular processes, leading to the
appearance of kidney diseases (38-40).
The presence of systemic inflammation was quantified
by measuring the acute phase reactants: ESR and CRP. Most often,
the normal values of these biological constants are associated with
low activity or remission of inflammatory joint diseases. Moreover,
they are directly proportional to the disease activity quantified
in our study by BASDAI and BASFI scores. Following analysis, ESR
and CRP were inversely correlated with the level of
Bacteroides and directly proportional to C. coccoides
and C. leptum.
Two studies that are part of the RISTOMED project
(Impact of personalized diet and probiotic supplementation on
inflammation, nutritional parameters and intestinal microbiota)
(41,42) analyzed the correlations between the
composition of the intestinal microbiota and various clinical
parameters, including inflammatory markers. The study included 125
cases divided into a group with low inflammation and a group with
moderate-high inflammation. At the level of intestinal microbiota,
the Bifidobacterium and Clostridium group IV species
were analyzed. The results showed, in the arm with low
inflammation, a decrease in Bifidobacterium followed by an
increase in Clostridium IV and the strong correlation with
the level of acute phase reactants.
Regarding disease activity, we did not find
significant correlations between BASDAI and BASFI scores and the
bacteria species. Our results are consistent with other studies
that have not shown an association between disease activity or
function and intestinal dysbiosis (43). Moreover, these studies showed
similarities regarding the highlighted bacterial species, namely an
increase in E. coli and a decrease in Clostridium, F.
prausnitzii and Bacteroides (4,44-46).
On the other hand, many data support the close link between the
activity of AS and the composition of the intestinal microbiota
(29,32) and between gut inflammation and
disease activity in AS (47,48).
Considering the genetic predisposition, significant
correlations were found between the antigen HLA-B27 and
Lactobacillus, respectively E. coli. The levels of
Lactobacillus and E. coli were decreased in patients
tested positive for the HLA B27 antigen. The ability of this
antigen to modulate the intestinal microbiota in transgenic
laboratory animals has been demonstrated and published recently
(13). Intestinal dysbiosis in
HLA-B27-positive laboratory animals was characterized by a decrease
in Firmicutes followed by a significant increase in
Proteobacteria group, of which E. coli takes part
(49). Moreover, Breban et
al (29) showed a different
intestinal dysbiosis in members of the same family depending on the
presence or absence of the HLA-B27 antigen.
Further correlations between gut microbiota
composition and clinical parameters included the following data:
The BMI, radiological sacroiliitis, smoking status, disease
phenotype and the treatment. No correlations were found between the
degree of radiological sacroiliitis and bacterial groups or between
BMI and gut microbiome. A study published in 2018 highlighted the
analysis of the intestinal microbiota in 61 cases of obese people.
Intestinal dysbiosis was characterized by a decrease in microbial
diversity which correlated with various metabolic parameters
(50). Many of the published
results are contradictory. Some support a decrease in diversity in
the phyla group, others do not show significant differences between
normal and obese people. Many authors have shown an increase in
Firmicutes followed by a decrease in Bacteroides in
cases of obesity (51,52). Kasai et al showed an increase
in the diversity of intestinal microbiota in cases of obesity
(53). Angelakis et al
observed a numerical increase in anaerobic bacteria in obese
patients (54). Hu and colleagues
found no significant differences between normal and obese
individuals in the Bacteroidetes, Firmicutes, and
Proteobacteria groups (55).
In the present study, significant data were recorded
in relation to smoker status. Correlations between C.
coccoides and Bifidobacterium were found in the arm of
smokers diagnosed with AS. The level of C. coccoides was
decreased, while Bifidobacterium was increased. On the other
hand, correlations with a decreased level of F. prausnitzii
were found for active smokers in the control group. The
relationship between smoking and IBD is well known. This connection
is not fully understood, but the alteration of the intestinal
microbiota, as well as of the innate and adaptative immune system,
has been incriminated (56).
In a recently published meta-analysis (57), the intestinal microbiota was
analyzed in healthy smokers. The authors noted a decrease in the
diversity of the intestinal microbiome characterized by a decrease
in Bifidobacterium and Lactobacillus species and an
increase in Bacteroides, Clostridium and Prevotella.
Intestinal dysbiosis caused by smoking was similar to IBD
dysbiosis.
In the present study, according to the form of the
disease, significant data were found only for the
Bifidobacterium bacterial group. In the peripheral form
there was a significant decrease in Bifidobacterium compared
to the axial form of AS. Differences in the intestinal microbiome
according to disease phenotype were also observed in a study by
Chen et al (58). They noted
an increase in Prevotella in axial disease and increased
Collinsella, Streptococcus and Comamonas in the
peripheral form of AS.
In the present study, the final data focused on the
correlations between the composition of the intestinal microbiome
and treatment. Significant data were recorded only for NSAID
therapy. Thus, these patients showed a decrease in bacterial
diversity, in Bifidobacterium and Lactobacillus and
an increase in Bacteroides, C. coccoides, C. leptum, F.
prausnitzii and E. coli.
Studies have shown that the intestinal microbiome
can predict the therapeutic response and can be considered a
‘biomarker’ for inflammation (59).
NSAID treatment may cause enteropathy (60,61) or
may aggravate intestinal inflammation in patients with IBD. Chronic
administration of NSAIDs (celecoxib) had the ability to modulate
intestinal microbiome, leading to a decrease in
Bifidobacterium and Lactobacillus followed by an
increase in Coriobacteriaceae, having also a chemoprotective
role by decreasing fecal metabolites (62). Montenegro et al (63) support the harmful effect of NSAIDs,
characterized by a marked reduction in Lactobacillus and by
modulation of local motility and immunity through the
Bifidobacteria group.
In the present study, in contrary, synthetic and
biological immunosuppressive drugs had a positive effect on the
intestinal microbiome, improving dysbiosis and decreasing systemic
inflammation. Anti-TNFα agents can act on the intestinal microbiome
by inhibiting the onset of vascular inflammation and by inducing T
cell apoptosis (64). Other authors
state that this improvement of the intestinal microbiome after TNFα
therapy is due to a decrease in bacterial arthritogenic peptides
(65).
A study which included proteoglycan-induced mice
treated with etanercept for 4 weeks demonstrated the efficacy of
anti-TNF therapy on articular manifestations and on gut microbiome
composition, leading even to a microbial composition similar to
that of the control group (66).
Another study (67) highlighted the
changes in the intestinal microbiome of patients with AS at 1, 3
and 6 months after initiating anti-TNFα therapy. Initially, a
decreased biodiversity was observed, which improved almost to
normal after the first month of treatment.
In conclusion, our findings indicate that the
intestinal microbiome in patients with AS has a special signature
characterized by an inflammatory status induced by the increase in
some bacterial species associated with the decrease in other
species. We demonstrated that the composition of the intestinal
microbiome is influenced by numerous factors, among which genetic
background, smoking status, inflammatory markers, disease phenotype
and treatment play the most important roles. Our results are
similar with those already published and participate in the
enrichment of knowledge in the field, bringing new data on
intestinal dysbiosis in patients with AS.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AC, ER and AMB conceived the study, the methodology
and drafted the manuscript. ER supervised and designed the study.
AC, SC, FP, AMB and CR contributed to the literature resources; and
AC, AMB, FP and ER validated the data and data analysis. AC, SC,
FP, AMB, CR and ER contributed to the review and editing; AC, AMB,
SC, CR, FP and ER contributed to the approval of the final version
of the manuscript. All authors have read and agreed to the
published version of the manuscript.
Ethics approval and consent to
participate
Approval was obtained from the Ethics Committees of
the Grigore T Popa University of Medicine and Pharmacy and
Rehabilitation Hospital Iasi from which the cases were selected.
All the included cases expressed their informed consent to
participate in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Taurog JD, Chhabra A and Colbert RA:
Ankylosing spondylitis and axial spondyloarthritis. N Engl J Med.
374:2563–2574. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Brewerton DA, Hart FD, Nicholls A, Caffrey
M, James DC and Sturrock RD: Ankylosing spondylitis and HL-A 27.
Lancet. 1:904–907. 1973.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Brown MA, Kennedy LG, MacGregor AJ, Darke
C, Duncan E, Shatford JL, Taylor A, Calin A and Wordsworth P:
Susceptibility to ankylosing spondylitis in twins: The role of
genes, HLA, and the environment. Arthritis Rheum. 40:1823–1828.
1997.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Manasson J, Shen N, Garcia Ferrer HR,
Ubeda C, Iraheta I, Heguy A, Von Feldt JM, Espinoza LR, Garcia
Kutzbach A, Segal LN, et al: Gut microbiota perturbations in
reactive arthritis and postinfectious spondyloarthritis. Arthritis
Rheumatol. 70:242–254. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang Y, Guerassimov A, Leroux JY, Cartman
A, Webber C, Lalic R, de Miguel E, Rosenberg LC and Poole AR:
Arthritis induced by proteoglycan aggrecan G1 domain in BALB/c
mice. Evidence for t cell involvement and the immunosuppressive
influence of keratan sulfate on recognition of t and b cell
epitopes. J Clin Invest. 101:1678–1686. 1998.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Capkova J, Hrncir T, Kubatova A and
Tlaskalova-Hogenova H: Lipopolysaccharide treatment suppresses
spontaneously developing ankylosing enthesopathy in B10.BR male
mice: The potential role of interleukin-10. BMC Musculoskelet
Disord. 13(110)2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Thjodleifsson B, Geirsson AJ, Björnsson S
and Bjarnason I: A common genetic background for inflammatory bowel
disease and ankylosing spondylitis: A genealogic study in Iceland.
Arthritis Rheum. 56:2633–2639. 2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Robinson PC, Leo PJ, Pointon JJ, Harris J,
Cremin K, Bradbury LA, Stebbings S and Harrison AA: Australian
Osteoporosis Genetics Consortium; Wellcome Trust Case Control
Consortium et al. Exome-wide study of ankylosing spondylitis
demonstrates additional shared genetic background with inflammatory
bowel disease. NPJ Genom Med. 1(16008)2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tsui FW, Tsui HW, Akram A, Haroon N and
Inman RD: The genetic basis of ankylosing spondylitis: New insights
into disease pathogenesis. Appl Clin Genet. 7:105–115.
2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Costello ME, Ciccia F, Willner D,
Warrington N, Robinson PC, Gardiner B, Marshall M, Kenna TJ, Triolo
G and Brown MA: Brief report: Intestinal dysbiosis in ankylosing
spondylitis. Arthritis Rheumatol. 67:686–691. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Taurog JD, Richardson JA, Croft JT,
Simmons WA, Zhou M, Fernández-Sueiro JL, Balish E and Hammer RE:
The germfree state prevents development of gut and joint
inflammatory disease in HLA-B27 transgenic rats. J Exp Med.
180:2359–2364. 1994.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Asquith MJ, Stauffer P, Davin S, Mitchell
C, Lin P and Rosenbaum JT: Perturbed mucosal immunity and dysbiosis
accompany clinical disease in a rat model of spondyloarthritis.
Arthritis Rheumatol. 68:2151–2162. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lin P, Bach M, Asquith M, Lee AY,
Akileswaran L, Stauffer P, Davin S, Pan Y, Cambronne ED, Dorris M,
et al: HLA-B27 and human β2-microglobulin affect the gut microbiota
of transgenic rats. PLoS One. 9(e105684)2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang L, Hu Y, Xu Y, Li P, Ma H, Li X and
Li M: The correlation between intestinal dysbiosis and the
development of ankylosing spondylitis. Microb Pathog. 132:188–192.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Turner MJ, Sowders DP, DeLay ML, Mohapatra
R, Bai S, Smith JA, Brandewie JR, Taurog JD and Colbert RA: HLA-B27
misfolding in transgenic rats is associated with activation of the
unfolded protein response. J Immunol. 175:2438–2448.
2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kaneko M and Nomura Y: ER signaling in
unfolded protein response. Life Sci. 74:199–205. 2003.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ciccia F, Accardo-Palumbo A, Rizzo A,
Guggino G, Raimondo S, Giardina A, Cannizzaro A, Colbert RA,
Alessandro R and Triolo G: Evidence that autophagy, but not the
unfolded protein response, regulates the expression of IL-23 in the
gut of patients with ankylosing spondylitis and subclinical gut
inflammation. Ann Rheum Dis. 73:1566–1574. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ciccia F, Rizzo A and Triolo G:
Subclinical gut inflammation in ankylosing spondylitis. Curr Opin
Rheumatol. 28:89–96. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tian P, Li B, He C, Song W, Hou A, Tian S,
Meng X, Li K and Shan Y: Antidiabetic (type 2) effects of
Lactobacillus G15 and Q14 in rats through regulation of
intestinal permeability and microbiota. Food Funct. 7:3789–3797.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Morris G, Berk M, Carvalho AF, Caso JR,
Sanz Y and Maes M: The role of microbiota and intestinal
permeability in the pathophysiology of autoimmune and neuroimmune
processes with an emphasis on inflammatory bowel disease type 1
diabetes and chronic fatigue syndrome. Curr Pharm Des.
22:6058–6075. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ebringer A and Wilson C: The use of a low
starch diet in the treatment of patients suffering from ankylosing
spondylitis. Clin Rheumatol. 15 (Suppl 1):S62–S66. 1996.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ugur M, Baygutalp NK, Melikoglu MA,
Baygutalp F, Altas EU and Seferoglu B: Elevated serum
interleukin-23 levels in ankylosing spondylitis patients and the
relationship with disease activity. Nagoya J Med Sci. 77:621–627.
2015.PubMed/NCBI
|
|
23
|
Smith JA and Colbert RA: Review: The
interleukin-23/interleukin-17 axis in spondyloarthritis
pathogenesis: Th17 and beyond. Arthritis Rheumatol. 66:231–241.
2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ciccia F, Guggino G, Rizzo A, Saieva L,
Peralta S, Giardina A, Cannizzaro A, Sireci G, De Leo G, Alessandro
R and Triolo G: Type 3 innate lymphoid cells producing IL-17 and
IL-22 are expanded in the gut, in the peripheral blood, synovial
fluid and bone marrow of patients with ankylosing spondylitis. Ann
Rheum Dis. 74:1739–1747. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Costello ME, Elewaut D, Kenna TJ and Brown
MA: Microbes, the gut and ankylosing spondylitis. Arthritis Res
Ther. 15(214)2013.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
van der Linden S, Valkenburg HA and Cats
A: Evaluation of diagnostic criteria for ankylosing spondylitis. A
proposal for modification of the New York criteria. Arthritis
Rheum. 27:361–368. 1984.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang W, Chen L, Zhou R, Wang X, Song L,
Huang S, Wang G and Xia B: Increased proportions of
Bifidobacterium and the Lactobacillus group and loss
of butyrate-producing bacteria in inflammatory bowel disease. J
Clin Microbiol. 52:398–406. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang L, Han R, Zhang X, Fang G, Chen J,
Li J, Xu S, Qian L, Chen W and Pan F: Fecal microbiota in patients
with ankylosing spondylitis: Correlation with dietary factors and
disease activity. Clin Chim Acta. 497:189–196. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Breban M, Tap J, Leboime A, Said-Nahal R,
Langella P, Chiocchia G, Furet JP and Sokol H: Faecal microbiota
study reveals specific dysbiosis in spondyloarthritis. Ann Rheum
Dis. 76:1614–1622. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Morgan XC, Tickle TL, Sokol H, Gevers D,
Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et
al: Dysfunction of the intestinal microbiome in inflammatory bowel
disease and treatment. Genome Biol. 13(R79)2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Scher JU, Ubeda C, Artacho A, Attur M,
Isaac S, Reddy SM, Marmon S, Neimann A, Brusca S, Patel T, et al:
Decreased bacterial diversity characterizes the altered gut
microbiota in patients with psoriatic arthritis, resembling
dysbiosis in inflammatory bowel disease. Arthritis Rheumatol.
67:128–139. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tito RY, Cypers H, Joossens M, Varkas G,
Van Praet L, Glorieus E, Van den Bosch F, De Vos M, Raes J and
Elewaut D: Brief report: Dialister as a microbial marker of disease
activity in spondyloarthritis. Arthritis Rheumatol. 69:114–121.
2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Haque TR and Barritt AS IV: Intestinal
microbiota in liver disease. Best Pract Res Clin Gastroenterol.
30:133–142. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Minemura M and Shimizu Y: Gut microbiota
and liver diseases. World J Gastroenterol. 21:1691–1702.
2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Morris A: Metabolism: New insights into
the BAT-liver-gut axis. Nat Rev Endocrinol. 13(438)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yip LY, Aw CC, Lee SH, Hong YS, Ku HC, Xu
WH, Chan JMX, Cheong EJY, Chng KR, Ng AHQ, et al: The liver-gut
microbiota axis modulates hepatotoxicity of tacrine in the rat.
Hepatology. 67:282–295. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Pallen MJ and Quraishi MN: The gut
microbiota and the hepatologist: Will our bugs prove to be the
missing link? Dig Dis. 35:377–383. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Rabb H, Pluznick J and Noel S: The
microbiome and acute kidney injury. Nephron. 140:120–123.
2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Noel S, Martina-Lingua MN, Bandapalle S,
Pluznick J, Hamad AR, Peterson DA and Rabb H: Intestinal
microbiota-kidney cross talk in acute kidney injury and chronic
kidney disease. Nephron Clin Pract. 127:139–143. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Sabatino A, Regolisti G, Brusasco I,
Cabassi A, Morabito S and Fiaccadori E: Alterations of intestinal
barrier and microbiota in chronic kidney disease. Nephrol Dial
Transplant. 30:924–933. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ostan R, Béné MC, Spazzafumo L, Pinto A,
Donini LM, Pryen F, Charrouf Z, Valentini L, Lochs H,
Bourdel-Marchasson I, et al: Impact of diet and nutraceutical
supplementation on inflammation in elderly people. Results from the
RISTOMED study, an open-label randomized control trial. Clin Nutr.
35:812–818. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Valentini L, Pinto A, Bourdel-Marchasson
I, Ostan R, Brigidi P, Turroni S, Hrelia S, Hrelia P, Bereswill S,
Fischer A, et al: Impact of personalized diet and probiotic
supplementation on inflammation, nutritional parameters and
intestinal microbiota-The ‘RISTOMED project’: Randomized controlled
trial in healthy older people. Clin Nutr. 34:593–602.
2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Klingberg E, Magnusson MK, Strid H,
Deminger A, Ståhl A, Sundin J, Simrén M, Carlsten H, Öhman L and
Forsblad-d'Elia H: A distinct gut microbiota composition in
patients with ankylosing spondylitis is associated with increased
levels of fecal calprotectin. Arthritis Res Ther.
21(248)2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Rizzatti G, Lopetuso LR, Gibiino G, Binda
C and Gasbarrini A: Proteobacteria: A common factor in human
diseases. Biomed Res Int. 2017(9351507)2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
McIlroy J, Ianiro G, Mukhopadhya I, Hansen
R and Hold GL: Review article: The gut microbiome in inflammatory
bowel disease-avenues for microbial management. Aliment Pharmacol
Ther. 47:26–42. 2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Ciccia F, Guggino G, Rizzo A, Alessandro
R, Luchetti MM, Milling S, Saieva L, Cypers H, Stampone T, Di
Benedetto P, et al: Dysbiosis and zonulin upregulation alter gut
epithelial and vascular barriers in patients with ankylosing
spondylitis. Ann Rheum Dis. 76:1123–1132. 2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Klingberg E, Strid H, Ståhl A, Deminger A,
Carlsten H, Öhman L and Forsblad-d'Elia H: A longitudinal study of
fecal calprotectin and the development of inflammatory bowel
disease in ankylosing spondylitis. Arthritis Res Ther.
19(21)2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Van Praet L, Jans L, Carron P, Jacques P,
Glorieus E, Colman R, Cypers H, Mielants H, De Vos M, Cuvelier C,
et al: Degree of bone marrow oedema in sacroiliac joints of
patients with axial spondyloarthritis is linked to gut inflammation
and male sex: Results from the GIANT cohort. Ann Rheum Dis.
73:1186–1189. 2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Onderdonk AB, Richardson JA, Hammer RE and
Taurog JD: Correlation of cecal microflora of HLA-B27 transgenic
rats with inflammatory bowel disease. Infect Immun. 66:6022–6023.
1998.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Aron-Wisnewsky J, Prifti E, Belda E, Ichou
F, Kayser BD, Dao MC, Verger EO, Hedjazi L, Bouillot JL, Chevallier
JM, et al: Major microbiota dysbiosis in severe obesity: Fate after
bariatric surgery. Gut. 68:70–82. 2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Ley RE, Turnbaugh PJ, Klein S and Gordon
JI: Microbial ecology: Human gut microbes associated with obesity.
Nature. 444:1022–1023. 2006.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ley RE, Bäckhed F, Turnbaugh P, Lozupone
CA, Knight RD and Gordon JI: Obesity alters gut microbial ecology.
Proc Natl Acad Sci USA. 102:11070–11075. 2005.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Kasai C, Sugimoto K, Moritani I, Tanaka J,
Oya Y, Inoue H, Tameda M, Shiraki K, Ito M, Takei Y and Takase K:
Comparison of the gut microbiota composition between obese and
non-obese individuals in a Japanese population, as analyzed by
terminal restriction fragment length polymorphism and
next-generation sequencing. BMC Gastroenterol.
15(100)2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Angelakis E, Armougom F, Carrière F,
Bachar D, Laugier R, Lagier JC, Robert C, Michelle C, Henrissat B
and Raoult D: A metagenomic investigation of the duodenal
microbiota reveals links with obesity. PLoS One.
10(e0137784)2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Hu HJ, Park SG, Jang HB, Choi MK, Park KH,
Kang JH, Park SI, Lee HJ and Cho SH: Obesity alters the microbial
community profile in Korean adolescents. PLoS One.
10(e0134333)2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Parkes GC, Whelan K and Lindsay JO:
Smoking in inflammatory bowel disease: Impact on disease course and
insights into the aetiology of its effect. J Crohn's Colitis.
8:717–725. 2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Savin Z, Kivity S, Yonath H and Yehuda S:
Smoking and the intestinal microbiome. Arch Microbiol. 200:677–684.
2018.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Chen Z, Qi J, Wei Q, Zheng X, Wu X, Li X,
Liao Z, Lin Z and Gu J: Variations in gut microbial profiles in
ankylosing spondylitis: Disease phenotype-related dysbiosis. Ann
Transl Med. 7(571)2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Kolho KL, Korpela K, Jaakkola T, Pichai
MV, Zoetendal EG, Salonen A and de Vos WM: Fecal microbiota in
pediatric inflammatory bowel disease and its relation to
inflammation. Am J Gastroenterol. 110:921–930. 2015.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Wallace JL: NSAID gastropathy and
enteropathy: Distinct pathogenesis likely necessitates distinct
prevention strategies. Br J Pharmacol. 165:67–74. 2012.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Wallace JL: Mechanisms, prevention and
clinical implications of nonsteroidal anti-inflammatory
drug-enteropathy. World J Gastroenterol. 19:1861–1876.
2013.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Montrose DC, Zhou XK, McNally EM, Sue E,
Yantiss RK, Gross SS, Leve ND, Karoly ED, Suen CS, Ling L, et al:
Celecoxib alters the intestinal microbiota and metabolome in
association with reducing polyp burden. Cancer Prev Res (Phila).
9:721–731. 2016.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Montenegro L, Losurdo G, Licinio R,
Zamparella M, Giorgio F, Ierardi E, Di Leo A and Principi M: Non
steroidal anti-inflammatory drug induced damage on lower
gastro-intestinal tract: Is there an involvement of microbiota?
Curr Drug Saf. 9:196–204. 2014.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Danese S, Sans M, Scaldaferri F, Sgambato
A, Rutella S, Cittadini A, Piqué JM, Panes J, Katz JA, Gasbarrini A
and Fiocchi C: TNF-alpha blockade down-regulates the CD40/CD40L
pathway in the mucosal microcirculation: A novel anti-inflammatory
mechanism of infliximab in Crohn's disease. J Immunol.
176:2617–2624. 2006.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Yin J, Sternes PR, Wang M, Song J,
Morrison M, Li T, Zhou L, Wu X, He F, Zhu J, et al: Shotgun
metagenomics reveals an enrichment of potentially cross-reactive
bacterial epitopes in ankylosing spondylitis patients, as well as
the effects of TNFi therapy upon microbiome composition. Ann Rheum
Dis. 79:132–140. 2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Liu B, Yang L, Cui Z, Zheng J, Huang J,
Zhao Q, Su Z, Wang M, Zhang W, Liu J, et al: Anti-TNF-α therapy
alters the gut microbiota in proteoglycan-induced ankylosing
spondylitis in mice. Microbiologyopen. 8(e927)2019.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Zhang F, Ma C and Zhang B: Dynamic
variations in gut microbiota in ankylosing spondylitis patients
treated with anti-TNF-α for six months. Ann Clin Lab Sci.
50:99–106. 2020.PubMed/NCBI
|