In the 1960s, researchers first observed that cells
degraded intracellular components by wrapping them in a membrane to
form a cystic structure and transporting the contents to a small
compartment (called the lysosomes) for recycling (1). Due to the difficulties in studying
this phenomenon, little was known about it until the early 1990s,
when the Japanese molecular cell biologist Ryoshi Ohsumi performed
a series of experiments. In the experiments, baker’s yeast was used
to identify key autophagy genes. The mechanism behind yeast
autophagy was further explained and it was demonstrated that human
cells use a similar mechanism, a tightly regulated cellular process
in which damaged proteins or organelles are wrapped in autophagic
vesicles with a bilayer membrane structure, which are transported
to lysosomes (mammals) or vacuoles (yeast and plants) for
degradation and recycling (2,3). By
contrast, autophagy occurs under normal conditions in response to
adverse stresses, including hypoxia and nutritional deficiency to
supply nutrients and energy and maintain cellular homeostasis.
Autophagy can also act as a collective scavenger to degrade
unwanted substances (including damaged organelles and misfolded
proteins). However, when excessive, autophagy can also lead to
autophagic programmed cell death, which leads to the occurrence of
several diseases (including neurodegenerative diseases, pathogenic
microbial infections and cancer) (4-6).
Cancer is a disease with serious metabolic disorders, and autophagy
serves dual roles in promoting and inhibiting the occurrence and
development of cancer. In addition, numerous signaling pathways and
factors are involved in cancer-related processes (7). Beclin1 is a major factor in autophagy,
but its expression is downregulated or absent in human ovarian
cancer and non-small cell lung cancer, suggesting that Beclin1 is a
tumor suppressor (8,9). Notably, Beclin1 is upregulated in
colon cancer (10). In addition,
numerous tumor suppressor factors (P53, PTEN, AMPK and LKB1) may
negatively regulate the mTOR pathway and activate autophagy
(11-14).
By contrast, oncogenes, including Akt and ERK, can activate the
mTOR signaling pathway to inhibit autophagy (15,16).
Therefore, these factors or their pathways are bridges between
autophagy and cancer. However, it is unclear whether tumor
suppressors promote or inhibit autophagy. It is necessary to
determine the advantages and disadvantages of autophagy is cancer,
as well as the roles served by autophagy-associated factors and
pathways in cancer in order to improve cancer diagnosis, treatment
and prognosis assessment. By reviewing the signaling pathways and
factors related to autophagy and cancer, the present review aimed
to further clarify the association between autophagy and cancer and
its mechanism to provide strategies for the prevention and
treatment of cancer.
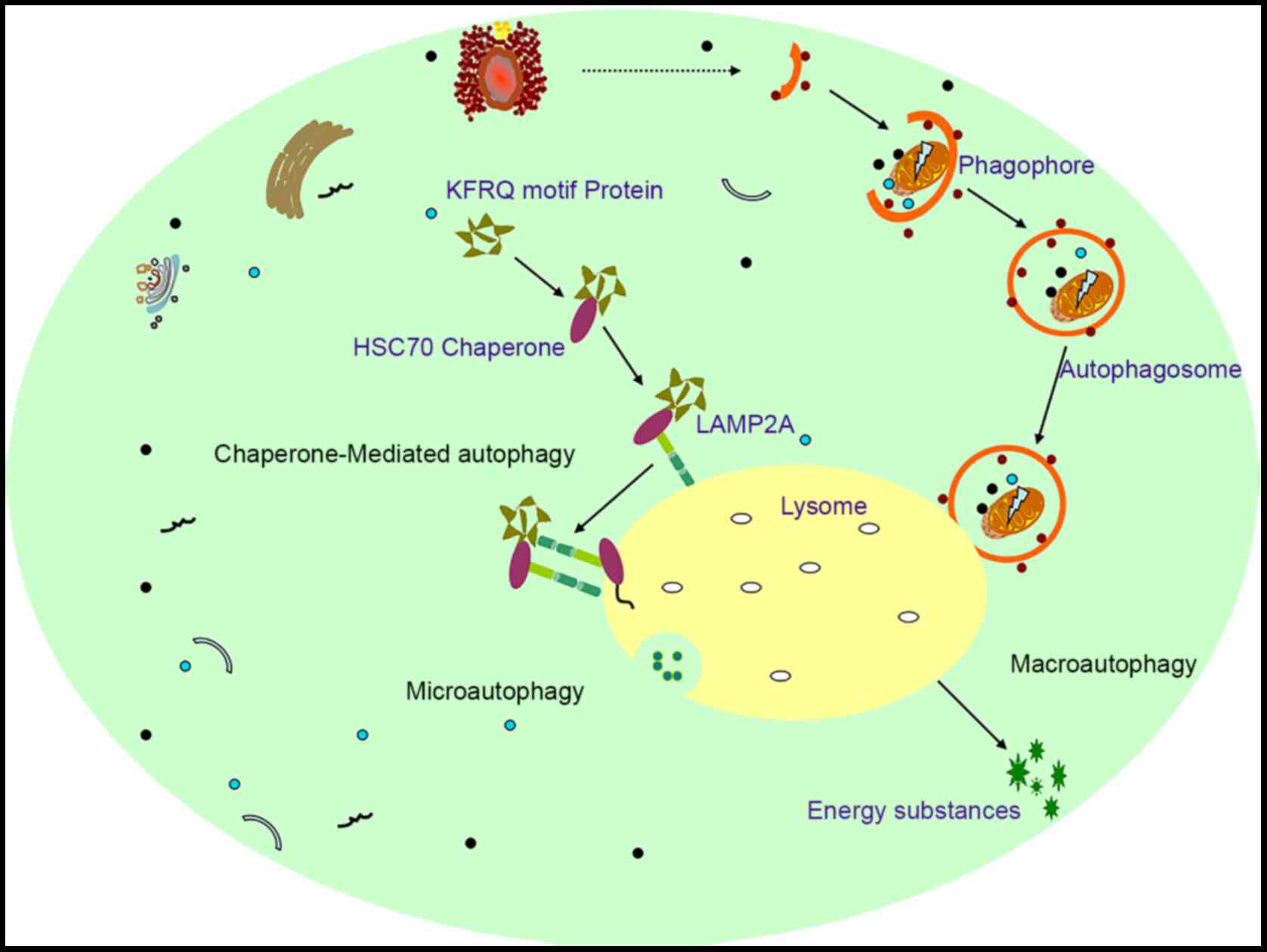
Autophagy is derived from the Greek prefix Auto and
is an intracellular catabolic process. At present, it has been
demonstrated that there are three types of autophagy,
microautophagy, chaperone-mediated autophagy (CMA) and
macroautophagy (MAC) (17).
Microautophagy is generally an invagination of lysosomal and
nuclear membranes, which directly engulf cytoplasmic contents
(Fig. 1). CMA selectively degrades
proteins containing soluble KFERQ-like motifs. These motifs are
recognized by 70-kD heat shock protein (Hsc70) and form complexes
with Hsc70 and chaperones proteins; these complexes are then
delivered to lysosomes through interactions with
lysosome-associated membrane protein (LAMP2A) receptors, resulting
in degradation (Fig. 1). MAC,
commonly known as autophagy (hereafter referred to as autophagy),
occurs through a free bilayer membrane structure derived from the
rough endoplasmic reticulum that wraps unwanted organelles/proteins
or cytosol to form a structure called an autophagosome, which
recognizes and isolates cellular contents that have been labeled by
autophagy receptors. Autophagic lysosomes are then fused with
lysosomes (mammals) or vacuoles (fungi or plants). During this
process, those engulfed organelles/proteins or other components are
degraded into amino acids or other raw materials for cell survival
or the maintenance of cell nutrition, thereby allowing cells to
maintain normal functions under various adverse conditions
(2,3) (Fig.
1).
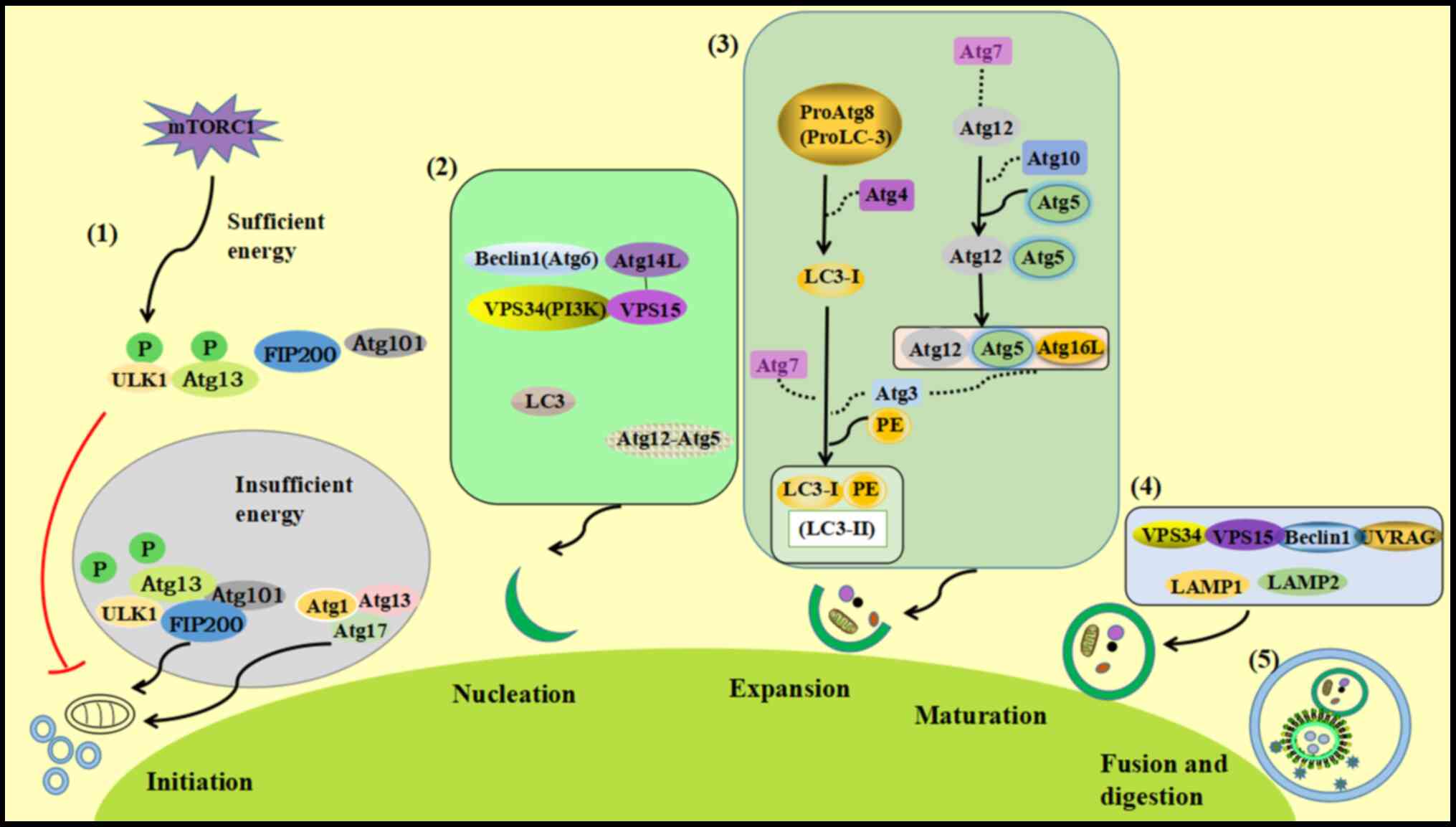
Autophagy includes five stages: autophagy induction,
nucleation, autophagosome extension, autophagosome maturation and
autophagosome lysis (Fig. 2)
(18).
Induction stage: The ULK1-Atg13-FIP200-Atg101
complex is involved in the induction of autophagy (19). Under conditions of sufficient
nutrition, mammalian rapamycin target protein complex 1 has a
certain activity and can phosphorylate unc-51 like kinase 1 (ULK1)
and Atg13 to prevent ULK1 binding to Atg13, FIP200 (FAK-family
interacting protein 200 kDa) and Atg101, thereby inhibiting
autophagy. During nutritional deficiency, the activity of mTORC1 on
the surface of the lysosome is inhibited, and ULK1 and Atg13 are
dephosphorylated, which leads to the activation of ULK1 kinase, and
the ULK complex localizes to phagosomes to form the
ULK1-Atg13-FIP200-Atg101 complex, which induces autophagy. When
mTORC1 activity is inhibited, Atg13-Atg1-Atg17 forms and initiates
autophagy.
Nucleation stage: The Beclin1
(Atg6)-Atg14L-VPS15-VPS34 (PI3K) complex mediates the formation of
pre-autophagosomes (PASs) and the initiation of autophagy, and may
also recruit associated autophagy proteins (including Atg12-Atg5,
Atg6 and LC3). Atg6 and LC3 may promote the expansion and extension
of phagosomes (20).
Autophagosome extension: This step is mainly
mediated by two ubiquitin-like systems, the Atg5-Atg12 system and
the LC3 system (21). Atg7 acts as
an E1-like ubiquitin-activating enzyme to activate Atg12; the E2
ubiquitin-transferase Atg10 transfers Atg12 to Atg5 to form an
Atg12-Atg5 complex, which then binds with Atg16L to form an E3-like
ligase-like Atg12-Atg5-Atg16L complex, which is located on the
outer membrane of autophagosomes and participates in membrane
expansion (22). At the same time,
Atg4 can cleave the C-terminal glycine of proAtg8 (pro-LC3) to form
cytoplasmic LC3-I and covalently bind to phosphatidylethanolamine
(PE) in response to Atg7 and Atg3 to form an Atg8-PE (LC3-II)
fat-soluble complex (22). In
addition, the Atg12-Atg5-Atg16L complex can activate the Atg3
enzyme, promote the transfer of Atg8 (LC3) from Atg3 to PE, and
promote the formation of the Atg8-PE (LC3-II) complex. These two
systems complement each other to promote the expansion and
extension of the autophagic membrane (23).
Autophagosome maturation: Soluble
N-ethylmaleimide-sensitive factor attachment protein receptor
(SNARE) serves an important role in cell material transport and
specific membrane fusion (24). The
PI3K complex (Vps34-Vps15-Beclin1) binds to the anti-ultraviolet
gene (ultraviolet radiation resistance-associated gene, UVRAG) to
form a Vps34-Vps15-Beclin1-UVRAG complex (class III
phosphatidylinositol kinase complex, PI3KC3), which participates in
autophagic maturation and transport. Cheng et al (25) reported that Pacer (protein
associated with UVRAG as an autophagy enhancer) is a
vertebrate-specific autophagy regulatory molecule. Pacer may
directly interact with UVRAG, one of the mammalian PIK3C3 subunits,
thereby alleviating Rubicon (complex subunit, autophagy negative
regulatory factor)-mediated inhibition of Vps34 kinase. By
contrast, Pacer may recruit PI3KC3 and HOPS complex subunits to
autophagic vesicles and promote the fusion of autophagosomes and
lysosomes. In addition, autophagic lysosome fusion requires the
participation of lysosome-associated membrane protein 1 (LAMP1) and
lysosome-associated membrane protein 2 (LAMP2) (26).
Autophagolysis: Acid hydrolase in autophagic
lysosomes degrades the contents of the vesicles.
Basal autophagy serves an important role in cellular
homeostasis by eliminating old or damaged organelles and aggregated
intracellular proteins. In the cancer microenvironment, when cancer
cells are subjected to stress (glucose/cytokine deficiency,
hypoxia, oxidative stress, radiation or anticancer drug therapy),
the level of autophagy increases, which leads to the enhancement of
cancer cell adaptability (cytoprotective response) and contributes
toward the development, proliferation and migration of cancer cells
(27). In addition, it has been
reported that the level of autophagy was decreased in a variety of
cancer types, indicating that autophagy also has a certain
anticancer effect (28). Therefore,
autophagy serves dual roles in the promotion and inhibition of
cancer. The occurrence and development of tumors involves numerous
signaling pathways and associated factors. Therefore, a more
comprehensive review of autophagy and tumor-related signaling
pathways is required to understand the pathogenesis and treatment
of cancer.
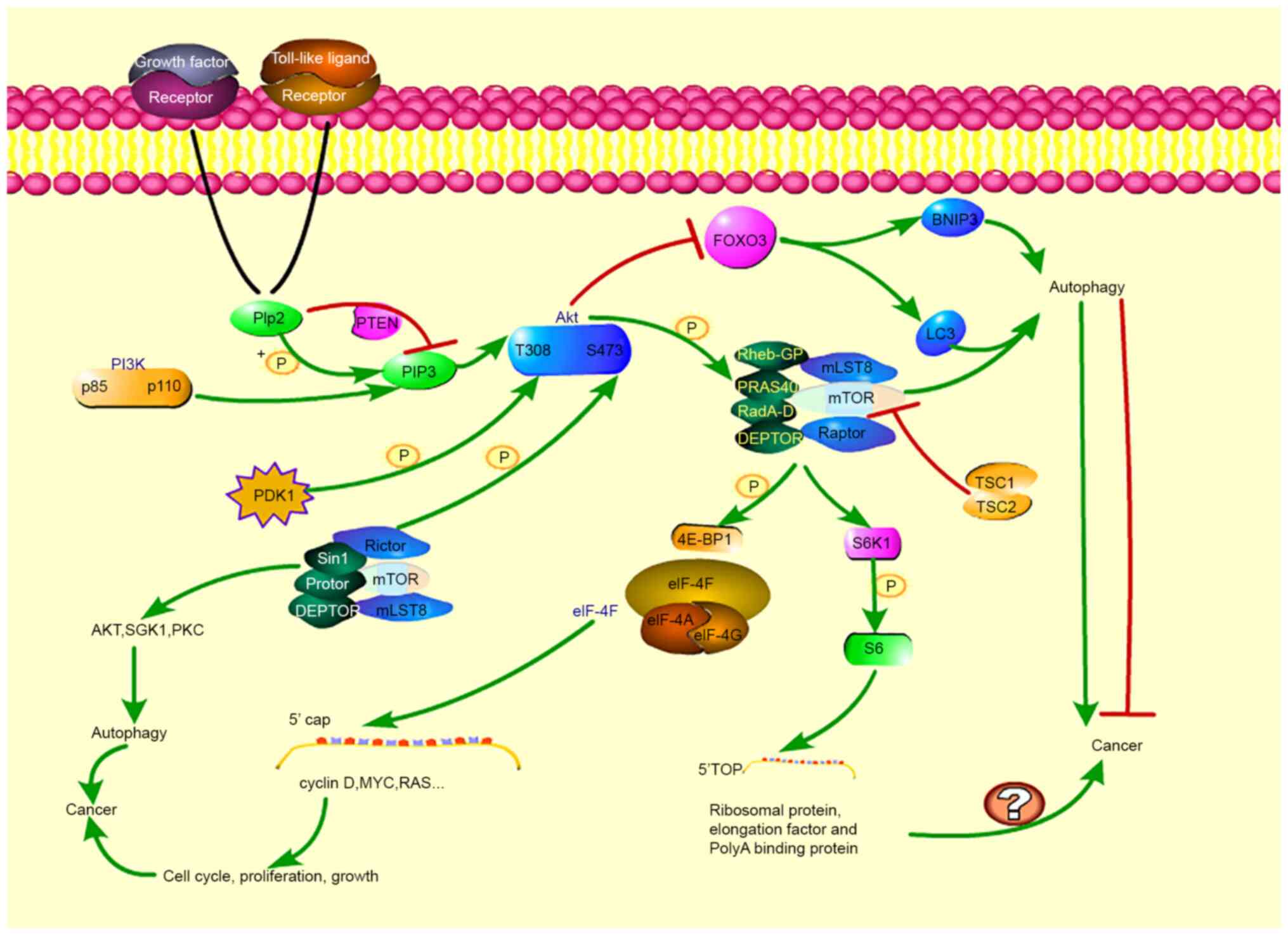
mTOR, a highly evolutionarily conserved kinase, is
important for cell proliferation, growth and metabolism (29). Early studies have demonstrated that
rapamycin, an mTOR inhibitor, may induce autophagy (30). A previous study reported that the
mTOR signal transduction pathway is associated with autophagy
regulation (31). There are two
types of mTOR complexes, rapamycin-sensitive mTORC1 and
rapamycin-insensitive mTORC2 (32,33).
Rapamycin-sensitive mTORC1 is composed of mTOR, mLST8, Raptor,
PRAS40, DEPTOR, RadA-D and Rheb-GP and mTORC2 is composed of mTOR,
mLST8, Rictor and related proteins (Sin1 and Protor, DEPTOR;
Fig. 3) (34). mTOR regulates autophagy through two
mechanisms. mTORC1 acts on 4E-BP1 (eIF-4E-binding protein 1) and
S6K1 through the signal transduction pathway and may initiate
transcription and translation of associated genes and regulate
autophagy (35). mTOR kinase may
also directly act on Atg and regulate autophagy (36). 4E-BP1 is a negative regulator of
translation and may be phosphorylated and inactivated by mTORC1.
The inactivated 4E-BP1 dissociates from eIF-4E, thereby activating
eIF-4E. EIF-4E binds with eIF-4A and eIF-4G to form an eIF-4F
complex. The EIF-4F complex binds to the cap structure at the 5'
end of target mRNA, which promotes the initiation of translation.
EIF-4E may regulate the translation of numerous proteins, including
cyclin D-MYC and RAS, which are closely associated with cancer cell
growth, proliferation and cell cycle regulation, thereby affecting
cancer progression (37,38). S6K1 is a serine/threonine kinase
with multiple phosphorylation sites that is directly or indirectly
regulated by mTORC1 and participates in the regulation of autophagy
(30). S6K1 may phosphorylate and
activate 40S ribosomal protein S6. Activated S6 may improve the
translation efficiency of 5' terminal oligopyrimidine bundle (TOPS)
RNA. 5'Top mRNA accounts for 15-20% of total intracellular RNA and
encodes numerous components required for protein translation,
including ribosomal proteins, elongation factors and polyA-binding
proteins. A previous study demonstrated that abnormal synthesis of
these proteins is associated with cancer (39). Members of the AGC protein kinase
family, including Akt, SGK1 and PKC, are known substrates of mTORC2
and serve important roles in regulating cytoskeletal remodeling and
autophagy (40). The expression of
the tumor suppressor FOXO3a is usually downregulated in cancer
(41). Activated FOXO3 may induce
autophagy by enhancing the transcription of autophagy-related genes
LC3 and BNIP3, while mTORC2 blocks the activation of FOXO3 by
activating Akt (42). Therefore,
FOXO3a is a key molecule connecting autophagy and cancer, and its
expression may be regulated by regulating autophagy-related
signaling pathways, suggesting that FOXO3a may be a target for the
treatment of cancer.
When a growth factor or a Toll-like ligand binds to
a growth factor receptor or a Toll-like receptor, p85 is activated,
and p110 is recruited, thereby activating PI3K and then catalyzing
PIP2 phosphorylation on the plasma membrane to produce the second
messenger PIP3, recruiting Akt (43). PTEN is a phosphatase that inhibits
the transition of PIP2 to PIP3, thereby preventing PIP3 from
recruiting Akt to the membrane, inhibiting Akt activation by
preventing phosphorylation (44).
PDK1 is a kinase. When Akt is recruited to the membrane, PDK1
phosphorylates Akt at threonine 308 (T308), and mTORC2 and other
kinases phosphorylate Akt at serine 473 (S473). When these two
sites are phosphorylated, Akt is fully activated, and PRAS40 is
phosphorylated, thereby alleviating PRAS40-mediated inhibition of
mTORC1(45). Following mTORC1
activation, some of the aforementioned regulatory responses take
place. In addition, the TSC2 and TSC1 complexes, which are encoded
by the Tsc1 and Tsc2 genes, respectively, may inactivate Rheb,
inhibiting the activity of mTORC1, and thereby promoting autophagy
(46). Activated Akt may
phosphorylate TSC2 and prevent the formation of the TSC complex,
which further leads to the activation of mTORC1, thereby inhibiting
autophagy (47). It has been
demonstrated that the mTOR/PI3K/Akt signaling pathway is triggered
in tumor cells and is considered to be a key therapeutic target for
the treatment of various cancer types (48). Han et al (49) demonstrated that β-rapazone may block
the mTOR/PI3K/Akt signaling pathway, induce autophagy, and
ultimately inhibit the migration and invasion of nasopharyngeal
carcinoma cells. In addition, angiogenesis may provide nutrition
and oxygen to tissues, which are essential for tumor growth and
metastasis. It has been demonstrated that dihydroartemisinin (DHA)
may induce autophagy in human umbilical vein endothelial cells by
inhibiting the Akt/mTOR signaling pathway (50). It has been reported that
angiogenesis is a major cause of cancer, and DHA is an effective
antiangiogenic agent (51). Recent
studies have demonstrated that angiogenesis is closely associated
with autophagy (52). Antitumor
therapy by inhibiting angiogenesis is also a promising strategy. In
numerous cancer types, protective autophagy induces chemical
resistance to various chemotherapeutic agents. Therefore, the
efficacy of chemotherapeutic agents in numerous cancer types is
limited by spontaneous protective autophagic induction, and
chemical resistance may be overcome by studying autophagy pathways
and properly regulating autophagy levels. Liu et al
(53) reported that the combination
of 9za with autophagy inhibitors (including chloroquine or
3-methyladenine) exhibited higher cytotoxicity and apoptosis than
9za alone and reported that the Akt/mTOR axis was associated with
9za-induced autophagy (53). In
addition, PDK1 overexpression leads to increased phosphorylation of
PDK1 and Akt, and blockade of 9za-mediated autophagy (53). The regulation of autophagy
contributes toward the expression of tumor suppressor proteins or
oncogenes. Tumor suppressors are negatively regulated by mTOR,
leading to autophagy induction and cancer initiation inhibition. By
contrast, oncogenes may be activated by mTOR, class I PI3K and Akt,
leading to autophagy inhibition and cancer development. The
induction of autophagy has a positive regulatory effect on cancer
chemotherapy resistance; therefore, reducing chemotherapy
resistance by inhibiting autophagy is one of the strategies that
may be considered in cancer treatment. Furthermore, studies have
reported that abnormal PI3K/Akt/mTOR signaling is associated with
cancer cell growth, survival, movement and resistance to targeted
therapy (54-56).
It was demonstrated that in head and neck squamous cell carcinoma,
the mutation rate of the mTOR genome reached 80-90% (57). In addition, previous studies have
reported that in gastric cancer, breast cancer, prostate cancer,
endometrial cancer, cholangiocarcinoma and bladder cancer,
inhibiting the mTOR signaling pathway may inhibit cancer
progression or has a synergistic effect with current conventional
treatments (including radiotherapy and chemotherapy), enhancing
sensitization and efficacy (58-63).
mTOR is a key factor in this pathway. A previous study has
demonstrated that there is inevitable resistance to first- and
second-generation mTOR inhibitors, which may be associated with
drug-resistant mutants and the induction of autophagy (64). However, the third-generation mTOR
inhibitor, Rapalink, (which inhibits both sites of mTOR at the same
time) may enter cancer cells, inactivate mTOR signaling, and
decrease resistance to first- or second-generation mTOR inhibitors
(64). Akt inhibitor VIII
(AKTi-1/2) may reversibly inhibit Akt, and when combined with other
inhibitors, it synergistically enhances the anticancer effect
(65). Therefore, the majority of
studies have focused on PI3K/Akt/mTOR signaling pathway inhibitors.
Unfortunately, the majority of mTOR signaling pathway inhibitors
are in clinical trials, and numerous challenges need to be overcome
if these treatments are to be used clinically. A previous study
reported that, in patients with HER2-positive cancer, inhibiting
the mTOR pathway alone may activate autophagy, causing cancer cells
to escape and develop drug resistance (66). Therefore, it is necessary to
investigate the inhibition of other signal transduction pathways,
dual pathway inhibition or multiple pathway inhibition to achieve
the inhibition of cancer progression. To date, no typical
biomarkers have been identified to predict the PI3K/Akt/mTOR
signaling pathway inhibitory responses or drug resistance.
Therefore, targeting the PI3K/Akt/mTOR signaling pathway in cancer
treatment requires comprehensive investigation.
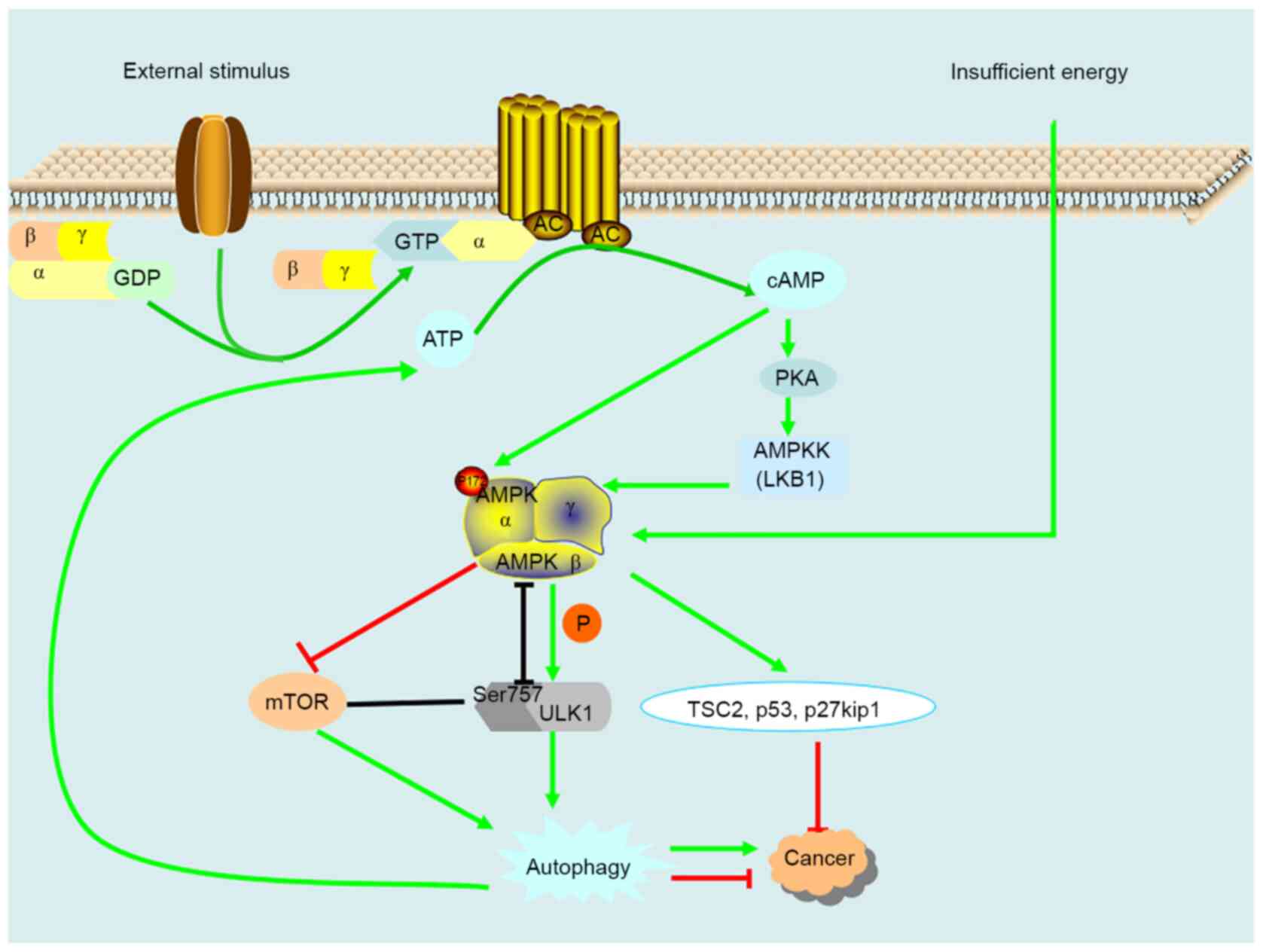
As a cellular energy receptor, AMPK serves a key
role, not only in the regulation of cellular energy homeostasis,
but also in carcinogenesis and anticancer drug resistance (67). AMPK may form a heterotrimeric
complex containing a catalytic α-subunit and regulatory β-and
γ-subunits. Following binding to the γ-subunit, AMP allosterically
activates the complex, resulting in substrate phosphorylation at
threonine 172 and further phosphorylation by upstream LKB1 in the
activation ring of the α-subunit (Fig.
4) (68). When cells are
stimulated, extracellular signaling molecules first bind to the
receptor to form a complex and then activate the Gs protein on the
cell membrane. The activated Gs protein reactivates adenylate
cyclase (AC) on the cell membrane to catalyze the removal of
pyrophosphate from ATP to produce cAMP. The decreased cellular
energy state enhances the phosphorylation and activation of AMPK by
LKB1, and following activation, AMPK catalyzes the phosphorylation
of ULK1, which induces autophagy (69). Induced autophagy removes the
accumulated unwanted components that cause inflammation, stimulate
ROS, trigger cell death and induce genomic instability, all of
which contribute toward cancer. Therefore, AMPK-induced autophagy
may prevent the occurrence and development of tumors. In addition,
induced autophagy may also provide nutrients for tumor cells and
aid tumors in adapting to adverse environments. It has been
demonstrated that in breast cancer cells, parthenolide activates
the apoptosis pathway and AMPK-autophagy survival pathway through
the production of ROS. Inhibition of AMPK or autophagy may
potentially enhance the anticancer effect of parthenolide on breast
cancer cells (70). In addition,
AMPK can also act on the downstream factors TSC2, p53 and
p27kip1(71). LKB1 is the upstream
tumor suppressor of AMPK, and TSC2, p53 and p27kip1 are downstream
tumor suppressor genes, indicating that AMPK has a certain role in
tumor inhibition.
Notably, AMPK is a key factor in the response to
metabolic stress and the maintenance of energy homeostasis. As AMPK
is able to support the proliferation of cancer cells by regulating
energy metabolism, the AMPK pathway has attracted much attention in
the treatment of cancer. In thyroid cancer, the AMPK pathway is
highly activated (72). It was
demonstrated that inhibiting the AMPK pathway may inhibit cancer.
JQ1 may induce autophagy by activating the LKB1/AMPK pathway,
thereby inhibiting cancer cell proliferation (73). In addition, NPC-26 kills human
colorectal cancer cells by activating AMPK signaling (74). Therefore, AMPK and its pathways are
expected to serve as therapeutic targets for the treatment of a
variety of cancer types. In summary, the role of the AMPK pathway
in autophagy and cancer is complicated, but AMPK pathway inhibition
is more conducive to cancer suppression than activation.
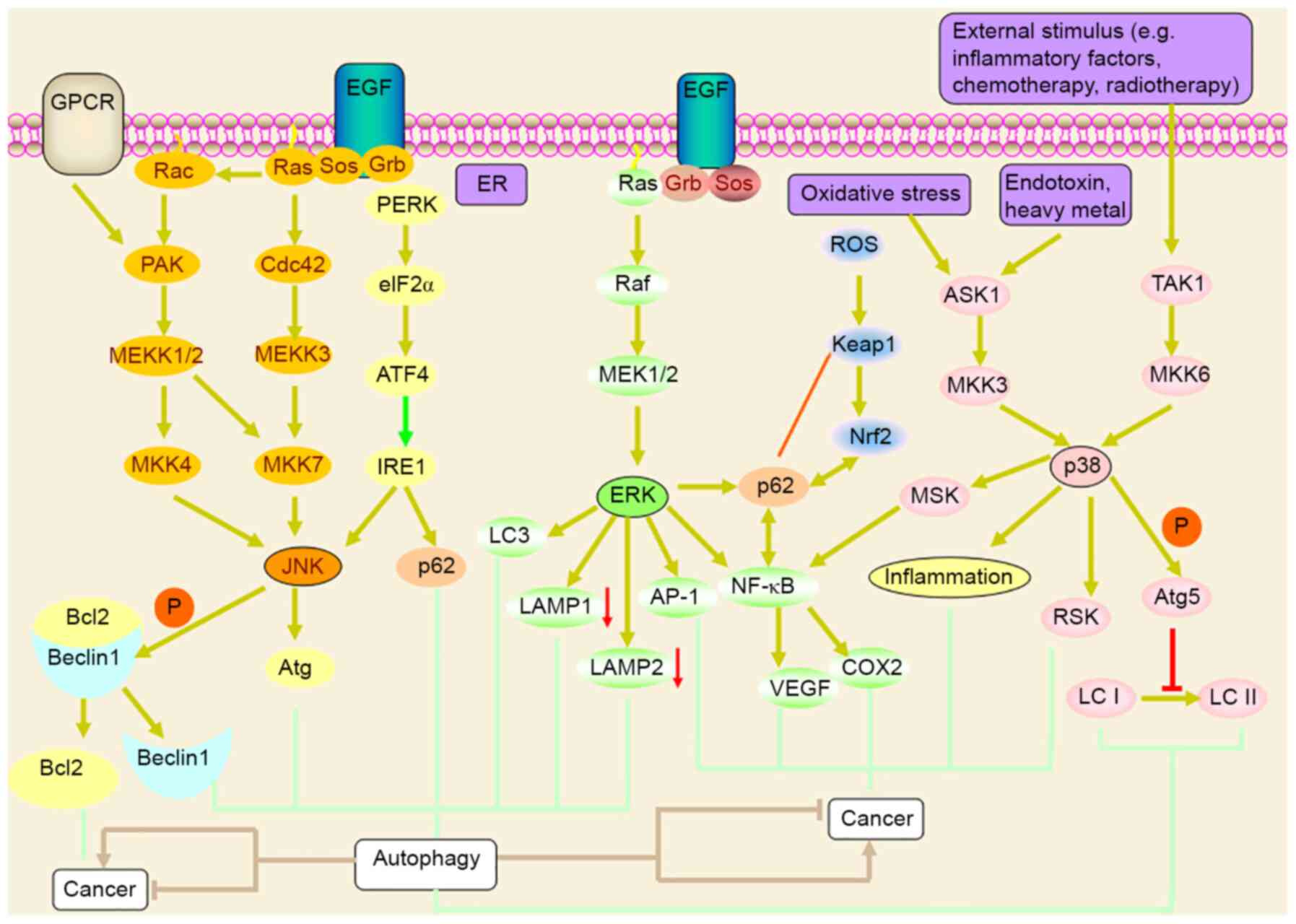
MAPK is a serine/threonine protein kinase that
primarily includes three subgroups: ERK, JNK/SAPKII and p38-MAPK
(Fig. 5). MAPK may regulate the
expression of downstream genes by regulating B lymphocyte tumor-2
gene (Bcl-2) family proteins. Bcl-2 family proteins are key
regulators in the disposal of abnormal cells and participate in
almost all cell pathological and physiological processes, including
cell cycle regulation, cell survival and death, gene expression,
and cell movement (75). JNK
regulates autophagy through Beclin-dependent and -independent
signaling (76). ER stress
upregulates p62 expression through the PERK-mediated eIF2 α-ATF4
pathway and activates the ER transmembrane sensor. Inositol
requires enzyme 1 (IRE1) to activate Jun N-terminal kinase (JNK),
and JNK activation may inhibit/phosphorylate the antiapoptotic
protein Bcl2, which leads to the dissociation of Bcl2 from
Beclin-1, resulting in autophagic flux. Pan et al (77) found that Prodigiosin-induced
endoplasmic reticulum stress could lead to breast cancer cell death
through the PERK-mediated eIF2 α-ATF4 pathway, in which IRE1-JNK
mediates Bcl2-induced cell death. However, JNK mediates the
increase in Bcl2 and protective autophagy, resulting in increased
autophagic flux and the induction of therapeutic resistance.
Therefore, the therapeutic agent Prodigiosin, could be used in
combination with autophagy inhibitors to inhibit associated
autophagic flux, which may improve treatment efficacy. P62 is a
ubiquitin-binding protein that is closely associated with the
ubiquitination of proteins. P62 is involved in the regulation of a
variety of cellular signal transduction pathways and autophagy
(78). In autophagy, p62 binds to
ubiquitin, forms a complex with LC3-II located on the autophagic
membrane, and is degraded in the autophagic lysosome. Therefore,
when autophagy occurs, p62 protein is continuously degraded in the
cytoplasm; when autophagic activity is weakened and autophagy is
deficient, p62 protein will accumulate in the cytoplasm. P62 is one
of the marker proteins that reflects autophagic activity. The P62
level indirectly reflects the level of autophagosome clearance. A
large number of studies have demonstrated that high expression of
p62 is associated with poor prognosis in colon cancer, non-small
cell lung cancer and breast cancer (79-81).
Therefore, p62 may be an important target for the treatment of
these cancer types. By contrast, JNK may upregulate the expression
of Atg and promote autophagy. Chen et al demonstrated that
tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)
promoted autophagy in A549 cells by regulating the expression of
ATG through the JNK pathway (82).
Autophagy inhibition may inhibit proliferation and enhance
apoptosis in A549 cells induced by TRAIL (82). A previous study have demonstrated
that JNK has a significant oncolytic effect, and autophagy inducers
acting on the JNK/Beclin-1 axis may enhance this oncolytic effect
(83). Notably, JNK is a potential
molecular target for tumor therapy. When cells are stimulated by
growth factors, mitogens and environmental stimuli, they activate
the ERK pathway through the Ras-Raf-MEK1/2-ERK cascade and act on
nuclear transcription factors, including AP-1 and NF-κB to regulate
gene expression (84). In addition,
the activation of ERK may promote the expression of LC3 and p62,
promote autophagy, and downregulate the expression of LAMP1 and
LAMP2, thereby inhibiting the binding of lysosomes to
autophagosomes and inhibiting the degradation of autophagosomes
(85). Mou et al (86) reported that Berbamine exhibited
anticancer effects on human HT-29 colon cancer cells by inhibiting
the MEK/ERK pathway and cell migration, and inducing autophagy and
apoptosis. Therefore, MEK/ERK is expected to be a therapeutic
target for colon cancer.
Autophagy exhibits dual effects on cells, not only
protecting cells, but also leading to cell death. The production of
tumor-associated ROS may be stimulated by endogenous and exogenous
factors. Endogenous factors generally include mitochondrial
products and nutrient deficiency, and exogenous factors include
chemotherapy and radiotherapy. ROS have been suggested to induce or
mediate the activation of MAPK family members and serve important
roles in autophagy and apoptosis (95). It is known that there is a series of
complex signal transduction pathways and interactions between ROS
and autophagy that regulate autophagy. In response to cellular
stress, there are checks and balances between the two responses;
ROS may participate in the induction of autophagy, and conversely,
autophagy may control the level of ROS (96). In addition, ROS-mediated autophagy
activation mainly serves a cytoprotective role in starvation
conditions. However, in certain cases, ROS may induce cell damage
and participate in certain signal transduction pathways, leading to
autophagic death. JNK activation is involved in the apoptosis- and
autophagy-mediated responses to various stress signals (97). JNK may mediate the phosphorylation
of the antiapoptotic protein, Bcl-2/Bcl-xL, to change the MMP. In
this respect, the mitochondrial dysfunction induced by lutein VI
may be associated with the JNK-mediated apoptotic response. JNK
activation also contributes toward autophagy induction. Lutein VI
induces G2/M arrest, apoptosis and autophagy in human osteosarcoma
cells by activating the ROS/JNK pathway (98). Chitooligosaccharide (COS) exerts
significant antitumor activity on human cervical cancer C33A cells
by activating oxidative stress and mitochondrial apoptosis and
autophagy signal transduction. Ubenimex may inhibit the
proliferation of pituitary adenoma GH3 and MMQ cells, and induce
apoptosis and autophagy, which may be associated with the
activation of the ROS/ERK1/2 pathway (99). These findings provide novel
perspectives for the possible application of Ubenimex in the
chemotherapeutic treatment of pituitary adenomas. In addition,
autophagy is associated with cancer treatment resistance. It has
been reported that breast cancer MCF-7 cells are resistant to
tamoxifen through autophagy induced by p38/c-JNK (100). A previous study reported that ECZ
induces apoptosis in mouse colon cancer CT-26 cells through a
caspase-dependent pathway and triggers autophagy by increasing the
formation of autophagosomes, and the production of ROS serves a key
role in this process (101).
Inhibition of autophagy enhances ECZ-induced apoptosis, which is
due to increased ROS production, suggesting that autophagy may
serve a cytoprotective role by resisting apoptosis (101). Therefore, ECZ may be used in
combination with autophagy inhibitors for the chemoprevention of
cancer or as a chemotherapeutic agent. By examining ROS production,
apoptosis and autophagy, it has been demonstrated that regulating
the apoptosis-autophagy balance in cells treated with ultralong
silver nanowires may protect A549 cells from ROS accumulation and
nutrient deficiency ultralong nanowire stimulation (102). Therefore, autophagy serves a role
in cytoprotection, which is also a factor leading to the
therapeutic resistance of cancer cells. In general, ROS serves
important roles in apoptosis and autophagy signal transduction, and
the production of ROS selectively induces cancer cell death.
Autophagic cell death leads to a significant decrease in the number
of cancer cells, but as autophagic cell death generally occurs in
normal cells rather than cancer cells, the inactivation of
autophagic cell death remains an important cause of tumor
formation.
The basal level of autophagy suppresses
tumorigenesis by decreasing damaged organelles and proteins and
other unwanted components to maintain cellular homeostasis.
However, autophagy disorders positively regulate the occurrence and
development of tumors. Due to the association between autophagy and
tumor occurrence and development, a series of autophagy regulators
have been developed to treat tumors (Table I).
A number of autophagy regulators, including
everolimus tablets, venezuela, AT-101, rapamycin, 3-MA, LY294002,
bafilomycin A1, autophinib, ROC-325, IITZ-01, daurisoline, 3BDO,
9f, chloroquine (CQ), hydroxychloroquine (HCQ) and spautin-1, are
used to treat cancer (103-117).
CQ and HCQ are lysosome inhibitors that inhibit autophagy and
induce the accumulation of autophagosomes by altering lysosomal pH
(110,117). Preclinical studies have
demonstrated that CQ or HCQ may inhibit the growth of cancer cells
by inhibiting autophagy in bladder cancer, pancreatic cancer, lung
cancer and glioma (110,116,118). In addition, a number of studies
have reported that these reagents enhance the therapeutic effects
of radiotherapy and chemotherapy by inhibiting autophagy-mediated
anticancer therapy (53,111,112). Therefore, the development of
autophagy-specific inhibitors is a novel therapeutic strategy for
anticancer therapy.
Under cellular stress conditions, including nutrient
deficiency or starvation, hypoxia, oxidative stress, pathogen
infection, radiation or anticancer drug therapy, the level of
autophagy increases, which enhances cell adaptability and viability
and serves a role in cell protection. For solid tumors with low
blood supply, autophagy provides energy for cancer cells, but this
is not always the case. Autophagy also serves a different role in
different types of cancer. AT-101 induces autophagy via
Beclin1-dependent or Beclin1-independent pathways; induces
survival-mediating protective autophagy in Burkitt lymphoma, breast
cancer, cervical cancer and non-small cell lung cancer; and induced
autophagic death in prostate cancer cells, malignant peripheral
schwannomas and gliomas (105,106). In addition, different types of
tumors have different autophagic activities. In ovarian cancer,
lung cancer, brain glioma and esophageal cancer, the protein
expression of Beclin1 and autophagic activity are decreased
(119,120). However, in nasopharyngeal
carcinoma, the prognosis of the group with high Beclin1 expression
combined with radiotherapy was worse than that of the group with
low Beclin1 expression. This effect may be due to the decrease in
microvessel density in tumor tissue following radiotherapy,
resulting in tissue hypoxia, thereby inducing autophagy and
promoting tumor survival in a hypoxic environment. Therefore, the
sensitivity of cancer cells to radiotherapy is decreased (121). Autophagy function was decreased
and cell necrosis was increased, promoting tumor progression.
Beclin1 deletion decreases autophagy, thereby promoting the
occurrence and development of tumors. Tumor cells degrade unwanted
substances through autophagy to generate nutrients and decrease
energy consumption, which is conducive to tumor survival; high
autophagic activity may confer a survival advantage in harsh
environments but may weaken the effect of radiotherapy. Therefore,
when developing tumor-associated autophagy inhibitors/activators,
comprehensive consideration should be given to different types of
tissues and different types of tumors. In numerous cancer types,
protective autophagy induces resistance to various chemotherapeutic
agents. Therefore, the efficacy of chemotherapeutic agents in
numerous cancer types is limited by spontaneous protective
autophagy induction, and chemoresistance may be overcome by
studying autophagic pathways and effectively regulating autophagy
levels.
In conclusion, the signaling pathways and factors
associated with autophagy and cancer are intertwined. Tumor
suppressors may serve a dual role in autophagy. The occurrence of
autophagy may promote cancer and inhibiting autophagy may inhibit
cancer. The occurrence of autophagy may also inhibit cancer
progression. Activation of autophagy may inhibit cancer, but at the
same time, autophagy is a key factor in the development of drug
resistance. Therefore, it is also necessary to prevent the
development of drug resistance, including through the inhibition of
multiple signal transduction pathways, to increase the inhibition
of cancer and inhibit drug resistance. The mTOR/PI3K/Akt, AMPK and
MAPK signaling pathways and their upstream and downstream factors
all serve important roles in cancer and autophagy, and no reliable
evidence has been collected for the reversal of these pathways.
Therefore, reasonable analysis, evaluation and identification of
cancer types, staging and the tumor microenvironment contribute
toward the selection of therapeutic targets and prognostic
markers.
Not applicable.
Funding: The present study was supported by grants from the
Inner Mongolia Autonomous Region Higher Education Research Project
of China (grant no. NJZY19106) and the National Natural Science
Foundation of China (grant no. 81660468).
Not applicable.
CC wrote the manuscript. HG constructed and edited
the figures. XS revised the manuscript. All authors read and
approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Klionsky DJ: Autophagy revisited: A
conversation with Christian de Duve. Autophagy. 4:740–743.
2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Nakatogawa H, Suzuki K, Kamada Y and
Ohsumi Y: Dynamics and diversity in autophagy mechanisms: Lessons
from yeast. Nat Rev Mol Cell Biol. 10:458–467. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Yamamoto H, Fujioka Y, Suzuki SW, Noshiro
D, Suzuki H, Kondo-Kakuta C, Kimura Y, Hirano H, Ando T, Noda NN,
et al: The intrinsically disordered protein Atg13 mediates
supramolecular assembly of autophagy initiation complexes. Dev
Cell. 38:86–99. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Byun S, Lee E and Lee KW: Therapeutic
implications of autophagy inducers in immunological disorders,
infection, and cancer. Int J Mol Sci. 18:1959–1980. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ma X, Yin X, Liu H, Chen Q, Feng Y, Ma X
and Liu W: Antiproliferative activity of plumbagin
(5-hydroxy-2-methyl-1,4-naphthoquinone) in human gastric carcinoma
cells is facilitated via activation of autophagic pathway,
mitochondrial-mediated programmed cell death and inhibition of cell
migration and invasion. J BUON. 24:2000–2005. 2019.PubMed/NCBI
|
|
6
|
Khan M, Imam H and Siddiqui A: Subversion
of cellular autophagy during virus infection: Insights from
hepatitis B and hepatitis C viruses. Liver Res. 2:146–156.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yan X, Zhou R and Ma Z: Autophagy-cell
survival and death. Adv Exp Med Biol. 1206:667–696. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Shen MX and Ding JB: Expression levels and
roles of EMC-6, Beclin1, and Rab5a in the cervical cancer. Eur Rev
Med Pharmacol Sci. 21:3038–3046. 2017.PubMed/NCBI
|
|
9
|
Yu S, Cheng C, Wang J, Wang J, Qu Z, Ren
H, Li Y, Ning Q, Chen M and Hu T: Loss of Beclin1 expression and
Nrf2 overexpression are associated with poor survival of patients
with non-small cell lung cancer. Anticancer Agents Med Chem.
18:1680–1687. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wu S, Sun C, Tian D, Li Y, Gao X, He S and
Li T: Expression and clinical significances of Beclin1, LC3 and
mTOR in colorectal cancer. Int J Clin Exp Pathol. 8:3882–3891.
2015.PubMed/NCBI
|
|
11
|
Chen JH, Zhang P, Chen WD, Li DD, Wu XQ,
Deng R, Jiao L, Li X, Ji J, Feng GK, et al: ATM-mediated PTEN
phosphorylation promotes PTEN nuclear translocation and autophagy
in response to DNA-damaging agents in cancer cells. Autophagy.
11:239–252. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang Z, Wang N, Liu P and Xie X: AMPK and
Cancer. Exp Suppl. 107:203–226. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Shaw RJ, Bardeesy N, Manning BD, Lopez L,
Kosmatka M, DePinho RA and Cantley LC: The LKB1 tumor suppressor
negatively regulates mTOR signaling. Cancer Cell. 6:91–99.
2004.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li H, Cao X, Chen X, Yi X, Xia J, Chen J
and Yang L: Bufadienolides induce apoptosis and autophagy by
inhibiting the AKT signaling pathway in melanoma A-375 cells. Mol
Med Rep. 20:2347–2354. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhang J, Zhao D, Xie Z and Qi Y:
Down-regulation of AKT combined with radiation-induced autophagy
and apoptosis roles in MCF-7 cells. Biomed Mater Eng. 26 (Suppl
1):S2259–S2265. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jiao YN, Wu LN, Xue D, Liu XJ, Tian ZH,
Jiang ST, Han SY and Li PP: Marsdenia tenacissima extract
induces apoptosis and suppresses autophagy through ERK activation
in lung cancer cells. Cancer Cell Int. 18(149)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhou J, Chong SY, Lim A, Singh BK, Sinha
RA, Salmon AB and Yen PM: Changes in macroautophagy,
chaperone-mediated autophagy, and mitochondrial metabolism in
murine skeletal and cardiac muscle during aging. Aging (Albany NY).
9:583–599. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Oczypok EA, Oury TD and Chu CT: It's a
cell-eat-cell world: Autophagy and phagocytosis. Am J Pathol.
182:612–622. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jung CH, Jun CB, Ro SH, Kim YM, Otto NM,
Cao J, Kundu M and Kim DH: ULK-Atg13-FIP200 complexes mediate mTOR
signaling to the autophagy machinery. Mol Biol Cell. 20:1992–2003.
2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhan ZZ, Chen X and Zhang Y: Autophagy and
the relativity of its function. Di Er Jun Yi Da Xue Xue Bao.
37:1189–1194. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kim SH, Yu HS, Park S, Park HG, Ahn YM,
Kang UG and Kim YS: Electroconvulsive seizures induce autophagy by
activating the AMPK signaling pathway in the rat frontal cortex.
Int J Neuropsychopharmacol. 23:45–52. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chang H, Peng X, Yan X, Zhang J, Xu S,
Wang H, Wang Z, Ma X and Gao Y: Autophagy and Akt-mTOR signaling
display periodic oscillations during torpor-arousal cycles in
oxidative skeletal muscle of Daurian ground squirrels
(Spermophilus dauricus). J Comp Physiol B 15: 2019.
https://doi.org/10.1007/s00360-019-01245-5.
|
|
23
|
Kabeya Y, Mizushima N, Yamamoto A,
Oshitani-Okamoto S, Ohsumi Y and Yoshimori T: LC3, GABARAP and
GATE16 localize to autophagosomal membrane depending on form-II
formation. J Cell Sci. 117:2805–2812. 2004.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang Y, Li L, Hou C, Lai Y, Long J, Liu J,
Zhong Q and Diao J: SNARE-mediated membrane fusion in autophagy.
Semin Cell Dev Biol. 60:97–104. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Cheng X, Ma X, Zhu Q, Song D, Ding X, Li
L, Jiang X, Wang X, Tian R, Su H, et al: Pacer is a mediator of
mTORC1 and GSK3-TIP60 signaling in regulation of autophagosome
maturation and lipid metabolism. Mol Cell. 73:788–802.e7.
2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Eskelinen EL: Roles of LAMP-1 and LAMP-2
in lysosome biogenesis and autophagy. Mol Aspects Med. 27:495–502.
2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang H: Targeting autophagy in lymphomas:
A double-edged sword? Int J Hematol. 107:502–512. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Nagar R: Autophagy: A brief overview in
perspective of dermatology. Indian J Dermatol Venereol Leprol.
83:290–297. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Rustin M: Postmodernism and antimodernism
in contemporary British architecture. Assemblage. 10:89–103.
1989.
|
|
30
|
Nakamura O, Hitora T, Yamagami Y, Mori M,
Nishimura H, Horie R, et al: The combination of rapamycin and MAPK
inhibitors enhances the growth inhibitory effect on Nara-H cells.
Int J Mol Med. 33:1491–1497. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Reddy D, Kumavath R, Tan TZ, Ampasala DR
and Kumar AP: Peruvoside targets apoptosis and autophagy through
MAPK Wnt/β-catenin and PI3K/AKT/mTOR signaling pathways in human
cancers. Life Sci. 241(117147)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bhaskar PT and Hay N: The two TORCs and
Akt. Dev Cell. 12:487–502. 2007.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Loewith R, Jacinto E, Wullschleger S,
Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P and Hall MN:
Two TOR complexes, only one of which is rapamycin sensitive, have
distinct roles in cell growth control. Mol Cell. 10:457–468.
2002.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Julien LA, Carriere A, Moreau J and Roux
PP: mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135
and regulates mTORC2 signaling. Mol Cell Biol. 30:908–921.
2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wang Y and Zhang H: Regulation of
autophagy by mTOR signaling pathway. Adv Exp Med Biol. 1206:67–83.
2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Petherick KJ, Conway OJ, Mpamhanga C,
Osborne SA, Kamal A, Saxty B and Ganley IG: Pharmacological
inhibition of ULK1 kinase blocks mammalian target of rapamycin
(mTOR)-dependent autophagy. J Biol Chem. 290:11376–11383.
2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Joseph B, Kumar RV, Champaka G, Shenoy A,
Sabitha KS, Lokesh V, Ramesh C and Vijay CR: Biological tailoring
of adjuvant radiotherapy in head and neck and oral malignancies -
The potential role of p53 and eIF4E as predictive parameters.
Indian J Cancer. 56:330–334. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Huang CI, Wang CC, Tai TS, Hwang TZ, Yang
CC, Hsu CM and Su YC: eIF4E and 4EBP1 are prognostic markers of
head and neck squamous cell carcinoma recurrence after definitive
surgery and adjuvant radiotherapy. PLoS One.
14(e0225537)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Alabdullah ML, Ahmad DA, Moseley P,
Madhusudan S, Chan S and Rakha E: The mTOR downstream regulator
(p-4EBP1) is a novel independent prognostic marker in ovarian
cancer. J Obstet Gynaecol. 39:522–528. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gleason CE, Oses-Prieto JA, Li KH, Saha B,
Situ G, Burlingame AL and Pearce D: Phosphorylation at distinct
subcellular locations underlies specificity in mTORC2-mediated
activation of SGK1 and Akt. J Cell Sci. 132:1–16. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liu Y, Ao X, Ding W, Ponnusamy M, Wu W,
Hao X, Yu W, Wang Y, Li P and Wang J: Critical role of FOXO3a in
carcinogenesis. Mol Cancer. 17:104–115. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Ni HM, Du K, You M and Ding WX: Critical
role of FoxO3a in alcohol-induced autophagy and hepatotoxicity. Am
J Pathol. 183:1815–1825. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Carnero A and Paramio JM: The
PTEN/PI3K/AKT pathway in vivo, cancer mouse models. Front Oncol.
4(252)2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Carnero A, Blanco-Aparicio C, Renner O,
Link W and Leal JF: The PTEN/PI3K/AKT signalling pathway in cancer,
therapeutic implications. Curr Cancer Drug Targets. 8:187–198.
2008.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Mangé A, Coyaud E, Desmetz C, Laurent E,
Béganton B, Coopman P, Raught B and Solassol J: FKBP4 connects
mTORC2 and PI3K to activate the PDK1/Akt-dependent cell
proliferation signaling in breast cancer. Theranostics.
9:7003–7015. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wang P, Gao W, Wang Y and Wang J:
Acetylshikonin inhibits in vitro and in vivo tumorigenesis in
cisplatin-resistant oral cancer cells by inducing autophagy,
programmed cell death and targeting m-TOR/PI3K/Akt signalling
pathway. J BUON. 24:2062–2067. 2019.PubMed/NCBI
|
|
47
|
Huang S, Xie T and Liu W: Icariin inhibits
the growth of human cervical cancer cells by inducing apoptosis and
autophagy by targeting mTOR/PI3K/AKT signalling pathway. J BUON.
24:990–996. 2019.PubMed/NCBI
|
|
48
|
Agostini D, Natalucci V, Baldelli G, De
Santi M, Donati Zeppa S, Vallorani L, Annibalini G, Lucertini F,
Federici A, Izzo R, Stocchi V and Barbieri E: New insights into the
role of exercise in inhibiting mTOR signaling in triple-negative
breast cancer. Oxid Med Cell Longev. 2018(5896786)2018.PubMed/NCBI View Article : Google Scholar : https://doi.org/10.1155/2018/5896786.
|
|
49
|
Han Y, Shi D and Li J: Inhibition of
nasopharyngeal carcinoma by beta-lapachone occurs by targeting the
mammalian target of rapamycin (mTOR)/PI3K/AKT pathway, reactive
oxygen species (ROS) production, and autophagy induction. Med Sci
Monit. 25:8995–9002. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Liu J, Ren Y, Hou Y, Zhang C, Wang B, Li
X, Sun R and Liu J: Dihydroartemisinin induces endothelial cell
autophagy through suppression of the Akt/mTOR pathway. J Cancer.
10:6057–6064. 2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Dong F, Han J, Jing G, Chen X, Yan S, Yue
L, Cao Z, Liu X, Ma G and Liu J: Dihydroartemisinin transiently
activates the JNK/SAPK signaling pathway in endothelial cells.
Oncol Lett. 12:4699–4704. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Li WD, Zhou DM, Sun LL, Xiao L, Liu Z,
Zhou M, Wang WB and Li XQ: LncRNA WTAPP1 promotes migration and
angiogenesis of endothelial progenitor cells via MMP1 through
microRNA 3120 and Akt/PI3K/autophagy pathways. Stem Cells.
36:1863–1874. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Liu R, Chen Z, Yi X, Huang F, Hu G, Liu D,
Li X, Zhou H and Liu Z: 9za plays cytotoxic and proapoptotic roles
and induces cytoprotective autophagy through the PDK1/Akt/mTOR axis
in non-small-cell lung cancer. J Cell Physiol. 234:20728–20741.
2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Dowling RJ, Topisirovic I, Fonseca BD and
Sonenberg N: Dissecting the role of mTOR: Lessons from mTOR
inhibitors. Biochim Biophys Acta. 1804:433–439. 2010.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Gera J and Lichtenstein A: The mammalian
target of rapamycin pathway as a therapeutic target in multiple
myeloma. Leuk Lymphoma. 52:1857–1866. 2011.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Tan FH, Bai Y, Saintigny P and Darido C:
mTOR signalling in head and neck cancer: Heads Up. Cells.
8(333)2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Bu Z and Ji J: Therapeutic implications of
mTOR inhibitors in the treatment of gastric cancer. Curr Cancer
Drug Targets. 13:121–125. 2013.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Alvarez RH, Bechara RI, Naughton MJ,
Adachi JA and Reuben JM: Emerging perspectives on mTOR
inhibitor-associated pneumonitis in breast cancer. Oncologist.
23:660–669. 2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Chang L, Graham PH, Ni J, Hao J, Bucci J,
Cozzi PJ and Li Y: Targeting PI3K/Akt/mTOR signaling pathway in the
treatment of prostate cancer radioresistance. Crit Rev Oncol
Hematol. 96:507–517. 2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Myers AP: New strategies in endometrial
cancer: Targeting the PI3K/mTOR pathway--the devil is in the
details. Clin Cancer Res. 19:5264–5274. 2013.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wu CE, Chen MH and Yeh CN: mTOR inhibitors
in advanced biliary tract cancers. Int J Mol Sci.
20(500)2019.PubMed/NCBI View Article : Google Scholar : https://doi.org/10.3390/ijms20030500.
|
|
63
|
Ching CB and Hansel DE: Expanding
therapeutic targets in bladder cancer: The PI3K/Akt/mTOR pathway.
Lab Invest. 90:1406–1414. 2010.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Rodrik-Outmezguine VS, Okaniwa M, Yao Z,
Novotny CJ, McWhirter C, Banaji A, Won H, Wong W, Berger M, de
Stanchina E, et al: Overcoming mTOR resistance mutations with a
new-generation mTOR inhibitor. Nature. 534:272–276. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Yu C, Sun P, Zhou Y, Shen B, Zhou M, Wu L
and Kong M: Inhibition of AKT enhances the anti-cancer effects of
Artemisinin in clear cell renal cell carcinoma. Biomed
Pharmacother. 118(109383)2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Vinayak S and Carlson RW: mTOR inhibitors
in the treatment of breast cancer. Oncology (Williston Park).
27:38–44, 46, 48 passim. 2013.PubMed/NCBI
|
|
67
|
Martisova A, Sommerova L, Kuricova K,
Podhorec J, Vojtesek B, Kankova K and Hrstka R: AGR2 silencing
contributes to metformin-dependent sensitization of colorectal
cancer cells to chemotherapy. Oncol Lett. 18:4964–4973.
2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Ciccarese F, Zulato E and Indraccolo S:
LKB1/AMPK pathway and drug response in cancer: A therapeutic
perspective. Oxid Med Cell Longev. 2019(8730816)2019.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Richani D, Lavea CF, Kanakkaparambil R,
Riepsamen AH, Bertoldo MJ, Bustamante S and Gilchrist RB:
Participation of the adenosine salvage pathway and cyclic AMP
modulation in oocyte energy metabolism. Sci Rep. 9:18395–18406.
2019.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Lu C, Wang W, Jia Y, Liu X, Tong Z and Li
B: Inhibition of AMPK/autophagy potentiates parthenolide-induced
apoptosis in human breast cancer cells. J Cell Biochem.
115:1458–1466. 2014.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Chen W, Pan Y, Wang S, Liu Y, Chen G, Zhou
L, Zhang C, Ni W, Wang A, Lu Y, et al: Correction to:
cryptotanshinone activates AMPK-TSC2 axis leading to inhibition of
mTORC1 signaling in cancer cells. BMC Cancer. 19:257–269.
2019.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Andrade BM and de Carvalho DP:
Perspectives of the AMP-activated kinase (AMPK) signalling pathway
in thyroid cancer. Biosci Rep. 34(e00105)2014.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Li F, Yang C, Zhang HB, Ma J, Jia J, Tang
X, Zeng J, Chong T, Wang X, He D, et al: BET inhibitor JQ1
suppresses cell proliferation via inducing autophagy and activating
LKB1/AMPK in bladder cancer cells. Cancer Med. 8:4792–4805.
2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Zhao Z, Feng L, Wang J, Cheng D, Liu M,
Ling M, Xu W and Sun K: NPC-26 kills human colorectal cancer cells
via activating AMPK signaling. Oncotarget. 8:18312–18321.
2017.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Chen L, Liu M, Luan Y and Liu Y, Zhang Z,
Ma B, Liu X and Liu Y: BMP-6 protects retinal pigment epithelial
cells from oxidative stress-induced injury by inhibiting the MAPK
signaling pathways. Int J Mol Med. 42:1096–1105. 2018.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Ma Z, Wang C, Liu C, Yan DY, Tan X, Liu K,
Jing MJ, Deng Y, Liu W and Xu B: Manganese induces autophagy
dysregulation: The role of S-nitrosylation in regulating autophagy
related proteins in vivo and in vitro. Sci Total Environ.
698(134294)2020.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Pan MY, Shen YC, Lu CH, Yang SY, Ho TF,
Peng YT and Chang CC: Prodigiosin activates endoplasmic reticulum
stress cell death pathway in human breast carcinoma cell lines.
Toxicol Appl Pharmacol. 265:325–334. 2012.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Kim JS, Bae GE, Kim KH, Lee SI, Chung C,
Lee D, Lee TH, Kwon IS and Yeo MK: Prognostic significance of LC3B
and p62/SQSTM1 expression in gastric adenocarcinoma. Anticancer
Res. 39:6711–6722. 2019.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Zhang J, Yang S, Xu B, Wang T, Zheng Y,
Liu F, Ren F, Jiang J, Shi H, Zou B, et al: p62 functions as an
oncogene in colorectal cancer through inhibiting apoptosis and
promoting cell proliferation by interacting with the vitamin D
receptor. Cell Prolif. 52(e12585)2019.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Tan P, Ye Y, He L, Xie J, Jing J, Ma G,
Pan H, Han L, Han W and Zhou Y: TRIM59 promotes breast cancer
motility by suppressing p62-selective autophagic degradation of
PDCD10. PLoS Biol. 16(e3000051)2018.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Schläfli AM, Adams O, Galván JA, Gugger M,
Savic S, Bubendorf L, Schmid RA, Becker KF, Tschan MP, Langer R, et
al: Prognostic value of the autophagy markers LC3 and p62/SQSTM1 in
early-stage non-small cell lung cancer. Oncotarget. 7:39544–39555.
2016.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Chen Y, Zhou X, Qiao J and Bao A:
Autophagy is a regulator of TRAIL-induced apoptosis in NSCLC A549
cells. J Cell Commun Signal. 11:219–226. 2017.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Wechman SL, Rao XM, Gomez-Gutierrez JG,
Zhou HS and McMasters KM: The role of JNK phosphorylation as a
molecular target to enhance adenovirus replication, oncolysis and
cancer therapeutic efficacy. Cancer Biol Ther. 19:1174–1184.
2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Wang Y, Xiong H, Liu D, Hill C, Ertay A,
Li J, Zou Y, Miller P, White E, Downward J, et al: Autophagy
inhibition specifically promotes epithelial-mesenchymal transition
and invasion in RAS-mutated cancer cells. Autophagy. 15:886–899.
2019.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Bryant KL, Stalnecker CA, Zeitouni D,
Klomp JE, Peng S, Tikunov AP, Gunda V, Pierobon M, Waters AM,
George SD, et al: Combination of ERK and autophagy inhibition as a
treatment approach for pancreatic cancer. Nat Med. 25:628–640.
2019.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Mou L, Liang B, Liu G, Jiang J, Liu J,
Zhou B, Huang J, Zang N, Liao Y, Ye L, et al: Berbamine exerts
anticancer effects on human colon cancer cells via induction of
autophagy and apoptosis, inhibition of cell migration and MEK/ERK
signalling pathway. J BUON. 24:1870–1875. 2019.PubMed/NCBI
|
|
87
|
Tian Y, Song W, Li D, Cai L and Zhao Y:
Resveratrol as a Natural regulator of autophagy for prevention and
treatment of cancer. OncoTargets Ther. 12:8601–8609.
2019.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Sánchez-Martín P, Saito T and Komatsu M:
p62/SQSTM1: ‘Jack of all trades’ in health and cancer. FEBS J.
286:8–23. 2019.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Li Y, Shi J, Qi S, Zhang J, Peng D, Chen
Z, Wang G, Wang Z and Wang L: IL-33 facilitates proliferation of
colorectal cancer dependent on COX2/PGE2. J Exp Clin Cancer Res.
37:196–227. 2018.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Panigrahi DP, Bhol CS, R N, Nagini S,
Patil S, Maiti TK and Bhutia SK: Abrus agglutinin inhibits oral
carcinogenesis through inactivation of NRF2 signaling pathway. Int
J Biol Macromol. 9:1–31. 2019.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Yang L, Sun X, Ye Y, Lu Y, Zuo J, Liu W,
Elcock A and Zhu S: p38α mitogen-activated protein kinase is a
druggable target in pancreatic adenocarcinoma. Front Oncol.
9:1294–1314. 2019.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Patel PH, Pénalva C, Kardorff M, Roca M,
Pavlović B, Thiel A, Teleman AA and Edgar BA: Damage sensing by a
Nox-Ask1-MKK3-p38 signaling pathway mediates regeneration in the
adult Drosophila midgut. Nat Commun. 10:4365–4378. 2019.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Del Barco Barrantes I, Stephan-Otto
Attolini C, Slobodnyuk K, Igea A, Gregorio S, Gawrzak S, Gomis RR
and Nebreda AR: Regulation of mammary luminal cell fate and
tumorigenesis by p38α. Stem Cell Reports. 10:257–271.
2018.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Mo'men YS, Hussein RM and Kandeil MA: A
novel chemoprotective effect of tiopronin against
diethylnitrosamine-induced hepatocellular carcinoma in rats: Role
of ASK1/P38 MAPK-P53 signalling cascade. Clin Exp Pharmacol
Physiol. 47:322–332. 2020.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Aggarwal V, Tuli HS, Varol A, Thakral F,
Yerer MB, Sak K, Varol M, Jain A, Khan MA and Sethi G: Role of
reactive oxygen species in cancer progression: Molecular mechanisms
and recent advancements. Biomolecules. 9:735–750. 2019.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Boldbaatar J, Gunarta IK, Suzuki R,
Erdenebaatar P, Davaakhuu G, Hohjoh H and Yoshioka K: Protective
role of c-Jun NH2-terminal kinase-associated leucine
zipper protein (JLP) in curcumin-induced cancer cell death. Biochem
Biophys Res Commun. 29:1–31. 2019.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Shi Y, Liu Y, Zheng Y, Tang Y, Zhu G, Qiu
W, Huang L, Han S, Yin J, Peng B, et al: Autophagy triggered by
MAVS inhibits Coxsackievirus A16 replication. Acta Virol.
63:392–402. 2019.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Yuan YL, Jiang N, Li ZY, Song ZZ, Yang ZH,
Xue WH, Zhang XJ and Du Y: Polyphyllin VI induces apoptosis and
autophagy in human osteosarcoma cells by modulation of ROS/JNK
activation. Drug Des Devel Ther. 13:3091–3103. 2019.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Zhao M, Gu L, Li Y, Chen S, You J, Fan L,
Wang Y and Zhao L: Chitooligosaccharides display anti-tumor effects
against human cervical cancer cells via the apoptotic and
autophagic pathways. Carbohydr Polym. 224:115171–115184.
2019.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Cheng X, Tan S, Duan F, Yuan Q, Li Q and
Deng G: Icariin induces apoptosis by suppressing autophagy in
tamoxifen-resistant breast cancer cell line MCF-7/TAM. Breast
Cancer. 26:766–775. 2019.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Kim KY, Oh TW, Yang HJ, Kim YW, Ma JY and
Park KI: Ethanol extract of Chrysanthemum zawadskii Herbich
induces autophagy and apoptosis in mouse colon cancer cells through
the regulation of reactive oxygen species. BMC Complement Altern
Med. 19:274–283. 2019.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Wang F, Chen Y, Wang Y, Yin Y, Qu G, Song
M and Wang H: Ultra-long silver nanowires induced mitotic
abnormalities and cytokinetic failure in A549 cells.
Nanotoxicology. 13:543–557. 2019.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Chen G, Ding XF, Bouamar H, Pressley K and
Sun LZ: Everolimus induces G1 cell cycle arrest through
autophagy-mediated protein degradation of cyclin D1 in breast
cancer cells. Am J Physiol Cell Physiol. 317:C244–C252.
2019.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Booth L, Roberts JL, Avogadri-Connors F,
Cutler RE Jr, Lalani AS, Poklepovic A and Dent P: The irreversible
ERBB1/2/4 inhibitor neratinib interacts with the BCL-2 inhibitor
venetoclax to kill mammary cancer cells. Cancer Biol Ther.
19:239–247. 2018.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Zhou D, Dai L, Liu X, Que F, Xu Y, Luo X,
Zhu Y, Liu S, Li Y and Yu L: Bortezomib and obatoclax for dual
blockade of protein degradation pathways show synergistic
anti-tumor effect in human acute T lymphoblastic leukemia cells.
Nan Fang Yi Ke Da Xue Xue Bao. 39:401–408. 2019.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
106
|
Antonietti P, Gessler F, Düssmann H,
Reimertz C, Mittelbronn M, Prehn JH and Kögel D: AT-101
simultaneously triggers apoptosis and a cytoprotective type of
autophagy irrespective of expression levels and the subcellular
localization of Bcl-xL and Bcl-2 in MCF7 cells. Biochim Biophys
Acta. 1863:499–509. 2016.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Tong H, Li T, Qiu W and Zhu Z: Claudin-1
silencing increases sensitivity of liver cancer HepG2 cells to
5-fluorouracil by inhibiting autophagy. Oncol Lett. 18:5709–5716.
2019.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Lee J, Jung JH, Hwang J, Park JE, Kim JH,
Park WY, Suh JY and Kim SH: CNOT2 is critically involved in
atorvastatin induced apoptotic and autophagic cell death in
non-small cell lung cancers. Cancers (Basel). 11:1470–1484.
2019.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Lee JE, Yoon SS and Moon EY:
Curcumin-induced autophagy augments its antitumor effect against
A172 human glioblastoma cells. Biomol Ther (Seoul). 27:484–491.
2019.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Silva VAO, Rosa MN, Tansini A, Martinho O,
Tanuri A, Evangelista AF, Cruvinel Carloni A, Lima JP, Pianowski LF
and Reis RM: Semi-synthetic ingenol derivative from euphorbia
tirucalli inhibits protein kinase C isotypes and promotes
autophagy and S-phase arrest on glioma cell lines. Molecules.
24:4265–4282. 2019.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Robke L, Laraia L, Carnero Corrales MA,
Konstantinidis G, Muroi M, Richters A, Winzker M, Engbring T,
Tomassi S, Watanabe N, et al: Phenotypic identification of a novel
autophagy inhibitor chemotype targeting lipid kinase VPS34. Angew
Chem Int Ed Engl. 56:8153–8157. 2017.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Carew JS, Espitia CM, Zhao W, Han Y,
Visconte V, Phillips J and Nawrocki ST: Disruption of autophagic
degradation with ROC-325 antagonizes renal cell carcinoma
pathogenesis. Clin Cancer Res. 23:2869–2879. 2017.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Guntuku L, Gangasani JK, Thummuri D,
Borkar RM, Manavathi B, Ragampeta S, Vaidya JR, Sistla R and Vegi
NGM: IITZ-01, a novel potent lysosomotropic autophagy inhibitor,
has single-agent antitumor efficacy in triple-negative breast
cancer in vitro and in vivo. Oncogene. 38:581–595. 2018.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Wu MY, Wang SF, Cai CZ, Tan JQ, Li M, Lu
JJ, Chen XP, Wang YT, Zheng W and Lu JH: Natural autophagy
blockers, dauricine (DAC) and daurisoline (DAS), sensitize cancer
cells to camptothecin-induced toxicity. Oncotarget. 8:77673–77684.
2017.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Zhao Y, Li K, Zhao B and Su L, Li K, Zhao
B and Su L: HSP90 inhibitor DPB induces autophagy and more
effectively apoptosis in A549 cells combined with autophagy
inhibitors. In Vitro Cell Dev Biol Anim. 55:349–354.
2019.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Han J, Lv W, Sheng H, Wang Y, Cao L, Huang
S, Zhu L and Hu J: Ecliptasaponin A induces apoptosis through the
activation of ASK1/JNK pathway and autophagy in human lung cancer
cells. Ann Transl Med. 7:539–560. 2019.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Polishchuk EV, Merolla A, Lichtmannegger
J, Romano A, Indrieri A, Ilyechova EY, Concilli M, De Cegli R,
Crispino R, Mariniello M, et al: Activation of autophagy, observed
in liver tissues from patients with Wilson disease and from
ATP7B-deficient animals, protects hepatocytes from copper-induced
apoptosis. Gastroenterology. 156:1173–1189.e5. 2019.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Klionsky DJ, Abdelmohsen K, Abe A, Abedin
MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD,
Adeli K, et al: Guidelines for the use and interpretation of assays
for monitoring autophagy (3rd edition). Autophagy. 12:1–222.
2016.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Xuan F, Huang M, Liu W, Ding H, Yang L and
Cui H: Homeobox C9 suppresses Beclin1-mediated autophagy in
glioblastoma by directly inhibiting the transcription of
death-associated protein kinase 1. Neuro-oncol. 18:819–829.
2016.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Du H, Che J, Shi M, Zhu L, Hang JB, Chen Z
and Li H: Beclin 1 expression is associated with the occurrence and
development of esophageal squamous cell carcinoma. Oncol Lett.
14:6823–6828. 2017.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Chu C, Niu X, Ou X and Hu C: LAPTM4B
knockdown increases the radiosensitivity of EGFR-overexpressing
radioresistant nasopharyngeal cancer cells by inhibiting autophagy.
OncoTargets Ther. 12:5661–5677. 2019.PubMed/NCBI View Article : Google Scholar
|