Introduction
Chronic obstructive pulmonary disease (COPD) is a
common respiratory disease with a high rate of morbidity and
mortality (1-3).
As the rates of COPD incidence have increased, so too has its
social and economic burden. It is estimated that by 2020, COPD will
rank fifth in the list of diseases that most impacts the world's
economy (4). The pathological
characteristics of COPD include chronic inflammation of the lung
parenchyma and the surrounding airways, emphysema and small airway
stenosis or remodeling (5-7).
A number of processes, including oxidative stress, inflammation and
mitochondrial dysfunction have all been reported to serve
mechanistical roles (8-11).
However, additional research is required to determine the molecular
mechanisms underlying COPD.
MicroRNAs (miRNA or miRs) are a group of non-coding
RNAs that consist of 18-25 nucleotides and serve to increase mRNA
degradation or repress target-specific mRNA translation (12). miRNAs can modulate almost all
biological processes, including proliferation, differentiation,
inflammatory response, autophagy, tissue remodeling, immune
regulation, angiogenesis, aging and tumorigenesis (13-15).
Previous reports have confirmed that miRNAs can also serve an
important role in the pathology of COPD (16-18).
Therefore, regulating miRNA expression may be a potential
therapeutic strategy for COPD.
Exosomes are nano-sized membrane-bound vesicles that
are released into the extracellular environment by almost all cell
types (19-21).
Once stimulated by inflammation, hypoxia, oxidative stress and
immune activation, macromolecules contained inside exosomes, such
as DNA and RNA, are transformed and exuded to affect the biology of
receptor cells (22-24).
In addition, exosomes can be successfully isolated from different
types of body fluids, including sputum, plasma and bronchoalveolar
lavage (25-27).
Exosomes contain non-coding RNA, protein, lipid and other
substances, which change with the state of diseases including COPD
(28). This implies that there may
be an association between exosomes and COPD pathogenesis (29).
Unfortunately, sensitive biomarkers that can
potentially be used for the prognosis of COPD remain lacking.
Previous studies have demonstrated that the levels of miR-29c and
miR-126 in the peripheral blood are potential biomarkers for the
diagnosis of COPD (30,31). However, the miRNA in the peripheral
circulation is susceptible to interference by other components,
making circulating miRNA results less consistent. Exosomes have a
dual-membrane structure that protects them from being degraded by
ribonucleases, rendering exosomal miRNAs (exo-miRNAs) ideal for use
as circulating biomarkers (32).
Therefore, the present study isolated exosomes from
the plasma samples of patients with COPD and healthy individuals.
High-throughput sequencing was performed to identify enriched
miRNAs in the exosomes of patients with COPD. The exo-miRNAs
closely associated with the clinical characteristics of COPD were
subsequently screened and their diagnostic values were
analyzed.
Materials and methods
Study subjects
The present study was approved by the Research
Ethics Committee of Taizhou Clinical Medical School of Nanjing
Medical University (approval no. KY201904701; Taizhou, China) and
conducted according to the Declaration of Helsinki. All the
selected patients voluntary participated and provided their written
informed consent.
Between January 2019 and August 2020, a total of 46
patients with COPD were selected for the current study at Taizhou
Clinical Medical School of Nanjing Medical University (Taizhou,
China). At the same time, 34 healthy non-smoking individuals
(range, 45-85 years) were recruited as the controls at Taizhou
Clinical Medical School of Nanjing Medical University (Taizhou,
China). To reduce heterogeneity, the age and sex of the healthy
controls were matched to the patients with COPD. The average age of
the patients with COPD was 62 years (age range, 45-85 years) with
moderate to severe pulmonary dysfunction [the post bronchodilator
ratio of the forced expiratory volume in the 1st second (FEV1) to
the forced vital capacity (FVC) value, <0.70; FEV1% predicted
value, <80%], were included in the present study. COPD diagnosis
and severity classification were established by the same
respiratory physician HL, according to the GOLD criteria (33). Patients with COPD were either
current or ex-smokers (lifetime smoking exposure, smoking index ≥10
pack-years; smoking cessation, <1 year). Prior to enrollment,
patients were included if they had a documented history of at ≥ two
COPD exacerbations leading to oral or intravenous glucocorticoid
therapy or antibiotic therapy, or at ≥ one COPD exacerbation
leading to hospitalization within the previous year. Patients were
excluded if they presented with other diseases, including severe
cardiovascular disease, uncontrolled high blood pressure, bleeding,
hepatic failure, renal failure, rheumatoid immune disease and
malignant tumors.
Study design
The present study was divided into two sections:
Discovery and validation. In the discovery stage (stage I),
high-throughput sequencing was performed to screen the
differentially expressed plasma-derived exo-miRNAs between samples
from the group of five patients with COPD and the group with five
healthy individuals. The function of differentially expressed
miRNAs was evaluated using Gene Ontology (GO) and Kyoto
Encyclopedia of Gene and Genomes (KEGG) pathway analysis. Exosomal
miRNAs exhibiting the most significant (according to the multiple
of differential expression calculated by log2FC;
P<0.05) differential expression were selected for further
validation.
In the validation stage (stage II), the expression
of the potential plasma-derived exo-miRNAs in 41 patients with COPD
and 29 healthy individuals were analyzed using reverse
transcription-quantitative PCR (RT-qPCR). Receiver operating
characteristic (ROC) curve-based risk assessment analysis was
performed to evaluate the sensitivity and specificity of using
exo-miRNAs to diagnose COPD.
Plasma sample collection
After overnight fasting, 20 ml whole blood was drawn
from each participant through the antecubital vein and injected
into an EDTA-coated vacutainer tube (lavender top EDTA tubes; BD
Biosciences). The tubes were incubated at 4˚C for 3-4 h and
centrifuged at 4,000 x g at 4˚C for 5 min. The supernatants
containing plasma were collected and stored in a 2-ml falcon tube
at -80˚C for further analysis.
Exosome isolation and
characterization
Plasma-derived exosomes were isolated using an
ExoQuick commercial kit Ribo™ Exosome Isolation Reagent
(Guangzhou RiboBio Co., Ltd.) according to the manufacturer's
protocol. Transmission electron microscopy (TEM) was performed to
observe the size and morphology of exosomes as described previously
(34,35). Exosomes were placed on a copper mesh
surface, fixed with 3% glutaraldehyde at room temperature for 5 min
and stained with 4% uranium acetate at room temperature for 1 min.
Finally, the copper mesh was transferred to 1% methylcellulose and
suspended at room temperature for 3 min. Excess liquid at the edge
of the copper mesh was dried using filter paper for ≥30 min.
Stained samples were observed using TEM (magnification, x80,000)
(Hitachi-7650; Hitachi, Ltd.) and imaged for preservation.
Nanoparticle tracking analysis (NTA) was used to evaluate the size
and concentration of plasma exosomes. Brownian particles were
tracked, following which their hydrodynamic diameters and
concentrations were calculated using the Stockes-Einstein equation
(36). NTA assay was performed
using the ZetaView PMX 110 (Particle Metrix GmbH). The data
obtained with the ZetaView instrument were analysed using the
corresponding software ZetaView v.8.02.28 (Particle Metrix
GmbH).
Western blotting
Western blotting was used to detect the specific
marker proteins (CD9, CD63, TSG101) on the surface of plasma
exosomes. Exosome samples were lysed using RIPA lysis buffer (cat.
no. 20-188; Sigma-Aldrich, Merck KGaA) on ice for 30 min. A
bicinchoninic acid (BCA) Protein Assay kit (cat. no. 23225; Pierce;
Thermo Fisher Scientific, Inc.) was used to detect protein
concentrations, and a 12% sodium dodecyl sulfate-polyacrylamide gel
was used for total protein (30 µg/lane) separation. The proteins in
the gel were transferred to a 0.45-µM pore size PVDF membrane (cat.
no. IPVH00010; EMD Millipore) via wet electrophoretic transfer. The
membranes were blocked with 5% skimmed milk powder for 1 h at room
temperature and incubated overnight at 4˚C with anti-CD9 (cat. no.
ab92726; Abcam), anti-CD63 (cat. no. ab216130), anti-TSG101 (cat.
no. ab125011) antibodies, all diluted to 1:1,000 in TBS-1%
Tween-20. Subsequently, the membranes were incubated with a
horseradish peroxidase-conjugated goat anti rabbit (1:3,000; cat.
no. A0208; Beyotime Institute of Biotechnology) or goat anti mouse
IgG secondary antibodies (1:3,000; cat. no. A0216; Beyotime
Institute of Biotechnology) for 1 h at room temperature. The
membranes were visualized using LumiBest enhanced chemiluminescence
(cat. no. SB-WB011; Shanghai Shenger Biotechnology Co., Ltd.).
Exosomal miRNA sequencing
A total of 4 ml plasma was mixed with
Ribo™ Exosome Isolation Reagent (Guangzhou RiboBio Co.,
Ltd.), after which exosomes were isolated. Exosomal RNA was
extracted using the HiPure Plasma miRNA kit (cat. no. R314;
Guangzhou Guangzhou Meiji Biological Technology Co., Ltd.). The
quantity and integrity of exosomal RNA was assessed using the
Qubit®2.0 (Thermo Fisher Scientific, Inc.) and Agilent
2200 TapeStation (Agilent Technologies, Inc.), respectively. For
each sample, 50 ng exosomal RNA was used to prepare small RNA
libraries using the NEBNext® Multiplex Small RNA Library
Prep Set for Illumina (cat. no. E7580; New England
BioLabs® Inc.). Libraries were sequenced using HiSeq
2500 (Illumina, Inc.) with single-end 50 bp at Guangzhou RiboBio
Co., Ltd.. The final loading concentration of each sample was above
2.5 nmol/l. The raw reads were processed by filtering out
containing adapter, poly ‘N’, low quality, smaller than 17 nt reads
by FastQC v.0.11.9 (https://directory.fsf.org/wiki/FastQC) to get clean
reads. Mapping reads were obtained by mapping clean reads to
reference genome of by BWA v.0.7.12(37). The differential expression between
two sets of samples was calculated using the edgeR algorithm
(38): Log2 (fold
change) ≥1 and P<0.05. R software v.3.1.3 (www.r-project.org) was used for data analysis.
RT-qPCR validation
As previously described (32), plasma samples were thawed and
centrifuged at 1,000 x g for 5 min at room temperature to form
granules. The absorbance of oxyhemoglobin was then determined via
spectrophotometry at 414 nm. Once hemolysis occurred, the specimen
was discarded. Total RNA was extracted from plasma exosomes using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The primers (One RT primer and a pair of qPCR primers for
each set) specific for miRNAs were designed by RiboBio (Guangzhou
RiboBio Co., Ltd.). The primer sequences were patented. cDNA was
synthesized from 500 ng RNA using a reverse transcriptase kit (cat.
no. R10031.8; Guangzhou RiboBio Co., Ltd.) with the following
temperature protocol: 42˚C for 60 min, and followed by 70˚C for 10
min. qPCR was carried out in accordance with the protocols of the
Bulge-Loop™ miRNA qRT-PCR Starter kit (cat. no.
c10211-1; Guangzhou RiboBio Co., Ltd.) and ABI Prism 7900HT
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used: Initial denaturation at 95˚C
for 20 sec, followed by 40 cycles of 95˚C for 10 sec, 60˚C for 20
sec and 70˚C for 10 sec. U6 small nuclear RNA and cel-miR-39 were
used as endogenous (39) and
exogenous controls, respectively. The expression of exo-miRNAs was
calculated using the 2-ΔΔCq method (40).
GO and KEGG pathway analysis
TargetScan (version 7.2, https://www.targetscan.org/), miRDB (version 5.0,
http://www.targetscan.org/), miRTarBase
(version 7.0, http://starbase.sysu.edu.cn/) and miRWalk (version
7.0, http://mirwalk.umm.uni-heidelberg.de/) databases were
used to predict the target genes of selected miRNAs. Those
simultaneously predicted by ≥ two of the tools aforementioned were
selected as candidate target genes. KOBAS release number 3.0
(http://kobas.cbi.pku.edu.cn/) was used
for further GO and KEGG pathway analyses (41). The significance of GO and KEGG
pathway analysis was then determined using Fisher's exact and
χ2 tests. The subsequent P-value was corrected using the
false discovery rate (FDR). GO and KEGG terms with an adjusted
P-value <0.05 and an FDR <0.05 were selected and considered
to be enriched.
miRNA-mRNA network construction
An exosome miRNA-mRNA network was constructed.
Pearson's correlation of each gene pair was calculated and pairs
with significant correlations were selected for network creation.
Exo-miRNAs and mRNAs with Pearson correlation coefficients ≥0.99
were selected to construct the network using Cytoscape
bioinformatics software v.3.8.0 (https://cytoscape.org/) (42). During network analysis, connectivity
was defined as the number of connections between nodes. In
addition, the degree (the number of target genes targeted by
miRNAs) was subsequently calculated, which is the simplest and most
important measure used to determine the relative importance of
genes in a network (43).
Statistical analysis
Demographic and clinical characteristics were
presented as the mean ± standard deviation. Experiments were
repeated at least 3 times. All analyses were performed using SPSS
24.0 software (IBM Corp.) and GraphPad Prism 5 software (GraphPad
Software, Inc.). Student's paired t-test and Mann-Whitney unpaired
test analysis were used to evaluate statistical differences between
patients with COPD and controls. Categorical variables were
analyzed using the χ2 test. The differential expression
of miRNAs was evaluated via the U Mann Whitney test. Furthermore,
Spearman's linear regression method, with Bonferroni's correction
for multiple comparisons, was used to analyze the correlation
between lung function parameters (FEV1/FVC) and the expression of
miRNAs. The ROC curve was used to analyze the efficiency of
exo-miRNA as a biomarker for COPD diagnosis. The sensitivity and
specificity of each exo-miRNA to diagnose COPD were calculated
using the area under the ROC curve (AUC) with 95% confidence
intervals (CI). The optimal cut off point was where the ‘true
positive rate’ was high and the ‘false positive rate’ was low.
Youden's index was determined to identify the optimal cut-off point
for calculating exact diagnostic indices. P<0.05 was considered
to indicate a statistically significant difference.
Results
Patient clinical characteristics
In the present study, a total of 80 individuals were
enrolled, including 46 patients with COPD and 34 healthy controls
(Table I). There was no difference
in age and sex between the two groups.
 | Table ICharacteristics of patients. |
Table I
Characteristics of patients.
| Parameters | Control | COPD | P-value |
|---|
| Number of
subjects | 34 | 46 | NA |
| Male/Female | 27/7 | 36/10 | NA |
| Age, years
(range) | 61.2±6.3 | 62.3±5.6 | >0.05 |
| Smoking, pack
years | N/A | 52.6±12.5 | <0.001 |
| Smoking/currently
smoking | N/A | 46 | NA |
| Predicted
FEV1% | 97.8±9.1 | 49.0±13.7 | <0.001 |
| FEV1/FVC% | 81.6±5.3 | 54.8±12.6 | <0.001 |
| Inhaled
corticosteroids | 0 | 46/46 | <0.001 |
Exosome isolation and
identification
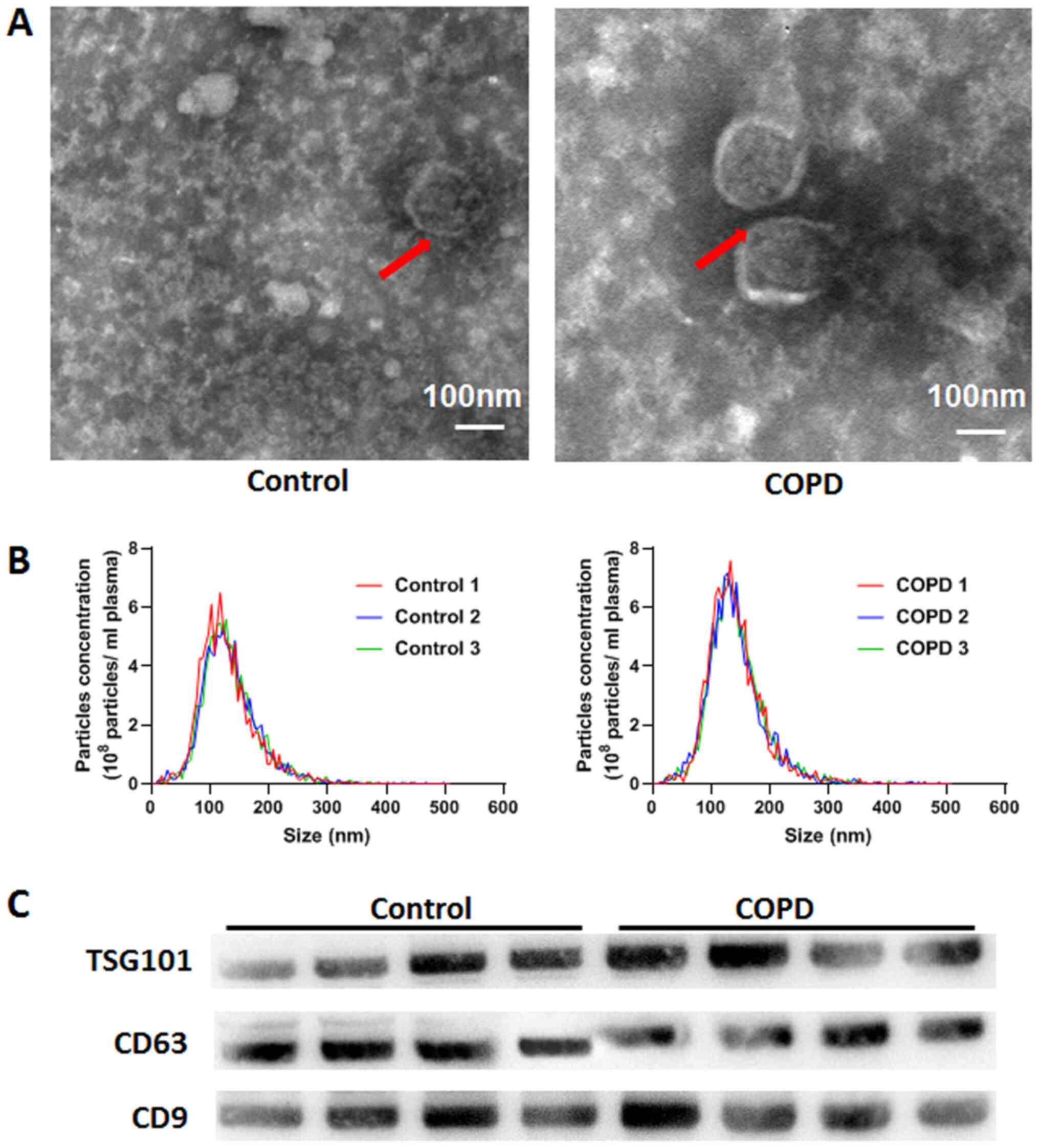
Extracted exosomes were found to be 50-150 nm in
size and exhibited round or spindle shapes (Fig. 1A). No significant difference was
observed in the size or total number of exosomes between patients
with COPD and healthy controls (Fig.
1B). Exosome markers TSG101, CD63 and CD9 were also highly
expressed in the exosomes isolated from the plasma samples of the
two groups (Fig. 1C).
Exosomal miRNAs are dysregulated in
patients with COPD
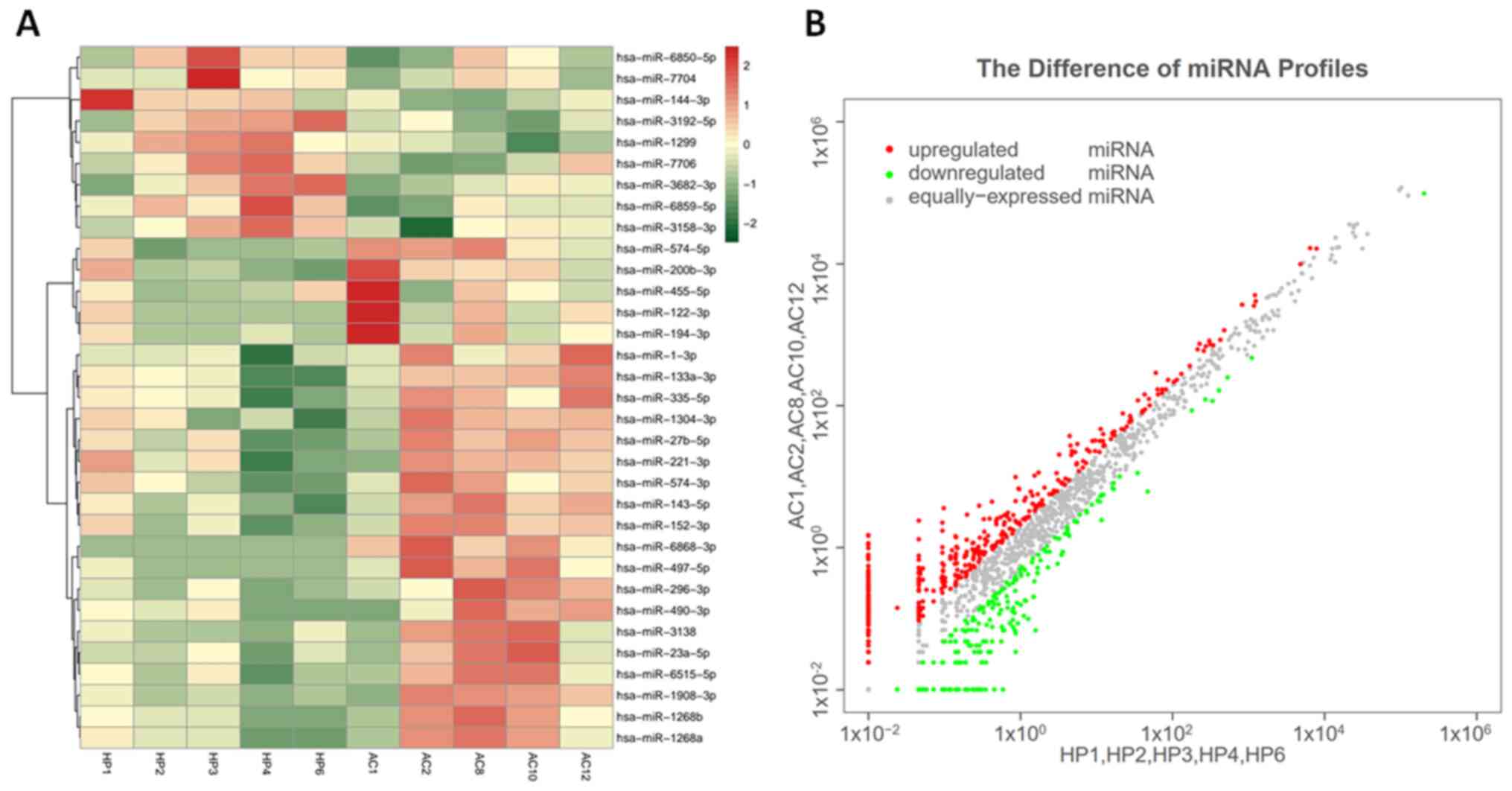
The exosomal miRNA profiles of five patients with
COPD and five healthy individuals were assessed (Fig. 2). Compared with those in healthy
individuals, 39 upregulated and 20 downregulated exo-miRNAs were
identified in patients with COPD. The top 20 miRNAs with the most
significant differential expression are presented in Table II. These miRNAs were selected for
further functional analysis.
 | Table IITop 10 miRNAs exhibiting the most
significant difference in expression levels. |
Table II
Top 10 miRNAs exhibiting the most
significant difference in expression levels.
| A, Upregulated |
|---|
| Rank | miRNAs | Log2
(FC) | P-value |
|---|
| 1 | hsa-miR-23a | 3.0915 | 0.0047 |
| 2 | hsa-miR-3138 | 2.8479 | 0.0214 |
| 3 | hsa-miR-1268a | 2.7234 | 0.0161 |
| 4 | hsa-miR-143 | 2.6790 | 0.0041 |
| 5 | hsa-miR-6515 | 2.5967 | 0.0084 |
| 6 | hsa-miR-1 | 2.2773 | 0.0094 |
| 7 | hsa-miR-221 | 2.0883 | 0.0493 |
| 8 | hsa-miR-574 | 2.0538 | 0.0271 |
| 9 | hsa-miR-335 | 1.7488 | 0.0204 |
| 10 | hsa-miR-152 | 1.7204 | 0.0364 |
| B,
Downregulated |
| Rank | miRNAs | Log2
(FC) | P-value |
| 1 | hsa-miR-6859 | -2.9372 | 0.0303 |
| 2 | hsa-miR-3682 | -2.8839 | 0.0365 |
| 3 | hsa-miR-3158 | -2.8433 | 0.0311 |
| 4 | hsa-miR-147b | -2.7802 | 0.0338 |
| 5 | hsa-miR-7706 | -2.7076 | 0.0300 |
| 6 | hsa-miR-6850 | -2.4224 | 0.0217 |
| 7 | hsa-miR-144 | -2.3069 | 0.0382 |
| 8 | hsa-miR-3192 | -2.1125 | 0.0307 |
| 9 | hsa-miR-7704 | -2.0504 | 0.0102 |
| 10 | hsa-miR-1299 | -1.9180 | 0.0017 |
GO and KEGG pathway analysis
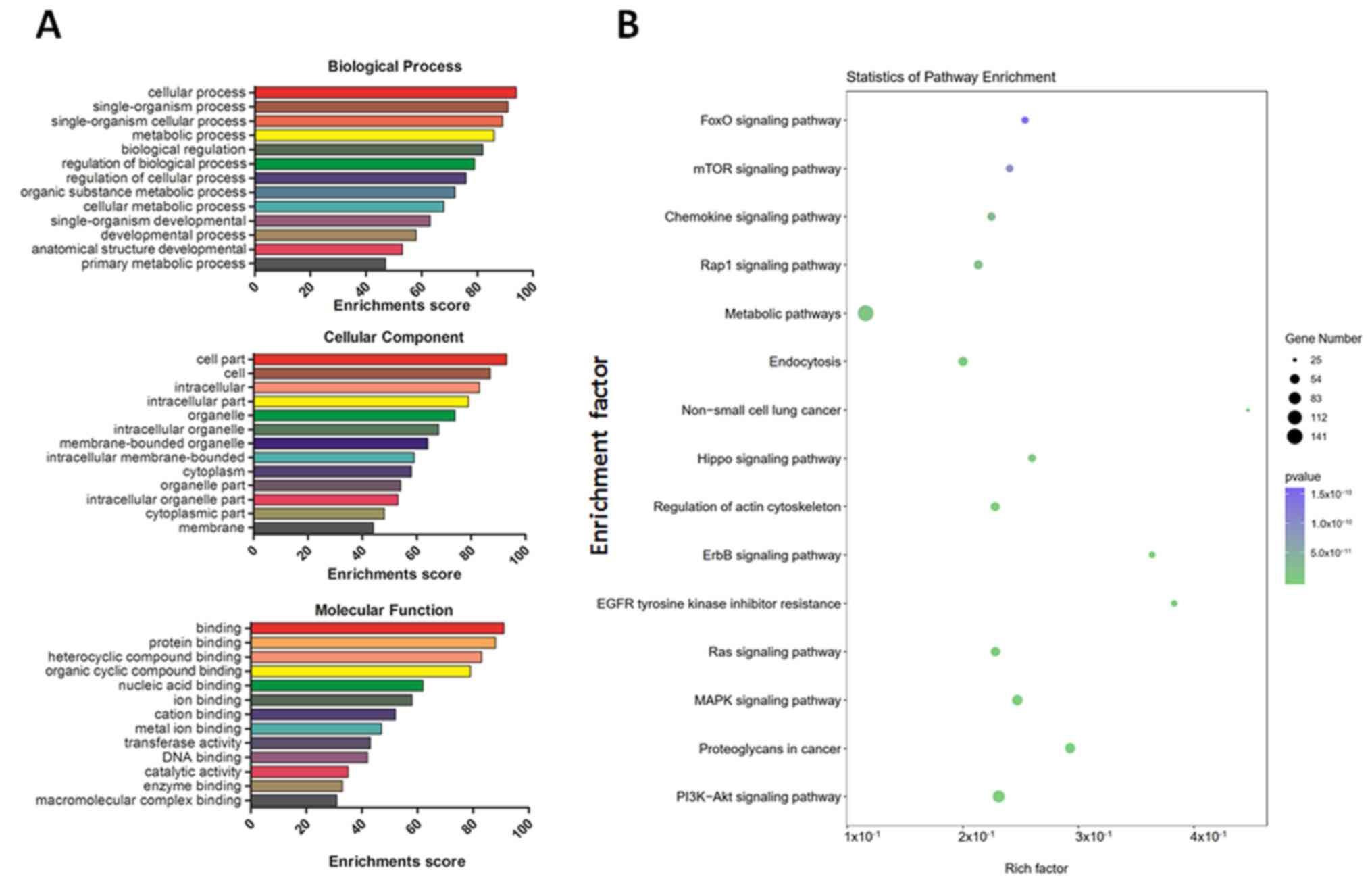
GO analysis revealed that the most enriched
biological processes were ‘single-organism process’, ‘metabolic
process’ and ‘biological regulation’. The top enriched cellular
components were ‘intracellular part’, ‘membrane-bounded organelle’,
‘cytoplasm’ and ‘cytoplasmic part’. Finally, the top enriched
molecular functions included ‘heterocyclic compound binding’,
‘nucleic acid binding’ and ‘transferase activity’ (Fig. 3A). The primarily enriched pathways
included the mTOR, chemokine, MAPK and PI3K-AKT signaling pathways
(Fig. 3B).
Validation of dysregulated miRNAs in
patients with COPD
The 20 exo-miRNAs with the most significant
differential expression in patients with COPD were further
validated via RT-qPCR. Nine of the exo-miRNAs were determined to be
significantly differentially expressed by exosome sequencing,
including five that were upregulated (miR-23a, miR-1, miR-574,
miR-152 and miR-221) and four that were downregulated (miR-3158,
miR-7706, miR-685 and miR-144) in patients with COPD compared with
those in healthy individuals. These results were consistent with
data obtained using RT-qPCR (Fig.
4). However, expression of the other 11 exo-miRNAs did not
significantly differ between patients with COPD and healthy
controls.
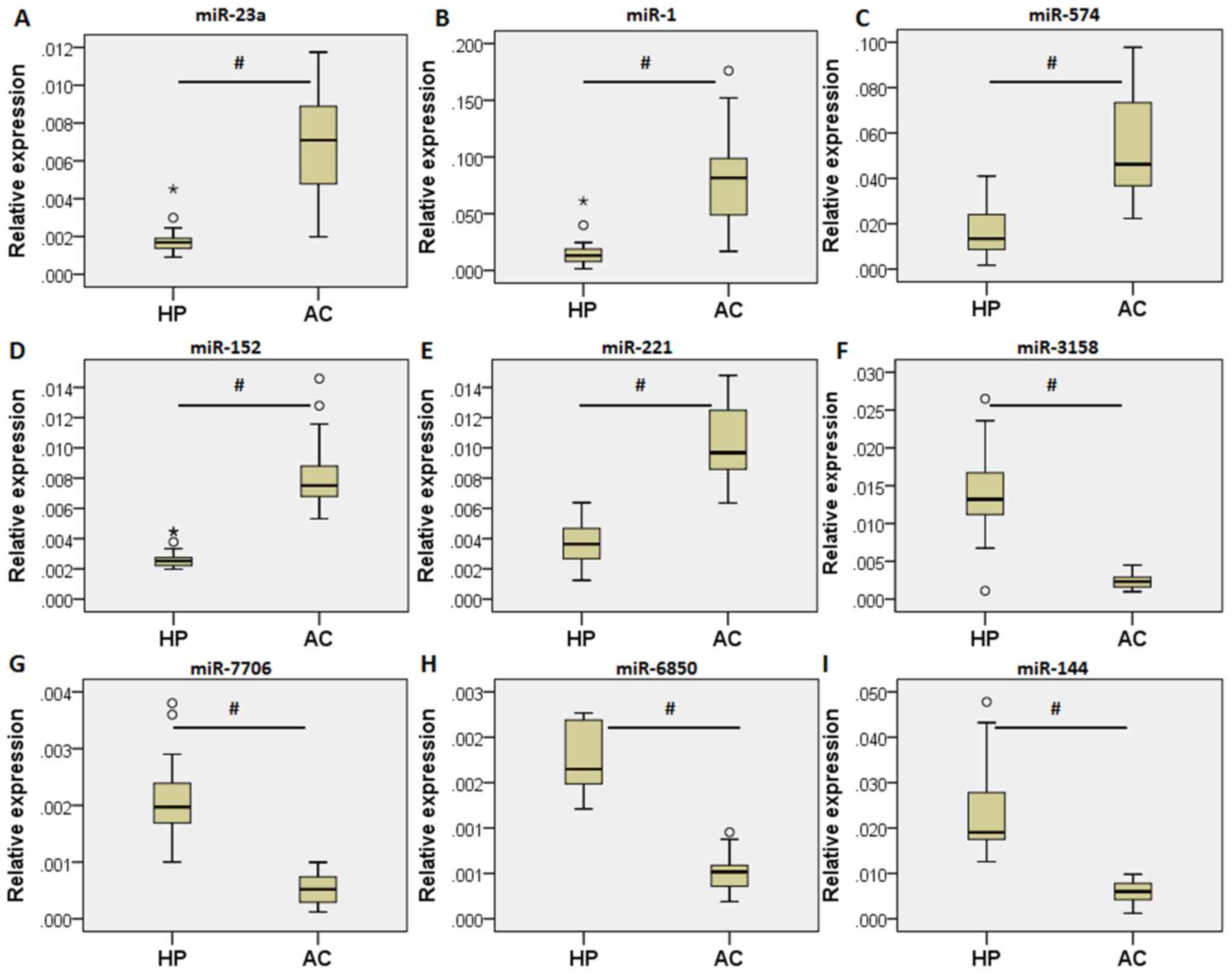 | Figure 4Validation of the differentially
expressed exosomal-miRNAs (41 patients with chronic obstructive
pulmonary disease and 29 healthy controls). Relative expression of
(A) miR-23a, (B) miR-1, (C) miR-574, (D) miR-152, (E) miR-221, (F)
miR-3158, (G) miR-7706, (H) miR-6850 and (I) miR-144 in the
validation cohort. Data are presented as median.
#P<0.05 vs. HP. °represents a mild outlier, and
*represents an extreme outlier. miRNA/miR, microRNA; HP,
healthy individuals; AC, patients with COPD. |
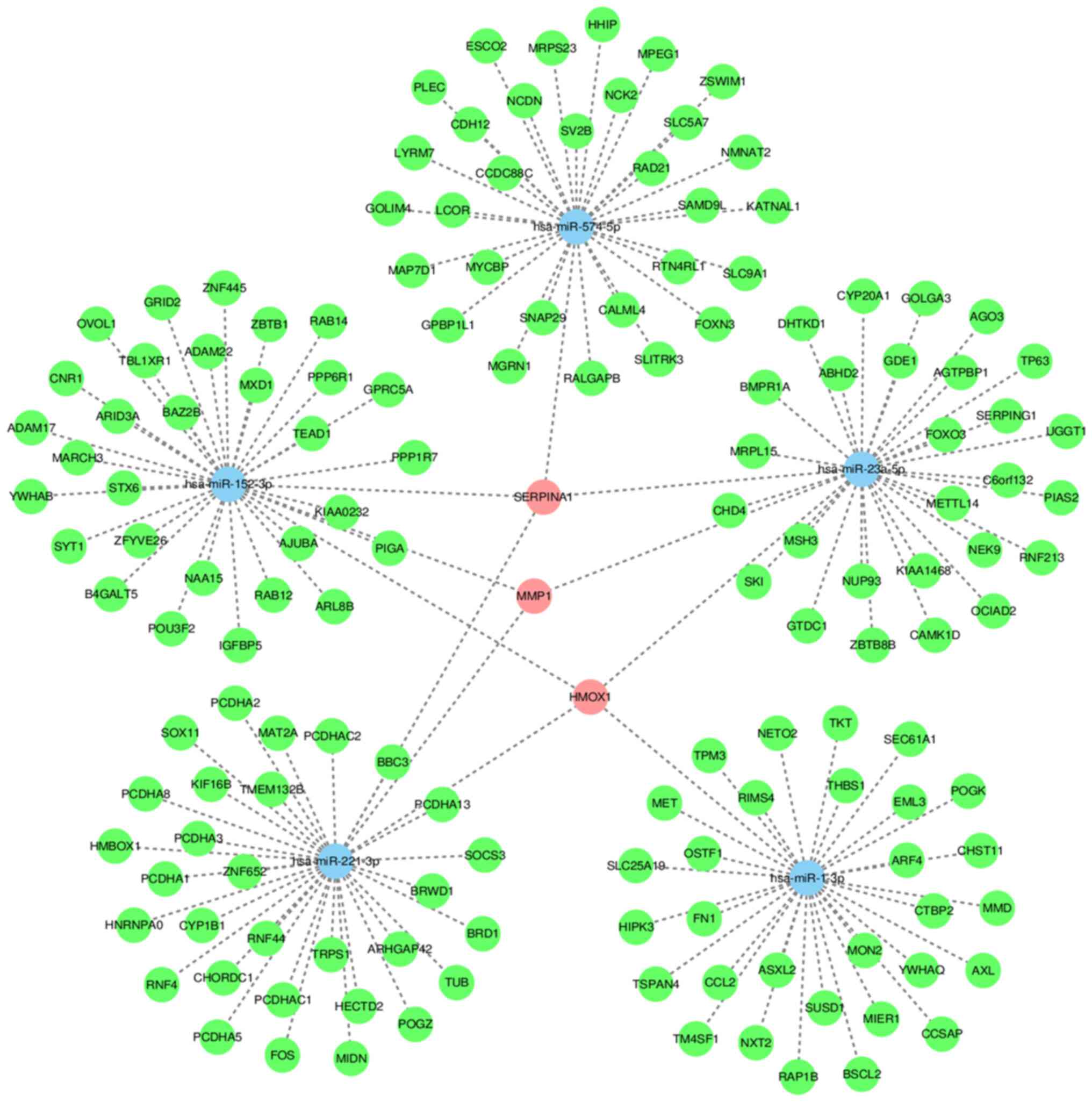
Construction of the exosomal
miRNA-mRNA network
A network was established based on the association
between the differentially expressed exo-miRNAs and mRNAs. The
network presented the interaction of the five miRNAs and 98 mRNAs
(Fig. 5). This network confirmed
that one exo-miRNA targeted one or two mRNAs, and one mRNA was
regulated by multiple miRNAs simultaneously, which suggested that a
regulatory mechanism existed between exo-miRNAs and mRNAs in
COPD.
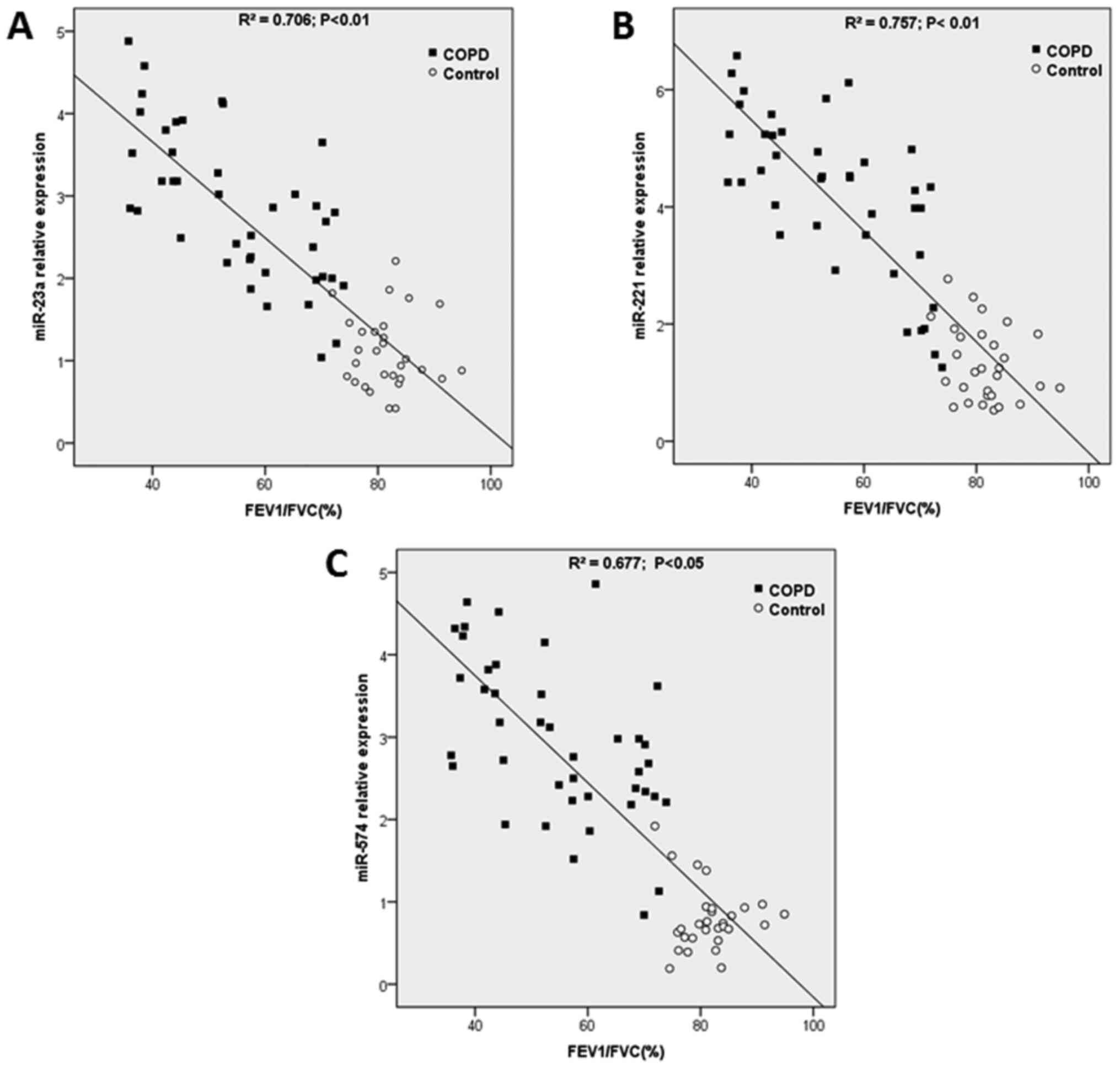
Correlation between exo-miRNA
expression with clinical parameters
The correlation between the exo-miRNA expression
levels and the FEV1/FVC value was further assessed using linear
regression. Potential confounding factors were adjusted, including
age, sex, smoking status and corticosteroid therapy were adjusted
using Bonferronis correction. The results indicated that the
expression levels of three upregulated exo-miRNAs (miR-23a, miR-221
and miR-574) correlated significantly with the FEV1/FVC values,
even after adjusting for the confounding factors. The correlation
analysis between exo-miRNA and FEV1/FVC is presented in Fig. 6. miR-23a (R2=0.706;
P<0.01; Fig. 6A), miR-221
(R2=0.757; P<0.01; Fig.
6B) and miR-574 (R2=0.677; P<0.01; Fig. 6C) were analyzed.
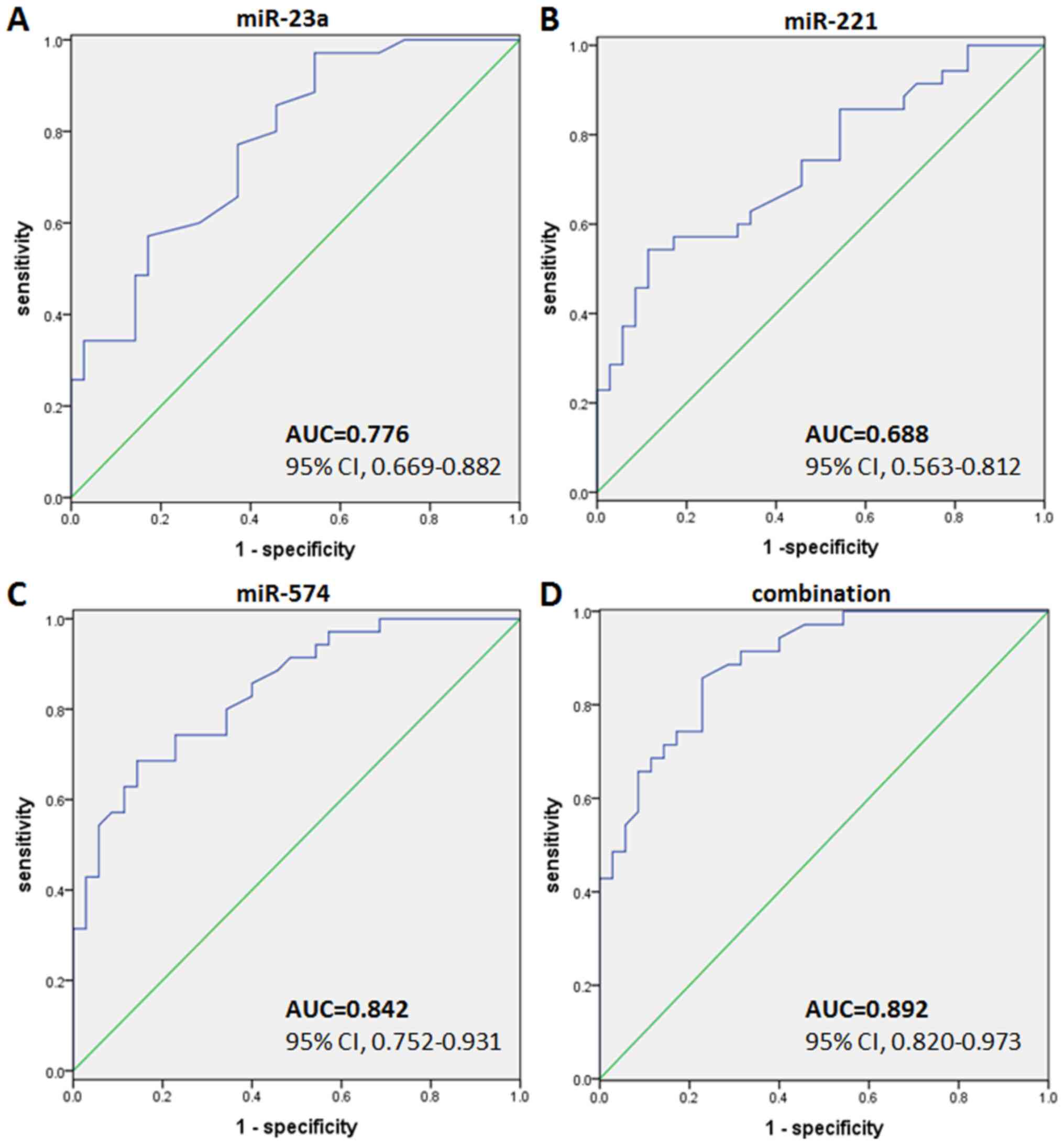
Diagnostic value of the
exo-miRNAs
ROC curves were plotted to evaluate the diagnostic
value of exo-miRNAs for COPD, which was expressed by sensitivity
and specificity. The AUC of ex-miR-23a was 0.776 (95% CI,
0.669-0.882; P<0.001). The sensitivity and specificity at the
optimal cut-off were 70.3 and 61.8%, respectively (Fig. 7A). The AUC of exo-miRNA-221 was
0.688 (95% CI, 0.563-0.812; P<0.001), where the sensitivity and
specificity at the optimal cut-off were 69.4 and 60.2%,
respectively (Fig. 7B). Finally,
the AUC of exo-miRNA-574 was 0.842 (95% CI, 0.752-0.931;
P<0.001) with a sensitivity and specificity value at the optimal
cut-off of 74.3 and 65.7%, respectively (Fig. 7C). Combining all three miRNAs
together, the AUC increased to 0.892 (95% CI, 0.820-0.973;
P<0.001). The sensitivity and specificity at the optimal cut-off
were 85.7 and 77.1%, respectively (Fig.
7D).
Discussion
Accumulating evidence has demonstrated that exosomes
mediate a variety of molecular and cellular events by facilitating
intercellular signaling (13,15,44,45).
Next-generation sequencing is a powerful tool that can identify
novel circulating biomarkers in various diseases, such as heart
disease (46,47), cancer (48) and polycystic ovary syndrome
(49). In recent years, the
relative distribution of miRNAs in different body fluids, including
blood, plasma, serum, saliva and urine have been determined
(50). In addition, it was
hypothesized that the components of exosomes may be altered during
the development of COPD (51).
Therefore, the present study performed a comprehensive analysis of
exo-miRNAs that were aberrantly expressed in patients with COPD.
The results suggested that three exo-miRNAs, miR-23a, miR-221 and
miR-574, may serve as potential diagnostic biomarkers of COPD.
Previous studies have verified the crucial role of
miRNAs in modulating gene expression under diverse conditions
(15,44). Dysregulation of miRNA expression in
airway epithelial cells, peripheral blood and lung tissues has been
associated with the pathogenesis of COPD (17,52-54).
Ezzie et al (52) compared
the expression of miRNAs in the lung tissue samples of COPD and
smokers without COPD, which elucidated 70 miRNAs that were
differentially expressed. In another study, Van Pottelberge et
al (55) revealed that 34
miRNAs were differentially expressed between non-smokers and
smokers without airflow restriction, where the expression of eight
miRNAs in smokers with COPD was significantly lower compared with
that in non-smokers. The results of the present study differed from
the aforementioned studies. This was mainly as miRNAs were
extracted from different samples, namely airway epithelial cells,
serum or lung tissues, difficulty remains in obtaining a
comprehensive comparison with previous experimental results. In
addition, these results may demonstrate discrepancy, arising from
other factors, including different sample sizes, analytical tools
and statistical methods.
Results from the current study suggested that
exosomal miRNAs were differentially expressed in patients with
COPD. Furthermore, 59 differentially expressed exo-miRNAs were
identified, including 39 that were upregulated and 20 that were
downregulated. GO enrichment analysis provides a unified vocabulary
to elaborate gene and gene product properties in various organisms
(56). GO analysis in the present
study indicated that the identified exo-miRNAs were enriched in
various annotations associated with COPD, as demonstrated by the
top enriched biological processes and molecular functions,
including ‘metabolic process’, ‘biological regulation’,
‘intracellular membrane-bounded organelle’, ‘transferase activity’,
‘catalytic activity’ and ‘enzyme binding’.
High-throughput sequencing, such as small RNA
sequencing technology makes it possible to measure the expression
of almost all coding genes, which assists in identifying genes and
pathways related to the development of diseases (51). In the present study, based on the
KEGG pathway result, the target genes of dysregulated exo-miRNAs
were involved in pathways that are closely associated with the
pathophysiology of COPD, such as the mTOR, chemokine, MAPK and
PI3K-Akt signaling pathways. Additionally, the present network
analysis indicated a potential association between miRNAs and their
target genes, suggesting that exo-miRNAs and their target genes
cooperate to regulate the pathogenesis of COPD. Furthermore, to
determine the function of the identified exo-miRNAs, interactions
between exo-miRNAs and their target mRNAs were theoretically
predicted using conserved seed-matching sequences with software for
miRNA target prediction, such as TargetScan and miRDB. This network
suggested the potential associations between exo-miRNAs and their
target genes. The network also provided an important reference
value for studying the interaction of other differentially
expressed exo-miRNAs with their potential targets. The current
study predicted that the interaction of exo-miRNAs and their target
genes was associated with COPD. The present study found 3 common
target genes [encoding α1-antitrypsin (SERPINA1), matrix
metalloproteinase 1 (MMP1), heme oxygenase-1 (HOMX1)] of the 5
miRNAs, which are closely related to the pathogenesis of COPD.
SERPINA1 has been shown to affect the susceptibility of COPD
(57). Dysregulation in the
production of MMP has been associated with lung matrix destruction
and small airways disease in COPD (58). HOMX1 (induction attenuated
senescence in chronic obstructive pulmonary disease lung
fibroblasts by protecting against mitochondria dysfunction
(59).
Exo-miRNAs that were differentially expressed
between patients with COPD and healthy controls were screened and
analyzed in an independent validation cohort. Through this method,
nine differentially expressed exo-miRNAs were identified, of which
five were upregulated and four were downregulated. To remove
various confounding factors, linear regression analysis was
performed to assess the relationship between plasma miRNA
expression and FEV1/FVC. As a result, three exo-miRNAs (miR-23a,
miR-221 and miR-574) were significantly correlated with FEV1/FVC
after adjusting for age, sex and treatment with corticosteroids.
These results suggested that these 3 exo-miRNAs may be used to
assess the severity of COPD lung function.
Exosomal molecules have the potential to serve as
disease biomarkers for a number of reasons. Exosomes contain
specific proteins and nucleic acids that carry information
regarding the physiology and microenvironment of their cells of
origin (60-62).
In addition, exosomes can exist in various biological fluids,
including blood (25), urine
(63), sputum, bronchoalveolar
lavage (26), synovial fluid
(64), pleural fluid and ascites
(65). Due to the bilayer structure
of phospholipids, exosomes are highly stable in the extracellular
environment (66). Therefore, many
exosomal proteins and miRNAs have been reported to be potential
biomarkers of multiple diseases, particularly in cancer (22,23).
There is considerable evidence to support the notion that
exo-miRNAs can serve an important role in multiple pulmonary
diseases, such as COPD (67). ROC
analysis in the present study revealed that three candidate miRNAs
(miR-23a, miR-221 and miR-574) can be used as new biomarkers of
COPD. Furthermore, when the three exo-miRNAs were combined
together, the diagnostic efficiency was improved further.
The present study provided genome-wide profiles of
exosomal miRNAs from human blood samples, demonstrating the
feasibility of identifying COPD biomarkers based on exosomal miRNA
profiling. Additionally, due to the double membrane structure of
exosomes, exosomal miRNAs can overcome certain limitations of
current biomarkers due to increased stability. However, the present
study also had a number of limitations. A single center, small
sample size, various forms of bias in the present study may lead to
inaccurate conclusions. Therefore, a large-scale, multi-center and
prospective validation study should be performed in future studies.
Additionally, the underlining mechanism of the association between
exosomal miRNAs and COPD remain unclear and should be investigated
further.
In summary, to the best of our knowledge, the
present study determined for the first time that miR-23a, miR-221
and miR-574 may serve as novel diagnostic biomarkers of COPD. The
results indicated that the evaluation of exosomal miRNA expression
could provide useful information regarding the diagnosis of
patients with COPD.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81870039),
the Jiangsu Provincial Medical Youth Talent (grant no. QNRC2016512)
and the Taizhou Municipal Science and Technology Bureau (grant no.
TS201729).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available in the National Center for Sequence Read
Archive (SRA) data, (SRA; https://www.ncbi.nlm.nih.gov/sra/; accession no.
PRJNA703816).
Authors' contributions
YS and XY designed the study and drafted the
manuscript. YS and LW performed the experiments. YW and YO
performed the statistical analysis. YO and HL performed sample
collection. HL diagnosed the patients and revised the manuscript
for important intellectual content. YS and XY confirm all the
authenticity of the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the research
ethics committee of Taizhou People's Hospital (Taizhou clinical
Medical School of Nanjing Medical university; Taizhou, China). All
individuals provided informed consent for the use of their samples
for clinical research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang C, Xu J, Yang L, Xu Y, Zhang X, Bai
C, Kang J, Ran P, Shen H, Wen F, et al: Prevalence and risk factors
of chronic obstructive pulmonary disease in China [the China
pulmonary health (CPH) study]: A national cross-sectional study.
Lancet. 391:1706–1717. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
GBD 2015 Chronic Respiratory Disease
Collaborators. Global, regional, and national deaths, prevalence,
disability-adjusted life years, and years lived with disability for
chronic obstructive pulmonary disease and asthma, 1990-2015: A
systematic analysis for the global burden of disease study 2015.
Lancet Respir Med. 5:691–706. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Fang X, Wang X and Bai C: COPD in China:
The burden and importance of proper management. Chest. 139:920–929.
2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Neumeier A and Keith R: Clinical guideline
highlights for the hospitalist: The GOLD and NICE guidelines for
the management of COPD. J Hosp Med. 15:240–241. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rabe KF, Hurd S, Anzueto A, Barnes PJ,
Buist SA, Calverley P, Fukuchi Y, Jenkins C, Rodriguez-Roisin R,
van Weel C, et al: Global strategy for the diagnosis, management,
and prevention of chronic obstructive pulmonary disease: GOLD
executive summary. Am J Respir Crit Care Med. 176:532–555.
2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Pauwels RA and Rabe KF: Burden and
clinical features of chronic obstructive pulmonary disease (COPD).
Lancet. 364:613–620. 2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lopez AD and Mathers CD: Measuring the
global burden of disease and epidemiological transitions:
2002-2030. Ann Trop Med Parasitol. 100:481–499. 2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ito K and Barnes PJ: COPD as a disease of
accelerated lung aging. Chest. 135:173–180. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Demedts IK, Demoor T, Bracke KR, Joos GF
and Brusselle GG: Role of apoptosis in the pathogenesis of COPD and
pulmonary emphysema. Respir Res. 7(53)2006.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Adcock IM, Tsaprouni L, Bhavsar P and Ito
K: Epigenetic regulation of airway inflammation. Curr Opin Immunol.
19:694–700. 2007.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wan ES and Silverman EK: Genetics of COPD
and emphysema. Chest. 136:859–866. 2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hough KP, Chanda D, Duncan SR, Thannickal
VJ and Deshane JS: Exosomes in immunoregulation of chronic lung
diseases. Allergy. 72:534–544. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Murgoci AN, Duhamel M, Raffo-Romero A,
Mallah K, Aboulouard S, Lefebvre C, Kobeissy F, Fournier I, Zilkova
M, Maderova D, et al: Location of neonatal microglia drives small
extracellular vesicles content and biological functions in vitro. J
Extracell Vesicles. 9(1727637)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Nana-Sinkam SP, Acunzo M, Croce CM and
Wang K: Extracellular vesicle biology in the pathogenesis of lung
disease. Am J Respir Crit Care Med. 196:1510–1518. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Stolzenburg LR and Harris A: The role of
microRNAs in chronic respiratory disease: Recent insights. Biol
Chem. 399:219–234. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
De Smet EG, Mestdagh P, Vandesompele J,
Brusselle GG and Bracke KR: Non-coding RNAs in the pathogenesis of
COPD. Thorax. 70:782–791. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Molina-Pinelo S, Pastor MD, Suarez R,
Romero-Romero B, González De la Peña M, Salinas A, García-Carbonero
R, De Miguel MJ, Rodríguez-Panadero F, Carnero A and Paz-Ares L:
MicroRNA clusters: Dysregulation in lung adenocarcinoma and COPD.
Eur Respir J. 43:1740–1749. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Robbins PD and Morelli AE: Regulation of
immune responses by extracellular vesicles. Nat Rev Immunol.
14:195–208. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Colombo M, Raposo G and Théry C:
Biogenesis, secretion, and intercellular interactions of exosomes
and other extracellular vesicles. Annu Rev Cell Dev Biol.
30:255–289. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yoshioka Y, Konishi Y, Kosaka N, Katsuda
T, Kato T and Ochiya T: Comparative marker analysis of
extracellular vesicles in different human cancer types. J Extracell
Vesicles. 2:2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mirzaei H, Sahebkar A, Jaafari MR,
Goodarzi M and Mirzaei HR: Diagnostic and therapeutic potential of
exosomes in cancer: The beginning of a new tale? J Cell Physiol.
232:3251–3260. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Saadatpour L, Fadaee E, Fadaei S, Nassiri
Mansour R, Mohammadi M, Mousavi SM, Goodarzi M, Verdi J and Mirzaei
H: Glioblastoma: Exosome and microRNA as novel diagnosis
biomarkers. Cancer Gene Ther. 23:415–418. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sun L, Zhu W, Zhao P, Wang Q, Fan B, Zhu
Y, Lu Y, Chen Q, Zhang J and Zhang F: Long noncoding RNA UCA1 from
hypoxia-conditioned hMSC-derived exosomes: A novel molecular target
for cardioprotection through miR-873-5p/XIAP axis. Cell Death Dis.
11(696)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Caby MP, Lankar D, Vincendeau-Scherrer C,
Raposo G and Bonnerot C: Exosomal-like vesicles are present in
human blood plasma. Int Immunol. 17:879–887. 2005.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Admyre C, Grunewald J, Thyberg J,
Gripenbäck S, Tornling G, Eklund A, Scheynius A and Gabrielsson S:
Exosomes with major histocompatibility complex class II and
co-stimulatory molecules are present in human BAL fluid. Eur Respir
J. 22:578–583. 2003.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Porro C, Lepore S, Trotta T, Castellani S,
Ratclif L, Battaglino A, Di Gioia S, Martínez MC, Conese M and
Maffione AB: Isolation and characterization of microparticles in
sputum from cystic fibrosis patients. Respir Res.
11(94)2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Salimian J, Mirzaei H, Moridikia A,
Harchegani AB, Sahebkar A and Salehi H: Chronic obstructive
pulmonary disease: MicroRNAs and exosomes as new diagnostic and
therapeutic biomarkers. J Res Med Sci. 23(27)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lener T, Gimona M, Aigner L, Börger V,
Buzas E, Camussi G, Chaput N, Chatterjee D, Court FA, Del Portillo
HA, et al: Applying extracellular vesicles based therapeutics in
clinical trials-an ISEV position paper. J Extracell Vesicles.
4(30087)2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Dang X, Qu X, Wang W, Liao C, Li Y, Zhang
X, Xu D, Baglole CJ, Shang D and Chang Y: Bioinformatic analysis of
microRNA and mRNA regulation in peripheral blood mononuclear cells
of patients with chronic obstructive pulmonary disease. Respir Res.
18(4)2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kara M, Kirkil G and Kalemci S:
Differential expression of MicroRNAs in chronic obstructive
pulmonary disease. Adv Clin Exp Med. 25:21–26. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang L and Zhang L: Circulating exosomal
miRNA as diagnostic biomarkers of neurodegenerative diseases. Front
Mol Neurosci. 13(53)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Albitar HAH and Iyer VN: Adherence to
global initiative for chronic obstructive lung disease guidelines
in the real world: Current understanding, barriers, and solutions.
Curr Opin Pulm Med. 26:149–154. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sundar IK, Li D and Rahman I: Small
RNA-sequence analysis of plasma-derived extracellular vesicle
miRNAs in smokers and patients with chronic obstructive pulmonary
disease as circulating biomarkers. J Extracell Vesicles.
8(1684816)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Théry C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol Chapter.
3(Unit 3 22)2006.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Mehdiani A, Maier A, Pinto A, Barth M,
Akhyari P and Lichtenberg A: An innovative method for exosome
quantification and size measurement. J Vis Exp: 50974, 2015.
|
|
37
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Parker VL, Cushen BF, Gavriil E, Marshall
B, Waite S, Pacey A and Heath PR: Comparison and optimisation of
microRNA extraction from the plasma of healthy pregnant women. Mol
Med Rep. 23(1)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39 (Web Server Issue):W316–W322. 2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Barabási AL and Oltvai ZN: Network
biology: Understanding the cell's functional organization. Nat Rev
Genet. 5:101–113. 2004.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Yáñez-MóM Siljander PR, Andreu Z, Zavec
AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J,
et al: Biological properties of extracellular vesicles and their
physiological functions. J Extracell Vesicles.
4(27066)2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Fujita Y, Kosaka N, Araya J, Kuwano K and
Ochiya T: Extracellular vesicles in lung microenvironment and
pathogenesis. Trends Mol Med. 21:533–542. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wang C, Song C, Liu Q, Zhang R, Fu R, Wang
H, Yin D, Song W, Zhang H and Dou K: Gene expression analysis
suggests immunological changes of peripheral blood monocytes in the
progression of patients with coronary artery disease. Front Genet.
12(641117)2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hou Z, Qin X, Hu Y, Zhang X, Li G, Wu J,
Li J, Sha J, Chen J, Xia J, et al: Longterm exercise-derived
exosomal miR-342-5p: A novel exerkine for cardioprotection. Circ
Res. 124:1386–1400. 2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Han Y, Chen J, Zhao X, Liang C, Wang Y,
Sun L, Jiang Z, Zhang Z, Yang R, Chen J, et al: MicroRNA expression
signatures of bladder cancer revealed by deep sequencing. PLoS One.
6(e18286)2011.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wang LP, Peng XY, Lv XQ, Liu L, Li XL, He
X, Lv F, Pan Y, Wang L, Liu KF and Zhang XM: High throughput
circRNAs sequencing profile of follicle fluid exosomes of
polycystic ovary syndrome patients. J Cell Physiol, Feb 18, 2019
(Epub ahead of print). doi: https://doi.org/10.1002/jcp.28201.
|
|
50
|
El-Mogy M, Lam B, Haj-Ahmad TA, McGowan S,
Yu D, Nosal L, Rghei N, Roberts P and Haj-Ahmad Y: Diversity and
signature of small RNA in different bodily fluids using next
generation sequencing. BMC Genomics. 19(408)2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
‘t Hoen PA, Ariyurek Y, Thygesen HH,
Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ and
den Dunnen JT: Deep sequencing-based expression analysis shows
major advances in robustness, resolution and inter-lab portability
over five microarray platforms. Nucleic Acids Res.
36(e141)2008.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ezzie ME, Crawford M, Cho JH, Orellana R,
Zhang S, Gelinas R, Batte K, Yu L, Nuovo G, Galas D, et al: Gene
expression networks in COPD: microRNA and mRNA regulation. Thorax.
67:122–131. 2012.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Akbas F, Coskunpinar E, Aynaci E, Oltulu
YM and Yildiz P: Analysis of serum micro-RNAs as potential
biomarker in chronic obstructive pulmonary disease. Exp Lung Res.
38:286–294. 2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Schembri F, Sridhar S, Perdomo C,
Gustafson AM, Zhang X, Ergun A, Lu J, Liu G, Zhang X, Bowers J, et
al: MicroRNAs as modulators of smoking-induced gene expression
changes in human airway epithelium. Proc Natl Acad Sci USA.
106:2319–2324. 2009.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Van Pottelberge GR, Mestdagh P, Bracke KR,
Thas O, van Durme YM, Joos GF, Vandesompele J and Brusselle GG:
MicroRNA expression in induced sputum of smokers and patients with
chronic obstructive pulmonary disease. Am J Respir Crit Care Med.
183:898–906. 2011.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Nakamura H: Genetics of COPD. Allergol
Int. 60:253–258. 2011.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Ostridge K, Williams N, Kim V, Bennett M,
Harden S, Welch L, Bourne S, Coombs NA, Elkington PT, Staples KJ
and Wilkinson TM: Relationship between pulmonary matrix
metalloproteinases and quantitative CT markers of small airways
disease and emphysema in COPD. Thorax. 71:126–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Even B, Fayad-Kobeissi S, Gagliolo JM,
Motterlini R, Boczkowski J, Foresti R and Dagouassat M: Heme
oxygenase-1 induction attenuates senescence in chronic obstructive
pulmonary disease lung fibroblasts by protecting against
mitochondria dysfunction. Aging Cell. 17(e12837)2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
de Jong OG, Verhaar MC, Chen Y, Vader P,
Gremmels H, Posthuma G, Schiffelers RM, Gucek M and van Balkom BW:
Cellular stress conditions are reflected in the protein and RNA
content of endothelial cell-derived exosomes. J Extracell Vesicles.
1:2012.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Beninson LA and Fleshner M: Exosomes: An
emerging factor in stress-induced immunomodulation. Semin Immunol.
26:394–401. 2014.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Iraci N, Leonardi T, Gessler F, Vega B and
Pluchino S: Focus on extracellular vesicles: Physiological role and
signalling properties of extracellular membrane vesicles. Int J Mol
Sci. 17(171)2016.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Pisitkun T, Shen RF and Knepper MA:
Identification and proteomic profiling of exosomes in human urine.
Proc Natl Acad Sci USA. 101:13368–13373. 2004.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Song JE, Kim JS, Shin JH, Moon KW, Park
JK, Park KS and Lee EY: Role of synovial exosomes in osteoclast
differentiation in inflammatory arthritis. Cells.
10(120)2021.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Hu Y, Qi C, Liu X, Zhang C, Gao J, Wu Y,
Yang J, Zhao Q, Li J, Wang X and Shen L: Malignant ascites-derived
exosomes promote peritoneal tumor cell dissemination and reveal a
distinct miRNA signature in advanced gastric cancer. Cancer Lett.
457:142–150. 2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Sun L, Zhu W, Zhao P, Zhang J, Lu Y, Zhu
Y, Zhao W, Liu Y, Chen Q and Zhang F: Down-regulated exosomal
MicroRNA-221-3p derived from senescent mesenchymal stem cells
impairs heart repair. Front Cell Dev Biol. 8(263)2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Chen J, Hu C and Pan P: Extracellular
vesicle MicroRNA transfer in lung diseases. Front Physiol.
8(1028)2017.PubMed/NCBI View Article : Google Scholar
|