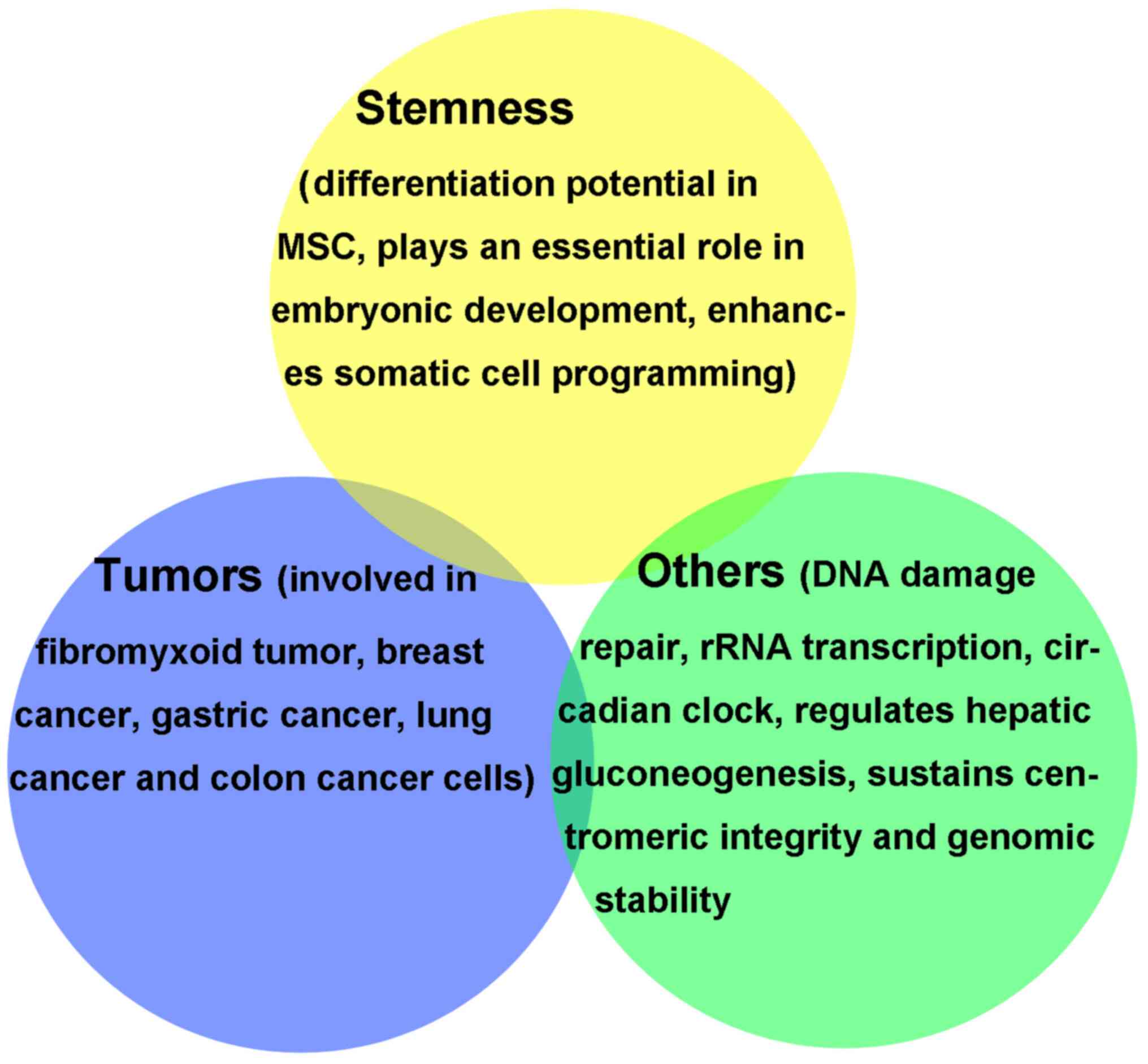
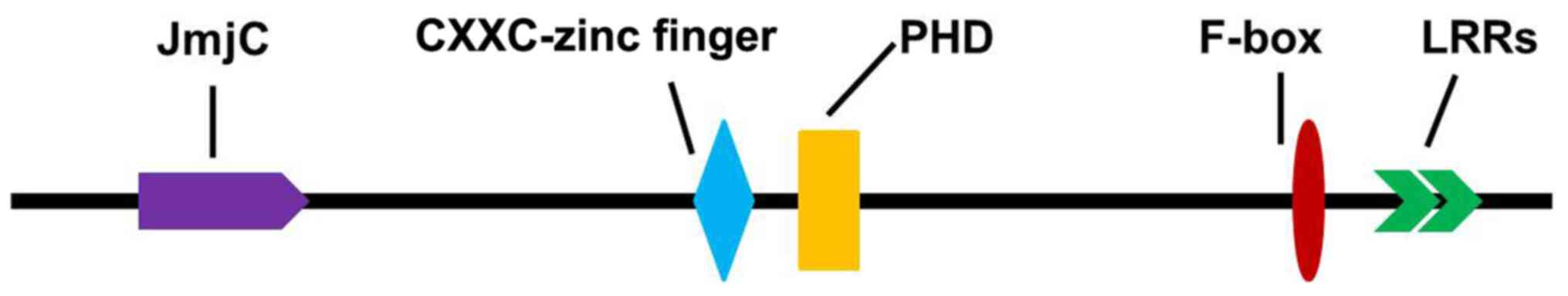
|
1
|
Luger K, Mader AW, Richmond RK, Sargent DF
and Richmond TJ: Crystal structure of the nucleosome core particle
at 2.8 a resolution. Nature. 389:251–260. 1997.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Richmond TJ, Finch JT, Rushton B, Rhodes D
and Klug A: Structure of the nucleosome core particle at 7 a
resolution. Nature. 311:532–537. 1984.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Hyun K, Jeon J, Park K and Kim J: Writing,
erasing and reading histone lysine methylations. Exp Mol Med.
49(e324)2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Black JC, Van Rechem C and Whetstine JR:
Histone lysine methylation dynamics: Establishment, regulation, and
biological impact. Mol Cell. 48:491–507. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hojfeldt JW, Agger K and Helin K: Histone
lysine demethylases as targets for anticancer therapy. Nat Rev Drug
Discov. 12:917–930. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Fang Y, Liao G and Yu B: Targeting histone
lysine demethylase LSD1/KDM1A as a new avenue for cancer therapy.
Curr Top Med Chem. 19:889–891. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tsukada Y, Fang J, Erdjument-Bromage H,
Warren ME, Borchers CH, Tempst P and Zhang Y: Histone demethylation
by a family of JmjC domain-containing proteins. Nature.
439:811–816. 2006.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z,
Ma Y, Yu Y, Lin H, Chen AP and Chen CD: JARID1B is a histone H3
lysine 4 demethylase up-regulated in prostate cancer. Proc Natl
Acad Sci USA. 104:19226–19231. 2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Klose RJ, Kallin EM and Zhang Y:
JmjC-domain-containing proteins and histone demethylation. Nat Rev
Genet. 7:715–727. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Cloos PA, Christensen J, Agger K and Helin
K: Erasing the methyl mark: Histone demethylases at the center of
cellular differentiation and disease. Genes Dev. 22:1115–1140.
2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Vacik T, Ladinovic D and Raska I: KDM2A/B
lysine demethylases and their alternative isoforms in development
and disease. Nucleus. 9:431–441. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zlotorynski E: Protein folding:
Structure-function analysis of KDM2A. Nat Rev Mol Cell Biol.
15(630)2014.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Blackledge NP, Zhou JC, Tolstorukov MY,
Farcas AM, Park PJ and Klose RJ: CpG islands recruit a histone H3
lysine 36 demethylase. Mol Cell. 38:179–190. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhou JC, Blackledge NP, Farcas AM and
Klose RJ: Recognition of CpG island chromatin by KDM2A requires
direct and specific interaction with linker DNA. Mol Cell Biol.
32:479–489. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Iuchi S and Paulo JA: Lysine-specific
demethylase 2A enhances binding of various nuclear factors to
CpG-rich genomic DNAs by action of its CXXC-PHD domain. Sci Rep.
9(5496)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu H, Liu L, Holowatyj A, Jiang Y and
Yang ZQ: Integrated genomic and functional analyses of histone
demethylases identify oncogenic KDM2A isoform in breast cancer. Mol
Carcinog. 55:977–990. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Cheng Z, Cheung P, Kuo AJ, Yukl ET, Wilmot
CM, Gozani O and Patel DJ: A molecular threading mechanism
underlies Jumonji lysine demethylase KDM2A regulation of methylated
H3K36. Genes Dev. 28:1758–1771. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Iuchi S and Green H: Lysine-specific
demethylase 2A (KDM2A) normalizes human embryonic stem cell derived
keratinocytes. Proc Natl Acad Sci USA. 109:9442–9447.
2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ladinovic D, Pinkas D, Šopin T, Raška O,
Liška F, Raška I and Vacík T: Alternative isoforms of KDM2A and
KDM2B lysine demethylases negatively regulate canonical Wnt
signaling. PLoS One. 15(e236612)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Frescas D, Guardavaccaro D, Kuchay SM,
Kato H, Poleshko A, Basrur V, Elenitoba-Johnson KS, Katz RA and
Pagano M: KDM2A represses transcription of centromeric satellite
repeats and maintains the heterochromatic state. Cell Cycle.
7:3539–3547. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Nagase T, Ishikawa K, Suyama M, Kikuno R,
Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N and Ohara O:
Prediction of the coding sequences of unidentified human genes.
XIII. The complete sequences of 100 new cDNA clones from brain
which code for large proteins in vitro. DNA Res. 6:63–70.
1999.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chen JY, Luo CW, Lai YS, Wu CC and Hung
WC: Lysine demethylase KDM2A inhibits TET2 to promote DNA
methylation and silencing of tumor suppressor genes in breast
cancer. Oncogenesis. 6(e369)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kong Y, Zou S, Yang F, Xu X, Bu W, Jia J
and Liu Z: RUNX3-mediated up-regulation of miR-29b suppresses the
proliferation and migration of gastric cancer cells by targeting
KDM2A. Cancer Lett. 381:138–148. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Dhar SS, Alam H, Li N, Wagner KW, Chung J,
Ahn YW and Lee MG: Transcriptional repression of histone
deacetylase 3 by the histone demethylase KDM2A is coupled to
tumorigenicity of lung cancer cells. J Biol Chem. 289:7483–7496.
2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tanaka Y, Obinata H, Konishi A, Yamagiwa N
and Tsuneoka M: Production of ROS by Gallic acid activates KDM2A to
reduce rRNA transcription. Cells. 9(2266)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tanaka Y, Konishi A, Obinata H and
Tsuneoka M: Metformin activates KDM2A to reduce rRNA transcription
and cell proliferation by dual regulation of AMPK activity and
intracellular succinate level. Sci Rep. 9(18694)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang F, Liang S, Liu X, Han L, Wang J and
Du Q: LINC00460 modulates KDM2A to promote cell proliferation and
migration by targeting miR-342-3p in gastric cancer. Onco Targets
Ther. 11:6383–6394. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Shou T, Yang H, Lv J, Liu D and Sun X:
MicroRNA3666 suppresses the growth and migration of glioblastoma
cells by targeting KDM2A. Mol Med Rep. 19:1049–1055.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li X, Wei C, Zhang Z, Jin Q and Xiao X:
MiR-134-5p regulates myocardial apoptosis and angiogenesis by
directly targeting KDM2A After myocardial infarction. Int Heart J.
61:815–821. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhao Y, Chen X, Jiang J, Wan X, Wang Y and
Xu P: Epigallocatechin gallate reverses gastric cancer by
regulating the long noncoding RNA LINC00511/miR-29b/KDM2A axis.
Biochim Biophys Acta Mol Basis Dis. 1866(165856)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yang H, Li G, Han N, Zhang X, Cao Y, Cao Y
and Fan Z: Secreted frizzled-related protein 2 promotes the
osteo/odontogenic differentiation and paracrine potentials of stem
cells from apical papilla under inflammation and hypoxia
conditions. Cell Prolif. 53(e12694)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ou R, Zhu L, Zhao L, Li W, Tao F, Lu Y, He
Q, Li J, Ren Y and Xu Y: HPV16 E7-induced upregulation of KDM2A
promotes cervical cancer progression by regulating miR-132-radixin
pathway. J Cell Physiol. 234:2659–2671. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang Y, Sun B, Zhang Q, Dong H and Zhang
J: p300 Acetylates JHDM1A to inhibit osteosarcoma carcinogenesis.
Artif Cells Nanomed Biotechnol. 47:2891–2899. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Tanaka Y, Yano H, Ogasawara S, Yoshioka S,
Imamura H, Okamoto K and Tsuneoka M: Mild glucose starvation
induces KDM2A-mediated H3K36me2 demethylation through AMPK To
reduce rRNA transcription and cell proliferation. Mol Cell Biol.
35:4170–4184. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lu S, Yang Y, Du Y, Cao LL, Li M, Shen C,
Hou T, Zhao Y, Wang H, Deng D, et al: The transcription factor
c-Fos coordinates with histone lysine-specific demethylase 2A to
activate the expression of cyclooxygenase-2. Oncotarget.
6:34704–34717. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
McAllister TE, England KS, Hopkinson RJ,
Brennan PE, Kawamura A and Schofield CJ: Recent progress in histone
demethylase inhibitors. J Med Chem. 59:1308–1329. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Suzuki T, Ozasa H, Itoh Y, Zhan P, Sawada
H, Mino K, Walport L, Ohkubo R, Kawamura A, Yonezawa M, et al:
Identification of the KDM2/7 histone lysine demethylase subfamily
inhibitor and its antiproliferative activity. J Med Chem.
56:7222–7231. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gerken PA, Wolstenhulme JR, Tumber A,
Hatch SB, Zhang Y, Müller S, Chandler SA, Mair B, Li F, Nijman SMB,
et al: Discovery of a highly selective cell-active inhibitor of the
histone lysine demethylases KDM2/7. Angew Chem Int Ed Engl.
56:15555–15559. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhou Q, Chen L, Song Y, Ma L, Xiao P, Chen
L, Zhen H, Han R, Chen X, Sun S, et al: Induction of co-inhibitory
molecule CTLA-4 by human papillomavirus E7 protein through
downregulation of histone methyltransferase JHDM1B expression.
Virology. 538:111–118. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Abdel-Hameed EA, Ji H and Shata MT:
HIV-induced epigenetic alterations in host cells. Adv Exp Med Biol.
879:27–38. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sheng W, LaFleur MW, Nguyen TH, Chen S,
Chakravarthy A, Conway JR, Li Y, Chen H, Yang H, Hsu PH, et al:
LSD1 ablation stimulates anti-tumor immunity and enables checkpoint
blockade. Cell. 174:549–563.e19. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Boehm D and Ott M: Host methyltransferases
and demethylases: Potential new epigenetic targets for HIV cure
strategies and beyond. AIDS Res Hum Retroviruses. 33
(Suppl):S8–S22. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Rose NR, Woon EC, Tumber A, Walport LJ,
Chowdhury R, Li XS, King ON, Lejeune C, Ng SS, Krojer T, et al:
Plant growth regulator daminozide is a selective inhibitor of human
KDM2/7 histone demethylases. J Med Chem. 55:6639–6643.
2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen JY, Li CF, Chu PY, Lai YS, Chen CH,
Jiang SS, Hou MF and Hung WC: Lysine demethylase 2A promotes
stemness and angiogenesis of breast cancer by upregulating Jagged1.
Oncotarget. 7:27689–27710. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wagner KW, Alam H, Dhar SS, Giri U, Li N,
Wei Y, Giri D, Cascone T, Kim JH, Ye Y, et al: KDM2A promotes lung
tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin
Invest. 123:5231–5246. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Huang Y, Liu Y, Yu L, Chen J, Hou J, Cui
L, Ma D and Lu W: Histone demethylase KDM2A promotes tumor cell
growth and migration in gastric cancer. Tumour Biol. 36:271–278.
2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lu DH, Yang J, Gao LK, Min J, Tang JM, Hu
M, Li Y, Li ST, Chen J and Hong L: Lysine demethylase 2A promotes
the progression of ovarian cancer by regulating the PI3K pathway
and reversing epithelialmesenchymal transition. Oncol Rep.
41:917–927. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Xi C, Ye NY and Wang YB: LncRNA LINC01278
accelerates colorectal cancer progression via miR-134-5p/KDM2A
axis. Eur Rev Med Pharmacol Sci. 24:10526–10534. 2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Cao LL, Du C, Liu H, Pei L, Qin L, Jia M
and Wang H: Lysine-specific demethylase 2A expression is associated
with cell growth and cyclin D1 expression in colorectal
adenocarcinoma. Int J Biol Markers: Apr 1, 2018 (Epub ahead of
print). doi: 10.1177/1724600818764069.
|
|
51
|
Rizwani W, Schaal C, Kunigal S, Coppola D
and Chellappan S: Mammalian lysine histone demethylase KDM2A
regulates E2F1-mediated gene transcription in breast cancer cells.
PLoS One. 9(e100888)2014.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chen JY, Li CF, Lai YS and Hung WC: Lysine
demethylase 2A expression in cancer-associated fibroblasts promotes
breast tumour growth. Br J Cancer. 124:484–493. 2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
De Nicola I, Guerrieri AN, Penzo M,
Ceccarelli C, De Leo A, Trerè D and Montanaro L: Combined
expression levels of KDM2A and KDM2B correlate with nucleolar size
and prognosis in primary breast carcinomas. Histol Histopathol.
35:1181–1187. 2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lin Q, Wu Z, Yue X, Yu X, Wang Z, Song X,
Xu L, He Y, Ge Y, Tan S, et al: ZHX2 restricts hepatocellular
carcinoma by suppressing stem cell-like traits through
KDM2A-mediated H3K36 demethylation. Ebiomedicine.
53(102676)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Scahill CM, Digby Z, Sealy IM,
Wojciechowska S, White RJ, Collins JE, Stemple DL, Bartke T,
Mathers ME, Patton EE and Busch-Nentwich EM: Loss of the chromatin
modifier Kdm2aa causes BrafV600E-independent spontaneous melanoma
in zebrafish. PLoS Genet. 13(e1006959)2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lu T, Jackson MW, Singhi AD, Kandel ES,
Yang M, Zhang Y, Gudkov AV and Stark GR: Validation-based
insertional mutagenesis identifies lysine demethylase FBXL11 as a
negative regulator of NFkappaB. Proc Natl Acad Sci USA.
106:16339–16344. 2009.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Lu T, Jackson MW, Wang B, Yang M, Chance
MR, Miyagi M, Gudkov AV and Stark GR: Regulation of NF-kappaB by
NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc
Natl Acad Sci USA. 107:46–51. 2010.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Gao R, Dong R, Du J, Ma P, Wang S and Fan
Z: Depletion of histone demethylase KDM2A inhibited cell
proliferation of stem cells from apical papilla by de-repression of
p15INK4B and p27Kip1. Mol Cell Biochem. 379:115–122.
2013.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Dong R, Yao R, Du J, Wang S and Fan Z:
Depletion of histone demethylase KDM2A enhanced the adipogenic and
chondrogenic differentiation potentials of stem cells from apical
papilla. Exp Cell Res. 319:2874–2882. 2013.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Yu G, Wang J, Lin X, Diao S, Cao Y, Dong
R, Wang L, Wang S and Fan Z: Demethylation of SFRP2 by histone
demethylase KDM2A regulated osteo-/dentinogenic differentiation of
stem cells of the apical papilla. Cell Prolif. 49:330–340.
2016.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Du J, Ma Y, Ma P, Wang S and Fan Z:
Demethylation of epiregulin gene by histone demethylase FBXL11 and
BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem
Cells. 31:126–136. 2013.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi
X, Qin B, Zeng L, Esteban MA, Pan G and Pei D: The histone
demethylases Jhdm1a/1b enhance somatic cell reprogramming in a
vitamin-C-dependent manner. Cell Stem Cell. 9:575–587.
2011.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Yi Q, Cao Y, Liu OS, Lu YQ, Wang JS, Wang
SL, Yao R and Fan ZP: Spatial and temporal expression of histone
demethylase, Kdm2a, during murine molar development. Biotech
Histochem. 91:137–144. 2016.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Kawakami E, Tokunaga A, Ozawa M, Sakamoto
R and Yoshida N: The histone demethylase Fbxl11/Kdm2a plays an
essential role in embryonic development by repressing cell-cycle
regulators. Mech Dev. 135:31–42. 2015.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Fu E, Shen J, Dong Z, Zhang W, Zhang Y,
Chen F, Cheng Z, Zhao X, Shuai L and Lu X: Histone demethylase
Kdm2a regulates germ cell genes and endogenous retroviruses in
embryonic stem cells. Epigenomics. 11:751–766. 2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Lu L, Gao Y, Zhang Z, Cao Q, Zhang X, Zou
J and Cao Y: Kdm2a/b lysine demethylases regulate canonical Wnt
signaling by modulating the stability of nuclear β-catenin. Dev
Cell. 33:660–674. 2015.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Acevedo N, Reinius LE, Vitezic M, Fortino
V, Söderhäll C, Honkanen H, Veijola R, Simell O, Toppari J, Ilonen
J, et al: Age-associated DNA methylation changes in immune genes,
histone modifiers and chromatin remodeling factors within 5 years
after birth in human blood leukocytes. Clin Epigenetics.
7(34)2015.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Salminen A, Kauppinen A and Kaarniranta K:
2-Oxoglutarate-dependent dioxygenases are sensors of energy
metabolism, oxygen availability, and iron homeostasis: Potential
role in the regulation of aging process. Cell Mol Life Sci.
72:3897–3914. 2015.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Pan D, Mao C, Zou T, Yao AY, Cooper MP,
Boyartchuk V and Wang YX: The histone demethylase Jhdm1a regulates
hepatic gluconeogenesis. PLoS Genet. 8(e1002761)2012.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Tanaka Y, Okamoto K, Teye K, Umata T,
Yamagiwa N, Suto Y, Zhang Y and Tsuneoka M: JmjC enzyme KDM2A is a
regulator of rRNA transcription in response to starvation. EMBO J.
29:1510–1522. 2010.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Tanaka Y, Umata T, Okamoto K, Obuse C and
Tsuneoka M: CxxC-ZF domain is needed for KDM2A to demethylate
histone in rDNA promoter in response to starvation. Cell Struct
Funct. 39:79–92. 2014.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Tsuneoka M, Tanaka Y and Okamoto K: A CxxC
domain that binds to unmethylated CpG is required for KDM2A to
control rDNA transcription. Yakugaku Zasshi. 135:11–21.
2015.PubMed/NCBI View Article : Google Scholar : (In Japanese).
|
|
73
|
Reischl S and Kramer A: Fbxl11 is a novel
negative element of the mammalian circadian clock. J Biol Rhythms.
30:291–301. 2015.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Jin MH and Oh DY: ATM in DNA repair in
cancer. Pharmacol Ther. 203(107391)2019.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Cao LL, Wei F, Du Y, Song B, Wang D, Shen
C, Lu X, Cao Z, Yang Q, Gao Y, et al: ATM-mediated KDM2A
phosphorylation is required for the DNA damage repair. Oncogene.
35(402)2016.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Bueno M, Baldascini M, Richard S and
Lowndes NF: Recruitment of lysine demethylase 2A to DNA double
strand breaks and its interaction with 53BP1 ensures genome
stability. Oncotarget. 9:15915–15930. 2018.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Rezazadeh S, Yang D, Biashad SA, Firsanov
D, Takasugi M, Gilbert M, Tombline G, Bhanu NV, Garcia BA, Seluanov
A and Gorbunova V: SIRT6 mono-ADP ribosylates KDM2A to locally
increase H3K36me2 at DNA damage sites to inhibit transcription and
promote repair. Aging (Albany NY). 12:11165–11184. 2020.PubMed/NCBI View Article : Google Scholar
|