Introduction
With the increasing amount of pressure on society
and the stress of everyday living, the number of individuals
suffering from depression is increasing (1). At present, depression is a common
disease which is recognized by the World Health Organization and it
is one of the main causes of psychiatric disability worldwide
(2). Establishing trust and rapport
is the primary intervention strategy for depression, followed by
specific interventions according to each diagnosis and the
individualized treatment care plan (3,4).
Researchers have also highlighted the need for nursing training in
order for nurses to perform initial assessment procedures and
acquire appropriate evidence-based intervention skills and
techniques (3). It has been
reported that hippocampal volume and nerve density are markedly
reduced in patients with depression through pathophysiological
studies (4-6).
In addition, a number of studies have demonstrated that the
pathogenesis of depression is affected by several factors, such as
neural and structural plasticity, neurotransmitter systems, and
epigenetic and genetic susceptibility (7-11).
Therefore, the study of depression has generally attracted the
attention of researchers (12,13).
MicroRNAs (miRNAs/miRs) are a group of endogenous
small and non-coding RNAs that regulate gene expression by
partially binding to the 3'-untranslated region (3'-UTR) of
targeted mRNAs (14-16).
Studies have indicated that miRNAs are involved in a number of
cellular biological processes in normal physiology and
pathogenesis, such as differentiation, cell growth, apoptosis and
inflammation (17). Previous
research has indicated that miRNAs are highly expressed in the
central nervous system and play a crucial role in brain functions,
such as neurogenesis, neuronal metabolism, proliferation and
apoptosis (18,19).
miR-135a has been found to be upregulated in
hepatoma cells, and to facilitate cell proliferation, migration and
invasion by targeting FOXO1 and TGFB1 (20,21).
Moreover, miR-135a has been found to promote cell proliferation by
targeting FOXO1 in malignant melanoma (22). In addition, a previous study
demonstrated that miR-135a is related to prostate cancer (23). It has been reported that miR-135a is
downregulated in the serum of patients with depression and may
serve as a potential marker for the diagnosis of depression
(24). In addition, Gheysarzadeh
et al (24) demonstrated
that miR-16 and miR-1202 expression was decreased in the serum of
patients with depression. However, the mechanism of action of
miR-135a in the development of depression remains unclear.
Currently, chronic unpredictable mild stress
(CUMS)-induced mouse model of depression has been widely used to
investigate depression in vivo (25-27).
Therefore, the aim of the present study was to explore the role of
miR-135a in CUMS-induced depression and to analyze its molecular
mechanisms of action.
Materials and methods
Clinical samples
In the present study, peripheral blood was obtained
from 50 patients with depression (male, 22; female, 28; age range,
37-53 years) and 50 healthy volunteers (male, 24; female, 26; age
range, 38-51 years) between January 2018 and December 2019 at
Binzhou Youfu Hospital (Binzhou, Shandong, China). Exclusion
criteria were a history of bipolar or any psychotic disorder, the
use of lithium or an antipsychotic within the prior 2 weeks;
substance-use disorder within 3 months; pregnancy or lactation. The
healthy controls had no lifetime history of any mental disorder.
All patients signed informed consent forms. The present study was
approved by the Ethics Committee of Binzhou Youfu Hospital.
Animals and experimental design
A total of 75 male C57BL/6 mice (age, 8-10 weeks;
weight, 18-22 g) were purchased from the Nanjing University Animal
Research Center. The mice were housed in a standard environment
(temperature, 22±2˚C; humidity, 55±5%; light/dark cycle, 12 h) with
free access to food and water. The mice were randomly divided into
five groups (n=15 per group) as follows: i) Unstressed control; ii)
CUMS; iii) CUMS + 0.5 nmol mimic control injected intraperitoneally
(5'-UUUGUACUACACAAAAGUACUG-3'; Guangzhou RiboBio Co., Ltd.); iv)
CUMS + 0.5 nmol miR-135a mimic injected intraperitoneally
(5'-UAUGGCUUUUUA UUCCUAUGUGA-3'; Guangzhou RiboBio Co., Ltd.); and
v) positive control group injected intraperitoneally with 20
mg/kg/day fluoxetine (Sigma-Aldrich; Merck KGaA) for 21 days (CUMS
+ FLU). All animal experiments were performed according to a
protocol approved by the Committee on Care and Use of the
Laboratory Animal Committee of Binzhou Youfu Hospital.
A total of 3 weeks after mimic control/miR-135a
mimic/FLU treatment, the mice were anesthetized with an
intraperitoneal injection of pentobarbital (Sigma-Aldrich; Merck
KGaA; 50 mg/kg) and sacrificed by cervical dislocation. Death was
defined as the lack of heartbeat and breathing. The peripheral
blood and hippocampus tissue were subsequently harvested following
euthanasia as previously described (28).
Mouse model of CUMS
A mouse model of depression was established by
inducing CUMS, as previously described (25). In brief, the CUMS procedures
included the following various mild stress factors: Continuous
night illumination (overnight), cage tilt (7 h), water and food
deprivation (24 h), swimming in cold water, noise, wet pads,
foreign object exposure (6 h), tail clamp (1 min), hanging of the
mice on the balance bar with a rope (10 min), physical restraint
for 3 h and a 5-min oscillation. The mice were subjected to 2-3
kinds of stimuli each day; however, the same stressor was not
applied again within less than 3 consecutive days. The CUMS
procedure lasted for 6 weeks, and treatments with miR-135a
mimic/mimic control/FLU were performed daily from the 4th to the
6th week. The body weights of the mice were monitored from the
beginning of the experiment and measured each week.
Sucrose preference test (SPT)
The SPT was performed using the method previously
described by Iñiguez et al (29). SPT was performed every week.
Briefly, the mice were deprived of water and food for 24 h and then
tested for sucrose preference. Each mouse was given free access to
two bottles for 12 h: One bottle contained 1% sucrose solution
(w/v) and the other bottle contained tap water. To avoid the
influence of the positioning, the two bottles were placed opposite
each other. After 12 h, the consumed volumes of tap water and
sucrose solution were recorded. The sucrose preference was
calculated as (sucrose solution intake)/(sucrose solution intake +
tap water intake) x100%.
Forced swimming test (FST)
After 3 weeks of mimic control/miR-135a mimic/FLU
treatment, FST was performed. The FST was carried out in a
cylindrical container (height, 65 cm; diameter, 30 cm), which was
filled to a height of 40 cm with water (temperature, 22-23˚C). The
FST lasted for 6 min and the immobility time (in sec) was recorded
during the last 4 min. The immobility time was defined as the time
when the mouse remained still and not struggling, using only the
basic motion to maintain its head on the water, or touching the
bottom for >1 sec.
Tail suspension test (TST)
After 3 weeks of mimic control/miR-135a mimic/FLU
treatment, the TST was performed as previously described (30). Briefly, the tape was placed at a
distance of 1 cm from the extremity of the tail of the mouse to fix
the position. The mice were suspended for 6 min and immobility was
recorded during the last 4 min.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the murine hippocampus
tissue or human and murine peripheral blood using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. Total RNA concentration was
detected using a NanoDrop 2000 spectrophotometer (NanoDrop; Thermo
Fisher Scientific, Inc.). Total RNA was stored at -80˚C until use.
The synthesis of cDNA was performed using the RevertAid™ First
Strand cDNA Synthesis kit (Vazyme Biotech Co., Ltd.). The reaction
conditions were as follows: 70˚C for 5 min, 37˚C for 5 min and 42˚C
for 60 min. SYBR-Green (Vazyme Biotech Co., Ltd.) qPCR assay was
performed to measure the expression level of the target gene. The
thermocycling conditions were as follows: Initial denaturation at
95˚C for 5 min; 40 cycles of denaturation at 95˚C for 10 sec,
annealing at 60˚C for 20 sec and extension at 72˚C for 30 sec.
Relative expression levels were calculated using the
2-ΔΔCq method following normalization with reference to
the expression of GAPDH or U6(31).
All experiments were performed in triplicate to ensure minimum
deviation. The following primer sequences were used: GAPDH forward,
5'-TTTGGTATCGTGGAAGGACTC-3' and reverse, 5'-GTA
GAGGCAGGGATGATGTTCT-3'; U6 forward, 5'-GCTTCG
GCAGCACATATACTAAAAT-3' and reverse, 5'-CGCTTC ACGAATTTGCGTGTCAT-3';
Toll-like receptor 4 (TLR4) forward, 5'-CCTGACACCAGGAAGCTTGAA-3'
and reverse, 5'-TCTGATCCATGCATTGGTAGGT-3'; miR-135a forward,
5'-ACACTCCAGCTCAGTATGGCTTTT TATTCCTATGT-3' and reverse,
5'-CTCAACTGGTGTCGTGGAGTCGGCAAT TCAG-3'.
Western blot analysis
Protein was extracted using RIPA buffer with 1 mM
protease inhibitor PMSF (Beijing Solarbio Science & Technology
Co., Ltd.). A bicinchoninic acid protein assay kit (Sigma-Aldrich;
Merck KGaA) was applied to determine protein concentration. Protein
(30 mg per lane) was separated on 10% SDS gels and transferred onto
PVDF membranes (EMD Millipore). The membranes were blocked with 5%
skim milk with TBS containing 0.1% Tween-20 for 1 h at room
temperature and incubated with the following primary antibodies:
Anti-Bcl-2 (cat. no. 3498; 1:1,000), anti-Bax (cat. no. 2772;
1:1,000), anti-TLR4 (cat. no. 14358; 1:1,000) and anti-GAPDH (cat.
no. 5174; 1:1,000) (all purchased from Cell Signaling Technology,
Inc.) at 4˚C overnight. The membranes were then incubated with
horseradish peroxidase-conjugated secondary antibody (goat
anti-rabbit; cat. no. 7074; 1:2,000; Cell Signaling Technology,
Inc.) for 2 h at room temperature. Subsequently, the protein bands
were detected and visualized by RapidStep™ ECL Reagent (EMD
Millipore). Band densities were quantified using Gel-Pro Analyzer
Densitometry software (version 6.3; Media Cybernetics, Inc.).
ELISA
After treatment, mice were sacrificed and
hippocampus were immediately dissected and then stored at -80˚C.
After the samples were homogenized and centrifuged at 5,000 x g at
4˚C for 15 min, the supernatant was collected. ELISA was performed
to examine the expression levels of TNF-α (cat. no. PT518), IL-6
(cat. no. PI330) and IL-1β (cat. no. PI305) in the mouse
hippocampus using ELISA kits according to the manufacturer's
instructions (all manufactured by Beyotime Institute of
Biotechnology).
Flow cytometric analysis
Cell apoptosis was analyzed using the Annexin V/PI
Apoptosis Detection kit (BD Biosciences). The hippocampus was
dissected as aforementioned and dissociated into a single cell
suspension by enzymatic degradation using a neural tissue
dissociation kit (Miltenyi Biotec, Inc.). The cells were then
collected, centrifuged at low temperature and high speed (1,000 x g
at 4˚C for 5 min), and re-suspended in 100 µl FITC binding buffer.
Subsequently, the buffer was supplemented with ~5 µl ready-to-use
Annexin V-FITC and 5 µl PI. In the dark, the cells were incubated
for 30 min at room temperature. Annexin V-FITC and PI fluorescence
were assessed using a BD FACSCalibur flow cytometer (BD
Biosciences). Data were analyzed using FlowJo software (version
7.2.4; FlowJo LLC).
Statistical analysis
Data are presented as the mean ± SD from at least
three independent experiments. GraphPad 6.0 software (GraphPad
Software, Inc.) was used for statistical analysis and unpaired
Student's t-test was performed to determine whether differences
between two groups were significant. Differences among multiple
groups were analyzed by one-way ANOVA followed by a Tukey's post
hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of miR-135a in patients
with depression and in the mouse model of depression
To explore the role of miR-135a in patients with
depression and in the mouse model of depression, peripheral blood
was collected from 50 patients with depression and 50 healthy
subjects, and the expression of miR-135a was detected by RT-qPCR.
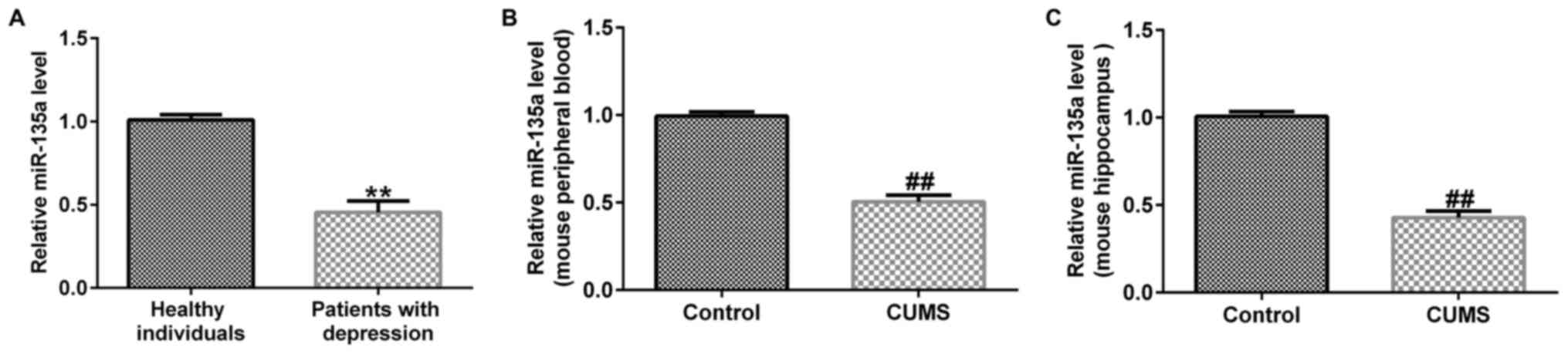
The results of RT-qPCR revealed that compared with the healthy
individuals, miR-135a expression was significantly reduced in
patients with depression (Fig. 1A).
Subsequently, a mouse model of depression was established by
inducing CUMS for 6 weeks. Peripheral blood and hippocampal tissue
samples were then collected from the mice, and the expression of
miR-135a was detected by RT-qPCR. The results revealed that
compared with the control group, the expression of miR-135a was
decreased in mouse peripheral blood (Fig. 1B) and in the hippocampus (Fig. 1C) in the CUMS group. These results
indicated that miR-135a expression was downregulated in patients
and mice with depression.
Effect of miR-135a on the body weights
of mice and depressive-like behavior
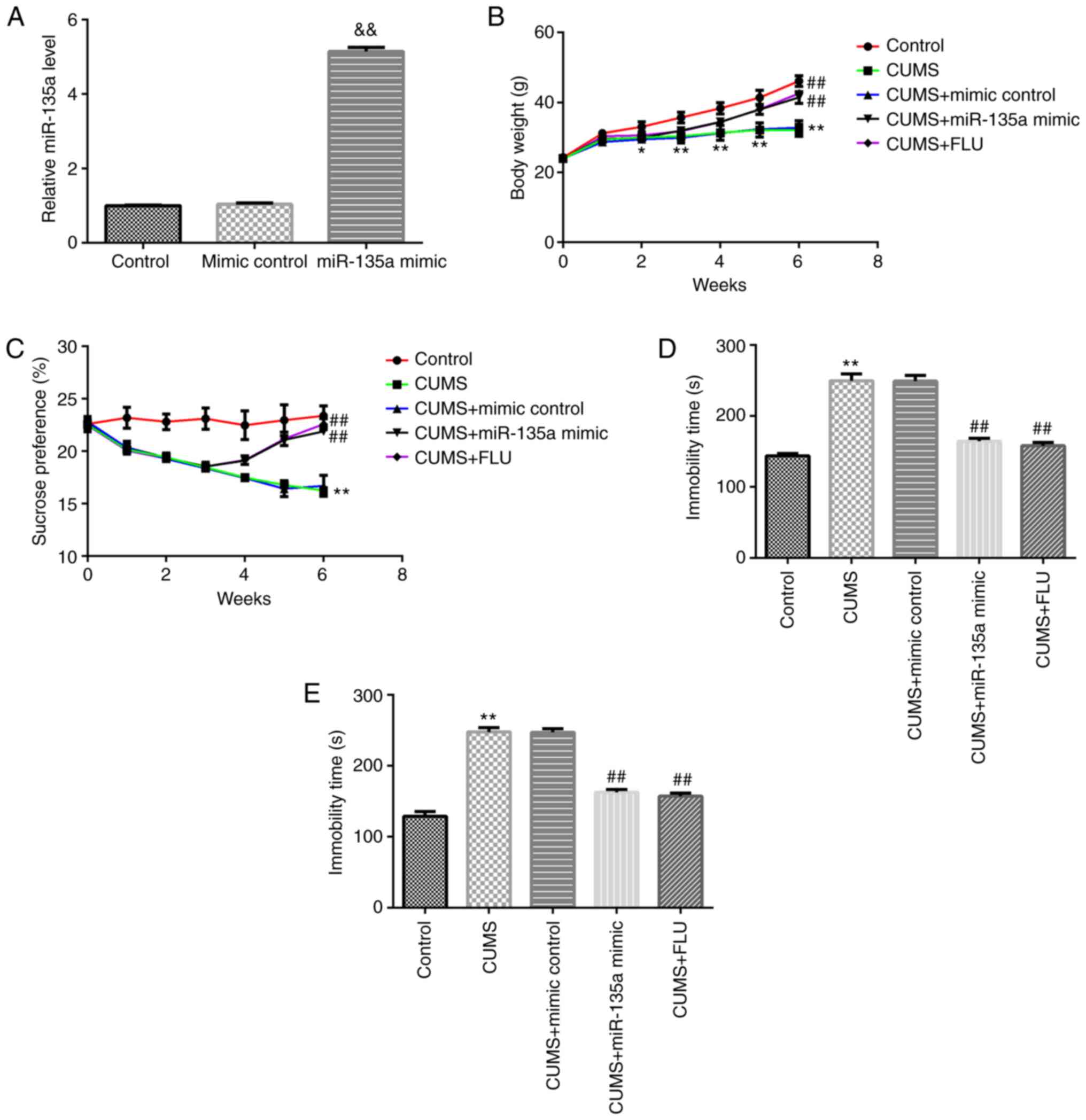
The present study first confirmed that compared with
the mimic control group, miR-135a mimic significantly enhanced the
level of miR-135a in the hippocampal tissue of mice (Fig. 2A). At the beginning of the
experiment, the weights of the mice in the different groups were
similar. Following 2 weeks of CUMS, the body weights of the mice in
the model group were significantly lower than those in the control
group. Treatment with miR-135a mimic and FLU significantly
attenuated the CUMS-induced reduction in weight gain (Fig. 2B). The effects of miR-135a on
depression-related symptoms in mice were then examined. The SPT was
determined every week. And following treatment of the mice
subjected to CUMS with miR-135a mimic for 3 weeks, the FST and TST
were used to evaluate the anti-depressant effects of miR-135a
mimic. Compared with CUMS + mimic control group, treatment with
miR-135a mimic and FLU markedly decreased CUMS-induced
depression-like behavior at 3 weeks after treatment. Compared with
the mice in the CUMS + mimic control group, miR-135a mimic
significantly increased SPT (1, 2 and 3 weeks after treatment) in
mice and reduced the immobility time in the FST and TST (Fig. 2C-E). These results indicated that
miR-135a treatment relieved CUMS-induced depressive-like
behavior.
Effect of miR-135a on CUMS-induced
hippocampal cell apoptosis
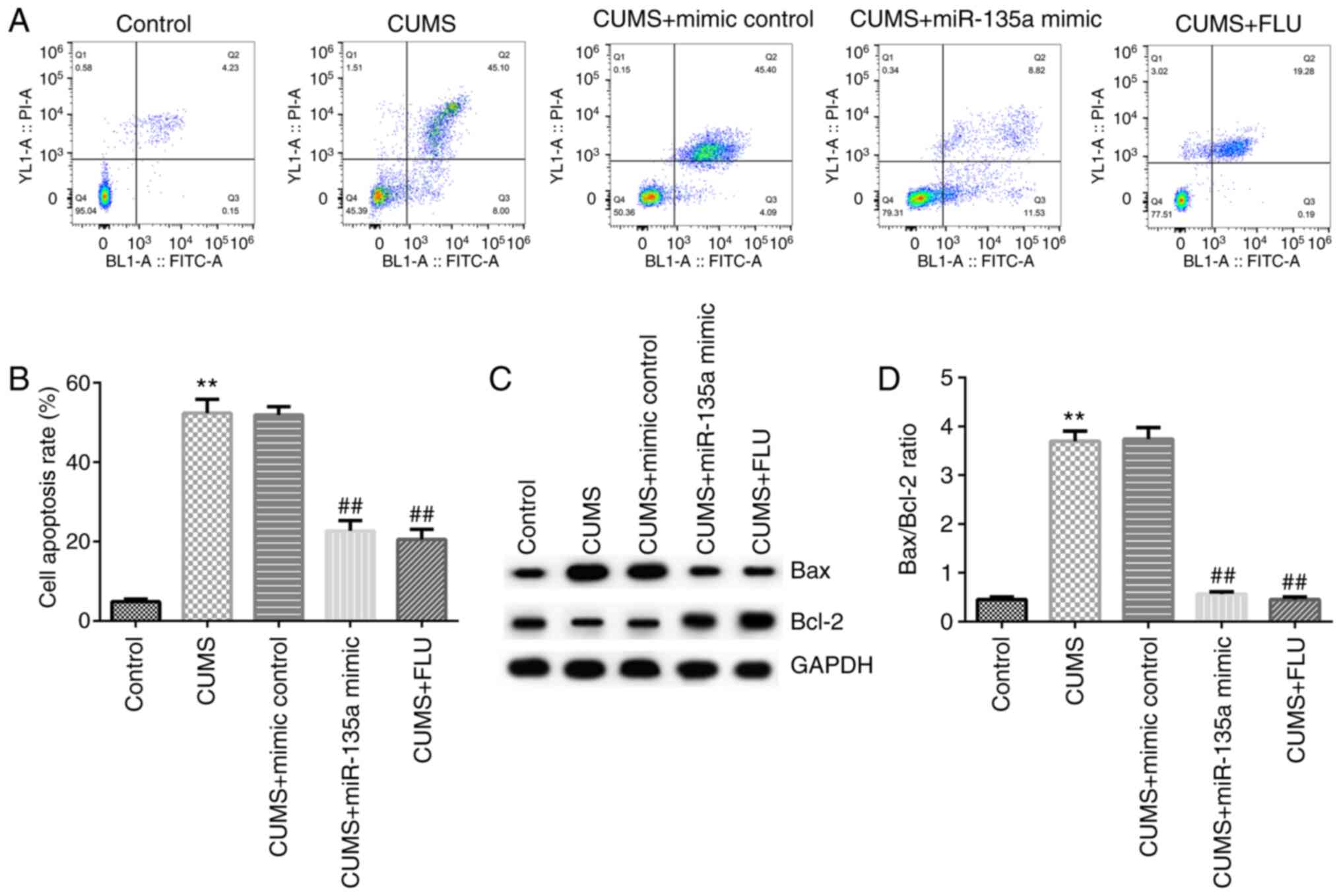
Subsequently, the effects of miR-135a on the
apoptosis of neural cells in the mouse hippocampus were examined by
flow cytometry. Compared with the control group, hippocampal
neuronal apoptosis was significantly increased in the CUMS group
(Fig. 3A and B). However, the apoptotic rate in the
hippocampus was decreased in the miR-135a mimic- and FLU-treated
group compared with the CUMS group (Fig. 3A and B). In addition, compared with the control
group, Bax protein expression was increased, Bcl-2 protein
expression was decreased, and the Bax/Bcl-2 ratio was increased in
the CUMS group (Fig. 3C and
D). However, miR-135a mimic and FLU
treatment significantly reduced the expression of Bax, increased
Bcl-2 expression and decreased the Bax/Bcl-2 ratio (Fig. 3C and D). Taken together, these results indicate
that miR-135a inhibited CUMS-induced hippocampal cell
apoptosis.
Effects of miR-135a on the
CUMS-induced hippocampal inflammatory response
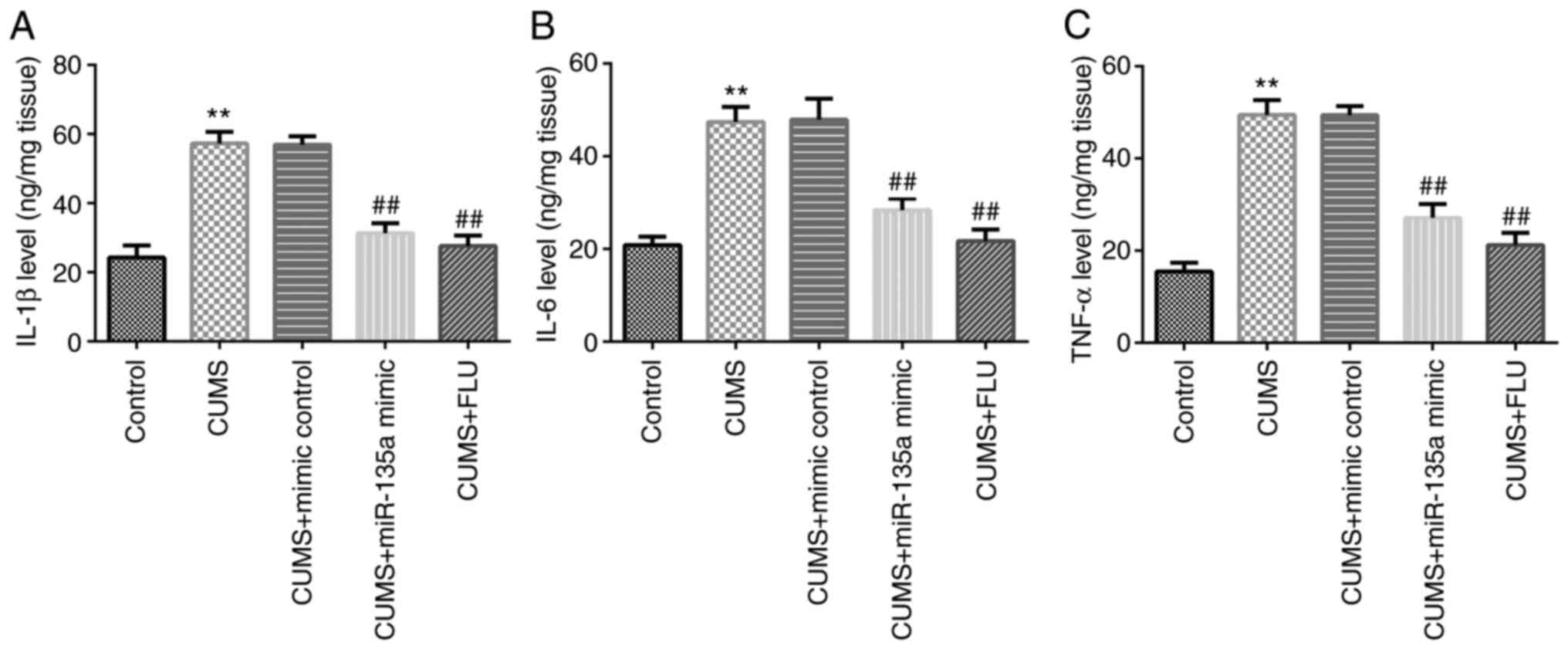
Subsequently, to examine the effects of miR-135a
treatment on neuroinflammation, ELISA was performed to detect the
levels of inflammatory factors in the hippocampus. The results
revealed that the levels of IL-1β, IL-6 and TNF-α were increased in
the CUMS group compared with the control group (Fig. 4A-C). However, miR-135a mimic and FLU
treatment significantly attenuated the CUMS-induced increase in the
levels of inflammatory factors in the hippocampus of mice (Fig. 4A-C). Therefore, miR-135a decreased
the CUMS-induced hippocampal inflammatory response.
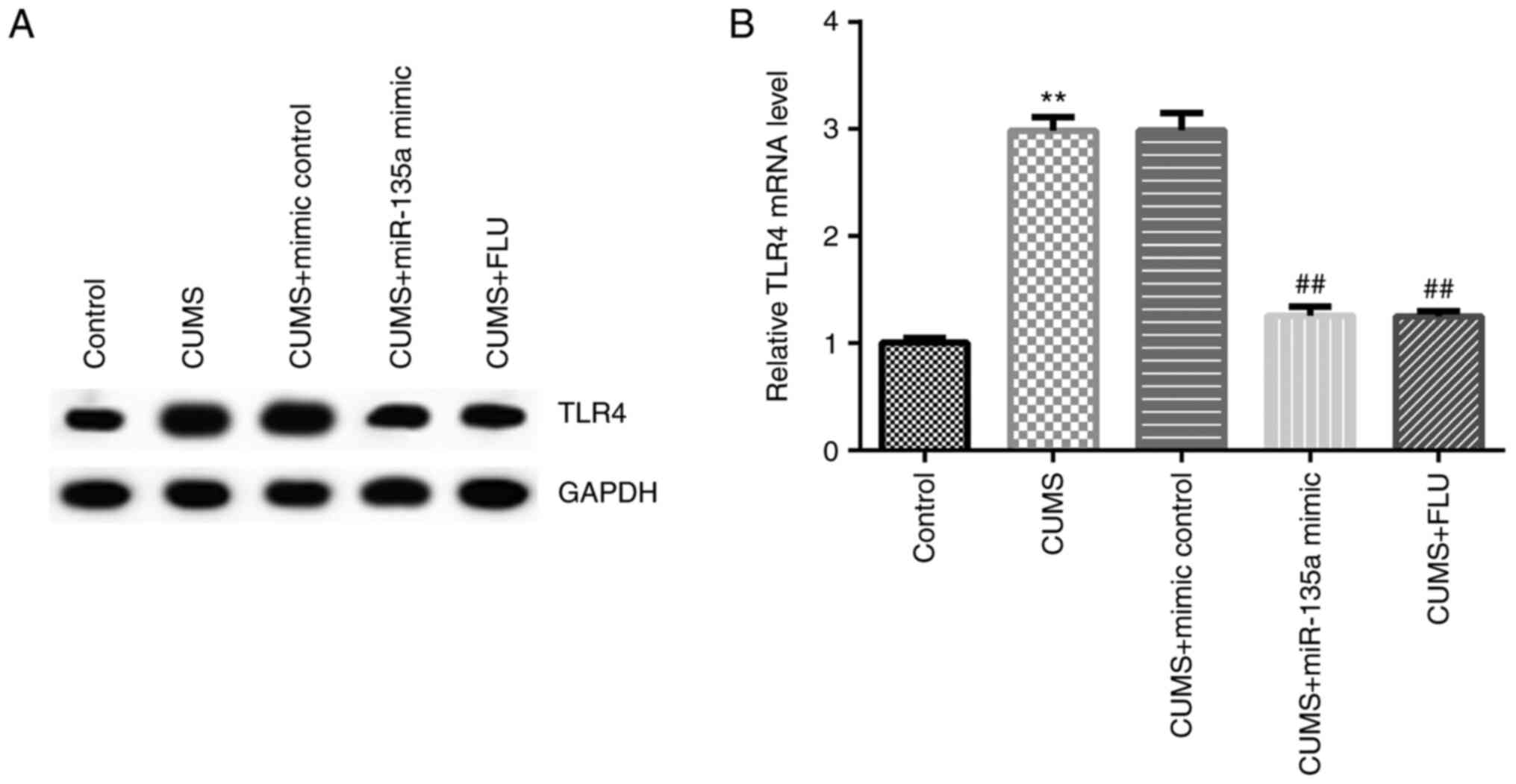
miR-135a may protect against
depression in mice by regulating TLR4 expression
A previous study demonstrated that TLR4 was a target
gene of miR-135a (32). Therefore,
in order to elucidate whether miR-135a affects depression by
regulating the expression of TLR4, the expression of TLR4 was
examined in the mice following treatment. After 3 weeks, the
expression of TLR4 was detected in the hippocampus of the mice in
each group by RT-qPCR and western blot analysis. The results
revealed that compared with the control group, the mRNA expression
of TLR4 was significantly increased in the CUMS group, and this
increase was significantly inhibited by miR-135a mimic (Fig. 5A and B).
Discussion
Depression is a common illness that severely
influences the quality of life. In recent years, an increasing
number of studies have focused on the exploration of the role of
miRNA in the pathological mechanism of depression (33-35).
It has previously been reported that miR-135a exerts an inhibitory
effect in prostate cancer and lung cancer (36,37).
Furthermore, miR-135a has been reported to function as an oncogene
in gastric and colorectal cancer (38,39).
In addition, Shi et al (40)
indicated that miR-135a expression was downregulated in glioma
tissues and cells. However, to the best of our knowledge, there are
few studies available on the expression and mechanisms of action of
miR-135a in patients with depression. Gheysarzadeh et al
(24) demonstrated that miR-135a
expression was decreased in patients with depression. CUMS-induced
animal models are currently considered as one of the most
appropriate animal models for studying depression (25-28,41).
CUMS exposure can induce depression-like behavior in animals, such
as behavioral despair, a lack of pleasure and reduced exercise
tolerance (42). In the present
study, it was found that the expression of miR-135a was
downregulated in patients with depression and in a mouse model of
CUMS-induced depression.
Wan et al (42) demonstrated that miR-29b-3p
overexpression improved depressive-like behaviors in rats with
CUMS. In the current study, the effects of miR-135a on
depression-related symptoms in mice with depression were examined.
It was found that miR-135a attenuated CUMS-induced in mice. In
addition, miR-135a mimic significantly increased SPT in mice and
shortened the immobility time in the FST and TST. The results of
the present study also demonstrated that miR-135a inhibited
CUMS-induced hippocampal cell apoptosis. There is increasing
evidence to indicate that inflammation is the primary pathological
mechanism of depression (43).
Psychological and physical stressors can stimulate immune and
inflammatory processes (44).
Previous studies have demonstrated that the increase in the levels
of pro-inflammatory cytokines (IL-1β, IL-6 and TNF-α) in certain
regions of the brain, such as the hippocampus of depressed mice, is
associated with the pathophysiology of depression (45,46).
In the present study, miR-135a attenuated the CUMS-induced
hippocampal inflammatory response and reduced the CUMS-induced
increase in the levels of inflammatory factors, including IL-1β,
IL-6 and TNF-α in the hippocampus of mice.
All the aforementioned results indicate that
miR-135a has a certain alleviating effect on depression. Du and Lu
(47) demonstrated that TLR4 was
the direct target gene of miR-135a, and miR-135a suppressed
oxidative stress and vascular inflammatory events through TLR4 in
atherogenesis. TLR4 plays an important role in the development
process of atherosclerosis (48)
and acts as a potential target of miR-590(49). The findings of the present study
demonstrated that miR-135a may protect mice against depression by
regulating TLR4 expression.
In conclusion, the present study demonstrated that
miR-135a regulated the apoptosis and inflammatory response in mouse
hippocampal neurons by regulating the expression of TLR4, thereby
alleviating the depressive behavior of mice and playing a
protective role in depression.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YXD conceived and designed the current study,
acquired, analyzed and interpreted the data, and prepared the
manuscript. MZ, BJQ, CPL and JFW contributed to the acquisition and
analysis of the data. JL contributed to acquisition and analysis of
the data and prepared the manuscript. YXD and JL confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All patients signed informed consent forms. The
present study was approved by the Ethics Committee of Binzhou Youfu
Hospital (Binzhou, China). All animal experiments were performed
according to a protocol approved by the Committee on Care and Use
of the Laboratory Animal Committee of Binzhou Youfu Hospital.
Patient consent for publication
All patients consented to the publication of their
data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bortolato B, Carvalho AF, Soczynska JK,
Perini GI and McIntyre RS: The involvement of TNF-α in cognitive
dysfunction associated with major depressive disorder: An
opportunity for domain specific treatments. Curr Neuropharmacol.
13:558–576. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Belmaker RH and Agam G: Major depressive
disorder. N Engl J Med. 358:55–68. 2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Prokofieva M, Koukia E and Dikeos D:
Mental health nursing in Greece: Nursing diagnoses and
interventions in major depression. Issues Ment Health Nurs.
37:556–562. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chuang YH and Kuo LM: Nurses' confidence
in providing and managing care for older persons with depressive
symptoms or depression in long-term care facilities: A national
survey. Int J Ment Health Nurs. 27:1767–1775. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Xie X, Shi Y and Zhang J: Structural
network connectivity impairment and depressive symptoms in cerebral
small vessel disease. J Affect Disord. 220:8–14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Suzuki H, Matsumoto Y, Ota H, Sugimura K,
Takahashi J, Ito K, Miyata S, Furukawa K, Arai H, Fukumoto Y, et
al: Hippocampal blood flow abnormality associated with depressive
symptoms and cognitive impairment in patients with chronic heart
failure. Circ J. 80:1773–1780. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Nabavi SM, Daglia M, Braidy N and Nabavi
SF: Natural products, micronutrients, and nutraceuticals for the
treatment of depression: A short review. Nutr Neurosci. 20:180–194.
2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Weil-Malherbe H: The biochemistry of
affective disorders. In: Handbook of Neurochemistry. Springer,
Boston, MA, pp371-416, 1972.
|
|
9
|
Lee S, Jeong J, Kwak Y and Park SK:
Depression research: Where are we now? Mol Brain. 3:8–0.
2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ebmeier KP, Donaghey C and Steele JD:
Recent developments and current controversies in depression.
Lancet. 367:153–167. 2006.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Leistedt SJ and Linkowski P: Brain,
networks, depression, and more. Eur Neuropsychopharmacol. 23:55–62.
2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kessler RC: The costs of depression.
Psychiatr Clin North Am. 35:1–14. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ota KT and Duman RS: Environmental and
pharmacological modulations of cellular plasticity: Role in the
pathophysiology and treatment of depression. Neurobiol Dis.
57:28–37. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M,
et al: Human microRNA genes are frequently located at fragile sites
and genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Mendell J and Olson E: . MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187.
2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Maes OC, Chertkow HM, Wang E and Schipper
HM: MicroRNA: Implications for Alzheimer disease and other human
CNS disorders. Curr Genomics. 10:154–168. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kocerha J, Kauppinen S and Wahlestedt C:
MicroRNAs in CNS disorders. Neuromolecular Med. 11:162–172.
2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zeng YB, Liang XH, Zhang GX, Jiang N,
Zhang T, Huang JY, Zhang L and Zeng XC: miRNA-135a promotes
hepatocellular carcinoma cell migration and invasion by targeting
forkhead box O1. Cancer Cell Int. 16(63)2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yao S, Tian C, Ding Y, Ye Q, Gao Y, Yang N
and Li Q: Down-regulation of Krüppel-like factor-4 by
microRNA-135a-5p promotes proliferation and metastasis in
hepatocellular carcinoma by transforming growth factor-β1.
Oncotarget. 7:42566–42578. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ren JW, Li ZJ and Tu C: miR-135
post-transcriptionally regulates FOXO1 expression and promotes cell
proliferation in human malignant melanoma cells. Int J Clin Exp
Pathol. 8:6356–6366. 2015.PubMed/NCBI
|
|
23
|
Xu B, Lu X, Zhao Y, Liu C, Huang X, Chen
S, Zhu W, Zhang L and Chen M: MicroRNA-135a induces prostate cancer
cell apoptosis via inhibition of STAT6. Oncol Lett. 17:1889–1895.
2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gheysarzadeh A, Sadeghifard N, Afraidooni
L, Pooyan F, Mofid MR, Valadbeigi H, Bakhtiari H and Keikhavani S:
Serum-based microRNA biomarkers for major depression: miR-16,
miR-135a, and miR-1202. J Res Med Sci. 23(69)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Deng XY, Li HY, Chen JJ, Li RP, Qu R, Fu Q
and Ma SP: Thymol produces an antidepressant-like effect in a
chronic unpredictable mild stress model of depression in mice.
Behav Brain Res. 291:12–19. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chen YP, Wang C and Xu JP: Chronic
unpredictable mild stress induced depression-like behaviours and
glutamate-glutamine cycling dysfunctions in both blood and brain of
mice. Pharm Biol. 57:280–286. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Duan CM, Zhang JR, Wan TF, Wang Y, Chen HS
and Liu L: SRT2104 attenuates chronic unpredictable mild
stress-induced depressive-like behaviors and imbalance between
microglial M1 and M2 phenotypes in the mice. Behav Brain Res.
378(112296)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lian N, Niu Q, Lei Y, Li X, Li Y and Song
X: miR-221 is involved in depression by regulating Wnt2/CREB/BDNF
axis in hippocampal neurons. Cell Cycle. 17:2745–2755.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Iñiguez SD, Riggs LM, Nieto SJ, Dayrit G,
Zamora NN, Shawhan KL, Cruz B and Warren BL: Social defeat stress
induces a depression-like phenotype in adolescent male c57BL/6
mice. Stress. 17:247–255. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Steru L, Chermat R, Thierry B and Simon P:
The tail suspension test: A new method for screening
antidepressants in mice. Psychopharmacology (Berl). 85:367–370.
1985.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Xie B, Lu C, Chen C, Zhou J and Deng Z:
miR-135a alleviates silica-induced pulmonary fibrosis by targeting
NF-κB/Inflammatory signaling pathway. Mediators Inflamm.
2020(1231243)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ferrúa CP, Giorgi R, da Rosa LC, do Amaral
CC, Ghisleni GC, Pinheiro RT and Nedel F: MicroRNAs expressed in
depression and their associated pathways: A systematic review and a
bioinformatics analysis. J Chem Neuroanat.
100(101650)2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Allen L and Dwivedi Y: MicroRNA mediators
of early life stress vulnerability to depression and suicidal
behavior. Mol Psychiatry. 25:308–320. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lou D, Wang J and Wang X: miR-124
ameliorates depressive-like behavior by targeting STAT3 to regulate
microglial activation. Mol Cell Probes. 48(101470)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Xu B, Tao T, Wang Y, Fang F, Huang Y, Chen
S, Zhu W and Chen M: hsa-miR-135a-1 inhibits prostate cancer cell
growth and migration by targeting EGFR. Tumour Biol.
37:14141–14151. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Shi H, Ji Y, Zhang D, Liu Y and Fang P:
miR-135a inhibits migration and invasion and regulates EMT-related
marker genes by targeting KLF8 in lung cancer cells. Biochem
Biophys Res Commun. 465:125–130. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yan LH, Chen ZN, Li-Li Chen J, Wei WE, Mo
XW, Qin YZ, Lin Y and Chen JS: miR-135a promotes gastric cancer
progression and resistance to oxaliplatin. Oncotarget.
7:70699–70714. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhou W, Li X, Liu F, Xiao Z, He M, Shen S
and Liu S: miR-135a promotes growth and invasion of colorectal
cancer via metastasis suppressor 1 in vitro. Acta Biochim Biophys
Sin (Shanghai). 44:838–846. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Shi HZ, Wang DN, Xu F, Teng JH and Wang
YL: miR-135a inhibits glioma cell proliferation and invasion by
directly targeting FOXO1. Eur Rev Med Pharmacol Sci. 22:4215–4223.
2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Henn FA and Vollmayr B: Stress models of
depression: Forming genetically vulnerable strains. Neurosci
Biobehav Rev. 29:799–804. 2005.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wan YQ, Feng JG, Li M, Wang MZ, Liu L, Liu
X, Duan XX, Zhang CX and Wang XB: Prefrontal cortex miR-29b-3p
plays a key role in the antidepressant-like effect of ketamine in
rats. Exp Mol Med. 50:1–14. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Dey A and Hankey Giblin PA: Insights into
macrophage heterogeneity and cytokineinduced neuroinflammation in
major depressive disorder. Pharmaceuticals (Basel).
11(E64)2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Iwata M, Ota KT and Duman RS: The
inflammasome: Pathways linking psychological stress, depression,
and systemic illnesses. Brain Behav Immun. 31:105–114.
2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Rush G, O'Donovan A, Nagle L, Conway C,
McCrohan A, O'Farrelly C, Lucey JV and Malone KM: Alteration of
immune markers in a group of melancholic depressed patients and
their response to electroconvulsive therapy. J Affect Disord.
205:60–68. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Shen Z, Xu Y, Jiang X, Wang Z, Guo Y, Pan
W and Hou J: Avicularin relieves depressive-like behaviors induced
by chronic unpredictable mild stress in mice. Med Sci Monit.
24:1643–3750. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Du XJ and Lu JM: miR-135a represses
oxidative stress and vascular inflammatory events via targeting
toll-like receptor 4 in atherogenesis. J Cell Biochem.
119:6154–6161. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
den Dekker WK, Cheng C, Pasterkamp G and
Duckers HJ: Toll like receptor 4 in atherosclerosis and plaque
destabilization. Atherosclerosis. 209:314–320. 2010.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yang L and Gao C: miR-590 inhibits
endothelial cell apoptosis by inactivating the TLR4/NF-κB pathway
in atherosclerosis. Yonsei Med J. 60:298–307. 2019.PubMed/NCBI View Article : Google Scholar
|