Introduction
Contrast-induced acute kidney injury (CI-AKI) is a
frequent complication following intravascular administration of
iodinated contrast medium (ICM), which is generally characterized
by an increase in serum creatinine (SCr) of 0.5 mg/dl or a 50%
relative elevation over baseline within 48 h following contrast
medium exposure, in the absence of an alternative etiology
(1). The development of this
iatrogenic syndrome is associated with adverse early and long-term
clinical outcomes (2,3). The introduction of safer contrast
agents and optimized hydration strategies have led to a decline in
the incidence of CI-AKI in the general population to 0.6-2.0%
(4). However, the morbidity rate
still remains as high as 20-50% in vulnerable subgroups, including
those with chronic kidney disease, acute myocardial infarction or
diabetes mellitus (1,5). Accumulating evidence has revealed a
variety of pathways implicated in CI-AKI, which involve direct
tubular cell toxicity, outer medullary ischemia, oxidative stress
and inflammation (6,7). However, the precise mechanisms
underlying the pathogenesis of CI-AKI remain largely unknown,
leading to a lack of early diagnostic biomarkers or more efficient
prevention strategies.
Long non-coding RNAs (lncRNAs) are a heterogeneous
class of RNAs that lack a protein-coding capability and are >200
nucleotides in length (8).
High-throughput RNA sequencing (RNA-seq) has led to the continuous
discovery of lncRNAs, which are emerging as important regulators in
a variety of physiological and pathological conditions, with a
relatively tissue-specific expression manner (9-13).
Mechanistically, lncRNAs have been proposed to function through
cis or trans transcriptional regulation, organization
of nuclear domains, and by acting as competing endogenous RNAs
(ceRNAs) (14,15). The complicated regulatory mechanisms
finally result in the formation of a large-scale regulatory network
across the transcriptome, and have provided useful explanations of
the pathological processes in various diseases. Over the past
decades, several studies have discovered the vital roles of lncRNAs
in acute kidney injury induced by a variety of etiologies (16-18).
For instance, NEAT1 was identified to be involved in
sepsis-induced kidney injury by targeting microRNA (miRNA) miR-204
and subsequently activating the nuclear factor-κB pathway (17). Recently, Cheng et al
(19) explored the potential link
between lncRNAs and CI-AKI. As reported, LNC_000343 potentially
regulates the expression of the Kielin/chordin-like protein by
acting as a ceRNA of rno-miR-1956-5p in CI-AKI rats (19). However, ICM was administered in an
intravenous manner in this model, which was slightly different from
the clinical practice of coronary angiography (20-22);
moreover, samples were collected at 24 h after iohexol injection in
that study (19). To the best of
our knowledge, the SCr level is significantly elevated at 24 h
following ICM intervention in rat models (23-25),
indicating that the regulation of the damage response might take
place even earlier. Thus, it is proposed that the lncRNA
transcriptome changes in CI-AKI might vary between the different
methods of modeling and time intervals of sample collection.
In the present study, a proven CI-AKI rat model with
significant elevation of SCr and remarkable histopathological
alterations was adopted (26). In
this model, ICM was administered in an intra-arterial manner, and
kidney samples were harvested at 12 h post ICM injection. By
performing deep RNA-seq analysis, the expression profiles of
lncRNAs were compared between the CI-AKI group and the control
group in kidney tissues. The potential function of these
differentially expressed (DE) lncRNAs (DElncRNAs) was then analyzed
using bioinformatic algorithms. Based on the expression profiles of
miRNAs and mRNAs identified previously in the same CI-AKI model
(GSE130796 for miRNA; and GSE130795 for mRNA) (26), lncRNA-mRNA co-expression analysis
was performed and an lncRNA-associated ceRNA network was
constructed to better understand the regulatory roles of the
DElncRNAs. Generally, the present study might provide new insights
of the dysregulated lncRNAs involved in the emergence of CI-AKI
complicated by clinical arteriography.
Materials and methods
Materials and animals
Indomethacin, N-ω nitro-L-arginine methyl ester
(L-NAME), and pentobarbital sodium were purchased from
Sigma-Aldrich; Merck KGaA. Iopromide (Ultravist 370; 370 mg/ml
iodine) was obtained from Bayer AG. Phosphate buffer (pH 8.4) was
synthesized by the experimental center. Indomethacin was dissolved
in phosphate buffer (5 mg/ml), and L-NAME was dissolved in 0.9%
normal saline (10 mg/ml), immediately before injection. In total,
18 3-month old male Sprague-Dawley rats, weighing ~300-400 g at the
start of the experiment, were obtained from Fujian Medical
University (Fuzhou, China). The 18 rats were kept in individual
cages under controlled conditions of light (12-h light/dark cycle),
temperature (21-23˚C) and humidity (50-60%), with free access to
tap water and standard rat chow for a 7-day adaptive period.
Ethics statement and establishment of
the CI-AKI rat model
The protocols of the animal experiments were
conducted in accordance with the Guiding Principles in the Use of
Animals in Toxicology (27), and
were approved by animal experiment ethics review committees of 900
Hospital of the Joint Logistics Team, Chinese People's Liberation
Army (approval no. IACUC-2017-17).
The study design was previously published (26). Briefly, rats were deprived of water
for 48 h and then anaesthetized using pentobarbital sodium (40
mg/kg, i.p.). Catheters were placed in the right femoral vein and
the common carotid artery (24 Gx21 mm; SPECATH) before a baseline
arterial blood sample was drawn (1 ml) to determine the SCr.
Indomethacin was then administered (10 mg/kg, i.v.), followed by
L-NAME (10 mg/kg, i.v.) after 15 min. After another 15 min, the
rats were randomized to receive iopromide (CI-AKI group, n=9) or
normal saline (control group, n=9) via the carotid artery
cannulation (7.8 ml/kg, i.a.). The rats were then allowed to
recover in individual cages with free access to tap water and
standard chow. A blood sample (1 ml) was obtained ~12 h after
iopromide injection from the abdominal aorta under pentobarbital
sodium anesthesia (40 mg/kg, i.p.) to measure postoperative SCr.
The kidney tissue for RNA-sequencing or reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
validation was then harvested immediately and stored in RNAsafety
Reagent (cat. no. 01901-50; http://www.shbio.com/products/3034) at 4˚C overnight,
before being transferred to a -20˚C refrigerator. For histological
analysis, part of the kidney tissue (~2-mm thick), containing both
the cortex and medulla, was fixed in 10% neutral formalin liquid
for 12-24 h at room temperature. The kidney tissue was then
dehydrated through an ascending series of ethanol, infiltrated with
acetone and embedded in paraffin at 60˚C. The paraffin blocks were
cut into 5-µm-thick sections and subsequently dewaxed in xylene and
rehydrated in a descending ethanol gradient. Hematoxylin and eosin
(H&E) staining of the sections was performed with hematoxylin
for 5 min and eosin for 2 min, both at room temperature. The Paller
scores were calculated to determine the severity of tubular injury
(13). Finally, the rats were
sacrificed with an overdose of pentobarbital anesthesia (200 mg/kg,
i.p.).
RNA extraction, qualification and
purification
According to the manufacturer's instructions, total
RNA was extracted using a mirVana™ miRNA Isolation kit (cat. no.
AM1561; Ambion, Thermo Fisher Scientific, Inc.). The RNA Integrity
Number (RIN) number was checked to inspect RNA integrity using an
Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.). Only those
samples with a RIN ≥7.0 and 28s/18s ≥0.7 were identified as
qualified and were selected for further analysis. The qualified
total RNA was then purified using an RNAClean XP kit (cat. no.
A63987; Beckman Coulter, Inc.) and RNase-Free DNase set (cat. no.
79254; Qiagen GmBH).
Library construction and RNA
sequencing
Library construction and RNA sequencing were
performed by Shanghai Biotechnology Corporation (http://www.shbio.com). Strand-specific cDNA libraries
were prepared from ribosomal RNA-depleted RNAs using a VAHTS Total
RNA-seq (H/M/R) Library PrepKit for Illumina (cat. no. NR603-02;
Vazyme Biotech Co., Ltd.). The RNAs were first interrupted into
shot fragments. Next, first-strand cDNA synthesis was performed
using Oligo(dT)12-18 primers and SuperScript™ II Reverse
Transcriptase kit (cat. no. 18064-014; Invitrogen, Thermo Fisher
Scientific, Inc.). The simplified steps were as follows: i) Heat
the mixture of total RNA, primers and dNTPs to 65˚C for 5 min; ii)
add First-Strand Buffer (5X) and incubate at 42˚C for 2 min; iii)
add SuperScript™ II Reverse Transcriptase and incubate at 42˚C for
50 min; iv) inactivate the reaction by heating at 70˚C for 15 min.
Double-strand cDNA synthesis was then performed and the cDNA
fragments were purified by Agencourt® AMPure XP Beads
(cat. no. A63881; Beckman Coulter, Inc.). After final PCR
enrichment, the cDNA libraries were quantified by Qubit®
2.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, Inc.) and
further validated by the Agilent 2100 system (Agilent Technologies,
Inc.) to calculate the library concentration. Cluster was generated
by the Illumina cBot system (version 02; Illumina, Inc.) with the
library diluted to a loading concentration to ~10 pM. Following
cluster generation, the libraries were sequenced in paired-end read
(2x150 bp) mode on the Illumina HiSeq 2500 platform (Illumina,
Inc.). TruSeq PE Cluster Kit v3 (cat. no. PE-401-3001; Illumina,
Inc.), TruSeq SR Cluster Kit v3 (cat. no. GD-401-3001; Illumina,
Inc.) and TruSeq Rapid Duo Dample Loading Kit (cat. no.
CT-402-4001; Illumina, Inc.) were used in library clustering and
sequencing. High-quality reads were aligned to the Rattus
norvegicus reference genome (ftp://ftp.ensembl.org/pub/release-83/fasta/rattus_norvegicus/dna/Rattus_norvegicus.Rnor_6.0.dna_rm.toplevel.fa.gz)
using the spliced mapping algorithm in the Hisat2 software (v
2.0.4) (28). The unmatched reads
were analyzed using the gffcompare software (v 0.9.8) to predict
novel lncRNAs (29). For the gene
fragment calculation, the fragments per kilobase of transcript per
million mapped reads (FPKM) value was determined using Stringtie
and trimmed mean of M values algorithm. Based on the FPKM data,
lncRNAs with a fold change ≥2 and P<0.05 were identified as
differentially expressed.
RT-qPCR verification of selective
DElncRNAs
In order to verify the reliability of the RNA-seq
results, several DElncRNAs from the RNA-seq data were selected for
validation in an independent cohort of 6 CI-AKI rats and 6 controls
using RT-qPCR. Total RNA was extracted from rat kidneys using
mirVana™ miRNA Isolation Kit (Ambion; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols, and reverse
transcribed using ReverTra Ace qPCR kit (Toyobo Life Science). The
reverse transcriptional conditions were as follows: 37˚C for 15
min, 98˚C for 5 min and then kept at 4˚C. Next, qPCR was performed
in triplicate using the 7500 Real-Time PCR System with Power
SYBR®-Green PCR Master Mix (Thermo Fisher Scientific,
Inc.). The thermocycling conditions were 50˚C for 2 min, then 95˚C
for 10 min, followed by 40 cycles at 95˚C for 15 sec and 60˚C for 1
min. The Gapdh gene (glyceraldehyde-3-phosphate
dehydrogenase) was used as internal control. The expression levels
of the lncRNAs was normalized and quantified using the
2-ΔΔCq method (30).
P<0.05 was considered to indicate a statistically significant
difference. The following reverse primers were used:
NONRATT027338.2 (forward, 5'-CAGGACAAAGGAACCCAGC-3' and reverse,
5'-CCGAGAGAGAGCAGCAATGA-3'); NONRATT027428.2 (forward,
5'-GCTGTAAATGTAGGCTATGGTGGTT-3' and reverse,
5'-AGCTCTCATGGGTTCTGTCATCTC-3'), NONRATT000173.2 (forward,
5'-ACCAAACAAGACCACCAGCAT-3' and reverse,
5'-GGAGGGACTGATGTGTACGAAAC-3'), NONRATT005775.2 (forward,
5'-CCTCCCACCCTCTGATGTAG-3' and reverse,
5'-AGAAAGTGCTCGTGGACAGG-3'), NONRATT016226.2 (forward,
5'-GAACCAGAGGATGGCGACA-3' and reverse, 5'-GATGGCATGAAGGGATGAAT-3'),
NONRATT018005.2 (forward, 5'-CCTTCTCCTTCCAGATAACTTACACA-3' and
reverse, 5'-GTGACTGCCAGGGTGCTAAAC-3'), NONRATT023682.2 (forward,
5'-CAGGTGCCTCCTCTCAGTCAA-3' and reverse,
5'-CCTCACCCCCCTAGTCTTCTTAA-3'), MSTRG.22041.2 (forward,
5'-TGCACTGAGCAGGACTGAAAA-3' and reverse,
5'-TTATCCCTTTGCATTCACTCCAA-3'), Gapdh (forward,
5'-TGGCCTCCAAGGAGTAAGAAAC-3' and reverse,
5'-GGCCTCTCTCTTGCTCTCAGTATC-3').
DElncRNA target gene prediction
Both the cis and trans regulation
analyses were applied to predict the target genes of lncRNAs. The
coding genes located within 10 kb upstream and downstream of the
lncRNAs were predicted as putative cis-targets. The
trans-prediction were performed using RNAplex (version
2.4.14) (31).
Co-expression analysis of DElncRNAs
and DEmRNAs
Pearson's correlation coefficient (PCC) between the
newly identified DElncRNAs and previously identified DEmRNAs was
calculated (P<0.05). The DEmRNAs were identified as having a
fold change ≥2 and P<0.05. The co-expression network of
DElncRNAs-DEmRNAs was presented using Cytoscape software (version
3.8.0) (32).
Construction of the lncRNA-associated
ceRNA network
The significant mature DElncRNAs (fold change ≥2;
q<0.05), DEmiRNAs; fold change ≥1.5; P<0.05), and DEmRNAs
(fold change ≥2; q<0.05), were selected for ceRNA analysis. The
lncRNA-miRNA interactions and miRNA-mRNA interactions were
predicted using the miRanda database according to their shared
miRNA-binding seed sequence sites (33). The lncRNA-miRNA pairs and miRNA-mRNA
pairs were then integrated into a ceRNA network in the
comprehensive analysis. After that, the PCCs of the expression
values between DElncRNAs and DEmRNAs, as well as the associations
among all the three kinds of RNA (DElncRNAs, DEmiRNAs and DEmRNAs),
were determined according to the expression levels. The
lncRNA-associated ceRNA network was further visualized using
Cytoscape (32).
Functional enrichment analysis
The functional enrichment analysis of putative
targets of lncRNAs were conducted using the Database for
Annotation, Visualization, and Integrated Discovery (DAVID)
software (34). Gene ontology (GO)
term analysis was performed in terms of functional classification,
including biological process, cellular component and molecular
function (35). Kyoto encyclopedia
of genes and genomes (KEGG) analysis was further conducted to
reveal the association of these genes with different pathways
(36). To account for multiple
comparisons, both Benjamini and Bonferroni corrections were used.
P<0.05 was considered to indicate a statistically significant
difference.
Statistical analysis
The SPSS 22.0 software (SPSS Inc.) was used for
statistical analysis. Data are presented as the mean ± SD. The
comparison between groups was conducted by unpaired Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
ICM exposure induced remarkable injury
to the kidneys
The CI-AKI rats exhibited a significantly increased
SCr, with an average level of 59.9±23.0%, as well as pronounced
histopathological changes to the kidney, as observed using H&E
staining, compared with those of the controls (26).
Characteristics of the RNA-seq
data
Total RNA in the kidney tissue derived from 3 paired
CI-AKI rats and 3 controls was isolated and analyzed. RNA-seq
produced over 90 million raw reads, with clean read ratios ranging
from 94.43 to 95.45%. Most of the clean reads could be mapped
perfectly to the Rattus reference genome (Table I). StringTie (v1.3.0) and gffcompare
(v0.9.8) were used to assemble and quantify the transcripts,
respectively (29,37). The identification of lncRNAs was
then performed using three tools (Pfam, CPC and CNCI) (38-40).
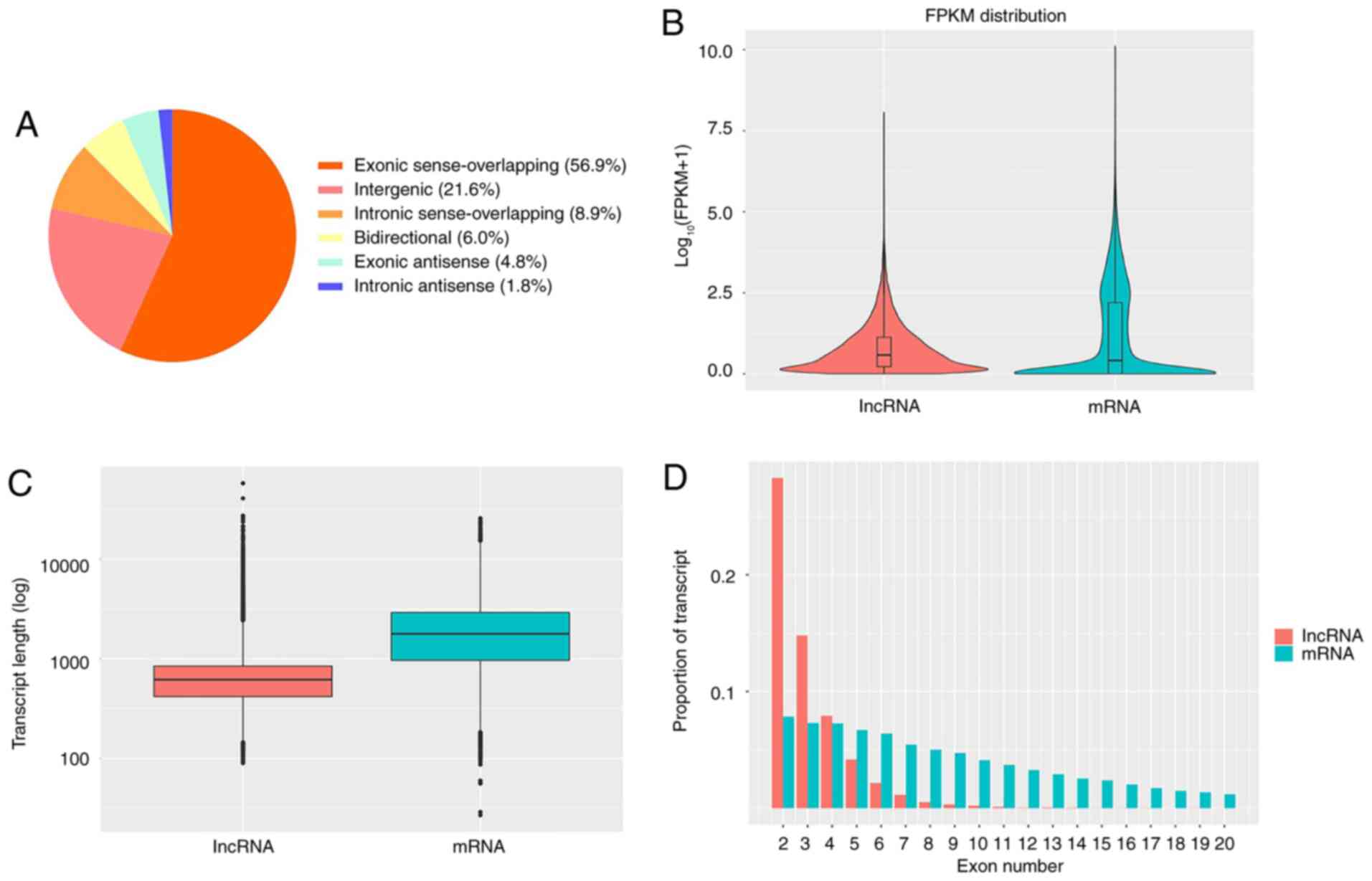
Finally, a total of 21,248 lncRNAs were identified (Table SI; Fig. S1), including 12,081 (56.9%) exonic
sense-overlapping lncRNAs, 4,584 (21.6%) intergenic lncRNAs, 1,900
(8.9%) intronic sense-overlapping lncRNAs, 1,281 (6.0%)
bidirectional lncRNAs, 1,012 (4.8%) exonic antisense lncRNAs and
390 (1.8%) intronic antisense lncRNAs (Fig. 1A). The features of the lncRNAs were
also analyzed. The lncRNAs tended to have a lower expression level
(calculated by the FPKM method) and a shorter transcript length
compared with those of the protein-coding transcripts (Fig. 1B and C). Moreover, the majority of lncRNAs were
identified to have fewer exons compared with mRNAs (Fig. 1D). The raw data has been uploaded to
the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134824).
 | Table ISummary of draft reads of cDNA
libraries. |
Table I
Summary of draft reads of cDNA
libraries.
| Sample | Raw reads | Clean reads | Clean ratio, % | Mapped reads | Mapping ratio,
% |
|---|
| Control_1 | 119,917,792 | 113,890,762 | 94.97 | 101,521,071 | 89.14 |
| Control_2 | 116,702,770 | 110,756,482 | 94.90 | 98,219,953 | 88.68 |
| Control_3 | 118,362,004 | 112,430,138 | 94.99 | 99,295,268 | 88.32 |
| Case_1 | 113,431,152 | 107,112,640 | 94.43 | 95,644,335 | 89.29 |
| Case_2 | 103,095,464 | 98,408,905 | 95.45 | 88,173,472 | 89.60 |
| Case_3 | 115,941,526 | 110,000,513 | 94.88 | 98,694,544 | 89.72 |
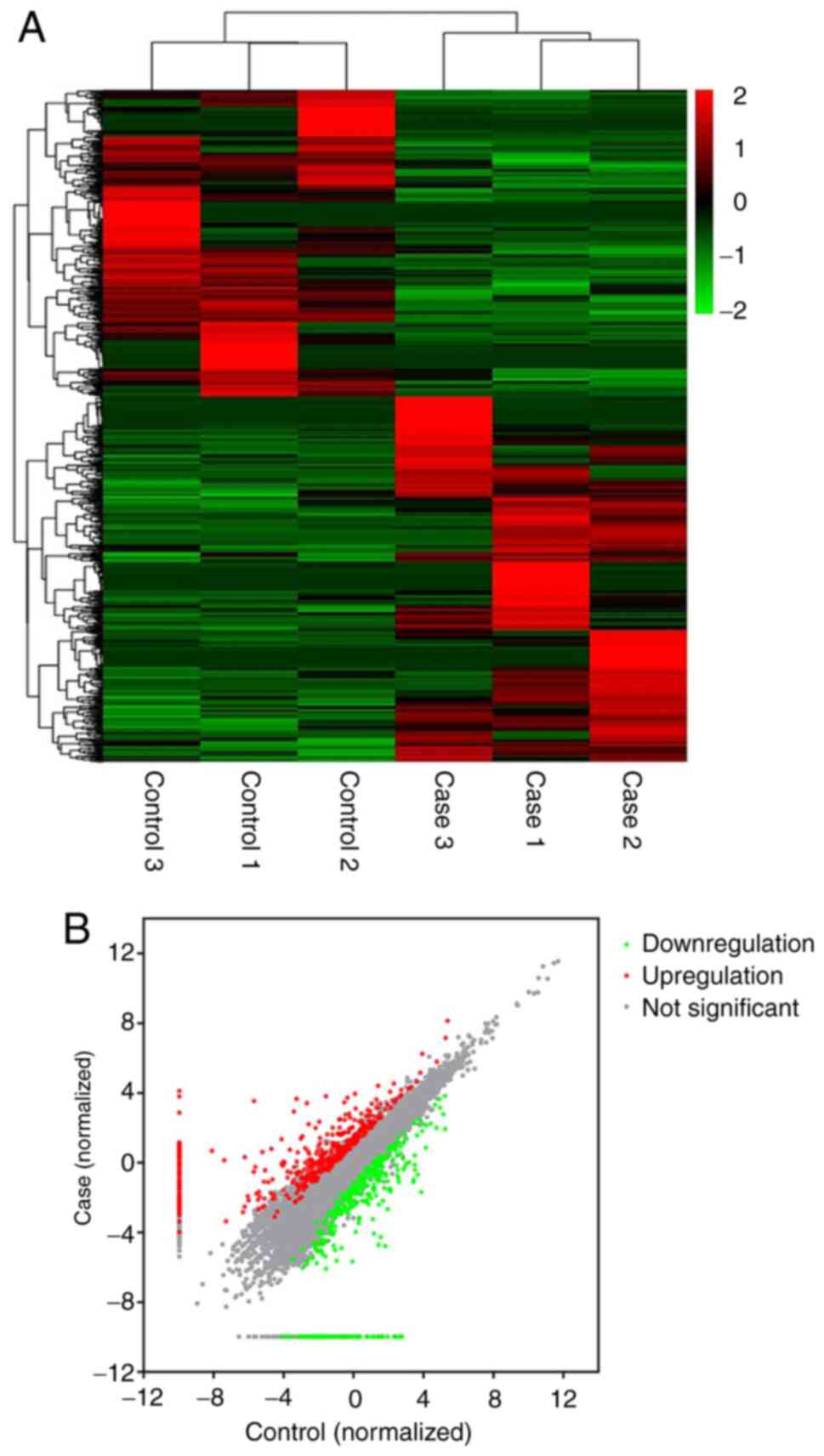
Differential expression analysis of
lncRNAs
According to the screening criteria, a total of 910
DElncRNAs, including 74 novel ones, were identified in the CI-AKI
rats compared with the controls (Table
SII). Among them, 495 DElncRNAs were upregulated and 415 were
downregulated. Notably, NONRATT027876.2 was the most highly
expressed, with a fold-change of 603.8, while NONRATT010468.2 was
the most downregulated, whose level decreased by 95.2-fold. Some of
the DElncRNAs had no expression in either the CI-AKI rats or the
controls, resulting in an infinite fold change value; for instance,
MSTRG.22041.2. Uniformly, these lncRNAs were considered as
dysregulated with a fold change of 2.0. The DElncRNAs were further
illustrated using a heatmap (Fig.
2).
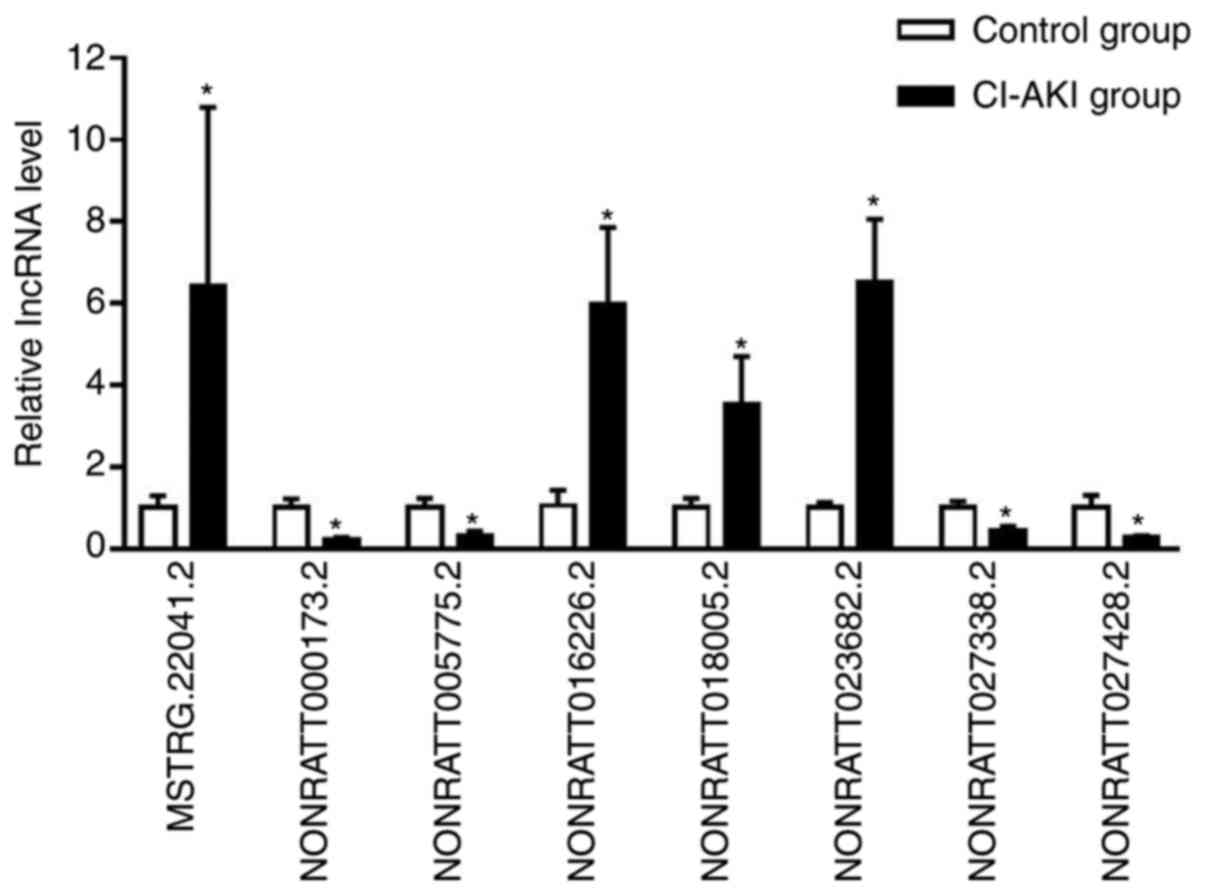
RT-qPCR validation of DElncRNAs
To validate the reliability of the RNA-seq data,
eight dysregulated lncRNAs were selected for further RT-qPCR
analysis in an independent cohort of six CI-AKI rats and six
controls. All the candidate lncRNAs were verified to be
differentially expressed (Fig. 3
and Table SIII; P<0.05), which
was consistent with the RNA-seq data. These results confirmed the
authenticity of the high-throughput sequencing data.
Trans and cis regulatory prediction
and functional enrichment analysis of DElncRNAs
The associations between DElncRNAs and their
potential targets were predicted according to both cis- and
trans-acting patterns (Tables
SIV and V). Location analysis
identified 805 DElncRNAs that were relatively close to 756
protein-coding genes, indicating of a cis-regulatory manner.
GO analysis was performed and 76 GO terms were significantly
enriched (Fig. S2A and Table SVI; P<0.05). KEGG analysis
revealed seven significant pathways (Fig. S2B and Table SVII; P<0.05). Notably, ‘p53
signaling pathway’ (rno04115), which plays a critical role in acute
kidney injury (41), was proposed
to be associated with CI-AKI. In the functional analysis of the
trans DElncRNA targets, 30 GO terms and 5 KEGG pathways were
significantly enriched (Fig. S3A
and B, Tables SVIII and SIX; P<0.05). Some of these GO terms
were associated with energy metabolism and apoptosis (GO:0005739,
GO:0006919, GO:0045335, GO:0005777 and GO:0045730).
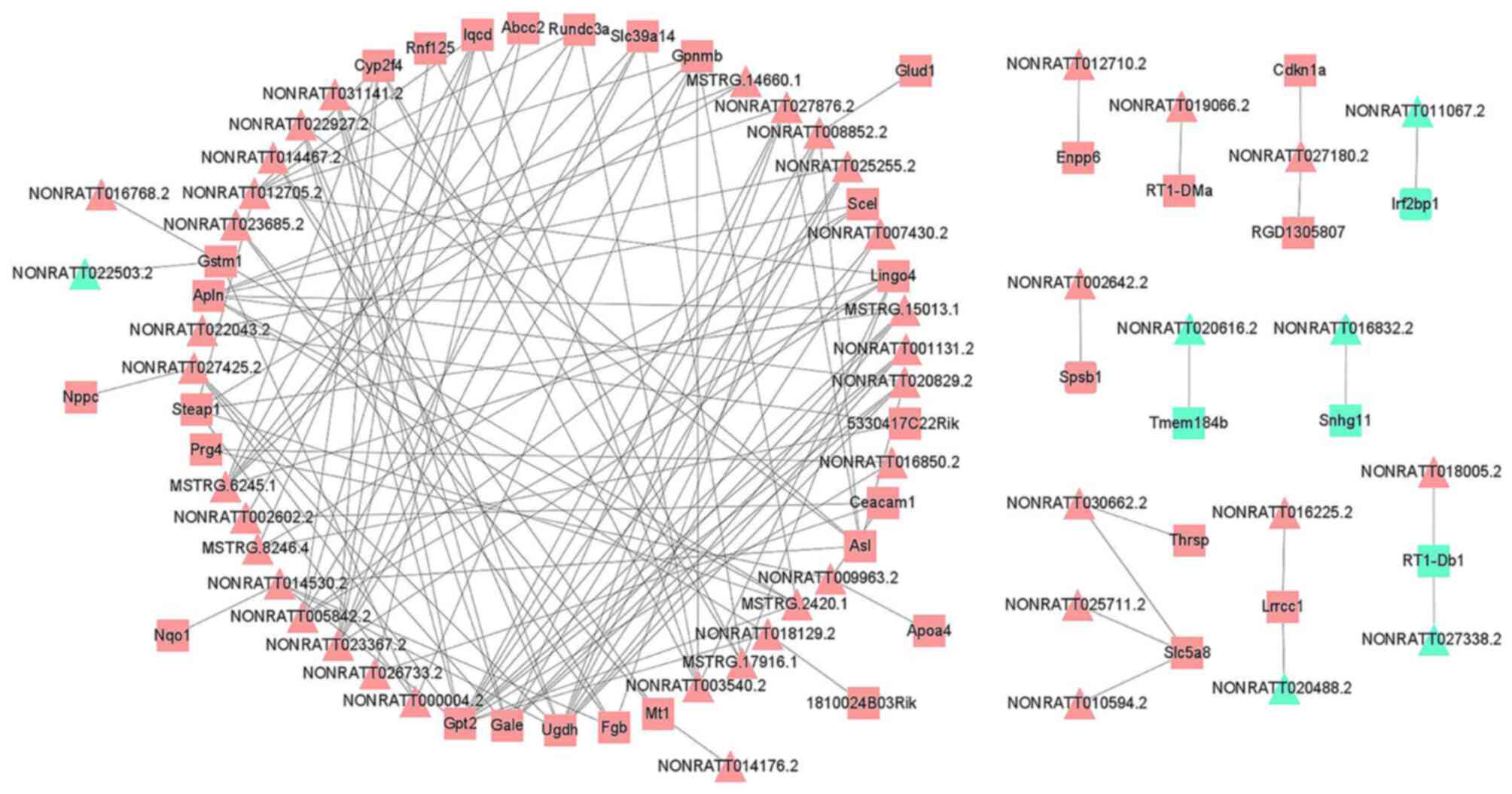
DElncRNA-DEmRNA co-expression
analysis
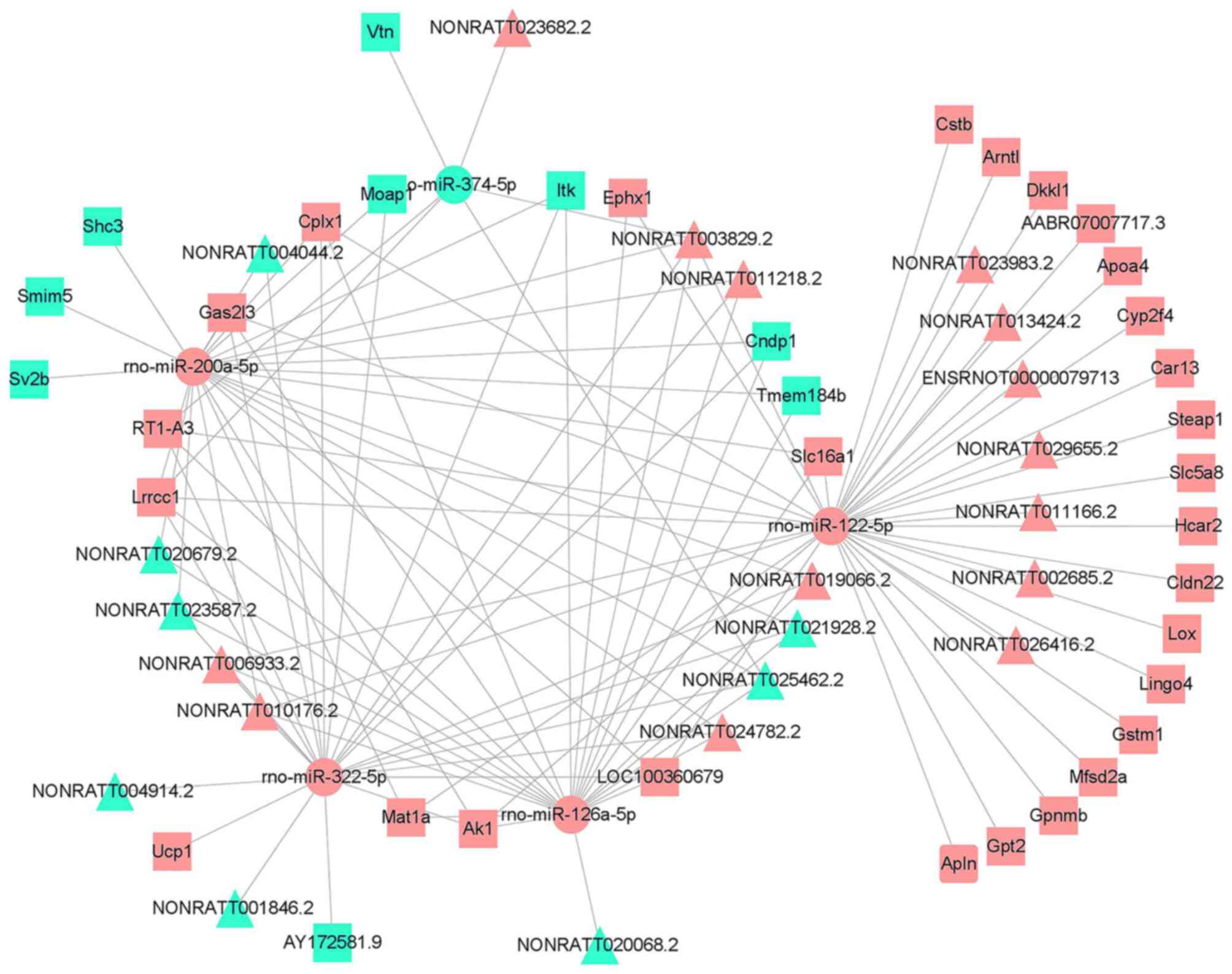
A co-expression network of the DElncRNAs and DEmRNAs
was constructed (Table SX). This
network comprised 1,632 lncRNA-mRNA interactions, containing 349
DElncRNA nodes and 203 DEmRNA nodes (PCC>0.990; P<0.05).
Finally, the DElncRNAs or DEmRNAs with significant expression
levels were selected and visualized, including 46 DElncRNAs and 38
DEmRNAs (Fig. 4). GO functional
enrichment analysis of the 203 differentially co-expressed genes
revealed 35 GO terms between the CI-AKI and control group. Among
them, 20 GO terms were biological process-associated, 9 were
cellular component-associated, and 6 were molecular
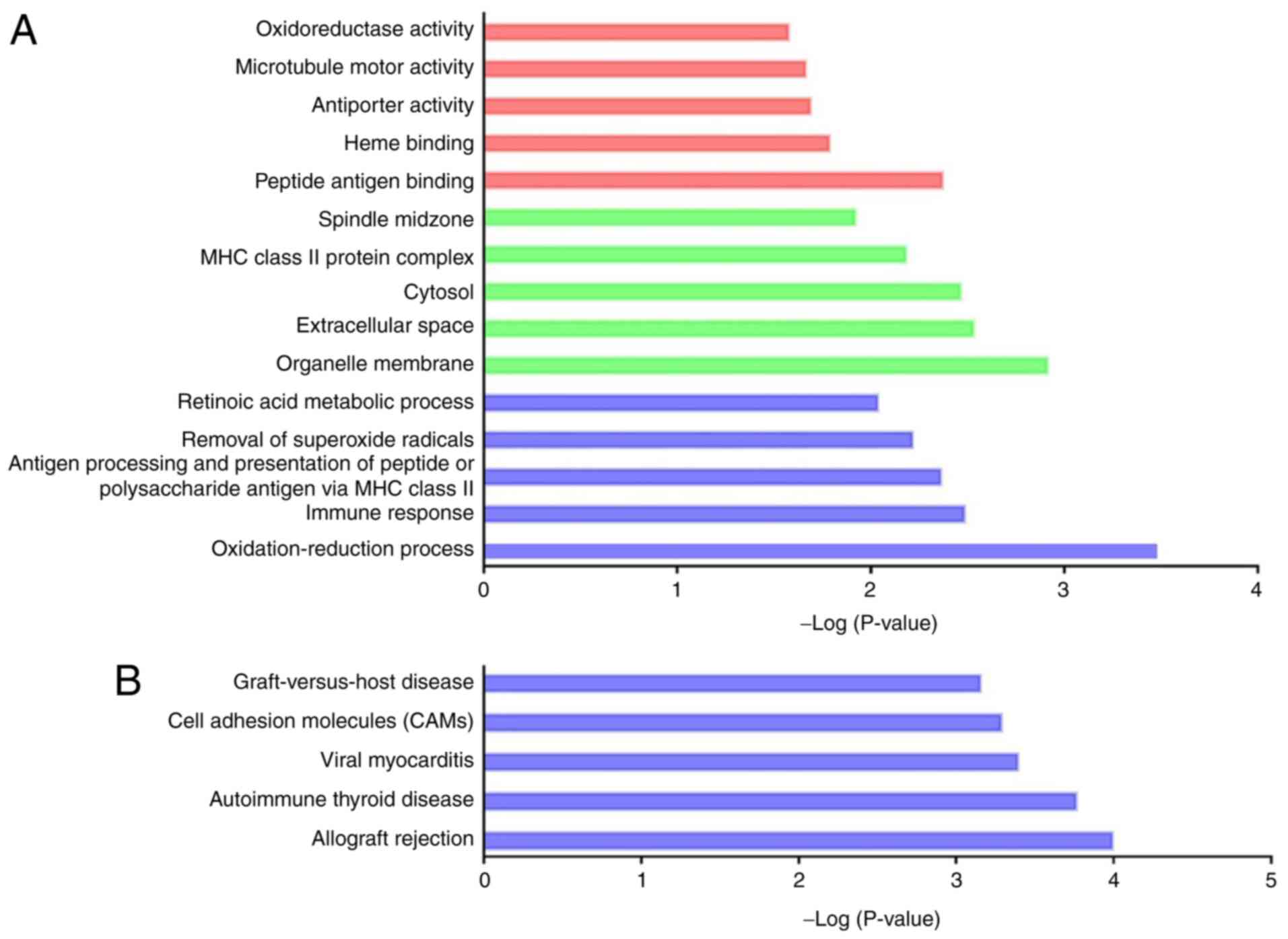
function-associated (Fig. 5A and
Table SXI; P<0.05).
Interestingly, a considerable number of significant oxidative
stress-related GO terms were identified, and oxidative stress has
been recognized as a hallmark of CI-AKI (6). Notably, 17 dysregulated genes were
remarkably enriched in the GO term ‘oxidation-reduction process’
(GO:0055114), including Ugdh (encoding UDP-glucose
6-dehydrogenase), Cmah (encoding cytidine
monophospho-N-acetylneuraminic acid hydroxylase), Glud1
(encoding glutamate dehydrogenase 1) and Cyp2f4 (encoding
cytochrome P450, family 2, subfamily f, polypeptide 4). The
aforementioned genes and associated DElncRNAs were expected to
modulate oxidative reactions during CI-AKI. In addition, several GO
terms were inflammation-associated (GO:0006955, GO:0002504,
GO:0034341, GO:0002474 and GO:0042130). KEGG analysis identified 19
remarkable pathways (Fig. 5B and
Table SXII; P<0.05).
Similarly, most of the significant pathways were relevant to the
pathological processes of inflammation. These results suggested
that the dysregulated lncRNAs might play a role in CI-AKI
pathogenesis by various mechanisms.
The DEmRNAs involved in the co-expression network
were further compared with predicted targets regulated by DElncRNAs
in a cis or trans manner. Significantly, 25
co-expressed DEmRNAs were found to be potentially targeted by 19
cis-acting DElncRNAs (Table
SXIII; P<0.05). The cis-acting DElncRNA-DEmRNA pairs
potentially involved in CI-AKI were displayed (Table II). It was also found that some
trans-acting DElncRNAs were predicted to target several
co-expressed differential genes. However, the majority of these
DElncRNAs and DEmRNAs had extremely low expression levels, making
them unsuitable for further studies.
 | Table IICis-regulatory DElncRNA-DEmRNA
co-expression pairs likely to be involved in CI-AKI. |
Table II
Cis-regulatory DElncRNA-DEmRNA
co-expression pairs likely to be involved in CI-AKI.
| DElncRNA | Fold change | Location | Strands | DEmRNA | Fold change | Location | Strands | r |
|---|
|
NONRATT025711.2 | 3.72 |
7:29479206-2948005 | + | Slc5a8 | 4.25 |
7:29435443-29477947 | + | 0.999 |
|
NONRATT016225.2 | 3.65 |
2:88404185-88408022 | - | Lrrcc1 | 2.36 |
2:88384795-88414012 | - | 0.992 |
|
NONRATT016226.2 | 4.72 |
2:88410568-88413958 | - | | | | | 0.970 |
|
NONRATT016224.2 | 3.66 |
2:88402887-88402723 | - | | | | | 0.967 |
|
NONRATT016768.2 | 5.00 |
2:210803868-210805276 | - | Gstm1 | 3.37 |
2:210803868-210809306 | - | 0.991 |
|
NONRATT014530.2 | 3.87 |
19:22621344-22632069 | - | Gpt2 | 3.89 |
19:22590880-22626653 | - | 0.986 |
|
NONRATT007430.2 | 2.24 |
12:30160774-30165923 | + | Asl | 2.03 |
12:30165693-30178341 | + | 0.985 |
|
NONRATT026655.2 | 2.53 |
7:29959585-29961154 | - | Gas2l3 | 2.41 |
7:29959596-29986163 | - | 0.971 |
|
NONRATT027381.2 | -2.19 |
7:145147281-145149879 | - | Ppp1r1a | -2.17 |
7:145146480-145154131 | - | 0.956 |
lncRNA-associated ceRNA network
LncRNAs can also function as molecular decoys for
miRNAs through miRNA response elements and thus regulate gene
expression as a ceRNA network (14). Thus, the lncRNA-associated ceRNA
network was constructed to further illustrate the role of DElncRNAs
in CI-AKI development. First, the candidate DElncRNAs and mature
DEmiRNAs with significant connections were matched (P<0.05).
Next, the selected DEmiRNAs were matched to their putative target
DEmRNAs (P<0.05). The DElncRNA-DEmiRNA network and the
DEmiRNA-DEmRNA network were then integrated into a ceRNA regulatory
network (Table SXIV). Meanwhile,
the associations among the expression levels of DElncRNAs, DEmiRNAs
and DEmRNAs were calculated to further test the possibilities of
these ceRNA pairs. The ceRNA pairs with negative associations
between DElncRNAs and DEmRNAs were selected and visualized in a
final ceRNA network, which covered 22 DElncRNAs, 5 DEmiRNAs and 37
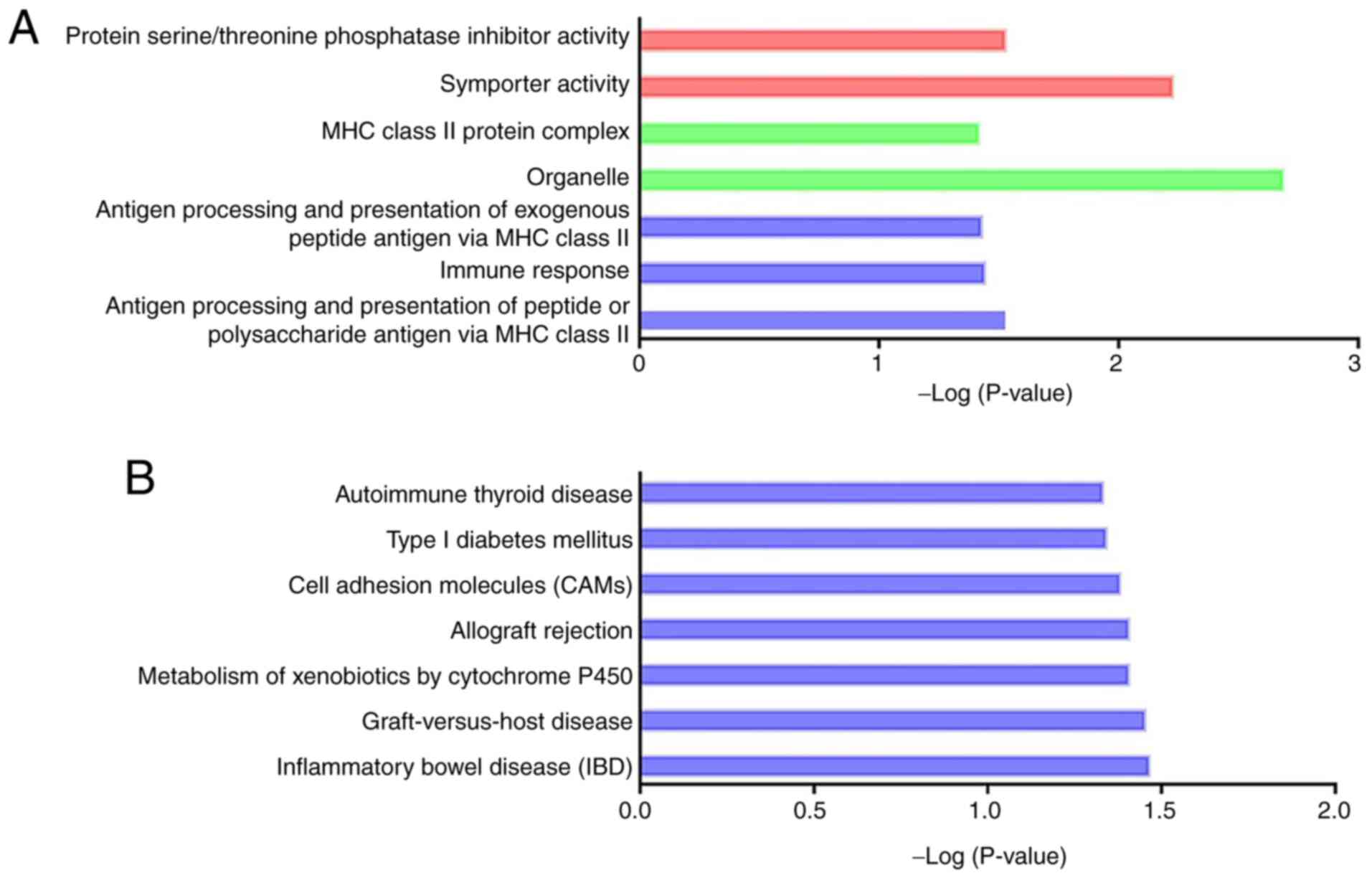
DEmRNAs (Fig. 6). As shown in
Fig. 7, the DEmRNAs within the
network were significantly enriched in seven GO terms and seven
KEGG pathways (Tables SXV and
SXVI; P<0.05), and most of
these processes were inflammation-associated. According to the
ceRNA hypothesis, lncRNA levels generally correlate negatively with
miRNA levels, and the miRNAs subsequently downregulate the
expression of target genes. The present study identified that the
downregulated lncRNAs, such as NONRATT021928.2 and NONRATT025462.2,
were ceRNAs of the upregulated rno-miR-126a-5p, rno-miR-200a-5p and
rno-miR-322-5p, which target Cndp1 (encoding carnosine
dipeptidase 1). The downregulated lncRNAs NONRATT020679.2 and
NONRATT023587.2 were ceRNAs of the upregulated rno-miR-126a-5p,
rno-miR-200a-5p and rno-miR-322-5p, which target Tmem184b
(encoding transmembrane protein 184B). The ceRNA pairs that are
likely to be associated with CI-AKI are shown in Table III.
 | Table IIISignificant ceRNA pairs likely to be
associated with CI-AKI. |
Table III
Significant ceRNA pairs likely to be
associated with CI-AKI.
| DElncRNA | Fold change | P-value | DEmiRNA | Fold change | P-value | DEmRNA | Fold change | P-value |
|---|
|
NONRATT025462.2 | -14.27 | <0.001 |
rno-miR-126a-5p | 2.60 | <0.001 | Cndp1 | -2.16 | <0.001 |
|
NONRATT021928.2 | -37.10 | <0.001 | | | | | | |
|
NONRATT025462.2 | -14.27 | <0.001 |
rno-miR-200a-5p | 2.24 | <0.001 | | | |
|
NONRATT021928.2 | -37.10 | <0.001 | | | | | | |
|
NONRATT025462.2 | -14.27 | <0.001 | rno-miR-322-5p | 2.41 | <0.001 | | | |
|
NONRATT021928.2 | -37.10 | <0.001 | | | | | | |
|
NONRATT004914.2 | -12.83 | <0.001 | | | | | | |
|
NONRATT001846.2 | -55.97 | <0.001 | | | | | | |
|
NONRATT020679.2 | -8.43 | <0.001 |
rno-miR-126a-5p | 2.60 | <0.001 |
Tmem184b | -2.08 | <0.001 |
|
NONRATT023587.2 | -12.2 | <0.001 | | | | | | |
|
NONRATT020679.2 | -8.43 | <0.001 |
rno-miR-200a-5p | 2.24 | <0.001 | | | |
|
NONRATT023587.2 | -12.2 | <0.001 | | | | | | |
Discussion
The present study was designed to compare the lncRNA
expression patterns of CI-AKI rats and controls from a global
perspective. To that end, the transcriptome of the kidney tissue
was generated using high throughput sequencing in the present
study. A total of 910 DElncRNAs were identified, including 74 novel
ones, that were dysregulated in CI-AKI rat kidney at 12 h following
intra-arterial iopromide exposure. Importantly, DElncRNA-DEmRNA
co-expression and functional enrichment analysis indicated that
these lncRNAs were mainly involved in oxidative stress and
inflammation reactions. Moreover, bioinformatic algorithms revealed
the CNDP1-specific and Tmem184b-specific networks that might be
associated with CI-AKI. The aforementioned results might provide
new insights into the lncRNA-associated regulatory mechanisms
underlying CI-AKI.
Depending on the CI-AKI model, striking differences
were observed in the expression profiles of lncRNAs between CI-AKI
rats and controls, as well as their functional characteristics.
DElncRNA-DEmRNA co-expression and functional enrichment analyses
implied that these DElncRNAs were likely to be associated with
several key aspects of CI-AKI, including oxidative stress and
inflammation. In particular, oxidative stress is recognized as one
of the most significant mechanisms of CI-AKI (42). Accumulating evidence has shown that
the injection of contrast medium might induce hypoxia and
consequently augment reactive oxygen species generation in the
kidney (43,44). Enhanced oxidative stress can cause
damage to membrane lipids, cellular proteins and DNA. Membrane
lipid peroxidation might change the cellular and mitochondrial
membrane permeability and activate specific cell signaling cascades
of cell death, ultimately resulting in renal parenchymal oxidative
injury (42,45). Several lncRNAs, including PRINS,
MIR210HG, linc-ATP13A4-8 and linc-KIAA1737-2, have been identified
to mediate hypoxic kidney injury (18,46).
In the present study, several significant oxidative stress-related
GO terms were identified in the functional analysis of the DEmRNAs
involved in the DElncRNA-DEmRNA co-expression network, including
‘oxidation-reduction process’ (GO:0055114), ‘removal of superoxide
radicals’ (GO:0019430), and ‘oxidoreductase activity’ (GO:0016491).
Moreover, several DElncRNA-DEmRNA co-expression pairs we found with
potential cis-regulatory relationships. In particular, the
NONRATT016768.2 locus was within the locus of the associated gene
Gstm1 (encoding glutathione S-transferase mu 1).
Gstm1 had been recognized as a target of nuclear factor
erythroid-derived 2-like 2 and played a protective role in response
to oxidative stress in kidney disease (47,48).
In addition, lncRNA-associated ceRNA analysis revealed a
significant Cndp1-specific network that was also likely to
be associated with the process of antioxidation. Cndp1
encodes a serum carnosinase that is associated with carnosine
degradation. A decreased level of CNDP1 might result in an
increased carnosine concentration, and could lead to renoprotective
effects by eliminating peroxyl and hydroxyl radicals (49,50).
These results suggested a vital role of lncRNAs in oxidative
stress-mediated injury in CI-AKI. The DElncRNAs implicated in
oxidative stress are worthy of further in-depth functional study to
help find indications regarding the important regulators of CI-AKI
pathogenesis.
Previous studies have also indicated a role of
inflammation in the pathogenesis of CI-AKI (51-55).
Andreucci et al (55) showed
that nuclear factor-κB (NF-κB) is activated together with an
increase in interleukin-8 in sodium diatrizoate-treated cells. An
in vivo study also suggested that NF-κB induced a
pro-inflammatory response, along with renal tubular damage, and
inhibition of NF-κB using sodium butyrate could protect the kidney
from contrast-induced injury (51).
In addition, the nucleotide-binding oligomerization domain-like
pyrin domain containing protein 3 inflammasome activated the
pro-inflammatory cytokines and apoptotic pathway, and mediated
kidney injury following contrast medium exposure, both in
vitro and in vivo (54).
Moreover, the inflammatory process of CI-AKI also involves the
regulatory effects of macrophage (7). In the present study, functional
analysis revealed that a fairly large number of DElncRNAs were
involved in the inflammation process. For instance, previous
studies had revealed that Gpnmb (encoding glycoprotein nmb)
was expressed in macrophages, negatively regulated inflammation,
and played a role in acute kidney injury (56,57).
The present study showed that a group of DElncRNAs (such as
MSTRG.11448.4, MSTRG.2420.1, MSTRG.6245.1 and NONRATT023367.2) were
linked to Gpnmb in the co-expression analysis. The
inflammation-associated DElncRNA-DEmRNA pairs further supported the
view that the CI-AKI pathological process is accompanied by
inflammation.
The mitogen-activated protein kinase (MAPK) pathway
regulates inflammation, apoptosis, and oxidative stress, and is
involved in kidney injury (58,59). A
previous study showed that Tmem184b might activate the MAPK
signaling pathway (60). The
present study identified a Tmem184b-specific ceRNA network
involving downregulated Tmem184b, two downregulated DElncRNAs
(NONRATT020679.2 and NONRATT023587.2) and two upregulated DEmiRNAs
(rno-miR-126a-5p and rno-miR-200a-5p). These aforementioned
dysregulated RNAs possessed remarkable expression levels and
significant fold changes; the functional roles of these RNAs in
CI-AKI should be further explored.
It should also be highlighted that the dysregulation
of several DElncRNAs and their putative targets likely resulted in
a reno-protective effect. Indeed, the activation of the reparative
response occurred at an early stage following injury. The recovery
process, especially tubular recovery, is vital to protect from
progression of chronic kidney disease after injury (61). Therein, the cross-talk between
tissue-reparative macrophages and tubular cells is an important
mechanism underlying tubular recovery after kidney injury. In
addition, Gpnmb is essential to switch macrophages towards a
wound-healing phenotype (57). The
upregulation of Gpnmb, together with the co-expressed
DElncRNAs, was potentially implicated in CI-AKI by modulating
macrophage polarization and tubular recovery. Besides, Gstm1
and Cndp1 in the bioinformatic networks were also associated
with a protective reaction. These results suggest that more
attention should be paid to the recovery process in research on
CI-AKI.
Despite the significant findings in the present
study, several limitations should be emphasized. Firstly, the
RNAseq analysis was performed using total RNA of kidneys, not on
specific cells. The specific roles of the distinct cells in kidney
tissue still remain unclear. Secondly, subcellular location is a
key feature for understanding a lncRNA's function; however, this
was not analyzed in the present study. Thirdly, these dysregulated
RNAs were identified in rats, the importance of these RNAs in
clinical samples is yet to be elucidated. Finally, the levels of
mRNAs do not necessarily associate with levels of their translation
products; however, the translation products of the critical DEmRNAs
had not been analyzed in animal models or clinical samples.
Intensive studies should be conducted to confirm the preliminary
results in the present study and further reveal the functional
characteristics of lncRNAs in CI-AKI development.
In conclusion, the present study identified the
lncRNA expression profile in vulnerable rats following
intra-arterial administration of contrast medium. The DElncRNAs and
their targets were likely to be associated with several key aspects
of CI-AKI, including oxidative stress and inflammation. The present
study provides an insight into the roles of lncRNAs in the
pathogenesis of CI-AKI.
Supplementary Material
PCA plot of long non-coding RNA
expression profiles for samples from CI-AKI and control groups.
Blue dots represent samples from CI-AKI group and red dots
represent control samples. The X and Y-axis represent PC1 and PC2,
respectively. The proportion of variance explained by PC1 is equal
to 27.44 and 21.02% for PC2. PCA, Principal component analysis;
CI-AKI, contrast-induced acute kidney injury; PC, principle
component.
Functional enrichment analysis of the
potential targets of differentially expressed long non-coding RNAs
regulated in a cis-regulatory manner. (A) Significant GO
terms enriched in the GO analysis. Red bars represent biological
process, green bars represent cell component, and blue bars
represent molecular function. (B) Significant pathways enriched in
the Kyoto encyclopedia of genes and genomes enrichment analysis.
GO, Gene Ontology.
Functional enrichment analysis of the
potential targets of differentially expressed long non-coding RNAs
regulated in a trans-regulatory manner. (A) Significant GO
terms enriched in the GO analysis. Red bars represent biological
process, green bars represent cell component, and blue bars
represent molecular function. (B) Significant pathways enriched in
the Kyoto encyclopedia of genes and genomes enrichment analysis.
GO, Gene Ontology.
lncRNA ide+A21250ntification.
Differentially expressed lncRNAs
between the CI-AKI group and the control group.
Relative expression of selected
differentially expressed long non-coding RNAs through reverse
transcription PCR.
Associations between differentially
expressed long non-coding RNAs and potential targets predicted
according to a cis-acting pattern.
Associations between differentially
expressed long non-coding RNAs and potential targets predicted
according to a trans-acting pattern.
Gene Ontology analysis of the
potential targets of differentially expressed long non-coding RNAs
in a cis regulatory manner.
Kyoto Encyclopedia of Genes and
Genomes analysis of the potential targets of differentially
expressed long non-coding RNAs regulated in a cis regulatory
manner.
Gene Ontology analysis of the
potential targets of differentially expressed long non-coding RNAs
regulated in a trans regulatory manner.
Kyoto Encyclopedia of Genes and
Genomes analysis of the potential targets of differentially
expressed long non-coding RNAs regulated in a trans regulatory
manner.
Co-expression analysis between
DElncRNAs and DEmRNAs.
Gene Ontology analysis of the DEmRNAs
involved in the differentially expressed long non-coding RNA-DEmRNA
co-expression network.
Kyoto Encyclopedia of Genes and
Genomes analysis of the DEmRNAs involved in the differentially
expressed long non-coding RNA-DEmRNA co-expression network.
Differentially expressed mRNA
potentially targeted by cis-acting co-expressed differentially
expressed long non-coding RNAs.
Long non-coding RNA-associated
competing endogenous RNA network.
Gene Ontology analysis of the
differentially expressed mRNAs involved in the long non-coding RNA
-associated competing endogenous RNA network.
Kyoto Encyclopedia of Genes and
Genomes analysis of the differentially expressed mRNA involved in
the long non-coding RNA -associated competing endogenous RNA
network.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the Natural Science
Foundation of Fujian province (grant no. 2016J01477), and the
Health Care Project of Chinese PLA (grant no. 16BJZ55).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the GEO repository [GSE130795
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130795);
GSE130796 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130796);
and GSE134824 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134824)].
Authors' contributions
MH and PT conceived and designed the experiments.
WB, ZX, ZW and DL performed the experiments. WB and ZW contributed
to the data analysis. WB and ZW wrote and reviewed the manuscript.
WB and MH confirmed the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocols were approved by animal
experiment ethics review committees of 900 Hospital of the Joint
Logistics Team, Chinese People's Liberation Army (approval no.
IACUC-2017-17).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ad-hoc working group of ERBP. Fliser D,
Laville M, Covic A, Fouque D, Vanholder R, Juillard L and Van
Biesen W: A European renal best practice (ERBP) position statement
on the Kidney disease improving Global Outcomes (KDIGO) clinical
practice guidelines on acute kidney injury: Part 1: Definitions,
conservative management and contrast-induced nephropathy. Nephrol
Dial Transplant. 27:4263–4272. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Solomon RJ, Mehran R, Natarajan MK, Doucet
S, Katholi RE, Staniloae CS, Sharma SK, Labinaz M, Gelormini JL and
Barrett BJ: Contrast-induced nephropathy and long-term adverse
events: Cause and effect? Clin J Am Soc Nephrol. 4:1162–1169.
2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
James MT, Samuel SM, Manning MA, Tonelli
M, Ghali WA, Faris P, Knudtson ML, Rannu N and Hemmelgarn BR:
Contrast-induced acute kidney injury and risk of adverse clinical
outcomes after coronary angiography: A systematic review and
meta-analysis. Circ Cardiovasc Interv. 6:37–43. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Caixeta A, Nikolsky E and Mehran R:
Prevention and treatment of contrast-associated nephropathy in
interventional cardiology. Curr Cardiol Rep. 11:377–383.
2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Finn WF: The clinical and renal
consequences of contrast-induced nephropathy. Nephrol Dial
Transplant. 21:i2–i10. 2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Azzalini L, Spagnoli V and Ly HQ:
Contrast-induced nephropathy: From pathophysiology to preventive
strategies. Can J Cardiol. 32:247–255. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lau A, Chung H, Komada T, Platnich JM,
Sandall CF, Choudhury SR, Chun J, Naumenko V, Surewaard BG, Nelson
MC, et al: Renal immune surveillance and dipeptidase-1 contribute
to contrast-induced acute kidney injury. J Clin Invest.
128:2894–2913. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sun W, Yang Y, Xu C and Guo J: Regulatory
mechanisms of long noncoding RNAs on gene expression in cancers.
Cancer Genet. 216-217:105–110. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sallam T, Sandhu J and Tontonoz P: Long
noncoding RNA discovery in cardiovascular disease: Decoding form to
function. Circ Res. 122:155–166. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Paller MS, Hoidal JR and Ferris TF: Oxygen
free radicals in ischemic acute renal failure in the rat. J Clin
Invest. 74:1156–1164. 1984.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46.
2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lorenzen JM, Schauerte C, Kielstein JT,
Hubner A, Martino F, Fiedler J, Gupta SK, Faulhaber-Walter R,
Kumarswamy R, Hafer C, et al: Circulating long noncoding RNATapSaki
is a predictor of mortality in critically ill patients with acute
kidney injury. Clin Chem. 61:191–201. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chen Y, Qiu J, Chen B, Lin Y, Chen Y, Xie
G, Qiu J, Tong H and Jiang D: Long non-coding RNA NEAT1 plays an
important role in sepsis-induced acute kidney injury by targeting
miR-204 and modulating the NF-κB pathway. Int Immunopharmacol.
59:252–260. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yu TM, Palanisamy K, Sun KT, Day YJ, Shu
KH, Wang IK, Shyu WC, Chen P, Chen YL and Li CY: RANTES mediates
kidney ischemia reperfusion injury through a possible role of
HIF-1α and LncRNA PRINS. Sci Rep. 6(18424)2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Cheng W, Li XW, Xiao YQ and Duan SB:
Non-coding RNA-Associated ceRNA networks in a new contrast-induced
acute kidney injury rat model. Mol Ther Nucleic Acids. 17:102–112.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Moore RD, Steinberg EP, Powe NR, Brinker
JA, Fishman EK, Graziano S and Gopalan R: Nephrotoxicity of
high-osmolality versus low-osmolality contrast media: Randomized
clinical trial. Radiology. 182:649–655. 1992.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Dong M, Jiao Z, Liu T, Guo F and Li G:
Effect of administration route on the renal safety of contrast
agents: A meta-analysis of randomized controlled trials. J Nephrol.
25:290–301. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Karlsberg RP, Dohad SY and Sheng R:
Iodixanol Peripheral Computed Tomographic Angiography Study
Investigator Panel. Contrast medium-induced acute kidney injury:
Comparison of intravenous and intraarterial administration of
iodinated contrast medium. J Vasc Interv Radiol. 22:1159–1165.
2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu TQ, Luo WL, Tan X, Fang Y, Chen J,
Zhang H, Yu XF, Cai JR and Ding XQ: A novel contrast-induced acute
kidney injury model based on the 5/6-nephrectomy rat and
nephrotoxicological evaluation of iohexol and iodixanol in vivo.
Oxid Med Cell Longev. 2014(427560)2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sun S, Zhang T, Nie P, Hu L, Yu Y, Cui M,
Cai Z, Shen L and He B: A novel rat model of contrast-induced acute
kidney injury. Int J Cardiol. 172:e48–e50. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yang D, Yang D, Jia R and Tan J: Na+/Ca2+
exchange inhibitor, KB-R7943, attenuates contrast-induced acute
kidney injury. J Nephrol. 26:877–885. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang Z, Bao W, Zou X, Tan P, Chen H, Lai
C, Liu D, Luo Z and Huang M: Co-expression analysis reveals
dysregulated miRNAs and miRNA-mRNA interactions in the development
of contrast-induced acute kidney injury. PLoS One.
14(e0218574)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Society of Toxicolgoy. Guiding principles
in the use of animals in toxicology. Adopted by the Society of
Toxicology in July 1989. Toxicol Appl Pharmacol.
178(4p)2002.PubMed/NCBI
|
|
28
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Tafer H and Hofacker IL: RNAplex: A fast
tool for RNA-RNA interaction search. Bioinformatics. 24:2657–2663.
2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The urihttp://microRNA.orgsimplemicroRNA.org resource:
Targets and expression. Nucleic Acids Res. 36 (Database
Issue):D149–D153. 2008.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Aoki-Kinoshita KF and Kanehisa M: Gene
annotation and pathway mapping in KEGG. Methods Mol Biol.
396:71–91. 2007.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sun L, Zhang Z, Bailey TL, Perkins AC,
Tallack MR, Xu Z and Liu H: Prediction of novel long non-coding
RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC
Bioinformatics. 13(331)2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ,
Wei L and Gao G: CPC: Assess the protein-coding potential of
transcripts using sequence features and support vector machine.
Nucleic Acids Res. 35:W345–W349. 2007.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C,
Liu Y, Chen R and Zhao Y: Utilizing sequence intrinsic composition
to classify protein-coding and long non-coding transcripts. Nucleic
Acids Res. 41(e166)2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Tang C, Ma Z, Zhu J, Liu Z, Liu Y, Liu Y,
Cai J and Dong Z: P53 in kidney injury and repair: Mechanism and
therapeutic potentials. Pharmacol Ther. 195:5–12. 2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Heyman SN, Rosen S, Khamaisi M, Idee JM
and Rosenberger C: Reactive oxygen species and the pathogenesis of
radiocontrast-induced nephropathy. Invest Radiol. 45:188–195.
2010.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Liss P, Nygren A, Erikson U and Ulfendahl
HR: Injection of low and iso-osmolar contrast medium decreases
oxygen tension in the renal medulla. Kidney Int. 53:698–702.
1998.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Rosenberger C, Rosen S and Heyman SN:
Renal parenchymal oxygenation and hypoxia adaptation in acute
kidney injury. Clin Exp Pharmacol Physiol. 33:980–988.
2006.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Yan M, Tang C, Ma Z, Huang S and Dong Z:
DNA damage response in nephrotoxic and ischemic kidney injury.
Toxicol Appl Pharmacol. 313:104–108. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lin J, Zhang X, Xue C, Zhang H, Shashaty
MG, Gosai SJ, Meyer N, Grazioli A, Hinkle C, Caughey J, et al: The
long noncoding RNA landscape in hypoxic and inflammatory renal
epithelial injury. Am J Physiol Renal Physiol. 309:F901–F913.
2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Chang J, Ma JZ, Zeng Q, Cechova S, Gantz
A, Nievergelt C, O'Connor D, Lipkowitz M and Le TH: Loss of GSTM1,
a NRF2 target, is associated with accelerated progression of
hypertensive kidney disease in the African American Study of Kidney
Disease (AASK). Am J Physiol Renal Physiol. 304:F348–F355.
2013.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Gigliotti JC, Tin A, Pourafshar S, Cechova
S, Wang YT, Sung SJ, Bodonyi-Kovacs G, Cross JV, Yang G, Nguyen N,
et al: GSTM1 deletion exaggerates kidney injury in experimental
mouse models and confers the protective effect of cruciferous
vegetables in mice and humans. J Am Soc Nephrol. 31:102–116.
2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Janssen B, Hohenadel D, Brinkkoetter P,
Peters V, Rind N, Fischer C, Rychlik I, Cerna M, Romzova M, de Heer
E, et al: Carnosine as a protective factor in diabetic nephropathy:
Association with a leucine repeat of the carnosinase gene CNDP1.
Diabetes. 54:2320–2327. 2005.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Kilis-Pstrusinska K: Carnosine and kidney
diseases: What we currently know? Curr Med Chem. 27:1764–1781.
2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Machado RA, Constantino Lde S, Tomasi CD,
Rojas HA, Vuolo FS, Vitto MF, Cesconetto PA, de Souza CT, Ritter C
and Dal-Pizzol F: Sodium butyrate decreases the activation of NF-κB
reducing inflammation and oxidative damage in the kidney of rats
subjected to contrast-induced nephropathy. Nephrol Dial Transplant.
27:3136–3140. 2012.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Toso A, Leoncini M, Maioli M, Tropeano F,
Di Vincenzo E, Villani S and Bellandi F: Relationship between
inflammation and benefits of early high-dose rosuvastatin on
contrast-induced nephropathy in patients with acute coronary
syndrome: The pathophysiological link in the PRATO-ACS study
(Protective Effect of Rosuvastatin and Antiplatelet Therapy on
Contrast-Induced Nephropathy and Myocardial Damage in Patients With
Acute Coronary Syndrome Undergoing Coronary Intervention). JACC
Cardiovasc Interv. 7:1421–1429. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Chen YH, Fu YC and Wu MJ: Does resveratrol
play a role in decreasing the inflammation associated with contrast
induced nephropathy in rat model? J Clin Med. 8(147)2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Shen J, Wang L, Jiang N, Mou S, Zhang M,
Gu L, Shao X, Wang Q, Qi C, Li S, et al: NLRP3 inflammasome
mediates contrast media-induced acute kidney injury by regulating
cell apoptosis. Sci Rep. 6(34682)2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Andreucci M, Lucisano G, Faga T, Bertucci
B, Tamburrini O, Pisani A, Sabbatini M, Salzano S, Vitale M, Fuiano
G, et al: Differential activation of signaling pathways involved in
cell death, survival and inflammation by radiocontrast media in
human renal proximal tubular cells. Toxicol Sci. 119:408–416.
2011.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ripoll VM, Irvine KM, Ravasi T, Sweet MJ
and Hume DA: Gpnmb is induced in macrophages by IFN-gamma and
lipopolysaccharide and acts as a feedback regulator of
proinflammatory responses. J Immunol. 178:6557–6566.
2007.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zhou L, Zhuo H, Ouyang H, Liu Y, Yuan F,
Sun L, Liu F and Liu H: Glycoprotein non-metastatic melanoma
protein b (Gpnmb) is highly expressed in macrophages of acute
injured kidney and promotes M2 macrophages polarization. Cell
Immunol. 316:53–60. 2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Chang L and Karin M: Mammalian MAP kinase
signalling cascades. Nature. 410:37–40. 2001.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Malik S, Suchal K, Bhatia J, Gamad N,
Dinda AK, Gupta YK and Arya DS: Molecular mechanisms underlying
attenuation of cisplatin-induced acute kidney injury by epicatechin
gallate. Lab Invest. 96:853–861. 2016.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Matsuda A, Suzuki Y, Honda G, Muramatsu S,
Matsuzaki O, Nagano Y, Doi T, Shimotohno K, Harada T, Nishida E, et
al: Large-scale identification and characterization of human genes
that activate NF-kappaB and MAPK signaling pathways. Oncogene.
22:3307–3318. 2003.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Basile DP, Bonventre JV, Mehta R, Nangaku
M, Unwin R, Rosner MH, Kellum JA and Ronco C: ADQI XIII Work Group.
Progression after AKI: Understanding maladaptive repair processes
to predict and identify therapeutic treatments. J Am Soc Nephrol.
27:687–697. 2016.PubMed/NCBI View Article : Google Scholar
|