Introduction
Kawasaki disease (KD) is an acute, self-limiting
form of vasculitis that affects infants and children, particularly
those aged <5 years. In developed countries, KD is the most
common cause of acquired heart disease in children (1). Up to 25% of patients develop coronary
artery aneurysm (CAA) if treatment is not provided in a timely
manner (2). Since 1983, Furusho
et al have used intravenous immunoglobulin (IVIG) to treat
patients with KD in the initial acute phase and the incidence of
CAA has decreased from 20-25 to 3-5% (3-5).
However, the mechanisms of action of IVIG treatment for KD have
remained elusive. There are several possible mechanisms, including
inhibition of inflammatory cytokines, blockade of Fc receptor on
macrophages, activation of augmenting T-cell suppressor, inhibition
of antibody formation and neutralization of bacterial superantigens
(6). Other mechanisms, such as
reduced platelet adhesion, decreased oxidative stress and
neutrophil apoptosis via a lectin-binding pathway have also been
reported (7-9).
In an animal model of KD, prevention of T-cell activation and
reduction of TNF-α production have been reported (10). In addition, adverse effects have
been revealed in the clinical application of IVIG, the most
frequent of which include flushing, fever, headache, chills and
hemolysis (11,12). In addition, not every family one of
patients who suffer from KD in poor countries can afford the
expense of treatment. Therefore, the aim of the present study was
to investigate the mechanisms of action of IVIG therapy for KD from
a genetic perspective. At present, there is a lack of research on
genetic changes in patients with KD prior to and after IVIG
treatment.
Currently, gene expression profiling microarrays and
proteome analysis are widely used to analyze the differential gene
expression profiles and enriched pathways in KD. A large amount of
gene chip expression profiling microarray data have been published
in public databases, such as the Gene Expression Omnibus (GEO)
database. In the present study, it was hypothesized that certain
genes are associated with IVIG treatment in patients with KD. To
test the hypothesis, bioinformatics methods were used to screen out
the differentially expressed genes (DEGs) in patients with KD after
vs. prior to IVIG therapy. The gene chip data were downloaded from
the GEO website and the R project was used to analyze DEGs.
Proteome analysis was used to assess the differentially expressed
proteins (DEPs). The DEGs and DEPs were combined for Gene Ontology
(GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analyses. The profiling changes in immune cells
were analyzed using xCell toll. A protein-protein interaction (PPI)
network was used to determine hub genes from DEGs and the
mRNA-microRNA (miRNA/miR) interaction network was established using
miRwalk. To further verify the aforementioned results, reverse
transcription-quantitative PCR (RT-qPCR) was performed. Potential
hub genes and their enriched pathways in patients with KD following
IVIG treatment were identified, which provided insight into the
mechanisms of action of IVIG in the treatment of KD.
Materials and methods
Patient data
A total of 50 patients with KD who received IVIG
therapy between January 2020 and June 2020 were included in the
present study. The whole blood cell samples from the 4 patients
with KD were sent for proteomic analysis, which was performed by
PTM Biolabs, Inc. They were randomly divided into two groups and
the mRNA levels detected in one group and miRNA levels in the
other. No significant differences in patient characteristics,
including sex, age and duration of fever, were identified between
the two groups. The characteristics of the cohort are provided in
Table I. Blood samples were
collected prior to and 36 h after IVIG. The study was performed in
accordance with the Declaration of Helsinki and approved by the
Ethics Committee of the Children's Hospital of Soochow University
Suzhou (Suzhou, China; approval no. 2020CS075). All participants
and their parents were informed of the study details by the Ethics
Committee and provided written informed consent.
 | Table IClinical characteristics. |
Table I
Clinical characteristics.
| | Subjects used for
RT-qPCR | |
|---|
| Characteristic | mRNA | miRNA | P-value |
|---|
| Male sex (n) | 15(25) | 18(25) | 0.157 |
| Age (months) | 29.95
(2.09-57.82) | 30.76
(6.8-54.7) | 0.427 |
| Duration of fever
(days) | 6.8
(5.57-8.03) | 6.76 (5.52-8) | 0.843 |
Microarray data
In order to identify the DEGs in the post-IVIG KD
samples, as compared with the pre-IVIG samples, the GES48498 gene
expression profile was retrieved from the National Center for
Biotechnology Information (NCBI) GEO database (https://www.ncbi.nlm.nih.gov/geo/). The GES48498
microarray dataset was contributed by Ogihara et al
(13), which included 6 whole blood
cell samples from patients that had or had not undergone IVIG
administration. Expression profiling arrays were generated using
the GPL570 Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix,
Inc.).
Identification of DEGs
The raw microarray dataset GES48498 in CEL format
was downloaded from the GEO database. The data were pre-processed
into expression values using the package Affy (version 1.64.0) of
the R project (14). Next, the data
were normalized to verify their quality. To screen out the DEGs,
the two groups of samples (pre-IVIG and post-IVIG) were analyzed
using the Limma package (version 3.42.2) in R (15). The fold-changes in the expression
profiling genes were calculated at the same time. |Log
fold-change|>1 and adjusted P<0.01 were set as the cut-off
criteria for the identification of DEGs. All significant DEGs were
demonstrated in a volcano plot generated using R.
Proteome analysis
In addition to microarray data, to further
understand the mechanism of IVIG in KD, proteome analysis was
performed on patients prior to and after IVIG treatment. The
American Heart Association defined fever for 36 h after the first
IVIG infusion as IVIG resistance (16). Therefore, the whole blood cell
samples from 2 patients with KD were collected prior to IVIG
treatment and those of 2 patients with KD were collected 36 h after
IVIG treatment and sent to PTM Biolabs, Inc. for high-throughput
quantitative proteome analysis. There was no change in the aspirin
dose in any of the patients' blood samples and no glucocorticoids
or antibiotics were used. Samples were sonicated three times on ice
using a high-intensity ultrasonic processor (Scientz) in lysis
buffer (8 M urea, 1% protease inhibitor cocktail). For PTM
experiments, inhibitors were also added to the lysis buffer, e.g. 3
µM trichostatin A and 50 mM nicotinamide for acetylation. The
remaining debris was removed by centrifugation at 12,000 x g at 4˚C
for 10 min. The protein sample was then diluted by adding 100 mM
tetraethylammonium bromide (TEAB) to achieve an urea concentration
of <2 M. Finally, trypsin (Promega Corp.) was added at a
trypsin-to-protein mass ratio of 1:50 for the first digestion at
37˚C overnight and 1:100 trypsin-to-protein mass ratio for a second
4-h digestion at room temperature. After trypsin digestion, the
peptide was desalted using a Strata X C18 SPE column (Phenomenex)
and vacuum-dried. The peptide was reconstituted in 0.5 M TEAB and
processed according to the manufacturer's protocol for the Tandem
Mass Tag (TMT) kit (Thermo Fisher Scientific, Inc.). In brief, one
unit of TMT/iTRAQ reagent was thawed and reconstituted in
acetonitrile. The peptide mixtures were then incubated for 2 h at
room temperature and pooled, desalted and dried by vacuum
centrifugation. The tryptic peptides were fractionated by high pH
reverse-phase high-performance liquid chromatography using a Thermo
Betasil C18 column (5-µm particles, 10 mm inner diameter, 250 mm in
length) (Thermo Fisher Scientific, Inc.). In brief, peptides were
first separated with a gradient of 8 to 32% acetonitrile (pH 9.0)
over 60 min and collected in 60 fractions. Subsequently, the
peptides were combined into 6 fractions and dried by vacuum
centrifugation. The tryptic peptides were dissolved in 0.1% formic
acid (solvent A) and directly loaded onto a home-made
reversed-phase analytical column (15-cm length, 75 µm inner
diameter). The gradient was comprised of an increase from 6 to 23%
solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, 23 to
35% in 8 min and climbing to 80% in 3 min and then a hold at 80%
for the last 3 min, all at a constant flow rate of 400 nl/min on an
EASY-nLC 1000 ultra-performance liquid chromatography (UPLC)
system. The peptides were subjected to nitrogen soluble index
source followed by tandem mass spectrometry (MS/MS) using a Q
Exactive™ Plus (Thermo Fisher Scientific, Inc.) coupled online to
the UPLC. The electrospray voltage applied was 2.0 kV. The mass to
charge (m/z) scan range was 350 to 1,800 for a full scan and intact
peptides were detected in the Orbitrap at a resolution of 70,000.
Peptides were then selected for MS/MS using the normalized
collision energy set to 28 and the fragments were detected in the
Orbitrap at a resolution of 17,500. A data-dependent procedure that
alternated between one MS scan followed by 20 MS/MS scans with
15.0-sec dynamic exclusion. Automatic gain control was set at 5E4.
The fixed first mass was set as 100 m/z. The raw data were
processed similar to microarray data using the R project. |Log
fold-change (FC)|>0.6 and adjusted P<0.05 were set as the
threshold for the identification of DEPs.
GO and KEGG pathway enrichment
analysis
To analyze the DEGs and significant proteins for GO
term and KEGG pathway enrichment analyses, the ClueGo package
(version 2.56) of Cytoscape software (version 3.8.0; https://cytoscape.org/) was used to process data and
the package ppglot2 (version 3.3.0) in R was utilized to visualize
the results. GO enrichment analysis included biological process
(BP), cellular component (CC) and molecular function (MF) terms. GO
annotation and KEGG pathway enrichment analysis of DEGs and DEPs
was performed. The intersection part of the pathway was selected to
analyze its enrichment in transcriptomic and proteomic analysis,
respectively. The adjusted P-value [false discovery rate
(FDR)<0.05] was considered to indicate a statistically
significant difference. The top 10 pathways with the greatest
difference in transcriptomics and proteomics were selected and the
intersection of the two groups was selected as an important GO and
KEGG pathway in the treatment of KD by IVIG.
Immune cell analysis
In order to further understand the changes in immune
cells prior to and after IVIG treatment, the webtool xCell was used
for cell type enrichment analysis of the GES48498 gene expression
profile for 64 immune and stroma cell types (17). Given that the distribution of the
present data was not normal, the Wilcoxon signed-rank test was used
to calculate the difference between two groups with P<0.05 set
as the cutoff value.
PPI network and hub gene
identification
In order to extract the hub genes from the DEGs in
the post-IVIG KD samples, the Search Tool for the Retrieval of
Interacting Genes and proteins (STRING) database (version 11.0) was
used to calculate and obtain the PPI network (18). A combined score of ≥0.4 for PPI
pairs was considered to indicate a statistically significant
difference. Visualization was performed with the MCODE (version
1.6.1) plug-in of Cytoscape software (19). The software parameters were set as
default. The genes contained in the gene cluster with the highest
scores were considered to be the hub genes (20).
Construction of the mRNA-miRNA
interaction network
The hub genes were selected and uploaded to the
miRWalk 2.0 database to predict its target miRNAs (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/).
To increase the accuracy of the results, four databases, including
TargetScan, miRanda, miRWalk and RNA22 (integrated in the miRWalk
2.0 database), were used to perform the intersection. These four
databases are the default databases used to analyse the mRNA-miRNA
interaction network. The mRNA-miRNA interaction network was
constructed with Cytoscape software (version 3.8.0). The miRNAs
that targeted >3 genes were screened out for further
verification.
Verification of mRNA and miRNA by
RT-qPCR
To verify the miRNAs and their target hub genes,
blood samples were collected from pediatric patients with KD
referred to the Children's Hospital of Soochow University (Suzhou,
China). mRNA and miRNA were tested in 25 patients. Clinical samples
were collected prior to and after treatment with IVIG. Total RNA
was extracted using TRIzol® reagent (Thermo Fisher
Scientific Inc.). Subsequently, miRNAs were isolated and purified
from total RNA using the miRNeasy Mini kit (Qiagen GmbH).
mRNA RT was performed using PrimeScript RT Master
mix (Takara Bio, Inc.) at 37˚C for 15 min and then at 85˚C for 10
sec. The miRNA RT was performed using the miScript II RT kit
(Qiagen GmbH) at 37˚C for 60 min and at 95˚C for 5 min. The
complementary DNA derived from mRNA and miRNA was diluted four
times with RNase-free water. Next, a LightCycler 480 II Real-Time
PCR system (Roche Diagnostics) was used for the detection of mRNA
and miRNA expression levels. qPCR for mRNA quantification was
performed with SYBR Green qPCR Master mix (Bimake.com)
at 50˚C for 2 min, followed by 40 cycles at 95˚C for 15 sec and
60˚C for 60 sec. PCR amplification of miRNA was conducted using the
miScript SYBR Green PCR kit (Qiagen GmbH) at 95˚C for 15 min,
followed by 40 cycles at 94˚C for 15 sec, 55˚C for 30 sec and 70˚C
for 30 sec. GAPDH and U6 were used as internal controls for mRNA
and miRNA, respectively. The relative expression of different
samples was calculated using the
2-ΔΔCt method (21). Primer sequences were designed by
Sangon Biotech Co., Ltd. and the universal primer was provided by
Qiagen GmbH. The primer sequences used are listed in Table II.
 | Table IIPrimers designed for real-time
PCR. |
Table II
Primers designed for real-time
PCR.
| Gene | Forward (5' to
3') | Reverse (5' to
3') |
|---|
| GAPDH |
GGAGAAAC-CTGCCAAGTATG |
TTACTCCTT-GGAGGCCATGTAG |
| RRM2 |
AGTG-GAAGGCATTTTCTTTTCC |
GCAAAATCACAGTG-TAAACCCT |
| TK1 |
GTTCTCAGGAAAAA-GCACAGAG |
GTCTTTGGCATACTT-GATCACC |
| TOP2A |
AAGATTCATTGAAGAC-GCTTCG |
GCTG-TAAAATGCCATTTCTTGC |
| GINS1 |
AGAGCACTCAGATGG-GAATATG |
ATCCTGTG-TAATGTCCAAACCT |
| ZWINT |
AATT-GCAGCTAAGGAACAATGG |
TTTCTCCATGGCCATTT-GTTTC |
| ICAM-1 |
ATGCCCAGA-CATCTGTGTCC |
GGGGTCTC-TATGCCCAACAA |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| miR-3929 |
GGAGGCTGATGTGAGTAGACCACT | |
| miR-30b-3p |
CGCTGGGAGGTGGATGTTTACTTC | |
| miR-3689b |
GCTGGGAGGTGTGATATTGTGGT | |
| miR-485-5p |
AGAGGCTGGCCGTGATGAATTC | |
| Universal reverse
of miRNAs | 10X miScript
Universal Primer (Qiagen GmbH) | |
Cell culture and treatment
Human coronary artery endothelial cells (HCAECs)
were purchased from Beijing Beina Chuanglian Biotechnology Research
Institute (cat. no. BNCC338441) and cultured with RPMI-1640 medium
(Biosharp Corp.) supplemented with 10% FBS (Biological Industries)
and antibiotics (100 U/ml penicillin-G and 100 mg/ml streptomycin)
at 37˚C in 5% CO2 and 95% humidified air. The HCAECs
were seeded in each well of a 12-well culture plate at a density of
5x104 cells and incubated for 18 h at 37˚C in 5% CO2 and
95% humidified air. The HCAECs were then subjected to 1 µg/ml
lipopolysaccharide (LPS; Merck KGaA) challenge for 3 h (22). Subsequently, the HCAECs were treated
with IVIGs (25 mg/ml) (Tonrol Bio-pharmaceutical Co., Ltd.) and
vehicle for 18 h (23). The
endothelial cells produce intercellular adhesion molecule 1 (ICAM1)
in response to inflammatory cytokines (23). The hub genes in HCAECs were
validated by RT-qPCR and imaged using an inverted phase-contrast
photo microscope at a magnification of x400 (Olympus IX73; Olympus
Corp.). All experiments were repeated at least three times with two
replicated each.
Statistical analysis
Data were analyzed using SPSS statistical software
for Windows (version 24.0; IBM Corp.) and experimental
non-parametric data and parametric data were presented as the mean
± standard deviation. The Shapiro-Wilk normality test was used to
assess the normal distribution of values. The Wilcoxon signed-rank
test was used for comparison of categorical data (age and days of
fever) and the χ2 test was used for the comparison of
categorical data (sex) in Table I
(P>0.05). The differences in mRNA/miRNA expression levels and
immune cells between the two groups (pre-IVIG group and post-IVIG
group) were analyzed using the Wilcoxon rank-sum test and those
between different time-points for the same group with the Wilcoxon
signed-rank test, based on the results of the Shapiro-Wilk
normality test. For RT-qPCR validation, each group contained 25
samples and each sample was measured by PCR and repeated three
times to obtain the mean value. P<0.05 was considered to
indicate a statistically significant difference.
Results
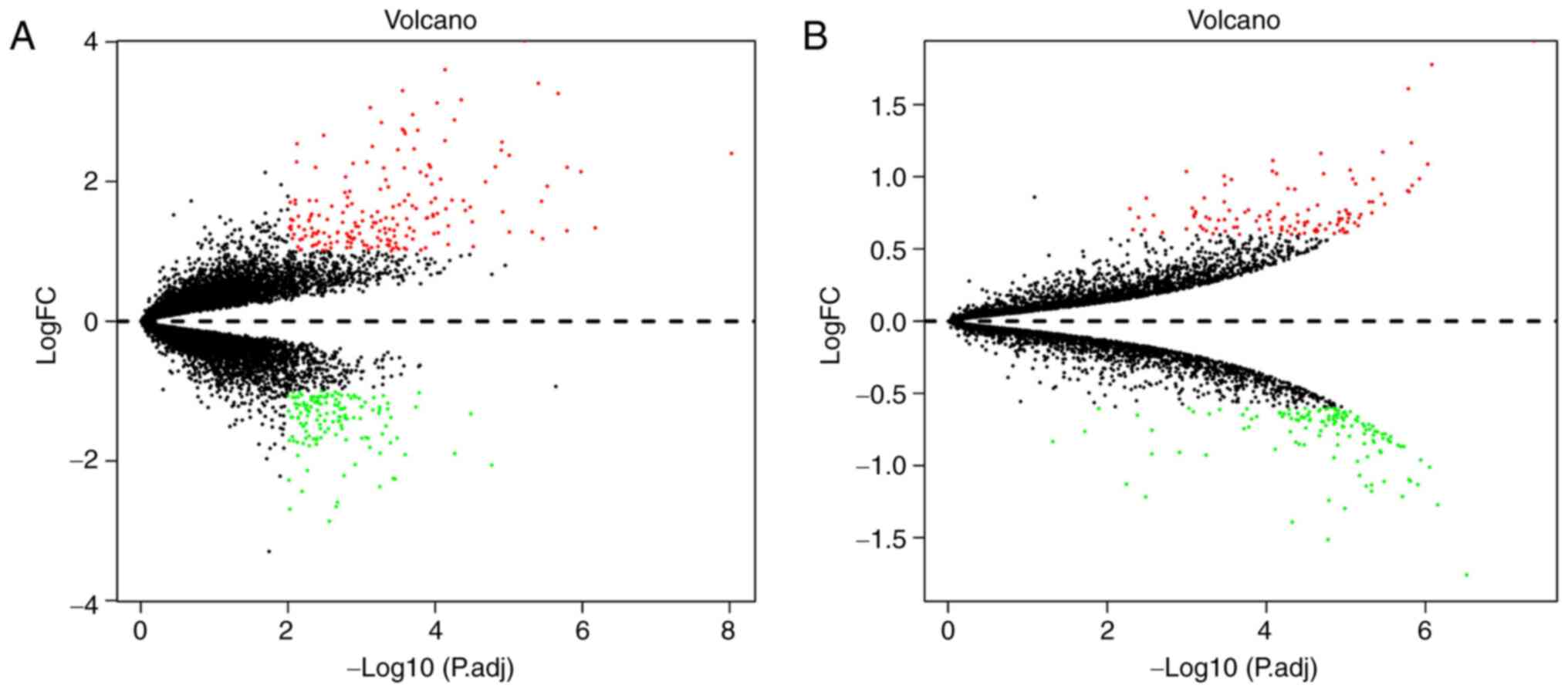
Identification of DEGs and DEPs
To identify DEGs in post-IVIG vs. pre-IVIG KD
samples, the raw microarray dataset GES48498 and the annotation of
the platform GPL570 were downloaded from the GEO database. DEGs
with |log2 FC|>1 and P<0.01 were determined. A total of 335
DEGs were selected, including 189 upregulated and 146 downregulated
genes. According to the results of the proteomic analysis, 253 DEPs
were identified, of which 117 were upregulated and 136
downregulated. A volcano plot of the DEGs and DEPs is presented in
Fig. 1A and B, respectively.
GO functional and KEGG pathway
enrichment analysis
To understand the functions and potential mechanisms
of IVIG treatment in KD, the results were analyzed and visualized
using Cytoscape software. In total, 335 DEGs and 253 DEPs were
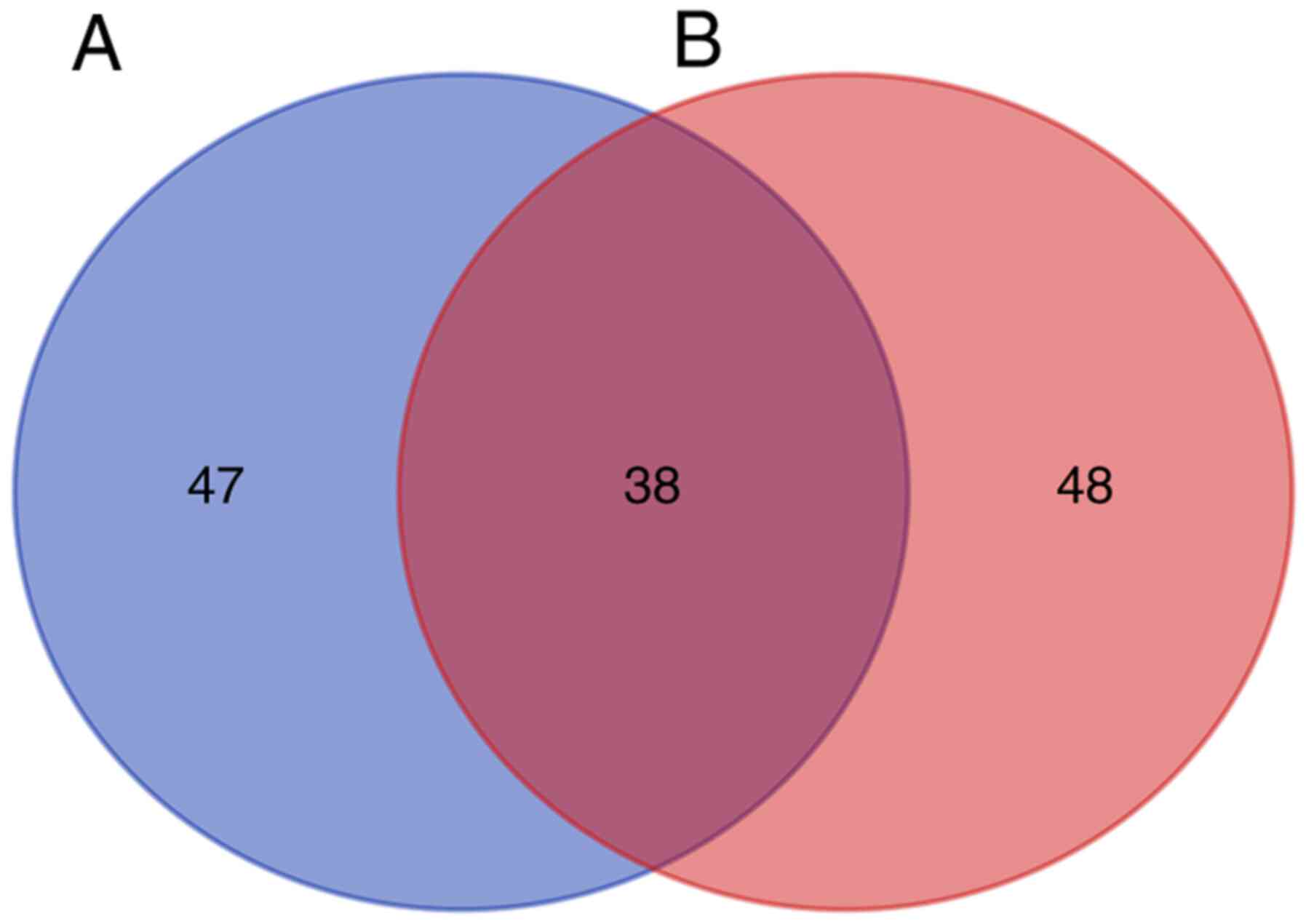
included in the analysis. The DEGs were enriched in 86 GO pathways
(FDR<0.05), including 68 BP, 17 CC and 1 MF terms. The DEPs were
enriched in 85 GO pathways, including 59 BP, 20 CC and 6 MF terms.
There were 38 pathways at the intersection of the two GO pathway
enrichments (Fig. 2). Common GO
pathways between DEGs and DEPs were further analyzed. The top 10
genes in each category of the GO pathway enrichment analysis of the
DEGs and DEPs according to the FDR are listed in Fig. 3A and B. DEGs enriched in 20 pathways and DEPs
enriched in 26 pathways were selected. Next, the intersection of
the two groups regarding important pathways was selected and a Venn
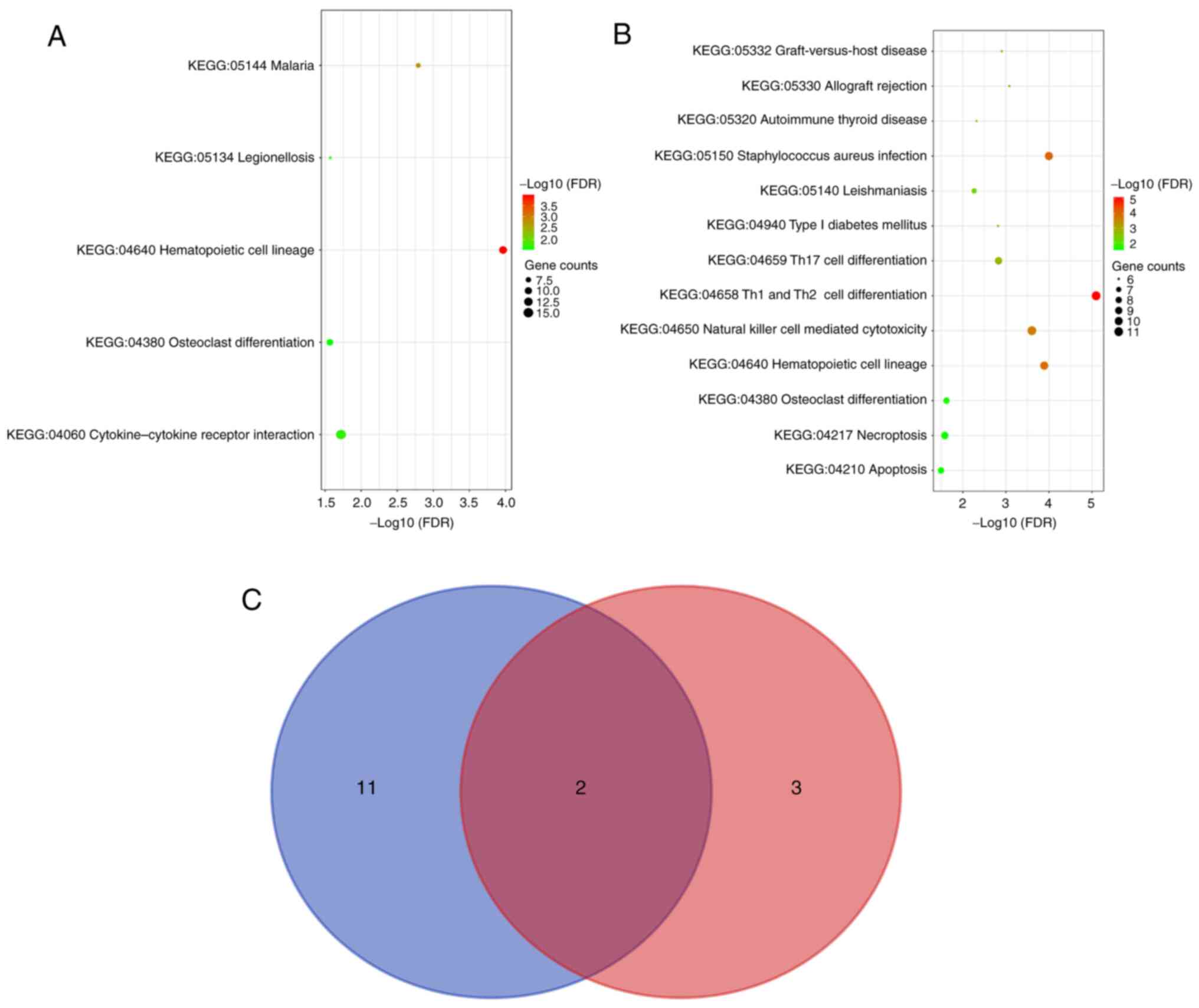
diagram was drawn to obtain 18 common pathways (Fig. 3C). The DEGs and DEPs were subjected
to enrichment analysis of the KEGG pathway and the top 10 pathways
are presented in Fig. 4A-C. The
results of the GO functional and KEGG pathway enrichment analyses
are presented in Table III,
Tables IV and Table V.
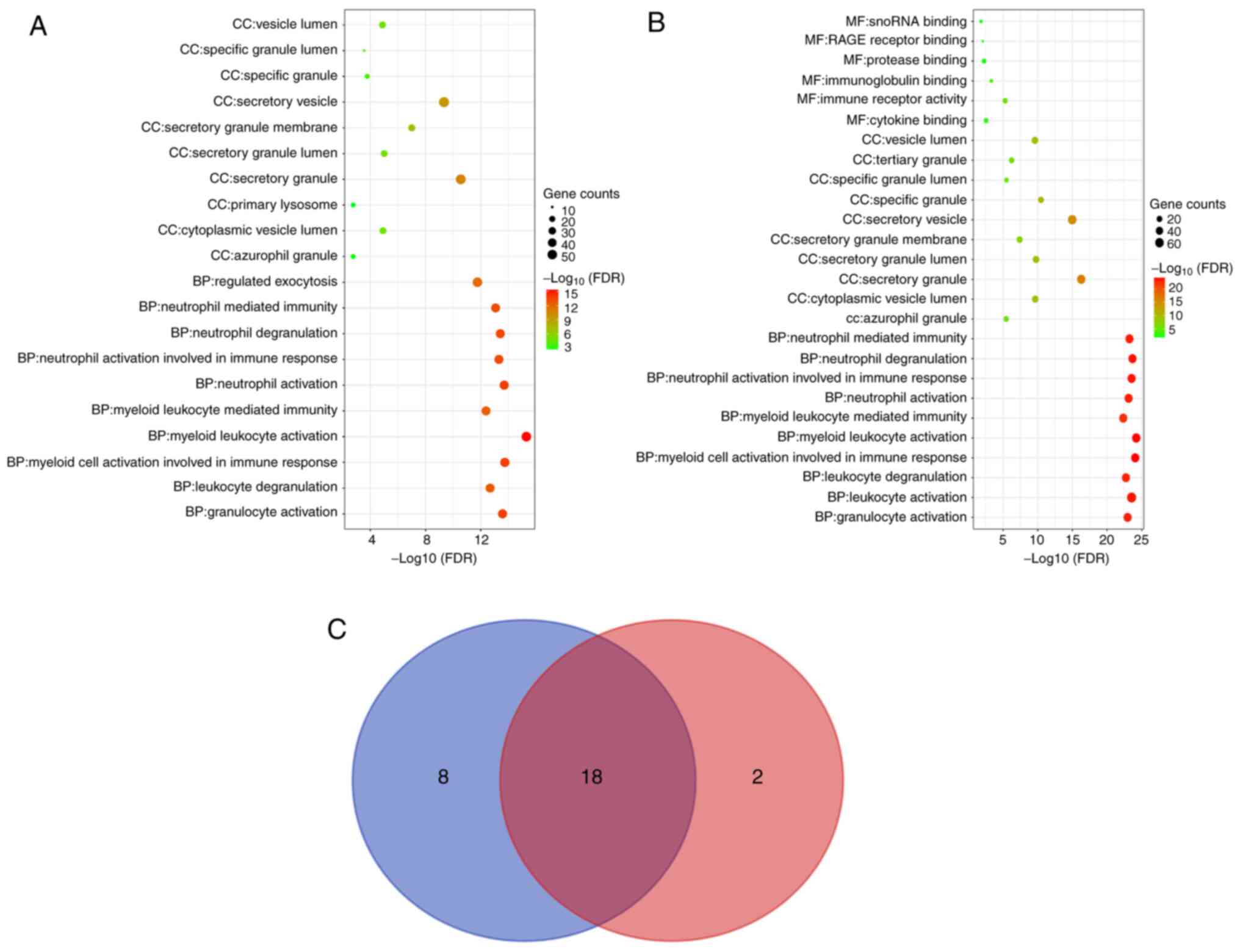 | Figure 3GO enrichment analysis of DEGs and
DEPs. For (A) DEGs and (B) DEPs, the top 10 pathways in each GO
category (MF, CC and BP) are presented in bubble charts. If the
analysis results yielded >10 pathways enriched in this category,
the top 10 pathways were listed according to the FDR. (C) Venn
diagram indicating the GO pathways for the DEGs and DEPs. The top
10 GO pathways for the DEPs in each GO category were determined and
represented by a blue circle, while those for the DEGs were
represented by a red circle. GO, gene ontology; MF, molecular
function; CC, cellular component; BP, biological process; FDR,
false discovery rate; DEGs, differentially expressed genes; DEPs,
differentially expressed proteins. |
 | Table IIIImportant common GO pathways in the
enrichment analyses of differentially expressed genes and
proteins. |
Table III
Important common GO pathways in the
enrichment analyses of differentially expressed genes and
proteins.
| A, Category BP |
|---|
| Pathway ID | Description |
|---|
| GO:0036230 | Granulocyte
activation |
| GO:0043299 | Leukocyte
degranulation |
| GO:0002275 | Myeloid cell
activation involved in immune response |
| GO:0002274 | Myeloid leukocyte
activation |
| GO:0002444 | Myeloid leukocyte
mediated immunity |
| GO:0042119 | Neutrophil
activation |
| GO:0002283 | Neutrophil
activation involved in immune response |
| GO:0043312 | Neutrophil
degranulation |
| GO:0002446 | Neutrophil mediated
immunity |
| B, Category CC |
| Pathway ID | Pathway ID |
| GO:0042582 | Azurophil
granule |
| GO:0060205 | Cytoplasmic vesicle
lumen |
| GO:0030141 | Secretory
granule |
| GO:0034774 | Secretory granule
lumen |
| GO:0030667 | Secretory granule
membrane |
| GO:0099503 | Secretory
vesicle |
| GO:0042581 | Specific
granule |
| GO:0035580 | Specific granule
lumen |
| GO:0031983 | Vesicle lumen |
 | Table IVKyoto Encyclopedia of Genes and
Genomes pathways in the enrichment analyses of dif-ferentially
expressed genes. |
Table IV
Kyoto Encyclopedia of Genes and
Genomes pathways in the enrichment analyses of dif-ferentially
expressed genes.
| Term | Description | Count | FDR |
|---|
| hsa04640 | Hematopoietic cell
lineage | 11 |
1.08x10-4 |
| hsa05144 | Malaria | 7 |
1.62x10-3 |
| hsa04060 | Cytokine-cytokine
receptor interaction | 15 |
1.91x10-2 |
| hsa05134 | Legionellosis | 6 |
2.68x10-2 |
| hsa04380 | Osteoclast
differentiation | 9 |
2.72x10-2 |
 | Table VTop five Kyoto Encyclopedia of Genes
and Genomes pathways in the enrichment anal-yses of differentially
expressed proteins. |
Table V
Top five Kyoto Encyclopedia of Genes
and Genomes pathways in the enrichment anal-yses of differentially
expressed proteins.
| Term | Description | Count | FDR |
|---|
| hsa04658 | Th1 and Th2 cell
differentiation | 11 |
8.03x10-6 |
| hsa05150 | Staphylococcus
aureus infection | 10 |
1.01x10-4 |
| hsa04640 | Hematopoietic cell
lineage | 10 |
1.29x10-4 |
| hsa04650 | Natural killer cell
mediated cytotoxicity | 11 |
2.49x10-4 |
| hsa05330 | Allograft
rejection | 6 |
8.33x10-4 |
The GO pathway analysis indicated that myeloid cells
and neutrophils have important roles in IVIG treatment of KD,
including the terms ‘granulocyte activation', ‘leukocyte
degranulation', ‘myeloid cell activation involved in the immune
response', ‘myeloid leukocyte activation',
‘myeloid-leukocyte-mediated immunity', ‘neutrophil activation',
‘neutrophil activation involved in the immune response',
‘neutrophil degranulation' and ‘neutrophil-mediated immunity'. CC
analysis indicated that the DEGs and DEPs were mainly enriched in
the terms of ‘azurophil granules', ‘cytoplasmic vesicle lumina',
‘secretory granules', ‘secretory granule lumina', ‘secretory
granule membranes', ‘secretory vesicles', ‘specific granules',
‘specific granule lumina' and ‘vesicle lumina'. Furthermore, KEGG
pathway analysis demonstrated that DEGs were enriched in
‘cytokine-cytokine receptor interaction', ‘hematopoietic cell
lineage', ‘osteoclast differentiation', ‘malaria' and
‘legionellosis'. The top five KEGG pathways in which DEPs were
enriched were ‘Type 1 T-helper (Th1) and Th2 cell differentiation',
‘Staphylococcus aureus infection', ‘hematopoietic cell
lineage', ‘natural killer cell-mediated cytotoxicity' and
‘allograft rejection'. The terms ‘hematopoietic cell lineage' and
‘osteoclast differentiation' were screened out for both DEGs and
DEPs.
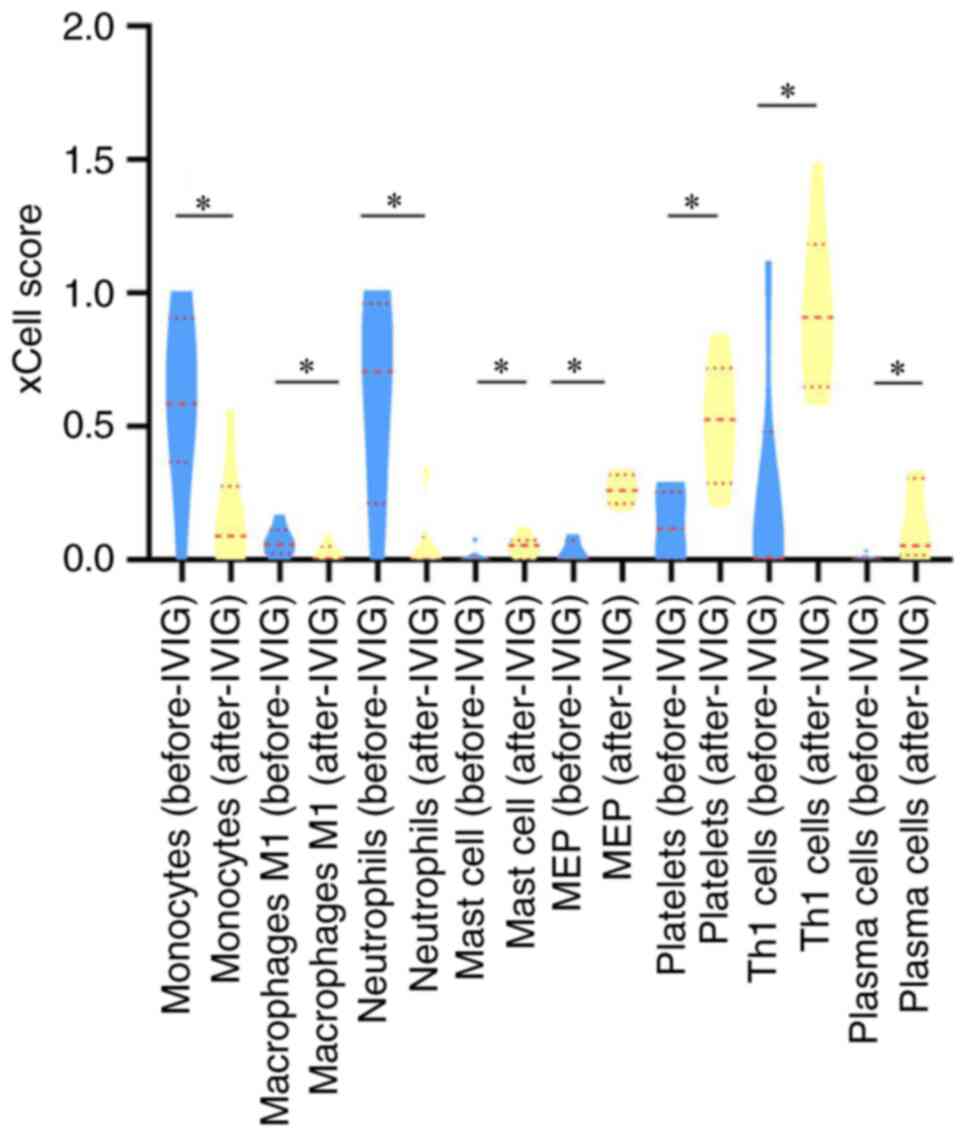
Immune cell deconvolution
analysis
To obtain a deeper understanding of the potential
mechanisms of IVIG treatment for KD, changes in immune cells were
analyzed. xCell was used to perform cell type enrichment based on
the gene microarray data. The types of cells that were
significantly different following calculation are presented in
Fig. 5. A total of eight
differential cell types were screened out: Monocytes (P=0.043), M1
macrophages (P=0.046), neutrophils (P=0.043), mast cells (P=0.043),
megakaryocyte/erythroid progenitor (MEP) cells (P=0.028), platelets
(P=0.028), Th1 cells (P=0.046) and plasma cells (P=0.043).
PPI network construction and hub gene
identification
To screen out the hub genes from a total of 335
DEGs, the online STRING database was used for analysis. The PPI
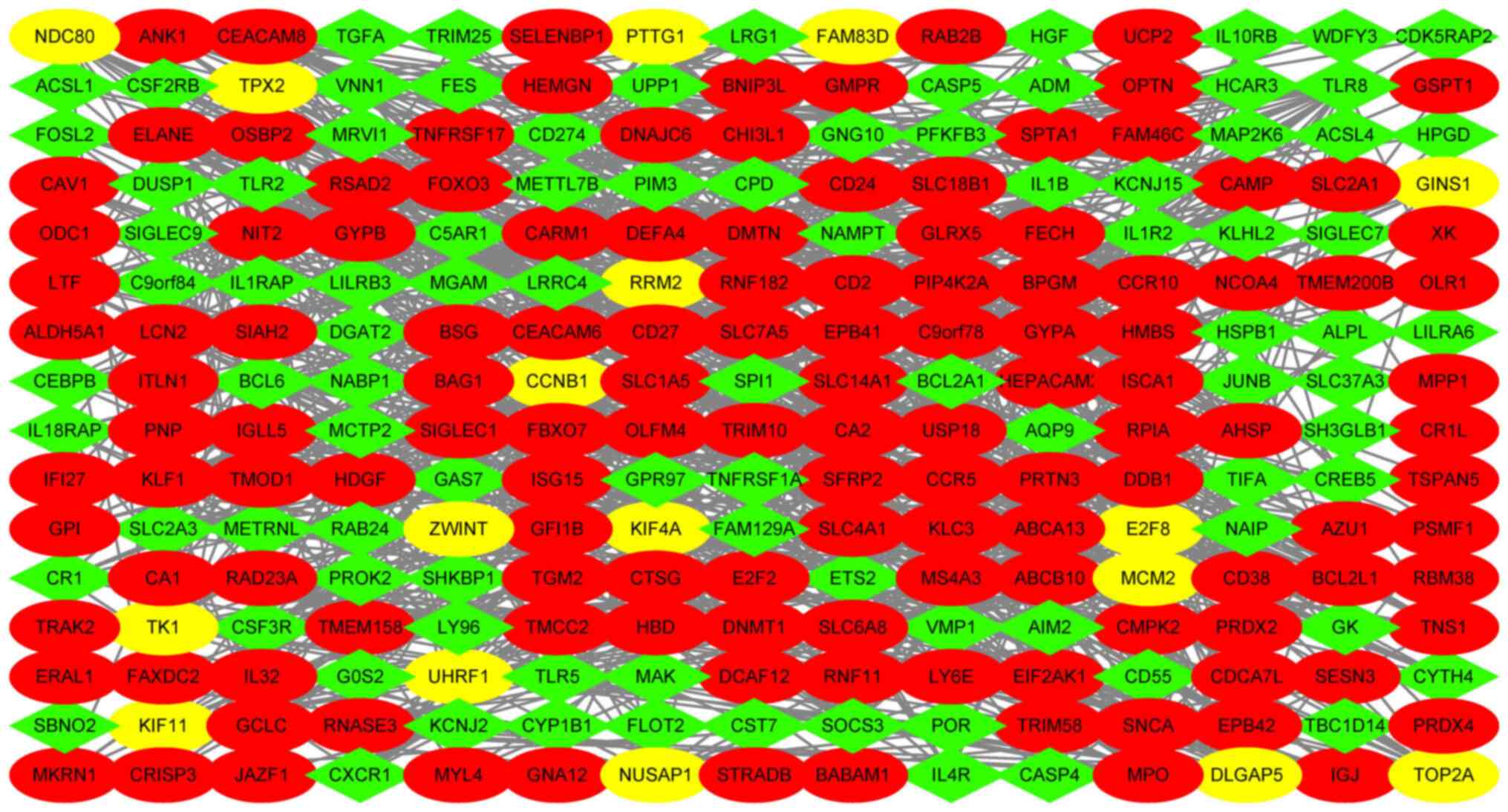
network of the DEGs was visualized using Cytoscape software. The
network contained 240 nodes and 932 edges and the average node
degree was 6.13. The data were analyzed using Cytoscape software
(Fig. 6). The MCODE plug-in of the
Cytoscape software was used to calculate the network data to
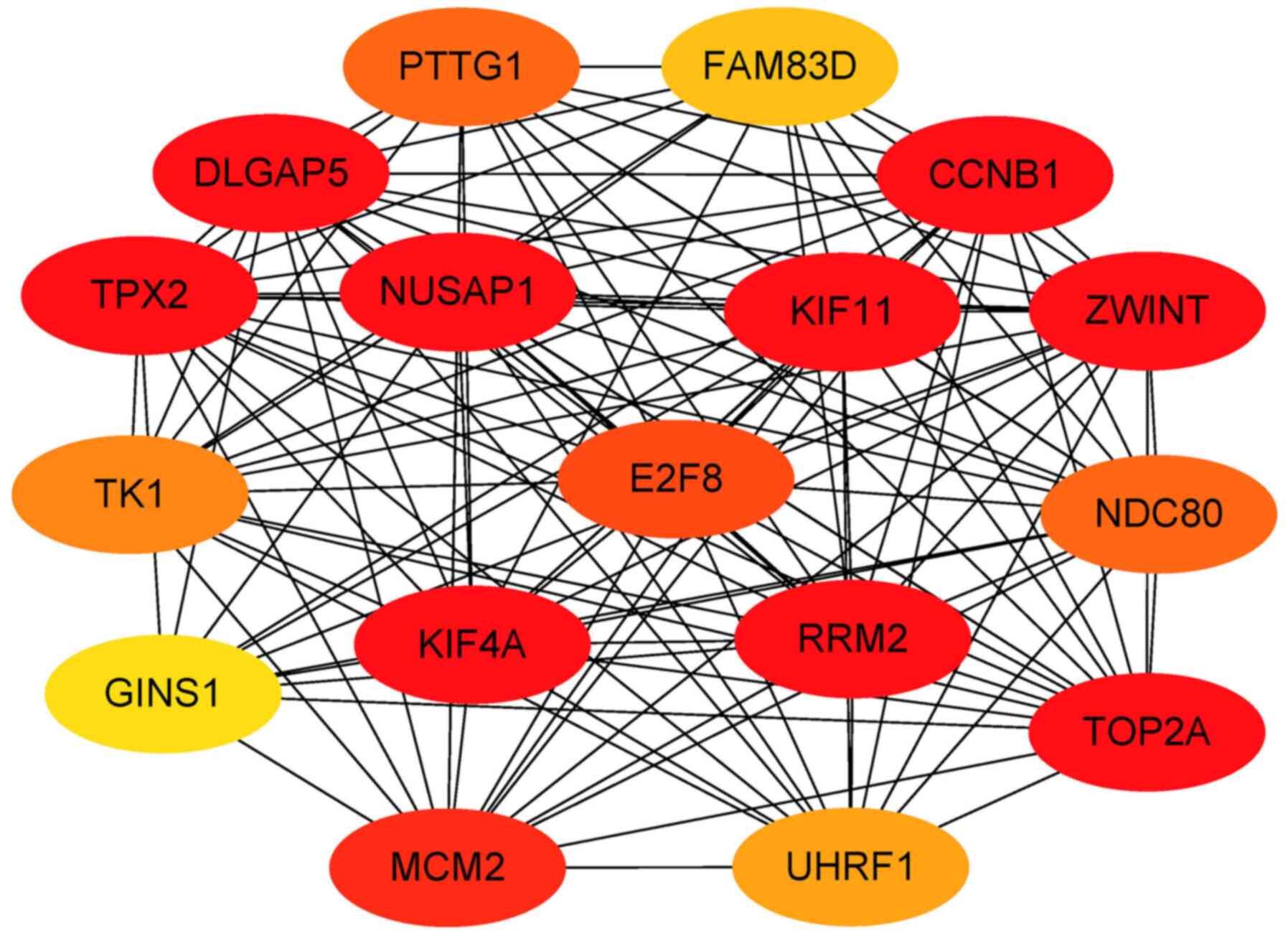
identify gene clusters. The genes in the highest-scoring gene
cluster were defined as hub genes. The results of all gene clusters
are presented in Table VI. A total
of 17 hub genes were screened out for further analysis. Those genes
are most likely to be essential genes for IVIG treatment of KD. The
hub genes are presented in Table
VII and Fig. 7.
 | Table VIMCODE was used to determine the
protein-protein interaction network downloaded from the Search Tool
for the Retrieval of Interacting Genes and Proteins database to
further mine gene clusters. |
Table VI
MCODE was used to determine the
protein-protein interaction network downloaded from the Search Tool
for the Retrieval of Interacting Genes and Proteins database to
further mine gene clusters.
| Cluster no. | Score
(density) | Nodes (n) | Edges (n) |
|---|
| 1 | 15.75 | 17 | 126 |
| 2 | 10 | 10 | 45 |
| 3 | 5.684 | 20 | 54 |
| 4 | 4.121 | 34 | 68 |
| 5 | 3.333 | 4 | 5 |
| 6 | 3 | 3 | 3 |
| 7 | 3 | 3 | 3 |
 | Table VIIGenes in the highest-scoring gene
cluster (Cluster 1) processed by MCODE. |
Table VII
Genes in the highest-scoring gene
cluster (Cluster 1) processed by MCODE.
| Gene/node ID | MCODE score | MCODE cluster |
|---|
| ZWINT | 12.752 | Cluster 1 |
| RRM2 | 12.752 | Cluster 1 |
| KIF11 | 12.752 | Cluster 1 |
| KIF4A | 12.752 | Cluster 1 |
| CCNB1 | 12.752 | Cluster 1 |
| E2F8 | 12.752 | Cluster 1 |
| TPX2 | 12.752 | Cluster 1 |
| TOP2A | 12.752 | Cluster 1 |
| NUSAP1 | 12.752 | Cluster 1 |
| MCM2 | 12.752 | Cluster 1 |
| DLGAP5 | 12.752 | Cluster 1 |
| TK1 | 11.868 | Cluster 1 |
| UHRF1 | 11.868 | Cluster 1 |
| NDC80 | 11.868 | Cluster 1 |
| PTTG1 | 11.657 | Cluster 1 |
| GINS1 | 11.000 | Cluster 1 |
| FAM83D | 10.859 | Cluster 1 |
Integrated network analysis of
mRNA-miRNA interactions
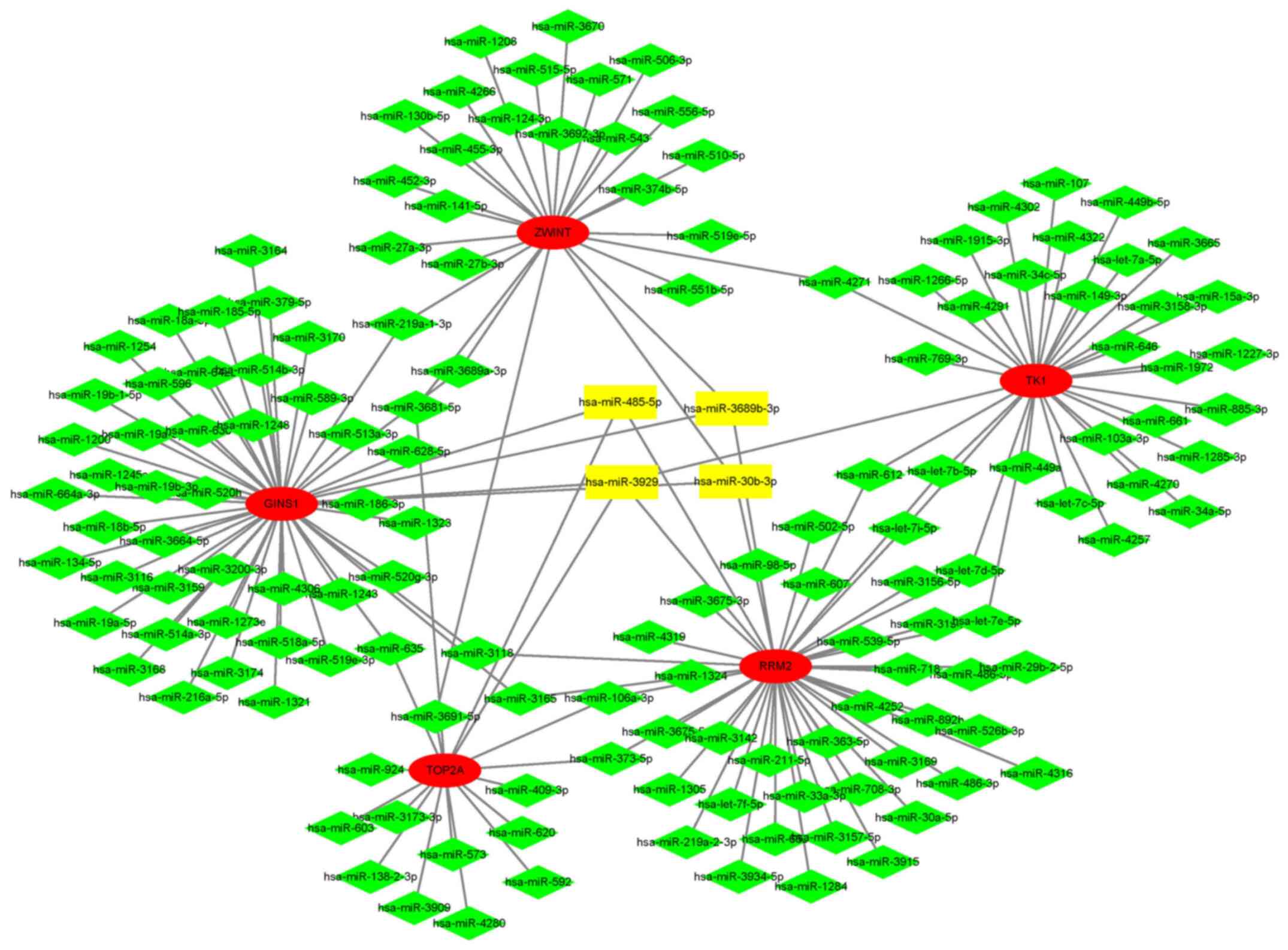
In total, 17 hub genes were uploaded to the online
miRWalk 2.0 database to predict the targeted miRNAs. The
intersection of miRNA results predicted using the TargetScan,
miRanda, miRWalk and RNA22 databases was defined as the prediction
result. The parameters were inputted as defaults. The miRNA-mRNA
interaction network was constructed and visualized with Cytoscape
software. A total of four miRNAs were screened out for further
verification, namely miR-3929, miR-30b-3p, miR-3689b-3p and
miR-485-5p. The results of the analysis and the interaction network
are presented in Table VIII and
Fig. 8. All four miRNAs targeted
ribonucleotide reductase regulatory subunit M2 and GINS complex
subunit 1 (GINS1).
 | Table VIIImiRNAs and their target hub
genes. |
Table VIII
miRNAs and their target hub
genes.
| miRNA | Genes targeted by
miRNA | Gene count |
|---|
| miR-3929 | RRM2, TK1, TOP2A,
GINS1 | 4 |
| miR-30b-3p | ZWINT, RRM2,
GINS1 | 3 |
| miR-3689b-3p | ZWINT, RRM2,
GINS1 | 3 |
| miR-485-5p | RRM2, TOP2A,
GINS1 | 3 |
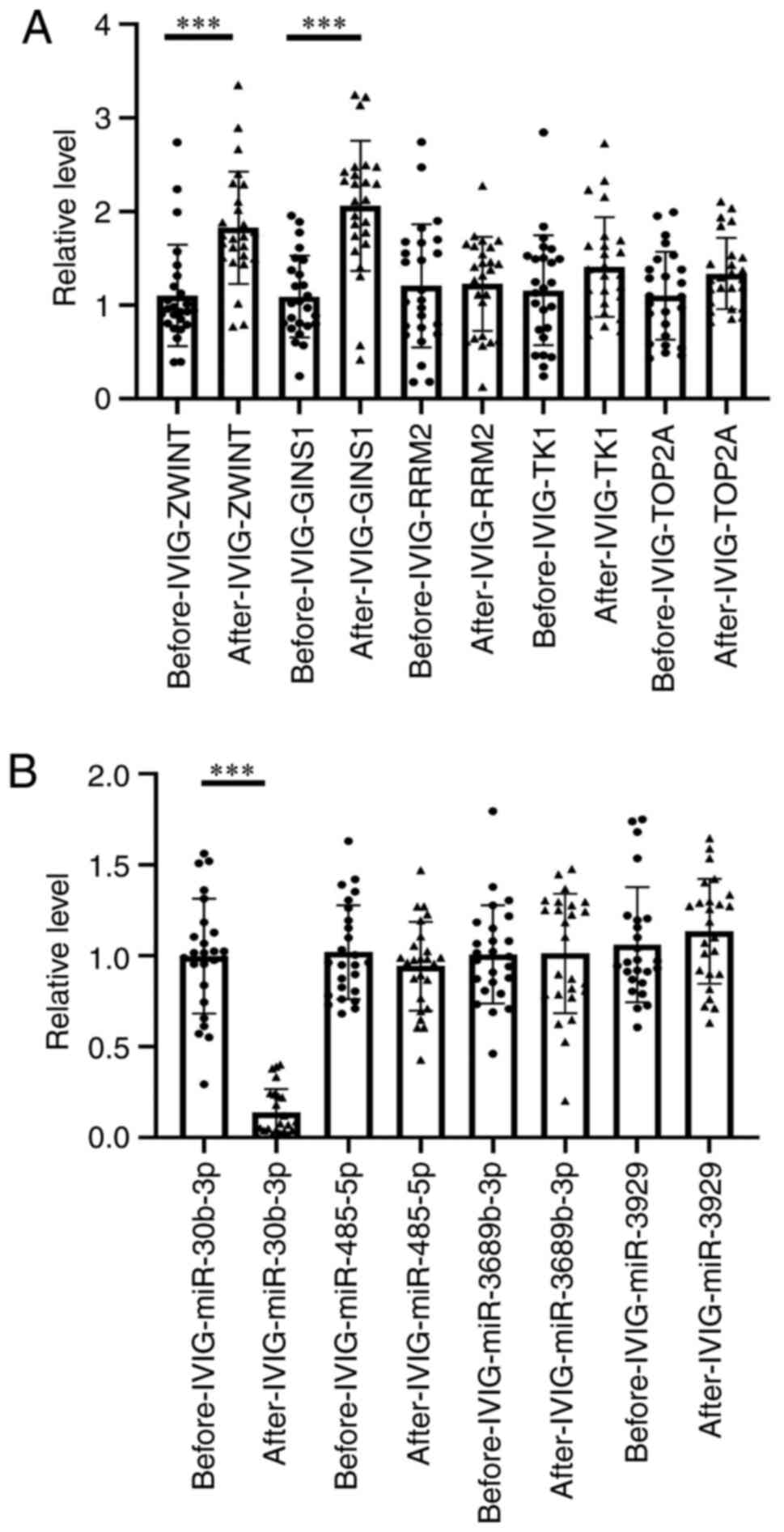
RT-qPCR validation
A total of five mRNAs and four miRNAs were detected
and compared in 50 patients with KD. All blood samples were
collected twice, namely prior to and 36 h after IVIG treatment. A
total of 50 (25 for mRNA and 25 for miRNA) samples were tested. Due
to the difficulty of collecting blood from children, one sample is
not adequate to detect miRNA and mRNA at the same time. The
expression levels of ZW10 interacting kinetochore protein (ZWINT),
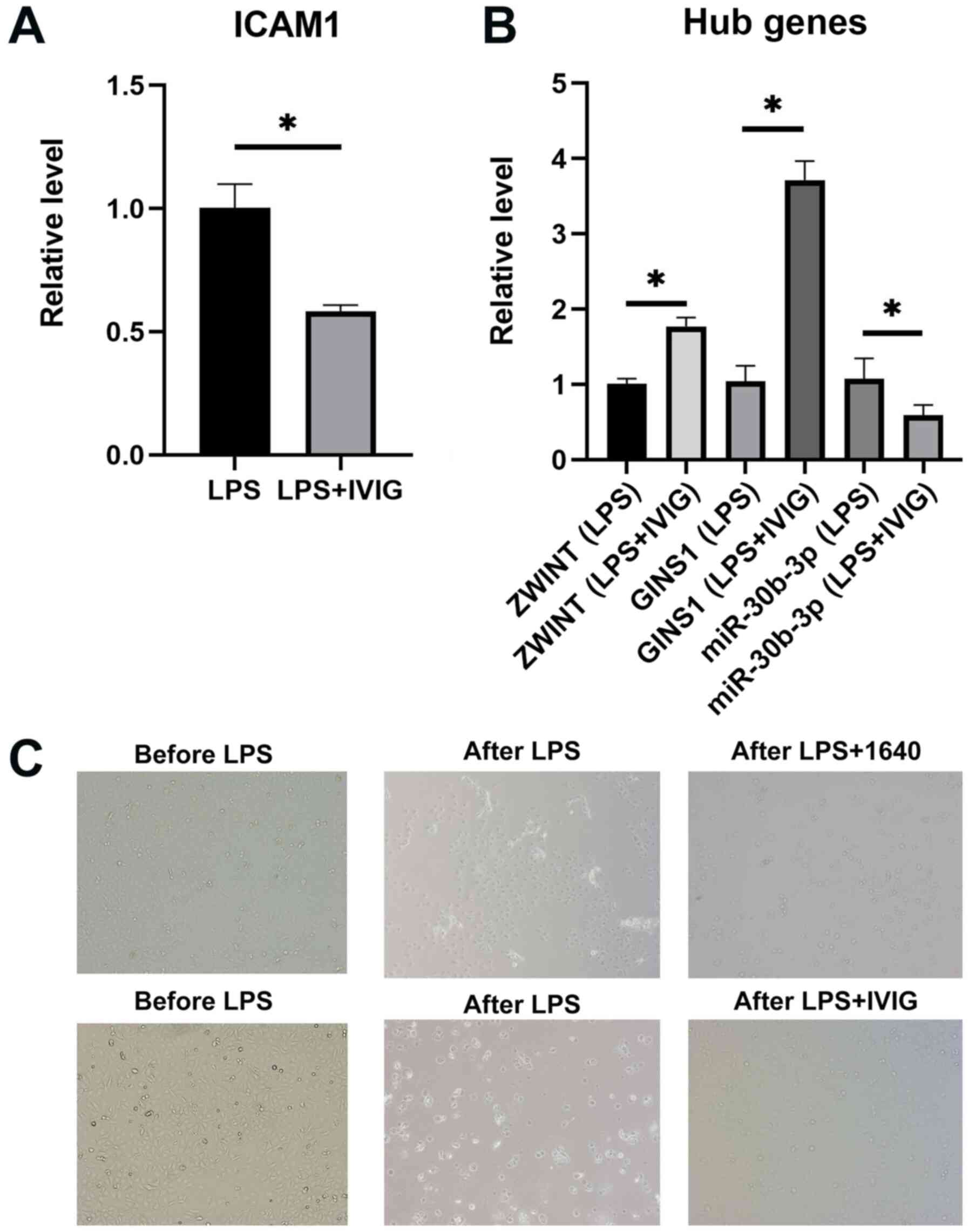
GINS1 and miR-30b-3p differed significantly post- vs. pre-IVIG
treatment among the patients with KD. As predicted, the relative
expression levels of ZWINT (P=0.00446) and GINS1 (P=0.00029) in the
pre-IVIG treatment group were lower than those in the post-IVIG
treatment group. Both ZWINT and GINS1 were targeted by miR-30b-3p,
whose expression level was higher in the pre-IVIG treatment group
as compared with that in the post-IVIG treatment group (P=0.00012)
(Fig. 9).
Cell experiment
The HCAECs were stimulated with LPS and underwent
IVIG treatment. ZWINT and GINS1 were upregulated, whereas
miR-30b-3p was significantly downregulated before and after IVIG
treatment in HCAEC. The expression level of ICAM1 was decreased
after IVIG treatment. Following LPS stimulation, certain HCAECs
exhibited apoptosis, which was assessed by observing the floating
cells in the medium. Following IVIG treatment, HCAECs had a faster
growth rate as compared with that in the control group (Fig. 10).
Discussion
KD is a form of vasculitis that frequently occurs in
childhood. IVIG, which may significantly reduce the incidence of
CAA in patients with KD, is used as initial therapy (24). In the present study, 335 DEGs and
253 DEPs were identified in patients with KD after vs. prior to
IVIG treatment. Enrichment analysis of GO terms and KEGG pathways
was performed for the selected DEGs and DEPs. Myeloid cell- and
neutrophil-related pathways have been indicated to have an
important role in IVIG treatment; these include myeloid
cell/neutrophil activation involved in the immune response, myeloid
leukocyte/neutrophil activation and myeloid
leukocyte/neutrophil-mediated immunity. In addition, through KEGG
enrichment pathway analysis, osteoclast differentiation and
hematopoietic cell lineage were determined for both DEGs and DEPs,
and it was suggested that cell differentiation may have a potential
key role in IVIG therapy. Similar to the results of the pathway
enrichment analysis, the changes in immune cells were analyzed
using xCell tools. A total of eight different types of cells were
screened out, including neutrophils and Th1 cells. The PPI and
mRNA-miRNA networks were used to screen out hub genes and their
target miRNAs of DEGs. Finally, the expression of ZWINT, GINS1 and
miR-30b-3p was confirmed to be significantly different between
clinical samples and HCAEC by using RT-qPCR analysis. These changes
in gene expression were consistent with the predicted results.
The PPI results were analyzed using MCODE in
Cytoscape. The gene cluster with the highest score in MCODE was
selected. The mRNA-miRNA network and RT-qPCR were used to identify
three genes, including two mRNAs and one miRNA. To the best of our
knowledge, the association among ZWINT, GINS1, miR-30b-3p and KD
has not been previously reported.
ZWINT encodes ZW10 interacting kinetochore protein,
which regulates centromere division (25). ZWINT has been reported to regulate
the cell proliferation cycle by regulating key mitotic nodes
(26). Chromosome instability is
also affected by ZWINT (27).
Therefore, the present study on ZWINT is mostly focused on tumors,
including breast cancer (25),
hepatocellular carcinoma (28) and
glioblastoma (29). Furthermore,
upregulation of ZWINT has a positive regulatory effect on the
antiviral signaling pathway (28).
Likewise, the relative expression levels of ZWINT were upregulated
in the present study. With the upregulation of ZWINT, the
proliferation of HCAEC was accelerated. Viral infections such as
coronavirus-19(30) and influenza
(31) are considered to be
associated with KD. Therefore, whether IVIG regulates the antiviral
signaling pathway through ZWINT requires further study.
GINS1 is also known as Go-Ichi-Ni-San complex
subunit 1 or PSF1. DNA replication is closely linked to GINS1 and
clinical studies have indicated that GINS1 deficiency may lead to
autoimmune disorders, such as neutrophil and NK-cell deficiency
(32). However, it has been
determined that GINS1 was upregulated and neutrophils were
decreased following IVIG treatment. The reason for this may that
GINS1 does not directly act on neutrophils. It has been reported
that GINS1 may regulate cell proliferation and apoptosis. GINS1
knockdown was indicated to suppress proliferation and accelerate
apoptosis in vitro (33),
and increased expression of GINS1 may be associated with poor
prognosis for patients with cancer, such as breast cancer (34). In the present study, increased
expression of GINS1 was indicated to be associated with the
proliferation of HCAEC.
miRNAs are short non-coding RNAs associated with
cell development, proliferation and apoptosis (35). miR-30b-3p has been reported to be
involved in cell inflammation and proliferation through the NF-κB,
T-cell receptor (36) and AKT
(37) signaling pathways.
miR-30b-3p is upregulated in viral diseases, such as
respiratory-syncytial-virus-associated pediatric pneumonia
(36). With regard to the
regulation of tumor cell proliferation, the results vary among
different types of tumor. Overexpression of miR-30b-3p promoted
apoptosis in ovarian cancer cells (38) and its knockdown suppressed the
proliferation of glioma cells (37). The role of miR-30b-3p in prostate
cancer remains controversial and the results are also conflicting
(39). These differences in
miR-30b-3p quantification may be associated with tissue processing,
RNA isolation methods, miRNA quantification or miRNA normalization
techniques. The present results indicated that these three genes
(ZWINT, GINS1 and miR-30b-3p) have a role in regulating the cell
proliferation cycle and participating in antiviral immunity. At
present, research on IVIG-induced changes in cell proliferation and
apoptosis in KD is still lacking. Kato et al (40) reported that immunoglobulin regulated
lymphocyte proliferation by suppressing superantigens. According to
the present results, ZWINT, GINS1 and miR-30b-3p may be key
molecules for the further study of the effect of IVIG in the
treatment of KD, since they may affect cell proliferation,
apoptosis and virus-related immune response.
Based on the GO analysis, nine BPs and nine CCs were
indicated to be significantly different between DEGs and DEPs.
Myeloid leukocytes and neutrophils were involved in almost all BPs.
The regulation of the activation involved in immune responses of
those two cell types may be associated with the mechanisms of
action of IVIG in KD. Andreozzi et al (41) indicated that neutrophils were
significantly increased in patients with acute-phase KD, since
neutrophils in the reserve pool of the bone marrow were released
into the peripheral circulation. A 12-year clinical study also
confirmed that neutrophils were significantly increased in the
acute phase of KD (42). Following
IVIG treatment, neutrophils were frequently significantly decreased
(42). Data on changes in
neutrophils were consistent with these reports (41,42).
This indicated that neutrophils may have an important role in the
therapeutic mechanisms of IVIG in KD. In addition, several
predictive models for IVIG resistance in KD have attempted to
identify potential risk factors; the percentage of neutrophils,
such as the Kobayashi and Formosa score and certain
neutrophil-related ratios, such as the neutrophil-to-lymphocyte
ratio, are considered to be associated with IVIG resistance
(43). It has also been suggested
that the control of neutrophil activation and neutrophil-mediated
immune response may be associated with the mechanism of IVIG
therapy. Furthermore, the prolonged lifespan of activated
neutrophils and prolonged activation of neutrophils may be
associated with the pathogenesis of KD and increase the possibility
of IVIG resistance (44,45). However, the exact mechanism remains
elusive. The results of the GO pathway analysis indicated that hub
genes may be involved in regulating the cell proliferation cycle.
However, whether IVIG regulates the neutrophil activation cycle
requires further study.
Through the KEGG database, the osteoclast
differentiation and hematopoietic cell lineage pathway were
calculated using ClueGo in Cytoscape. Most of the studies on
osteoclast differentiation have focused on rheumatoid arthritis and
osteoporosis. Certain transcription factors, such as Jun
dimerization protein 2, are able to regulate both osteoclast and
neutrophil differentiation. Neutrophils may be linked to the
therapeutic mechanism of IVIG (46). A bioinformatics analysis of other
microarray data also indicated that osteoclast differentiation was
associated with KD (47).
Blood-cell development progresses from hematopoietic stem cells.
Several cell types, such as neutrophils, T cells, monocytes and
macrophages, are derived from hematopoietic cells and are
associated with the pathogenesis of KD. The results of immune
infiltration analysis of changes in macrophages and monocytes using
xCell were consistent with those reported by Sugitani et al
(48) and Koizumi et al
(49), who confirmed that
macrophages and monocytes were upregulated in the acute phase of KD
and downregulated following treatment. Similar to macrophages and
monocytes, the changing trend in platelets was also consistent with
previous studies and the degree of platelet increase was associated
with the level of immunoglobulin (50). The role of B cells in the
pathogenesis of KD remains elusive. Unlike in the present study, a
previous trial reported no changes in the acute and convalescent
B-cell subgroups (51). Leung et
al (52) reported that the
activated Th1 cells were reduced following IVIG treatment in KD.
Since only 12 patients were examined and the changing trends in
plasma cells came from microarray calculations, the present results
may have been accidental and should be confirmed in a large study.
A small number of studies on mast and MEP cells in KD are available
(53). According to the present
analysis of the KEGG pathways and immune cell infiltration, the
aforementioned eight types of cells may participate in the
mechanism of action of IVIG. Further exploration of the therapeutic
mechanisms of IVIG in those cells would be a worthwhile first step
for future studies.
The present study had several limitations. First,
patients with KD who did not use IVIG were not included as a
control group in the study. Due to ethical issues, it is not
possible to treat patients with KD without IVIG. Furthermore, the
number of samples included in the study was small, in particular
the number of samples for proteome analysis. Proteome analysis from
4 different patients was also another limitation of the present
study. The results require to be confirmed in a larger study.
Finally, certain conclusions came from bioinformatics analyses and
require further experimental verification.
In conclusion, in the present study, the mechanisms
of action of IVIG in the treatment of KD were analyzed using an
integrated bioinformatics analysis. The results indicated that IVIG
treatment specifically increased the levels of ZWINT and GINS1, and
markedly decreased those of miR-30b-3p, and these changes are
mainly linked to cell cycle regulation and virus-related immune
response. The activation of myeloid leukocytes, neutrophils,
monocytes and the M1 type of macrophages, as well as neutrophils
and platelets, which are regulated by IVIG, may have an important
role in KD. Furthermore, osteoclast differentiation and the
hematopoietic cell lineage targeted by IVIG also have a critical
role in KD, although these points still require further
research.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81971477,
81870365, 82070512 and 81970436), Jiangsu Provincial Medical Young
Talents (grant no. QNRC2016756), the Applied Foundational Research
of Medical and Health Care of Suzhou City (grant no. SYS2019086),
Suzhou Medical Key Discipline - Pediatric Cardiology (grant no.
Szxk201507), Key Medical Talents in Jiangsu Province (grant no.
ZDRCA2016049), Second cycle key subjects of maternal and child
health in Jiangsu Province (grant no. FXK201740) and Introduction
Project of Clinical Medical Expert Team in Suzhou City (grant no.
SZYJTD201805).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HH and LX performed the experiments, analyzed the
data, prepared figures and/or tables, and authored and wrote drafts
of the paper. YD performed the experiments and prepared figures
and/or tables. JQ and CH performed the experiments. XL and YT
prepared figures and/or tables. XL performed the experiments like
qRT-PCR, analyzed the data with the software of SPSS. YT prepared
figures with Photoshop and helped design and implement cell
experiments. GQ and HL conceived and designed the project and
revised the final draft. GQ and HL confirm the authenticity of all
the raw data. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the Children's Hospital of Soochow University Suzhou (Suzhou,
China; approval no. 2020CS075). Written informed consent was
obtained from the guardians of each participant.
Patient consent for publication
Written informed consent for publication was
obtained from the guardians of each participant.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
de Graeff N, Groot N, Ozen S, Eleftheriou
D, Avcin T, Bader-Meunier B, Dolezalova P, Feldman BM, Kone-Paut I,
Lahdenne P, et al: European consensus-based recommendations for the
diagnosis and treatment of Kawasaki disease - the SHARE initiative.
Rheumatology (Oxford). 58:672–682. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lo MS and Newburger JW: Role of
intravenous immunoglobulin in the treatment of Kawasaki disease.
Int J Rheum Dis. 21:64–69. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Furusho K, Kamiya T, Nakano H, Kiyosawa N,
Shinomiya K, Hayashidera T, Tamura T, Hirose O, Manabe Y, Yokoyama
T, et al: High-dose intravenous gammaglobulin for Kawasaki disease.
Lancet. 2:1055–1058. 1984.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Newburger JW, Takahashi M, Beiser AS,
Burns JC, Bastian J, Chung KJ, Colan SD, Duffy CE, Fulton DR, Glode
MP, et al: A single intravenous infusion of gamma globulin as
compared with four infusions in the treatment of acute Kawasaki
syndrome. N Engl J Med. 324:1633–1639. 1991.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sosa T, Brower L and Divanovic A:
Diagnosis and Management of Kawasaki Disease. JAMA Pediatr.
173:278–279. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rodriguez MM and Wagner-Weiner L:
Intravenous Immunoglobulin in Pediatric Rheumatology: When to Use
It and What Is the Evidence. Pediatr Ann. 46:e19–e24.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Inagaki M and Yamada K: Inhibitory effects
of high doses of intravenous γ-globulin on platelet interaction
with the vessel wall in Kawasaki disease. Acta Paediatr Jpn.
33:791–798. 1991.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kaneko K, Takahashi M, Yoshimura K, Kitao
T, Yamanouchi S, Kimata T and Tsuji S: Intravenous immunoglobulin
counteracts oxidative stress in Kawasaki disease. Pediatr Cardiol.
33:1086–1088. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
von Gunten S, Schaub A, Vogel M, Stadler
BM, Miescher S and Simon H-U: Immunologic and functional evidence
for anti-Siglec-9 autoantibodies in intravenous immunoglobulin
preparations. Blood. 108:4255–4259. 2006.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lau AC, Duong TT, Ito S and Yeung RS:
Intravenous immunoglobulin and salicylate differentially modulate
pathogenic processes leading to vascular damage in a model of
Kawasaki disease. Arthritis Rheum. 60:2131–2141. 2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Guo Y, Tian X, Wang X and Xiao Z: Adverse
Effects of Immunoglobulin Therapy. Front Immunol.
9(1299)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang G, Xu S, Zhang Z, Zhang Y, Wu Y, An
J, Lin J, Yuan Z, Shen L and Si T: Identification of Key Genes and
the Pathophysiology Associated With Major Depressive Disorder
Patients Based on Integrated Bioinformatics Analysis. Front
Psychiatry. 11(192)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ogihara Y, Ogata S, Nomoto K, Ebato T,
Sato K, Kokubo K, Kobayashi H and Ishii M: Transcriptional
regulation by infliximab therapy in Kawasaki disease patients with
immunoglobulin resistance. Pediatr Res. 76:287–293. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121.
2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3(e3)2004.PubMed/NCBI View Article : Google Scholar
|
|
16
|
McCrindle BW, Rowley AH, Newburger JW,
Burns JC, Bolger AF, Gewitz M, Baker AL, Jackson MA, Takahashi M,
Shah PB, et al: American Heart Association Rheumatic Fever,
Endocarditis, and Kawasaki Disease Committee of the Council on
Cardiovascular Disease in the Young; Council on Cardiovascular and
Stroke Nursing; Council on Cardiovascular Surgery and Anesthesia;
and Council on Epidemiology and Prevention: Diagnosis, treatment,
and long-term management of Kawasaki disease: A scientific
statement for health professionals from the American Heart
Association. Circulation. 135:e927–e999. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Aran D, Hu Z and Butte AJ: xCell:
Digitally portraying the tissue cellular heterogeneity landscape.
Genome Biol. 18:220. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45D:D362–D368. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4(2)2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yi XH, Zhang B, Fu YR and Yi ZJ: STAT1 and
its related molecules as potential biomarkers in Mycobacterium
tuberculosis infection. J Cell Mol Med. 24:2866–2878.
2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods.
25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Uthman L, Kuschma M, Römer G, Boomsma M,
Kessler J, Hermanides J, Hollmann MW, Preckel B, Zuurbier CJ and
Weber NC: Novel Anti-inflammatory Effects of Canagliflozin
Involving Hexokinase II in Lipopolysaccharide-Stimulated Human
Coronary Artery Endothelial Cells. Cardiovasc Drugs Ther: Oct 13,
2020 (Epub ahead of print). doi: 10.1007/s10557-020-07083-w.
|
|
23
|
Armaroli G, Verweyen E, Pretzer C, Kessel
K, Hirono K, Ichida F, Okabe M, Cabral DA, Foell D, Brown KL, et
al: Monocyte-Derived Interleukin-1β As the Driver of
S100A12-Induced Sterile Inflammatory Activation of Human Coronary
Artery Endothelial Cells: Implications for the Pathogenesis of
Kawasaki Disease. Arthritis Rheumatol. 71:792–804. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen S, Dong Y, Kiuchi MG, Wang J, Li R,
Ling Z, Zhou T, Wang Z, Martinek M, Pürerfellner H, et al: Coronary
artery complication in Kawasaki disease and the importance of early
intervention: A systematic review and meta-analysis. JAMA Pediatr.
170:1156–1163. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Shao M-T, Hu Y-Z, Ding H, Wu Q, Pan J-H,
Zhao X-X and Pan Y-L: The overexpression of ZWINT in integrated
bioinformatics analysis forecasts poor prognosis in breast cancer.
Transl Cancer Res. 9:187–193. 2020.
|
|
26
|
Dou Z, Prifti DK, Gui P, Liu X, Elowe S
and Yao X: Recent progress on the localization of the spindle
assembly checkpoint machinery to kinetochores. Cells.
8(278)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Vargas-Rondón N, Villegas VE and
Rondón-Lagos M: The role of chromosomal instability in cancer and
therapeutic responses. Cancers (Basel). 10(4)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhu Z, Huang S, Zhang Y, Sun C, Tang Y,
Zhao Q, Zhou Q, Ju W and He X: Bioinformatics analysis on multiple
Gene Expression Omnibus datasets of the hepatitis B virus infection
and its response to the interferon-alpha therapy. BMC Infect Dis.
20(84)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yang L, Han N, Zhang X, Zhou Y, Chen R and
Zhang M: ZWINT: A potential therapeutic biomarker in patients with
glioblastoma correlates with cell proliferation and invasion. Oncol
Rep. 43:1831–1844. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jones VG, Mills M, Suarez D, Hogan CA, Yeh
D, Segal JB, Nguyen EL, Barsh GR, Maskatia S and Mathew R: COVID-19
and Kawasaki disease: Novel virus and novel case. Hosp Pediatr.
10:537–540. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Rowley AH and Shulman ST: The epidemiology
and pathogenesis of Kawasaki disease. Front Pediatr.
6(374)2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Cottineau J, Kottemann MC, Lach FP, Kang
YH, Vély F, Deenick EK, Lazarov T, Gineau L, Wang Y, Farina A, et
al: Inherited GINS1 deficiency underlies growth retardation along
with neutropenia and NK cell deficiency. J Clin Invest.
127:1991–2006. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tang L, Yu W, Wang Y, Li H and Shen Z:
Anlotinib inhibits synovial sarcoma by targeting GINS1: A novel
downstream target oncogene in progression of synovial sarcoma. Clin
Transl Oncol. 21:1624–1633. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fu Y, Zhou QZ, Zhang XL, Wang ZZ and Wang
P: Identification of Hub Genes Using Co-Expression Network Analysis
in Breast Cancer as a Tool to Predict Different Stages. Med Sci
Monit. 25:8873–8890. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang X, Huang F, Yang D, Peng T and Lu G:
Identification of miRNA-mRNA Crosstalk in Respiratory Syncytial
Virus- (RSV-) Associated Pediatric Pneumonia through Integrated
miRNAome and Transcriptome Analysis. Mediators Inflamm.
2020(8919534)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jian Y, Xu CH, Li YP, Tang B, Xie SH and
Zeng EM: Down-regulated microRNA-30b-3p inhibits proliferation,
invasion and migration of glioma cells via inactivation of the AKT
signaling pathway by up-regulating RECK. Biosci Rep.
39(BSR20182226)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li Y, Zhou J, Wang J, Chen X, Zhu Y and
Chen Y: miR-30b-3p affects the migration and invasion function of
ovarian cancer cells by targeting the CTHRC1 gene. Biol Res.
53(10)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kumar B, Khaleghzadegan S, Mears B, Hatano
K, Kudrolli TA, Chowdhury WH, Yeater DB, Ewing CM, Luo J, Isaacs
WB, et al: Identification of miR-30b-3p and miR-30d-5p as direct
regulators of androgen receptor signaling in prostate cancer by
complementary functional microRNA library screening. Oncotarget.
7:72593–72607. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kato K, Sakamoto T and Ito K:
Gamma-globulin inhibits superantigen-induced lymphocyte
proliferation and cytokine production. Allergol Int. 56:439–444.
2007.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Andreozzi L, Bracci B, D'Errico F and
Rigante D: A master role for neutrophils in Kawasaki syndrome.
Immunol Lett. 184:112–114. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sun L, Tang Y, Wang Y, Qian G, Yan W, Wang
B, Li X and Lv H: Changes in Profiles of Kawasaki Disease Noted
over Time in Suzhou, China. Cardiology. 141:25–31. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wu G, Yue P, Ma F, Zhang Y, Zheng X and Li
Y: Neutrophil-to-lymphocyte ratio as a biomarker for predicting the
intravenous immunoglobulin-resistant Kawasaki disease. Medicine
(Baltimore). 99(e18535)2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Tsujimoto H, Takeshita S, Nakatani K,
Kawamura Y, Tokutomi T and Sekine I: Delayed apoptosis of
circulating neutrophils in Kawasaki disease. Clin Exp Immunol.
126:355–364. 2001.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Inamo Y, Harada K, Okuni M, Kimoto K,
Takeuchi S and Sakurabayashi I: Immunoreactive polymorphonuclear
leukocyte elastase in complex with alpha 1-antitrypsin in Kawasaki
disease. Acta Paediatr Jpn. 29:202–205. 1987.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Maruyama K, Fukasaka M, Vandenbon A,
Saitoh T, Kawasaki T, Kondo T, Yokoyama KK, Kidoya H, Takakura N,
Standley D, et al: The transcription factor Jdp2 controls bone
homeostasis and antibacterial immunity by regulating osteoclast and
neutrophil differentiation. Immunity. 37:1024–1036. 2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Chang D, Qian C, Li H and Feng H:
Comprehensive analyses of DNA methylation and gene expression
profiles of Kawasaki disease. J Cell Biochem. 120:13001–13011.
2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Sugitani Y, Furuno K, Sueishi K and Hara
T: Macrophages and cytotoxic T cells infiltrate the destructed
mitral tissue in Kawasaki disease. BMJ Case Rep: Feb 22.
2018(bcr2017223584)2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Koizumi K, Hoshiai M, Moriguchi T,
Katsumata N, Toda T, Kise H, Hasebe Y, Kono Y, Sunaga Y, Yoshizawa
M, et al: Plasma Exchange Downregulates Activated Monocytes and
Restores Regulatory T Cells in Kawasaki Disease. Ther Apher Dial.
23:92–98. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Han JW, Oh JH, Rhim JW and Lee KY:
Correlation between elevated platelet count and immunoglobulin
levels in the early convalescent stage of Kawasaki disease.
Medicine (Baltimore). 96(e7583)2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Ikeda K, Yamaguchi K, Tanaka T, Mizuno Y,
Hijikata A, Ohara O, Takada H, Kusuhara K and Hara T: Unique
activation status of peripheral blood mononuclear cells at acute
phase of Kawasaki disease. Clin Exp Immunol. 160:246–255.
2010.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Leung DY, Burns JC, Newburger JW and Geha
RS: Reversal of lymphocyte activation in vivo in the Kawasaki
syndrome by intravenous gammaglobulin. J Clin Invest. 79:468–472.
1987.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ross R and Conti P: COVID-19 induced by
SARS-CoV-2 causes Kawasaki-like disease in children: Role of
pro-inflammatory and anti-inflammatory cytokines. J Biol Regul
Homeost Agents. 34:767–773. 2020.PubMed/NCBI View Article : Google Scholar
|