Introduction
Lung cancer ranks first in incidence and mortality
among malignant tumors worldwide. The incidence of malignant lung
tumors has increased over the last decades and this type of cancer
is responsible for >1.3 million deaths worldwide annually
(1). Lung tumors are one of the
most frequent malignant tumors types in China (2,3). Lung
cancer is mainly divided into non-small cell lung cancer (NSCLC)
and small cell lung cancer, with NSCLC accounting for ~80% of all
cases (4). The two main subtypes of
NSCLC are lung adenocarcinoma and lung squamous cell carcinoma
(LUSC), and LUSC is insidious and develops rapidly (5). A subset of patients with LUSC do not
have the opportunity to receive radical surgery and, consequently,
the survival rate of patients is low (6). It has been shown that <5% of
patients survive 5 years following chemotherapy treatment (7). Therefore, the identification of new
and effective treatments for this disease is crucial.
Basic calponin (CNN) or CNN1 (also referred to as
calmodulin 1) is a marker for the differentiation of cardiac and
smooth muscle (8). It is one of
three subtypes of CNN and is encoded by a gene on human chromosome
19 (19p13.2-p13.1) (8). The
expression levels of CNN1 are abnormal under various pathological
conditions, such as abnormal gastrointestinal motility (9) and hypoxia (10). In addition, a number of studies have
shown that CNN1 is expressed at low levels in a variety of tumor
tissue types, such as malignant melanoma (11), hepatocellular carcinoma (12), ovarian cancer (13) and breast cancer (14). However, to the best of our
knowledge, the association between CNN1 and LUSC has not been
reported previously. In the present study, the effects of CNN1 on
the invasive and migratory abilities of LUSC cells were
investigated.
The Wnt/β-catenin signaling pathway is involved in
various crucial cellular functions, such as stem cell regeneration
and organogenesis (15). Wnt
activation has been observed in various types of malignant tumor,
including in the breast, lung and hematopoietic system, and has
been shown to contribute to tumor recurrence (16). In NSCLC, it has been reported that
targeting the negative regulators of Wnt signaling for degradation
to increase β-catenin-mediated Wnt activity may lead to the
maintenance of lung cancer ‘stemness’ (17). The activation of the Wnt/β-catenin
signaling pathway promotes tumor growth, metastasis and
epithelial-to-mesenchymal transition (EMT) of NSCLC cells (18). Dickkopf-1 (DKK1) is a member of the
DKK protein family, which is a secretory protein that acts as an
inhibitor of the extracellular Wnt signal transduction pathway
(19). c-myc is one of the target
genes of Wnt/β-catenin pathway and can be activated by the
transcription factor, β-catenin, which enters the nucleus to
regulate target genes expression upon being stabilized by Wnt
binding (20). Notably, c-myc
functions as a critical oncogene and has been shown to be
implicated in enhancing the aggressiveness of various cancer types,
including lung cancer (21). The
present study aimed to investigate whether the
DKK1/Wnt/β-catenin/c-myc signaling pathway participated in the
effect of CNN1 on LUSC.
Materials and methods
Cell culture and transfection
The LUSC cell lines NCI-H520, SK-MES-1 and NCI-H2170
were obtained from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. All cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) and maintained at 37˚C and
5% CO2. Overexpression (OE) vectors for CNN1 and
knockdown vectors for DKK1 and tissue inhibitor of
metalloproteinases 2 (TIMP2) [OE-negative control (NC; empty
pcDNA3.1), OE-CNN1 (pcDNA3.1-CNN1), short hairpin RNA (shRNA)-NC,
shRNA-TIMP2 and shRNA-DKK1] were purchased from Biomics
Biotechnologies. Cell transfection in NCI-H2170 cells was performed
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Briefly, Lipofectamine 2000 was mixed with 20 µg plasmids, which
was then added to the cells at 70-80% confluence and incubated for
6 h at 37˚C. The transfection efficiency in cells was measured
using reverse transcription-quantitative PCR (RT-qPCR). At 48 h
post-transfection, the transfection efficacy was validated using
RT-qPCR and successfully transfected cells were selected for
subsequent experiments.
Dynamic analysis of GEPIA gene
expression profile data
GEPIA (http://gepia.cancer-pku.Cn/index.html) is a website
for cancer data mining. Using RNA sequencing data for CNN1 and
TIMP2 from tumor and normal samples, the database was used to
analyze the expression levels and association between CNN1 and
TIMP2 in tumor samples.
RT-qPCR
Total RNA was extracted using TRIzol®
(Invitrogen: Thermo Fisher Scientific, Inc.). The mRNA sequence was
reversed-transcribed into cDNA using a reverse transcriptase
(HiScript II Reverse Transcriptase; Vazyme Biotech Co., Ltd.) and
the reaction conditions were set as follows: 42˚C for ~1 h and 90˚C
for 5 min. The expression levels of CNN1, MMP2, MMP9, DKK1 and
TIMP2 were subsequently detected using qPCR by SYBR-Green ROX-mix
(ChamQ SYBR qPCR Master Mix; Vazyme Biotech Co., Ltd.). The PCR
reaction procedure was as follows: 5 min at 95˚C, with 40 cycles of
30 sec at 95˚C and 45 sec at 65˚C. Expression levels of target
genes were analyzed using the 2-ΔΔCq method (22) after being normalized to the
expression levels of GAPDH. The primer sequences used were as
follows: MMP2 forward, 5'-GATACCCCTTTGACGGTAAGGA-3' and reverse,
5'-CCTTCTCCCAAGGTCCATAGC-3'; MMP9 forward,
5'-GGGACGCAGACATCGTCATC-3' and reverse, 5'-TCGTCATCGTCGAAATGGGC-3';
CNN1 forward, 5'-TGAAGAAGATCAATGAGTCAACC-3' and reverse,
5'-CGTTCACCTTGTTTCCTTTCG-3'; DKK1 forward,
5'-GAAGAGTGTTAAAGGTTTTTTTTTATGTAT-3' and reverse,
5'-CCAAAATCCTAACTACAAAAAACACA-3'; TIMP2 forward,
5'-CTCTGATTTGGTCGTATTGGG-3' and reverse,
5'-TGGAAGATGGTGATGGGATT-3'; and GAPDH forward,
5'-GCAACCGGGAAGGAAATGAATG-3' and reverse,
5'-CCCAATACGACCAAATCAGAGA-3'.
Western blotting
The cells were harvested and total protein was
extracted using RIPA lysis buffer (Beyotime Institute of
Biotechnology). Protease inhibitors were added (Beyotime Institute
of Biotechnology) to the lysis buffer (1:100). The lysates were
centrifuged at 4˚C, 850 x g for 15 min. The supernatant was
collected and mixed with a loading buffer (Beyotime Institute of
Biotechnology) containing 100 mM dithiothreitol. Total protein was
quantified using a Protein Concentration Determination BCA kit
(Beyotime Institute of Biotechnology) and the proteins (30 µg/lane)
were separated via SDS-PAGE (15%). The separated proteins were
subsequently transferred onto PVDF membranes (EMD Millipore) and
blocked using 5% BSA (Beyotime Institute of Biotechnology) at room
temperature for 2 h. The membranes were incubated with the
following primary antibodies at 4˚C overnight: Anti-CNN1 (cat. no.
ab46794; 1:1,000 dilution), anti-MMP2 (cat. no. ab92536; 1:1,000
dilution), anti-MMP9 (cat. no. ab76003; 1:1,000 dilution),
anti-E-cadherin (E-cad; cat. no. ab40772; 1:1,000 dilution),
anti-N-cadherin (N-cad; cat. no. ab245117; 1:1,000 dilution),
anti-SLUG (cat. no. ab51772; 1:1,000 dilution), anti-DKK1 (cat. no.
ab109416; 1:1,000 dilution), anti-β-catenin (cat. no. ab32572;
1:1,000 dilution), anti-c-myc (cat. no. ab32072; 1:1,000 dilution)
and anti-GAPDH (cat. no. ab8245; 1:10,000 dilution). All antibodies
were purchased from Abcam. Following the primary incubation, the
membranes were washed with TBS containing Tween-20 (0.1%) and
incubated at room temperature for 1.5 h with a horseradish
peroxidase-conjugated donkey anti-rabbit IgG secondary antibody
(cat. no. SA00001-9; 1:5,000 dilution; ProteinTech Group, Inc.) or
a donkey anti-mouse IgG secondary antibody (SA00001-8; 1:5,000
dilution; ProteinTech Group, Inc.). The protein bands were
visualized using the Odyssey Western Blot Analysis system (LI-COR
Biosciences) and quantified using Image J version 7.6.5 (National
Institutes of Health).
Wound healing and Transwell
assays
NCI-H2170 cells were cultured in six-well plates
(6x104 cells/well) and transiently transfected with the
aforementioned specific plasmids. Following 24 h of transfection
(37˚C) the cells were ~100% confluent, and a linear scratch was
created in the cell monolayer using a 200-µl pipette tip. The cells
were subsequently cultured under standard conditions in serum-free
DMEM medium (Procell Life Science and Technology Co., Ltd.) at 37˚C
in the presence of 5% CO2. Image J software (version
1.8.0; National Institutes of Health) was used to calculate the
wound width. Images of the wound were captured at 24 h using a
light microscope (Nikon Corporation; magnification x100). The
migration distance was calculated as the width of the scratch at 24
h minus the width of the scratch at 0 h. The relative migration
rate was calculated by normalizing to the control group.
The invasive ability of the cells was observed using
a Transwell invasion assay. Following 48 h of treatment, the cells
were collected for detection. A total of 200 µl cell suspension in
serum-free DMEM containing 5x105 cells were inoculated
in the upper Transwell chamber which was coated with Matrigel at
37˚C for 30 min (Corning, Inc.), and cultured at 37˚C in the
presence of 5% CO2 for 12 h. DMEM medium containing 10%
FBS was filled into the lower chamber for 24 h. Subsequently, 4%
paraformaldehyde was added for 0.5 h for fixation at room
temperature and the cells were stained with 0.1% crystal violet for
30 min at 37˚C. The number of cells passing through the membrane in
three random fields was observed under a light microscope with a
100x magnification.
Immunofluorescence
The cells were fixed with 4˚C 4% polyphosphate
formaldehyde for 14 min and permeabilization with 0.1% Triton X-100
(Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) for 30 min at
room temperature. This process (permeabilization) was repeated 3
times and the cells were blocked with 10% goat serum (Beijing
Zhongshan Jinqiao Biotechnology Co., Ltd.) for 30 min at room
temperature. Following incubation overnight at 4˚C with primary
antibodies (anti-N-cad; cat. no. ab245117; Abcam; 1:200 dilution),
FITC-labeled secondary fluorescent antibodies (goat anti-rabbit
IgG; cat. no. ab6717; Abcam; 1:200 dilution) were added to the
cells. Following incubation for 0.5 h at 37˚C, DAPI was added in
the dark and the film was sealed and observed using a fluorescence
microscope (magnification, x200). Three random fields per sample
were analyzed.
Statistical analysis
All experiments were repeated three times.
Statistical analysis was performed using GraphPad Prism 5 software
(GraphPad Software, Inc.) and all data are presented as mean ± SEM,
unless otherwise specified. Statistical differences between two
groups were determined using an unpaired two-tailed Student's
t-test, while a one-way ANOVA followed by Tukey's post hoc test
were used to analyze data corresponding to more than two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Overexpression of CNN1 inhibits cell
migration, invasion and EMT
Initially, the GEPIA2 database was used to explore
the expression of CNN1 in lung cancer tissues. The results
indicated that the expression levels of CNN1 were decreased in LUSC
tissues (Fig. 1A). RT-qPCR and
western blot analyses were used to detect the expression levels of
CNN1 in the following LUSC cell lines: 16HBE, NCI-H520, SK-MES-1
and NCI-H2170 (Fig. 1B-D). The
results indicated that the expression levels of CNN1 were the
lowest in NCI-H2170 cells. Therefore, these cells were selected for
the following experiments. Following successful overexpression of
CNN1 (Fig. 1E), cell migratory
(Fig. 1F) and invasive (Fig. 1G) rate, protein expression of MMP2
and MMP9 (Fig. 1H) along with N-cad
and Slug expression levels (Fig. 1I
and J) were decreased, but E-cad
was increased, indicating the inhibitory effect of CNN1
overexpression on cell migration, invasion and EMT.
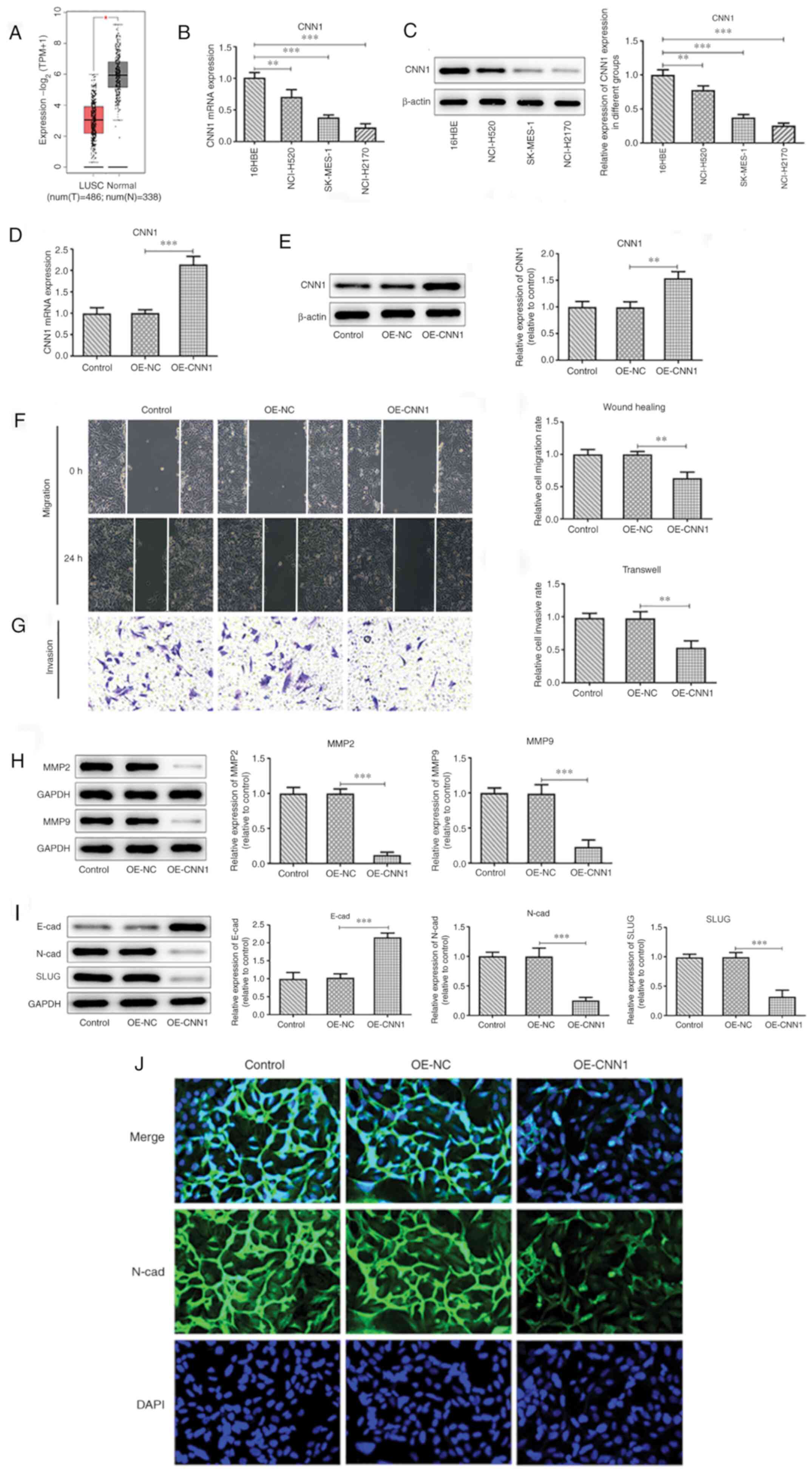 | Figure 1Overexpression of CNN1 inhibits the
migration, invasion and EMT of NCL-H2170 cells. (A) GEPIA database
predicted the decreased expression of CNN1 in LUSC. (B) CNN1
expression levels in cells were detected using RT-qPCR. (C) CNN1
protein expression levels in the various cell lines were detected
using western blotting. (D) Protein expression was quantified using
Image J. (E) CNN1 expression levels in NCL-H2170 cells before and
after CNN1 overexpression were detected using western blotting and
RT-qPCR. (F) Wound healing assays were conducted to evaluate the
migratory capacity of NCL-H2170 cells (magnification, x100). (G)
Transwell assays were performed to assess the invasive capacity of
NCL-H2170 cells (magnification, x100). (H) Western blot analysis
was used to detect migration-associated proteins in NCL-H2170
cells. (I) Western blot analysis was used to detect the levels of
EMT-associated proteins in NCL-H2170 cells. (J) Immunofluorescence
analysis was used to detect the expression levels of N-cad
(magnification, x200) in NCL-H2170 cells. *P<0.05,
**P<0.01, ***P<0.001. All experiments
were repeated three times. CNN1, calponin 1; E-cad; E-cadherin;
EMT, epithelial-to-mesenchymal transition; LUSC, lung squamous cell
carcinoma; N, normal; N-cad, N-cadherin; NC, negative control; OE,
overexpression; RT-qPCR, reverse-transcription quantitative PCR;
SLUG, zinc finger protein SNAI2; T, tumor; TPM, transcripts per
million. |
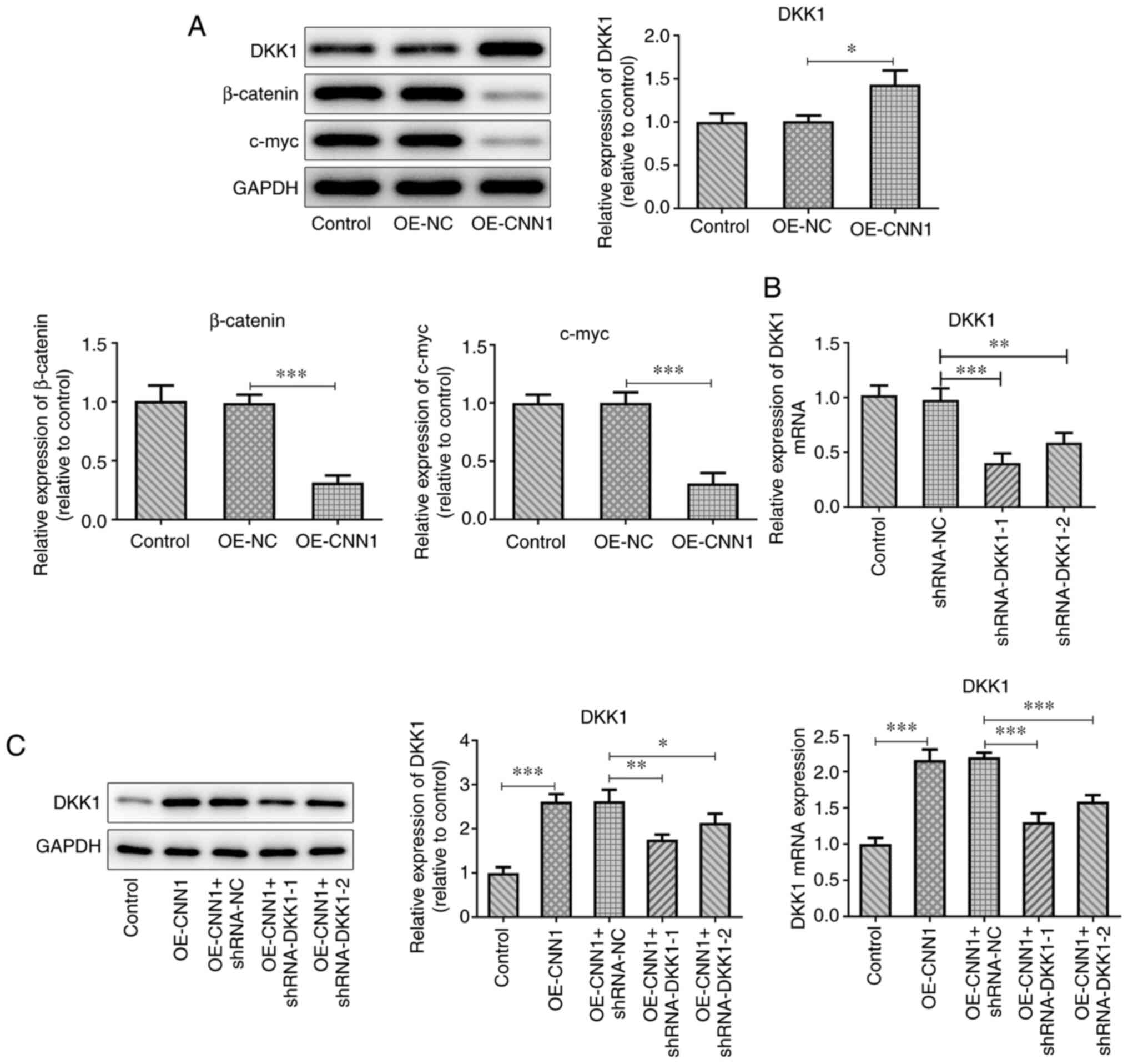
Overexpression of CNN1 inhibits the
expression levels of Wnt/β-catenin signaling proteins
The expression levels of DKK1 were upregulated, and
those of β-catenin and c-myc were downregulated, compared with
those of the OE-NC group following overexpression of CNN1 (Fig. 2A). An shRNA interference plasmid
targeting DKK1 was constructed and the transfection efficacy was
confirmed using RT-qPCR (Fig. 2B).
The expression levels of DKK1 in shRNA-DKK1 + OE-CNN1
co-transfected cells were detected using RT-qPCR and western blot
analyses (Fig. 2C). The mRNA and
protein expression levels of CNN1 in the OE-CNN1 + shRNA-DKK1-1
group were lower than those of the OE-CNN1 + shRNA-DKK1-2 group.
Therefore, the DKK1-1 group was selected for further
experiments.
Interference of DKK1 blocks CNN1
expression and inhibits the migration, invasion and EMT of lung
cancer cells
The scratch width, as determined using wound healing
assays, was reduced following interference of DKK1 expression
compared with that of the plasmid control group (Fig. 3A and C). Corresponding results were observed in
the Transwell experiment; interference with DKK1 expression
attenuated the ability of increased CNN1 expression levels to
inhibit cell invasion (Fig. 3B and
C). Following interference with
DKK1, the expression levels of MMP2 and MMP9 were increased
compared with the OE-CNN1 + shRNA-NC group, as demonstrated using
western blot analysis (Fig. 3D).
This further suggested that the migratory and invasive ability of
the cells in the OE-CNN1 + shRNA-DKK1-1 group was enhanced. The
inhibitory effects of CNN1 on the EMT process of lung cancer cells
also appeared to be blocked.
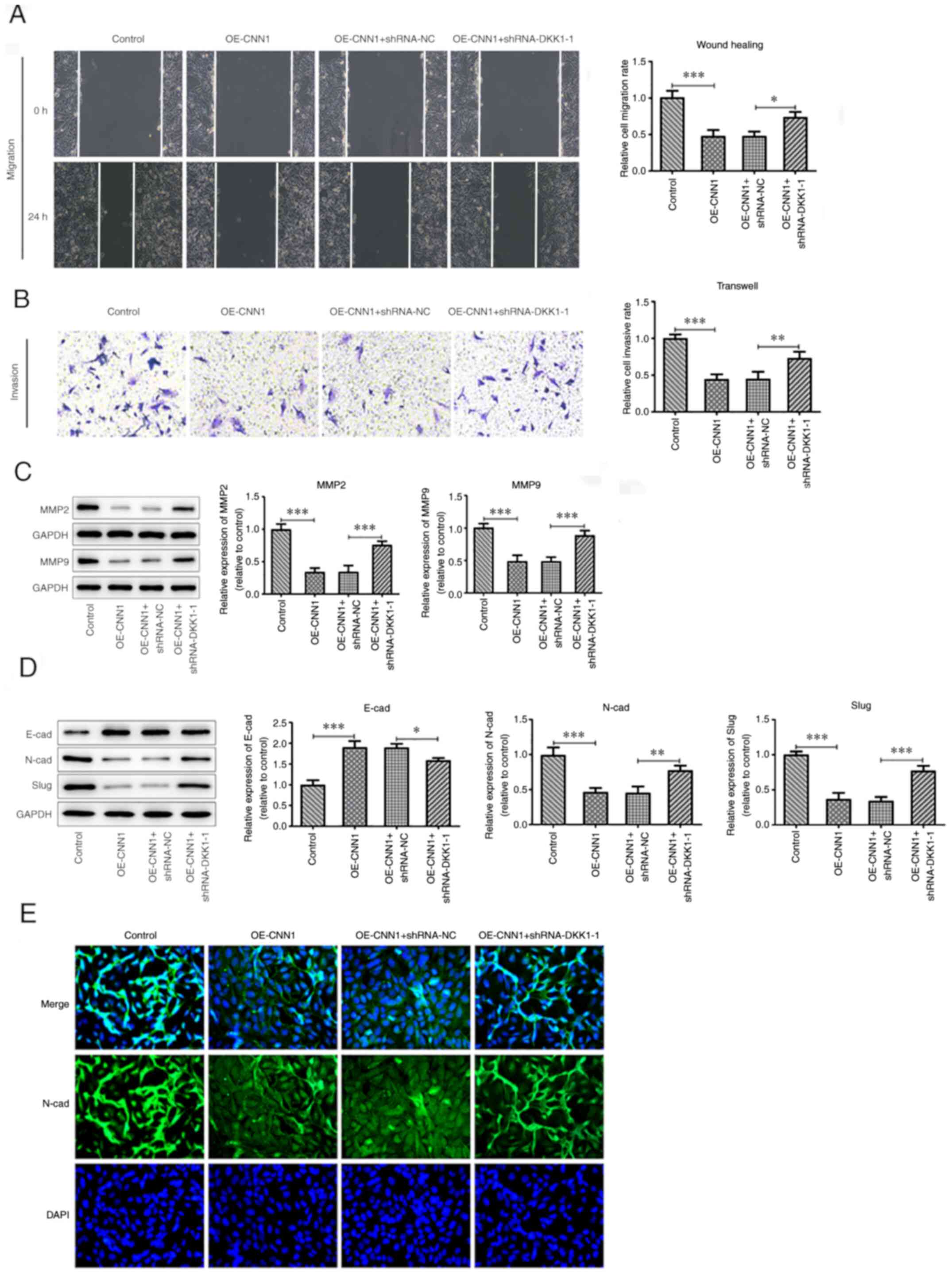 | Figure 3Interference of DKK1 inhibits the
migration, invasion and EMT of NCL-H2170 cells. (A) Wound healing
assays were conducted to evaluate the migratory capacity of
NCL-H2170 cells (magnification, x100). (B) Transwell assays were
performed to assess the invasive capacity of NCL-H2170 cells
(magni-fication, x100). (C) MMP2 and MMP9 protein expression levels
were detected using western blot analysis. (D) EMT-associated
protein expression was detected by western blotting. (E)
Immunofluorescence analysis was used to detect the expression
levels of N-cad (magnification, x200). *P<0.05,
**P<0.01, ***P<0.001. The experiments
were repeated three times. CNN1, calponin 1; DKK1, Dickkopf-1; EMT,
epithelial-to-mes-enchymal transition; N-cad, N-cadherin; NC,
negative control; OE, overexpression; shRNA, short hairpin RNA;
SLUG, zinc finger protein SNAI2. |
The expression levels of N-cad and SLUG along with
N-cad fluorescence were increased in the OE-CNN1 + shRNA-DKK1-1
group (Fig. 3E), whereas E-cad
expression was decreased (Fig. 3E),
when compared with OE-CNN1 + shRNA-NC group.
Interference of TIMP2 inhibits the
expression levels of the Wnt/β-catenin signaling pathway
proteins
GEPIA predicted a positive correlation between CNN1
and TIMP2, with a correlation coefficient of 0.68 (Fig. 4A). It was also predicted that TIMP2
expression was decreased in LUSC tissues (Fig. 4B). The mRNA expression levels of
TIMP2 were upregulated following CNN1 overexpression (Fig. 4C). The knockdown efficiency of TIMP2
in cells was detected using RT-qPCR (Fig. 4D and E), and shRNA-TIMP2-1, which was selected
for subsequent experiments, exhibited a better knockdown
efficiency. CNN1 overexpression increased DKK1 expression level,
but reduced β-catenin and c-myc expression levels (Fig. 4F). Following TIMP2 silencing, the
expression levels of DKK1 were downregulated, and those of
β-catenin and c-myc were upregulated compared with the OE-CNN1 +
shRNA-NC group (Fig. 4F).
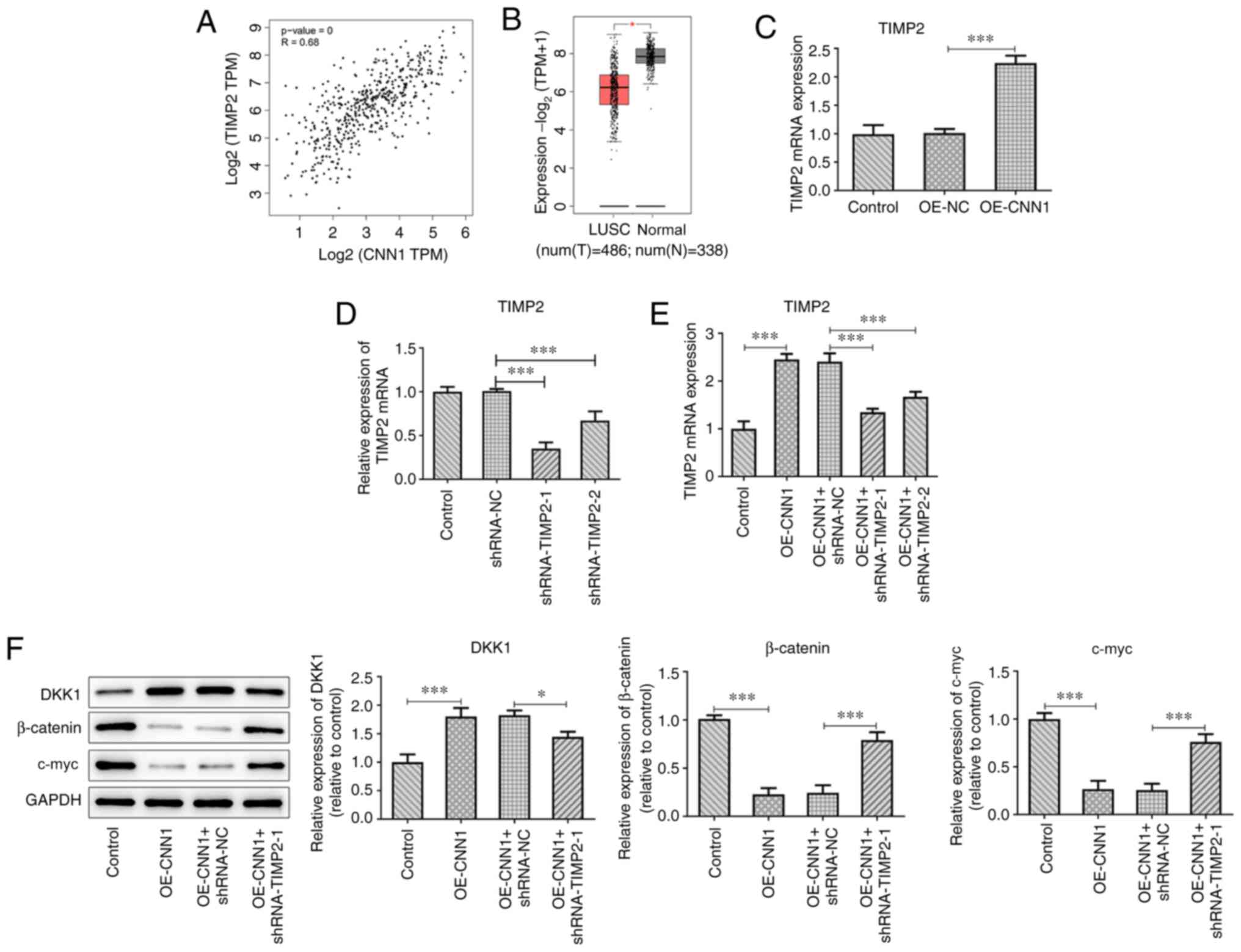 | Figure 4Interference of TIMP2 affects the
expression of DKK1-associated proteins in NCL-H2170 cells. (A)
GEPIA database prediction of the association between CNN1 and
TIMP2. (B) GEPIA database prediction of the expression of TIMP2 in
LUSC. (C) TIMP2 expression was detected using RT-qPCR following
CNN1 overexpression. (D and E) TIMP2 mRNA expression levels were
assessed following transfection with (D) shRNA-TIMP2, as well as
(E) OE-CNN1 and shRNA-TIMP2 using RT-qPCR. (F) Protein expression
levels of DKK1, β-catenin and c-myc were detected using western
blot analysis. *P<0.05 and ***P<0.001.
The experiments were repeated three times. CNN1, calponin 1; DKK1,
Dickkopf-1; LUSC, lung squamous cell carcinoma; N, normal; NC,
negative control; OE, overexpression; RT-qPCR,
reverse-transcription quantitative PCR; shRNA, short hairpin RNA;
T, tumor; TIMP2, tissue inhibitor of metalloproteinases 2; TPM,
transcripts per million. |
Discussion
Lung cancer is a malignant tumor with high morbidity
and mortality, which poses a serious threat to human health
worldwide (23). NSCLC accounts for
~85% of all lung cancer cases (24). Significant advances have been made
in the treatment of lung cancer (24). Although targeted therapy and
immunotherapy have significantly increased the 5-year survival
rates, high recurrence rates, metastasis and drug resistance are
characteristic of lung cancer (5).
Therefore, novel treatments with high efficiency, low cost and low
adverse reactions are urgently required.
CNN1 is a known marker of cardiac and smooth muscle
cell differentiation. CNN1 is also a member of the cysteine rich
61/connective tissue growth factor/nephroblastoma overexpressed
family of growth regulators (8,25).
CNN1 is characterized as a pro-angiogenic factor that mediates
diverse roles in cell development, proliferation and tumorigenesis
(26). However, the association
between CNN1 and lung cancer or squamous cell carcinoma development
has, to the best of our knowledge, not been previously reported. In
the present study, the GEPIA database was used to predict the
association between CNN1 and LUSC cells. The expression levels of
CNN1 in 16HBE, NCL-H520, SK-MES-1 and NCL-H2170 cells were also
detected using western blot analysis. Following overexpression of
CNN1, the cell migratory and invasive abilities decreased. Several
studies have shown that MMP2 and MMP9 are tumor promoter genes that
are highly expressed in a variety of malignant tumor tissue types,
such as glioma, lung, liver, colon, pancreatic, breast and cervical
cancers (27,28). They promote the formation of new
blood vessels in tumors, and accelerate the invasion and metastasis
of tumor cells (27,28). In the present study, MMP2 and MMP9
expression levels were downregulated, indicating that
overexpression of CNN1 inhibited cell migration. The EMT process
involves the transformation of epithelial cells into mesenchymal
cells under specific physiological and pathological conditions,
resulting in loss of polarity of epithelial cells and intercellular
transformation (29,30). The end-result of EMT is cells with
mesenchymal cell morphology and properties (29,30).
E-cad is a transmembrane glycoprotein widely present in human
epithelial cells (31). Its
physiological activity is to maintain the adhesion between cells
and preserve the integrity of the tissue structure (31). N-cad is a transmembrane protein,
which functions to transmit signals. A previous study has shown
that the N-cad protein is upregulated in a variety of tumor cell
types and that its abnormal expression is associated with the
survival, migration, invasion and proliferation of tumor cells
(32). The expression levels of
E-cad and N-cad were downregulated, indicating that overexpression
of CNN1 inhibited EMT.
DKK1 is a secretory protein that acts as an
inhibitor of the extracellular Wnt signal transduction pathway
(19). DKK1 is a member of the DKK
protein family and its abnormal regulation is closely associated
with the pathogenesis of various types of tumors (19). The Wnt/β-catenin pathway is
inhibited by degradation of proteasomal β-catenin, induction of
apoptosis and inhibition of cell proliferation (33). Following CNN1 overexpression, DKK1
was upregulated, and β-catenin and c-myc expression levels were
downregulated. Following DKK1 interference, increased cell
migratory, invasive and EMT abilities were observed. These results
demonstrated that CNN1 could inhibit the invasion, migration and
the EMT process of LUSC cells by inactivating the
Wnt/β-catenin/c-myc signaling pathway. However, the silencing of
DKK1 did not block the effects of CNN1 completely, indicating the
presence of at least one other pathway that mediates the actions of
CNN1 in LUSC, which need to be further explored.
DKK1 is a member of the TIMP family, which is
closely associated with lung cancer. TIMP2 can impair the activity
of the protease MMP2 to hydrolyze the extracellular matrix, which
impedes the process of cell migration and invasion (19,34).
The GEPIA database predicted a positive correlation between CNN1
and TIMP2, and TIMP2 expression was found to decrease in LUSC
tissues. TIMP2 has been reported to inhibit Wnt signaling to
suppress tumor growth and metastasis (18). Therefore, it was speculated that
CNN1 exerted its inhibitory effect on the Wnt/β-catenin signaling
pathway by upregulating TIMP2 expression. Consistent with this
hypothesis, the expression levels of TIMP2 were upregulated
following CNN1 overexpression. TIMP2 knockdown blocked the
inhibitory effect of CNN1 on the Wnt/β-catenin signaling.
In conclusion, CNN1 regulated the
Wnt/β-catenin/c-myc signaling pathway by activating TIMP2 to
inhibit the invasion, migration and the EMT process in LUSC cells.
However, other potential mechanisms need to be uncovered, as well
as the usage of other cell lines together with in vivo studies to
further clarify the conclusions of the present study.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
WSL contributed to study conception and design. WSL
and XGF contributed to acquisition of data. RML contributed to
analysis and interpretation of data. WSL drafted the initial
manuscript and revised it critically for important intellectual
content. WSL and XGF confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nasim F, Sabath BF and Eapen GA: Lung
Cancer. Med Clin North Am. 103:463–473. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The Eighth Edition AJCC Cancer Staging Manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jain D and Roy-Chowdhuri S: Molecular
Pathology of Lung Cancer Cytology Specimens: A Concise Review. Arch
Pathol Lab Med. 142:1127–1133. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhang R, Deng Y, Zhang Y, Zhai GQ, He RQ,
Hu XH, Wei DM, Feng ZB and Chen G: Upregulation of HOXA13 as a
potential tumorigenesis and progression promoter of LUSC based on
qRT-PCR and bioinformatics. Int J Clin Exp Pathol. 10:10650–10665.
2017.PubMed/NCBI
|
|
7
|
Drilon A, Rekhtman N, Ladanyi M and Paik
P: Squamous-cell carcinomas of the lung: Emerging biology,
controversies, and the promise of targeted therapy. Lancet Oncol.
13:e418–e426. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Miano JM, Krahe R, Garcia E, Elliott JM
and Olson EN: Expression, genomic structure and high resolution
mapping to 19p13.2 of the human smooth muscle cell calponin gene.
Gene. 197:215–224. 1997.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang X, Wu K, Zhang Z, Lan M, Jin J and
Fan D: The effect of calponin and caldesmon in regulation of the
gastrointestinal motility during pathophysiological adaptation.
Zhonghua Nei Ke Za Zhi. 40:459–462. 2001.PubMed/NCBI(In Chinese).
|
|
10
|
Lv B, Zhao J, Yang F, Huang X, Chen G,
Yang K, Liu S, Fan C, Fu H and Chen Z: Phenotypic transition of
corpus cavernosum smooth muscle cells subjected to hypoxia. Cell
Tissue Res. 357:823–833. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Koganehira Y, Takeoka M, Ehara T, Sasaki
K, Murata H, Saida T and Taniguchi S: Reduced expression of
actin-binding proteins, h-caldesmon and calponin h1, in the
vascular smooth muscle inside melanoma lesions: An adverse
prognostic factor for malignant melanoma. Br J Dermatol.
148:971–980. 2003.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sasaki Y, Yamamura H, Kawakami Y, Yamada
T, Hiratsuka M, Kameyama M, Ohigashi H, Ishikawa O, Imaoka S,
Ishiguro S, et al: Expression of smooth muscle calponin in tumor
vessels of human hepatocellular carcinoma and its possible
association with prognosis. Cancer. 94:1777–1786. 2002.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yamane T, Asanoma K, Kobayashi H, Liu G,
Yagi H, Ohgami T, Ichinoe A, Sonoda K, Wake N and Kato K:
Identification of the critical site of calponin 1 for suppression
of ovarian cancer properties. Anticancer Res. 35:5993–5999.
2015.PubMed/NCBI
|
|
14
|
Martín de las Mulas J, Reymundo C,
Espinosa de los Monteros A, Millán Y and Ordás J: Calponin
expression and myoepithelial cell differentiation in canine, feline
and human mammary simple carcinomas. Vet Comp Oncol. 2:24–35.
2004.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Krishnamurthy N and Kurzrock R: Targeting
the Wnt/beta-catenin pathway in cancer: Update on effectors and
inhibitors. Cancer Treat Rev. 62:50–60. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Teng Y, Wang X, Wang Y and Ma D:
Wnt/beta-catenin signaling regulates cancer stem cells in lung
cancer A549 cells. Biochem Biophys Res Commun. 392:373–379.
2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yang S, Liu Y, Li MY, Ng CSH, Yang SL,
Wang S, Zou C, Dong Y, Du J, Long X, et al: FOXP3 promotes tumor
growth and metastasis by activating Wnt/β-catenin signaling pathway
and EMT in non-small cell lung cancer. Mol Cancer.
16(124)2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Huang Y, Liu L and Liu A: Dickkopf-1:
Current knowledge and related diseases. Life Sci. 209:249–254.
2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Katoh M: Multi-layered prevention and
treatment of chronic inflammation, organ fibrosis and cancer
associated with canonical WNT/β-catenin signaling activation
(Review). Int J Mol Med. 42:713–725. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chanvorachote P, Sriratanasak N and
Nonpanya N: C-myc Contributes to Malignancy of Lung Cancer: A
potential anticancer drug target. Anticancer Res. 40:609–618.
2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Samarghandian S, Azimi-Nezhad M and
Farkhondeh T: Thymoquinone-induced antitumor and apoptosis in human
lung adenocarcinoma cells. J Cell Physiol. 234:10421–10431.
2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Cagle PT, Allen TC and Olsen RJ: Lung
cancer biomarkers: Present status and future developments. Arch
Pathol Lab Med. 137:1191–1198. 2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Williams JP, Micoli K and McDonald JM:
Calmodulin-an often-ignored signal in osteoclasts. Ann N Y Acad
Sci. 1192:358–364. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Menéndez JA, Mehmi I, Griggs DW and Lupu
R: The angiogenic factor CYR61 in breast cancer: Molecular
pathology and therapeutic perspectives. Endocr Relat Cancer.
10:141–152. 2003.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Jackson MT, Moradi B, Smith MM, Jackson CJ
and Little CB: Activation of matrix metalloproteinases 2, 9, and 13
by activated protein C in human osteoarthritic cartilage
chondrocytes. Arthritis Rheumatol. 66:1525–1536. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Milner JM, Rowan AD, Cawston TE and Young
DA: Metalloproteinase and inhibitor expression profiling of
resorbing cartilage reveals pro-collagenase activation as a
critical step for collagenolysis. Arthritis Res Ther.
8(R142)2006.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Lee HM, Hwang KA and Choi KC: Diverse
pathways of epithelial mesenchymal transition related with cancer
progression and metastasis and potential effects of endocrine
disrupting chemicals on epithelial mesenchymal transition process.
Mol Cell Endocrinol. 457:103–113. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hao Y, Baker D and Ten Dijke P:
TGF-β-mediated epithelial-mesenchymal transition and cancer
metastasis. Int J Mol Sci. 20(2767)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Luo T, Yan A, Liu L, Jiang H, Feng C, Liu
G, Liu F, Tang D and Zhou T: In vitro study of joint intervention
of E-cad and Bmi-1 mediated by transcription activator-like
effector nuclease in nasopharyngeal carcinoma. Zhong Nan Da Xue Xue
Bao Yi Xue Ban. 43:229–239. 2018.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
32
|
Liu Y, Yang F, Liang S, Liu Q, Fu S, Wang
Z, Yang C and Lin J: N-Cadherin Upregulation Promotes the
Neurogenic Differentiation of Menstrual Blood-Derived Endometrial
Stem Cells. Stem Cells Int. 2018(3250379)2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Igbinigie E, Guo F, Jiang SW, Kelley C and
Li J: Dkk1 involvement and its potential as a biomarker in
pancreatic ductal adenocarcinoma. Clin Chim Acta. 488:226–234.
2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jaschke N, Hofbauer LC, Göbel A and
Rachner TD: Evolving functions of Dickkopf-1 in cancer and
immunity. Cancer Lett. 482:1–7. 2020.PubMed/NCBI View Article : Google Scholar
|