Introduction
Diabetic nephropathy (DN) has long been considered
the most pervasive and serious diabetic chronic complication
(1). Currently, DN is mostly
treated with drugs, including rosiglitazone (2), tripterysium glycosides (3) and alprostadil (4). However, these drug treatments have
limitations and adverse reactions, including leukopenia,
gastrointestinal reactions, irregular menstruation and abnormal
liver function (5,6). Therefore, the investigation into novel
treatment strategies for DN remains necessary.
Long non-coding RNAs (lncRNAs) are important
regulators of cell proliferation, inflammation and fibrosis in DN
(7,8). For instance, lncRNA plasmacytoma
variant translocation 1 promotes mesangial cell (MC) proliferation
under high-glucose conditions in DN (9,10) and
lncRNA nuclear enriched abundant transcript 1 (NEAT1) accelerates
the occurrence and development of DN (11). Additionally, non-coding RNA
activated by DNA damage (NORAD) is a conserved and abundant lncRNA
that preserves genomic stability (12). NORAD has been demonstrated to be an
onco-lncRNA in various types of human cancer, including prostate
(13), ovarian (14), lung (15) and gastric (16) cancers. lncRNA NORAD is also involved
in DN progression (17). Qi et
al (17) demonstrated that the
NORAD/miR-520h/Toll-like receptor 4 regulatory loop promotes the
proliferation and inhibits the apoptosis of glomerular MCs, thereby
aggravating the progression of DN. However, the possible effects of
NORAD on the inflammation and fibrosis of DN and the underlying
regulatory mechanisms remain to be fully revealed.
Emerging evidence has indicated that lncRNAs serve
roles as competing endogenous RNAs (ceRNAs) for miRNAs and regulate
the expression of their target genes in certain diseases (18). miRNAs are involved in cellular
processes, including cell viability and apoptosis (19). Furthermore, miRNAs act as efficient
inhibitors in DN progression (20).
For example, miR-544 attenuates diabetic renal injury by
suppressing glomerulosclerosis and inflammation (21). miR-320a may be a potential curative
target in DN (22). miR-874
overexpression alleviated renal injury in DN rats (23). Notably, miR-485 may serve as a
regulator of inflammatory and fibrotic responses (24). In a recent study, miR-485 suppressed
MC inflammation and proliferation in an in vitro model of DN
(25). Additionally, lncRNAs may
act as competing endogenous RNAs or sponges of miRNAs. NORAD has
been reported to regulate numerous miRNAs in several types of human
disease, including miR-136-5p in retinoblastoma (26), miR-144-3p in hepatocellular
carcinoma (27), miR-520a-3p in
non-small cell lung cancer (28)
and miR-214 in gastric cancer (16). However, the regulatory mechanisms
between lncRNA NORAD and miR-485 have not yet been reported.
The present study investigated the effects of lncRNA
NORAD on human (H)MC proliferation, inflammation and fibrosis and
investigated the regulatory mechanisms between NORAD and
miR-485/nuclear respiratory factor 1 (NRF1). The present study
revealed that the NORAD/miR-485/NRF1 axis will be a theoretical
basis for DN-targeted therapy.
Materials and methods
Tissue collection
A total of 21 patients with DN without other
complications were selected in Shengli Oilfield Central Hospital
(Dongying, China) between March 2017 and June 2018. The patients
included 11 males and 10 females (age range, 46-64 years; mean age,
54.6±6.2 years). These patients had not received treatment within 3
months before admission. Pathological kidney and adjacent normal
tissues were obtained by biopsy. Each patient provided written
informed consent and agreed to the study being published. The
present study was approved by the Ethics Committee of Shengli
Oilfield Central Hospital (approval no.
Q/ZXYY-ZY-YWB-LL202037).
Cell grouping and transfection
HMCs were purchased from The Cell Bank of Type
Culture Collection of the Chinese Academy of Sciences. DMEM
containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was used
to culture the cells at 37˚C in 5% CO2. Small
interfering (siRNA)-negative control (si-NC,
5'-UUCUCCGAACGUGUCACGU-3') and siRNA-NORAD-1/-2 (si-NORAD-1,
5'-AAGCCACCUUUGUGAACAGUA-3'; si-NORAD-2, 5'-GAGAAAUGGUAGAAUGACA-3')
were obtained from Sangon Biotech Co., Ltd. NORAD overexpression
(ov-NORAD), NRF1 overexpression (ov-NRF1), miR-485 mimics
(5'-AGAGGCUGGCCGUGAUGAAUUC-3'), miR-NC
(5'-UUCUCCGAACGUGUCACGUTT-3'), miR-485 inhibitor
(5'-GUCAUACACGGCUCUCCUCUCU-3'), and inhibitor NC
(5'-CAGUACUUUUGUAGUACAAA-3') were procured from Guangzhou RiboBio
Co., Ltd. HMCs were transfected with the aforementioned agents (all
50 nM) using a Lipofectamine RNAiMAX kit (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h at 37˚C. In addition, the HMCs
(6x105 cells/well) were further divided into high
glucose (HG; 30 mM) and normal glucose (NG; 5.5 mM) groups. At 48 h
after treatment, the cells were harvested to perform the following
experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from HMCs using a
TRIzol® reagent kit (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols. In
accordance with the manufacturer's protocols of GoScript™ reverse
transcription system (Promega Corporation), the extracted RNA was
initially reverse transcribed into cDNA at 37˚C for 60 min,
followed by 85˚C for 5 min and then subjected to qPCR analyses with
the Applied Biosystems SYBR™ Green PCR Master mix (Thermo Fisher
Scientific, Inc.). The thermocycling conditions were: Initial
denaturation at 95˚C for 10 min, followed by 40 cycles of 95˚C for
10 sec, 60˚C for 20 sec and 72˚C for 34 sec. U6 or GAPDH was used
as the internal reference standard. The primers were designed as
follows: NORAD forward: 5'-GGAGAATCGCTTGAACT-3' and reverse,
5'-CAAACACCCAATGAATAG-3'; miR-485 forward,
5'-CCAAGCTTCACCCATTCCTAACAGGAC-3' and reverse,
5'-CGGGATCCGTAGGTCAGTTACATGCATC-3'; NRF1 forward,
5'-TTACTCTGCTGTGGCTGATGG-3' and reverse,
5'-CCTCTGATGCTTGCGTGGTCT-3'; U6 forward,
5'-GCTTCGGCAGCACATATACTAAAAT-3' and reverse,
5'-CGCTTCACGAATTTGCGTGTCAT-3' and GAPDH forward,
5'-GAAGGTGAAGGTCGGAGTC-3' and reverse, 5'-GAAGATGGTGATGGGATTTC-3'.
Gene expression was quantified using the 2-ΔΔCq method
(29).
Western blot analysis
Radioimmunoprecipitation assay buffer (Beyotime
Institute of Biotechnology) containing protease inhibitors was used
to extract proteins from cells. The protein concentrations were
determined using a bicinchoninic acid Protein assay kit (Thermo
Fisher Scientific, Inc.). A total of 50 µg of protein/lane was
separated via 10% SDS-PAGE (Boster Biological Technology). The
resolved proteins were then transferred onto polyvinylidene
fluoride membranes. Blocking was performed using 5% bovine serum
albumin (Thermo Fisher Scientific, Inc.) at room temperature for 2
h. Following blocking, the membranes were incubated overnight at
4˚C with primary antibodies against NRF1 (1:1,000; cat. no.
ab175932; Abcam) and GAPDH (1:1,000; cat. no. ab8245; Abcam). Next,
the membranes were washed three times in TBS-Tween-20 (0.05%). The
secondary antibody horseradish peroxidase-conjugated anti-mice
immunoglobulin G (1:2,000; cat. no. ab6728; Abcam) was added and
incubated for 1 h at room temperature. GADPH was used as the
internal reference. The membranes were developed using an ECL
reagent (Thermo Fisher Scientific, Inc.) under Gel-Pro analyzer
(version 4.0; Media Cybernetics, Inc.).
Cell viability assay
MTT assays were used to determine HMC viability.
Cells were seeded onto a 96-well plate with 2x105
cells/well and cultured in serum-free medium overnight at 37˚C.
Subsequently, cells were incubated under the designated glucose
conditions for 24, 48, 72 and 96 h at 37˚C. Next, 20 µl MTT (Merck
KGaA) was added to each well and the cells were incubated for
another 2 h at 37˚C. The supernatant was removed and the formazan
crystals were dissolved using DMSO (150 µl/well). The absorbance at
450 nm was analysed using a Multiskan Spectrum microplate reader
(Thermo Fisher Scientific, Inc.).
ELISA
According to manufacturer protocols, the levels of
inflammatory [tumour necrosis factor (TNF)-α (cat. no.
70-EK182HS-96), interleukin (IL)-1β (cat. no. 70-EK101BHS-96) and
IL-6 (cat. no. 70-EK106/2-96) and fibrotic [type IV collagen (Col.
IV) (cat. no. RK-009-001-106), fibronectin (FN; cat. no.
RK-KOA0169) and plasminogen activator inhibitor 1 (PAI-1) (cat. no.
70-EK1136-96)] factors in HMCs were measured using specific ELISA
kits purchased from Multisciences Biotech, Ltd. The absorbance at
450 nm was determined using a Multiskan Spectrum microplate reader
(Thermo Fisher Scientific, Inc.).
Target prediction
The miRNA targets of NORAD were predicted using
StarBase software version 2.0 (http://starbase.sysu.edu.cn/), and 272 targets were
predicted. Among these miRNA targets, miR-485 was selected for the
following assays due to its important role in DN (25) and unknown regulatory association
with NORAD. In addition, the mRNA targets of miR-485 were predicted
using miRDB software version 3.0 (http://mirdb.org/), and 1,646 targets were predicted.
NRF1 was selected for the following assays due to its important
role in renal diseases (30,31).
Dual-luciferase reporter assay
To verify the direct interactions between miR-485
and NORAD/NRF1, a dual-luciferase reporter assay was performed.
Briefly, the mutant type (MUT) and wild type (WT) of NORAD/NRF1
binding sequences were cloned into the pGL3-promoter (Promega
Corporation) to generate the recombinant vectors pGL3-NORAD-WT/-MUT
or pGL3-NRF1-WT/-MUT. Cells were co-transfected with the
aforementioned recombinant vectors and miR-485 mimic/miR-NC (all 50
nM) using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37˚C for 48 h. The supernatant was used to
measure relative luciferase activity on a Dual-Luciferase Reporter
assay system (Promega Corporation). The activity of firefly
luciferase was normalized to that of Renilla luciferase.
Statistical analysis
SPSS version 20.0 (IBM Corp.) and GraphPad Prism
version 5.01 (GraphPad Software, Inc.) were used to perform the
statistical analyses. Data are expressed as the mean ± standard
deviation. Student's t-tests were used to assess the differences
between groups (paired, Figs. 1A,
3D and 5C; unpaired, Figs. 1B, 3E and 5D).
One-way analysis of variance (ANOVA) was used to investigate the
differences among multiple groups. Following ANOVA, pairwise
comparisons were performed using Tukey's multiple comparisons
tests. P<0.05 was considered to indicate a statistically
significant difference. All experiments were conducted in
triplicate in at least three independent experiments.
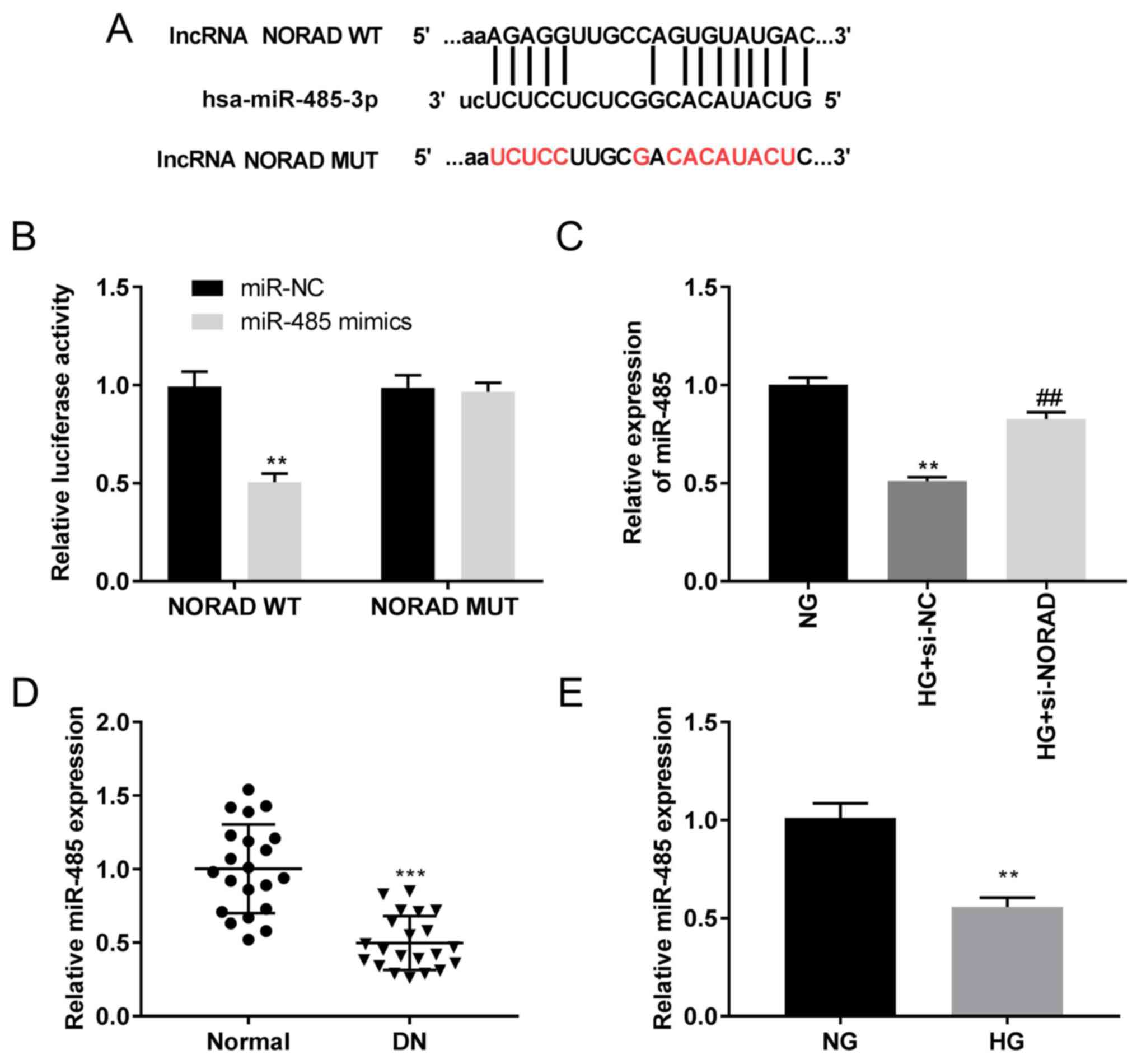 | Figure 3miR-485 is a direct target of NORAD.
(A) Predicted complementary binding site of NORAD and miR-485. (B)
Luciferase activity in HMCs co-transfected with pGL3-NORAD
WT/pGL3-NORAD MUT and miR-485 mimics/NC as determined by a dual
luciferase reporter assay. **P<0.01 vs. miR-NC. (C)
Expression of miR-485 in HMCs detected by RT-qPCR.
**P<0.01 vs. NG; ##P<0.01 vs. HG +
si-NC. (D) Expression of miR-485 in DN tissues (n=21) and adjacent
normal tissues (n=21) detected by RT-qPCR. ***P<0.001
vs. normal. (E) Expression of miR-485 in HG-induced HMCs and NG
HMCs detected by RT-qPCR. **P<0.01 vs. NG. miR,
microRNA; NORAD, non-coding RNA activated by DNA damage; HMCs,
human mesangial cells; si, small interfering RNA; NC, negative
control; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction; HG, high glucose; NG, normal glucose; DN, diabetic
nephropathy; WT, wild-type; MUT, mutant. |
Results
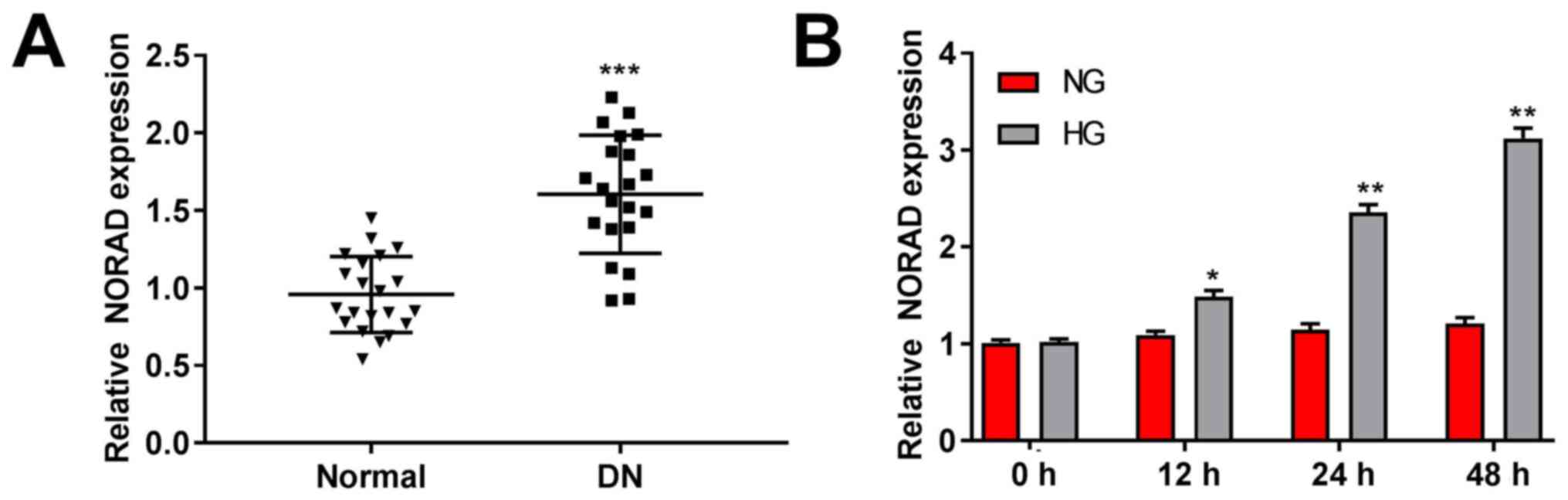
NORAD is highly expressed in DN
tissues and HG-stimulated HMCs
The NORAD expression in DN and normal tissues was
detected by RT-qPCR. The results demonstrated higher expression in
DN tissues compared with in normal tissues (P<0.001; Fig. 1A). Meanwhile, NORAD expression was
significantly increased in HG-stimulated HMCs compared with in the
NG group (P<0.05; Fig. 1B).
Knockdown of NORAD inhibits HG-induced
HMC proliferation, inflammation and fibrosis
To investigate the possible role of NORAD in DN
pathogenesis in vitro, si-NORAD-1/-2 was initially
transfected into HMCs to detect the silencing efficiency. As
illustrated in Fig. 2A, RT-qPCR
demonstrated significantly decreased NORAD expression following
transfection of si-NORAD-1 and si-NORAD-2 (P<0.01). Following
treatment with HG, NORAD expression was also significantly
decreased in the HG + si-NORAD-1 and HG + si-NORAD-2 groups
(P<0.01; Fig. 2B). si-NORAD-1
was selected in the following experiments due to its greater
silencing efficiency. The results of the MTT assay demonstrated
increased HMC viability in the HG + si-NC group compared with in
the NG group, whilst cell activity was partially inhibited in the
HG + si-NORAD group compared with in the HG + si-NC group
(P<0.05; Fig. 2C). Similarly,
the ELISA results demonstrated highly increased levels of
inflammatory factors (IL-6, IL-1β and TNF-α) in the HG + si-NC
group compared with those in the NG group, which was attenuated in
the HG + si-NORAD group (P<0.01; Fig. 2D-F). The levels of PAI-1, Col. IV
and FN were higher in the HG + si-NC group compared with in the NG
group, which was partially reversed in the HG + si-NORAD group
(P<0.01; Fig. 2G-I). These
results indicated that NORAD knockdown suppressed HMC
proliferation, inflammation and fibrosis in vitro.
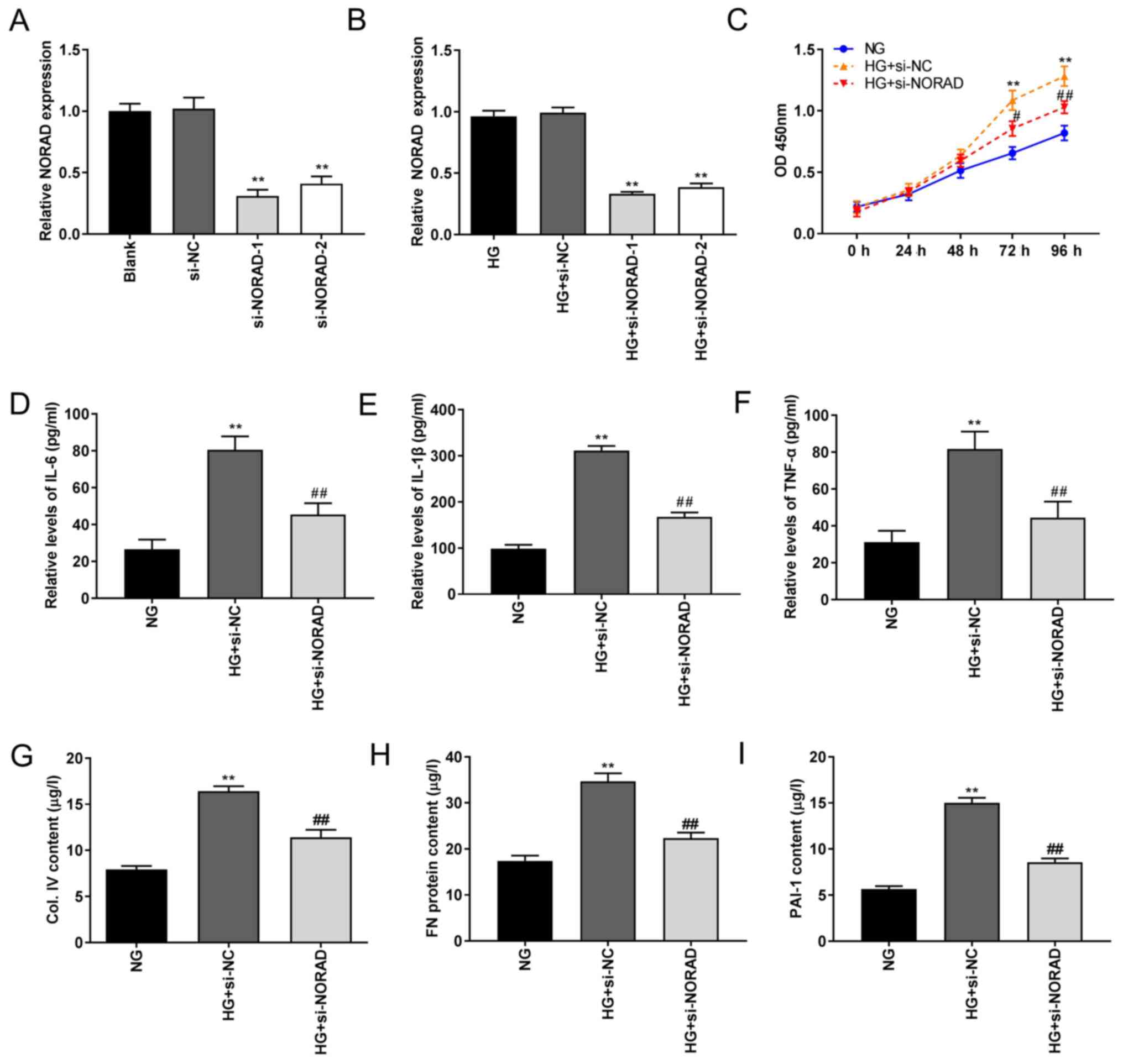 | Figure 2Knockdown of NORAD inhibits
proliferation, inflammation and fibrosis in HG-induced HMCs. (A)
Expression of NORAD determined by RT-qPCR following transfection of
si-NORD-1/-2. **P<0.01 vs. si-NC. (B) Expression of
NORAD detected by RT-qPCR following transfection of si-NORD-1/-2
under HG conditions. **P<0.01 vs. HG + si-NC. (C)
Viability of HMCs measured using MTT assay. **P<0.01
vs. NG; #P<0.05, ##P<0.01 vs. HG +
si-NC. Levels of (D) IL-6, (E) IL-1β, (F) TNF-α, (G) Col. IV, (H)
FN and (I) PAI-1 in HMCs measured via ELISA. **P<0.01
vs. NG; ##P<0.01 vs. HG + si-NC. NORAD, non-coding
RNA activated by DNA damage; NG, normal glucose; HG, high glucose;
HMCs, human mesangial cells; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; si, small
interfering RNA; NC, negative control; OD, optical density; TNF,
tumour necrosis factor; IL, interleukin; Col. IV, type IV collagen;
FN, fibronectin; PAI-1, plasminogen activator inhibitor 1. |
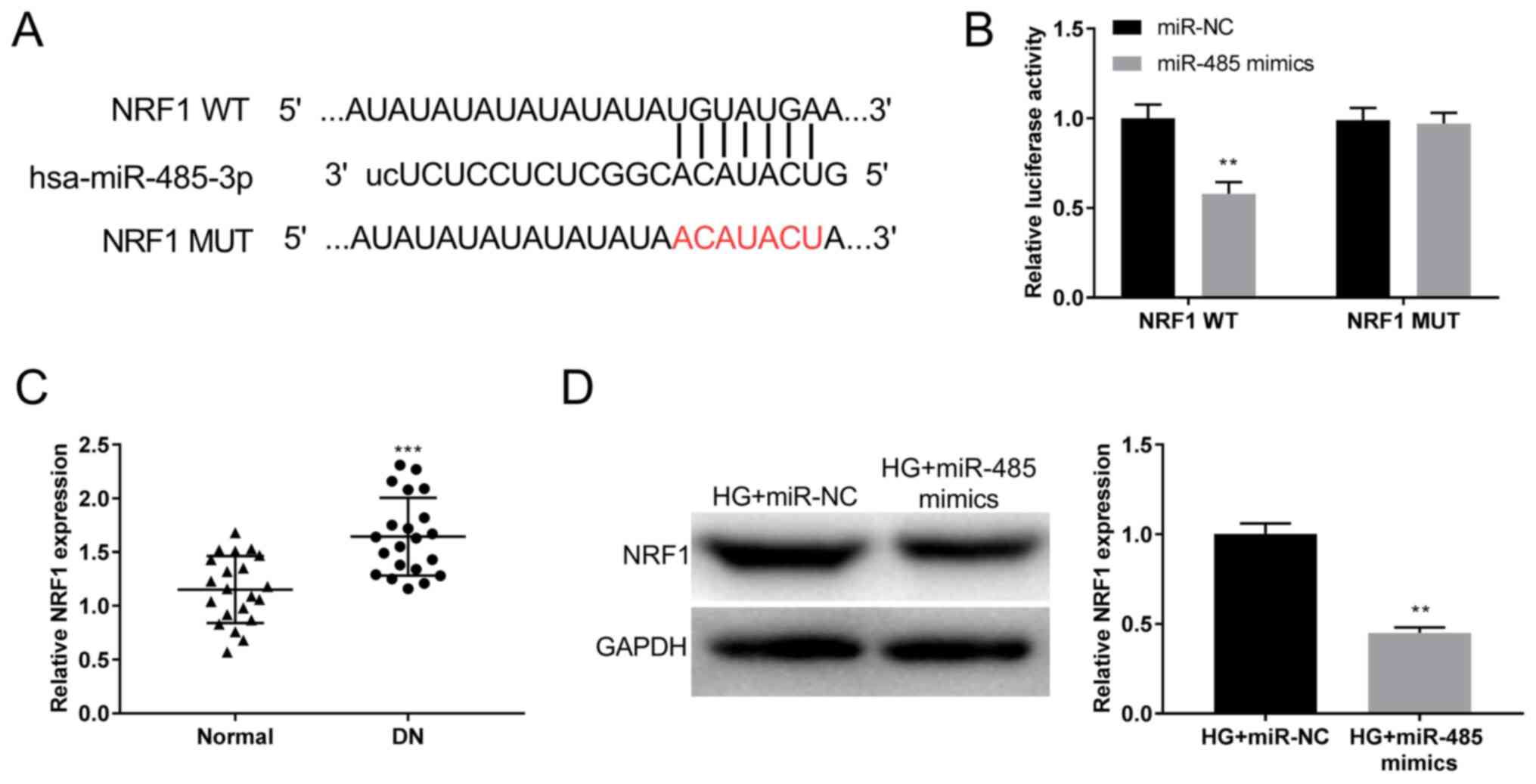
NORAD targets miR-485
Analysis using StarBase software demonstrated
potential binding sequences between lncRNA NORAD and miR-485
(Fig. 3A). The luciferase activity
in the NORAD WT/miR-485 mimics group was decreased compared with
that in the NORAD WT/miR-NC group (P<0.01; Fig. 3B). miR-485 expression was
significantly downregulated in the HG + si-NC group compared with
in the NG group, which was attenuated in the HG + si-NORAD group
(P<0.01; Fig. 3C). RT-qPCR
assays demonstrated decreased miR-485 expression in DN tissues
compared with in normal tissues (P<0.001; Fig. 3D). The expression of miR-485 was
also decreased in the HG group compared with in the NG group
(P<0.01; Fig. 3E). These data
indicated a negative regulatory association between NORAD and
miR-485.
NORAD reverses the inhibitory effects
of miR-485 on HG-induced HMCs
Next, miR-485 mimics, miR-485 inhibitor and miR-485
mimics + ov-NORAD were transfected into HMCs to detect the
transfection efficiency. The expression of miR-485 was upregulated
in the miR-485 mimics group compared with in the miR-NC group,
whilst the inverse was observed in the miR-485 inhibitor group
compared with the inhibitor NC group (P<0.01; Fig. 4A). Meanwhile, compared with the
miR-485 mimics + ov-NC group, miR-485 expression in the miR-485
mimics + ov-NORAD group was partially inhibited (P<0.01;
Fig. 4A). NORAD expression was then
determined following transfection. The results of RT-qPCR analysis
demonstrated that the expression of NORAD was upregulated by
ov-NORAD, but was partially suppressed by ov-NORAD + miR-485 mimics
(P<0.01; Fig. 4B). In addition,
it was also discovered that miR-485 expression was significantly
decreased in the HG group; its expression was upregulated in the HG
+ miR-485 mimics group and downregulated in the HG + miR-485
inhibitor group (P<0.01; Fig.
4C). All the results suggested that miR-485 mimics, miR-485
inhibitor, ov-NORAD and miR-485 mimics + ov-NORAD were successfully
transfected into HMCs. The results of the MTT assay demonstrated
significantly decreased HG-induced HMC viability in the miR-485
mimics group compared with in the miR-NC group; however, the
viability of HG-induced HMCs was partially promoted in the miR-485
mimics + ov-NORAD group compared with in the miR-485 mimics group
(P<0.05; Fig. 4D). Similarly,
the ELISA results demonstrated significantly decreased levels of
inflammatory (IL-6, IL-1β and TNF-α) and fibrotic (PAI-1, Col. IV
and FN) factors in the miR-485 mimics group compared with in the
miR-NC group. Furthermore, overexpression of NORAD attenuated the
effects of miR-485 on inflammation and fibrosis in HG-induced HMCs
(P<0.05; Fig. 4E and F). Therefore, the results suggested that
NORAD may affect the occurrence and development of DN in
vitro by regulating miR-485 expression.
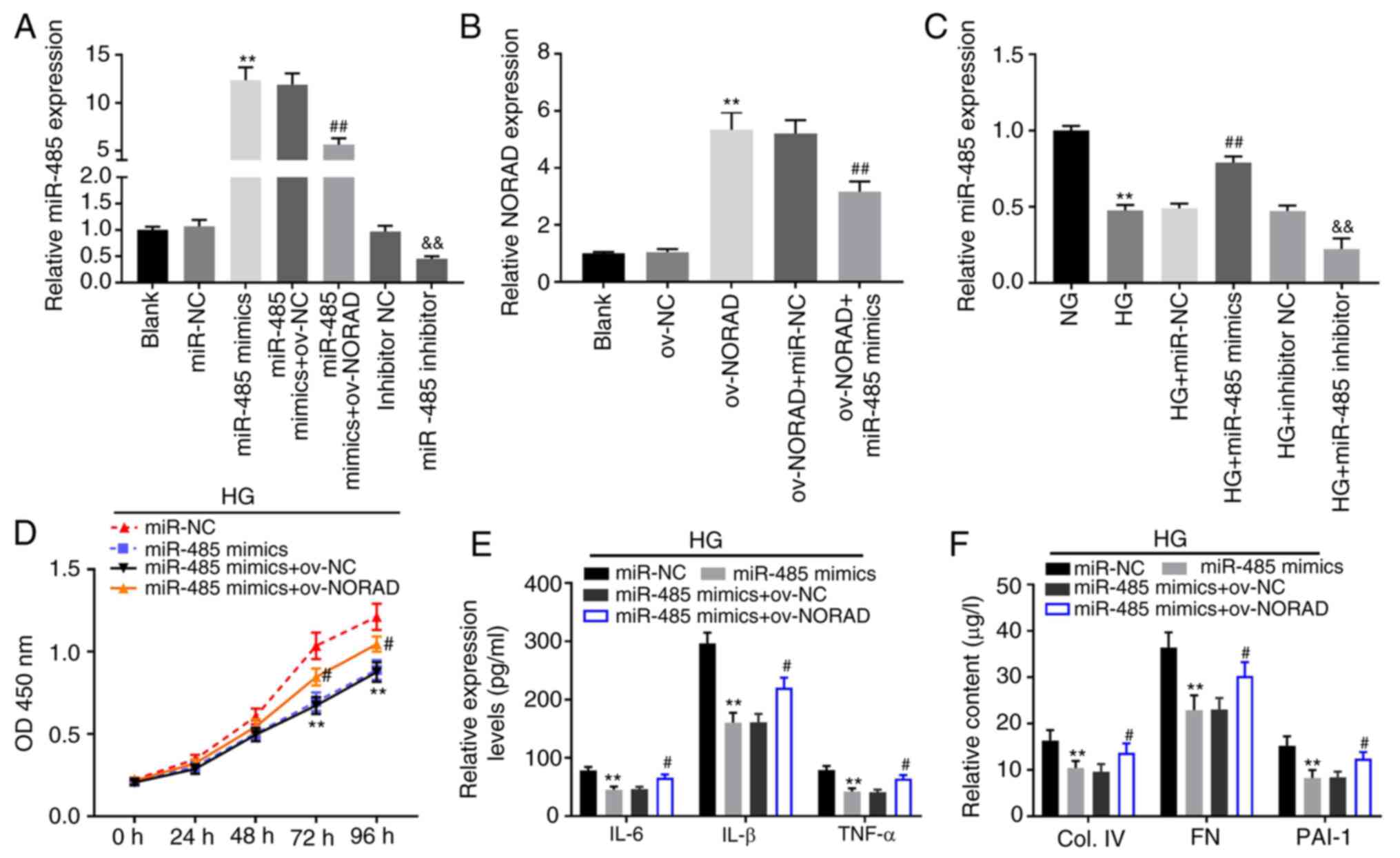 | Figure 4NORAD reverses the inhibitory effects
of miR-485 on HG-induced HMCs. (A) Expression of miR-485 following
transfection of miR-485 mimics/inhibitor or miR-485 mimics +
ov-NORAD detected by RT-qPCR. **P<0.01 vs. miR-NC;
##P<0.01 vs. miR-485 mimics + ov-NC;
&&P<0.01 vs. inhibitor NC. (B) Expression of
NORAD following transfection of ov-NORAD or ov-NORAD + miR-485
mimics detected by RT-qPCR. **P<0.01 vs. ov-NC;
##P<0.01 vs. ov-NORAD + miR-NC. (C) Expression of
miR-485 in HG-induced HMCs detected by RT-qPCR.
**P<0.01 vs. NG; ##P<0.01 vs. HG +
miR-NC; &&P<0.01 vs. HG + inhibitor NC. (D)
Viability of HG-induced HMCs measured by an MTT assay.
**P<0.01 vs. miR-NC; #P<0.05 vs.
miR-485 mimics. (E) Levels of IL-6, IL-1β and TNF-α in HG-induced
HMCs measured via ELISA. **P<0.01 vs. miR-NC;
#P<0.05 vs. miR-485 mimics. (F) Contents of Col. IV,
FN and PAI-1 in HG-induced HMCs measured via ELISA
**P<0.01 vs. miR-NC; #P<0.05 vs.
miR-485 mimics. miR, microRNA; NORAD, non-coding RNA activated by
DNA damage; NG, normal glucose; HG, high glucose; HMCs, human
mesangial cells; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; ov, overexpression vector; NC, negative
control; OD, optical density; TNF, tumour necrosis factor; IL,
interleukin; Col. IV, type IV collagen; FN, fibronectin; PAI-1,
plasminogen activator inhibitor 1. |
Identification of NRF1 as the target
gene of miR-485
Using miRDB software, the potential binding sites
between NRF1 and miR-485 were predicted (Fig. 5A). Dual-luciferase reporter assays
demonstrated significantly decreased luciferase activity in the
NRF1 WT/miR-485 mimics group compared with in the control group.
These results indicated that NRF1 was a direct target gene of
miR-485 (P<0.01; Fig. 5B). The
RT-qPCR results suggested that the expression of NRF1 was higher in
DN tissues compared with in normal tissues (P<0.001; Fig. 5C). Similar to the mRNA levels, the
results of the western blot analysis demonstrated downregulation of
the NRF1 protein level in the HG + miR-485 mimics group compared
with that in the HG + miR-NC group (P<0.01; Fig. 5D). These results suggested that
miR-485 inhibited NRF1 expression.
NORAD knockdown inhibits
proliferation, inflammation and fibrosis in HG-induced HMCs by
regulating the miR-485/NRF1 axis
The expression of NORAD and miR-485 following
co-transfection was detected. The results of RT-qPCR demonstrated
that NORAD expression was significantly increased in the si-NORAD +
miR-485 inhibitor and si-NORAD + ov-NRF1 groups compared with their
controls (si-NORAD + miR-NC and si-NORAD + ov-NC, respectively;
P<0.05; Fig. 6A). Furthermore,
the expression of miR-485 was inhibited by co-transfection with
si-NORAD + miR-485 inhibitor compared with si-NORAD + inhibitor NC
(P<0.01; Fig. 6B). The
transfection efficiency of ov-NRF1 was subsequently determined
using RT-qPCR, which demonstrated that NRF1 expression was
increased by ov-NRF1 (P<0.01; Fig.
6C). The aforementioned results indicated that si-NORAD +
miR-485 inhibitor or ov-NRF1 was successfully transfected into
HMCs. Western blot analysis demonstrated downregulated NRF1 protein
expression following transfection of si-NORAD, while
co-transfection with miR-485 inhibitor or ov-NRF1 reversed this
inhibitory effect (P<0.05; Fig.
6D). MTT assays demonstrated significantly inhibited HG-induced
HMC viability in the si-NORAD group compared with that in the si-NC
group. Meanwhile, cell viability was partially promoted in the
si-NORAD + miR-485 inhibitor and si-NORAD + ov-NRF1 groups compared
with in the si-NORAD group (P<0.05; Fig. 6E). Similarly, the ELISA results
indicated that NORAD knockdown downregulated the levels of
inflammatory (IL-6, IL-1β and TNF-α) and fibrotic (PAI-1, Col. IV
and FN) factors. However, downregulation of miR-485 and
upregulation of NRF1 reversed the effects of NORAD knockdown on
inflammation and fibrosis in HG-induced HMCs (P<0.05; Fig. 6F and G). These results indicated that knockdown
of NORAD may suppress HG-induced HMC proliferation, inflammation
and fibrosis by regulating miR-485/NRF1.
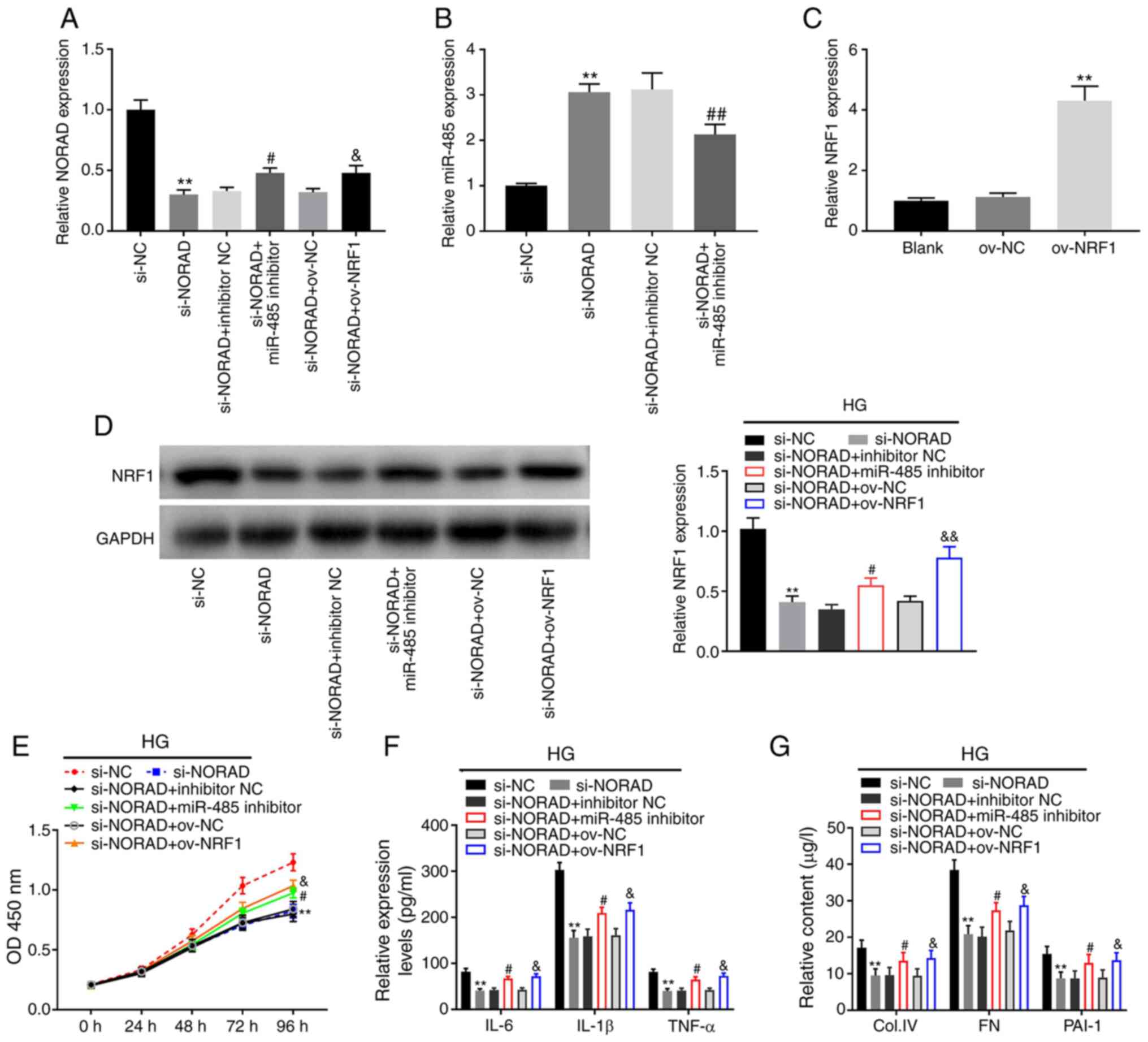 | Figure 6NORAD knockdown inhibits
proliferation, inflammation and fibrosis in HG-induced HMCs by
regulating the miR-485/NRF1 axis. (A) Expression of NORAD following
transfection of si-NORAD, si-NORAD + miR-485 inhibitor or si-NORAD
+ ov-NRF1 as determined via RT-qPCR. **P<0.01 vs.
si-NC; #P<0.05 vs. si-NORAD + inhibitor NC;
&P<0.05 vs. si-NORAD + ov-NC. (B) Expression of
miR-485 following transfection of si-NORAD or si-NORAD + miR-485
inhibitor as detected via RT-qPCR **P<0.01 vs. si-NC;
##P<0.01 vs. si-NORAD + inhibitor NC. (C) Expression
of NRF1 following transfection of ov-NRF1 detected by RT-qPCR.
**P<0.01 vs. ov-NC. (D) Protein expression of NRF1
was measured via western blot analysis. **P<0.01 vs.
si-NC; #P<0.05 vs. si-NORAD + inhibitor NC;
&&P<0.01 vs. si-NORAD + ov-NC. (E) Viability
of HG-induced HMCs measured by MTT assays. **P<0.01
vs. si-NC; #P<0.05 vs. si-NORAD + inhibitor NC;
&P<0.01 vs. si-NORAD + ov-NC. (F) Levels of IL-6,
IL-1β and TNF-α in HG-induced HMCs as determined via ELISA.
**P<0.01 vs. si-NC; #P<0.05 vs.
si-NORAD + inhibitor NC; &P<0.05 vs. si-NORAD +
ov-NC. (G) Contents of Col. IV, FN and PAI-1 in HG-induced HMCs as
determined via ELISA. **P<0.01 vs. si-NC;
#P<0.05 vs. si-NORAD + inhibitor NC;
&P<0.05 vs. si-NORAD + ov-NC. miR, microRNA;
NORAD, non-coding RNA activated by DNA damage; NRF1, nuclear
respiratory factor 1; HG, high glucose; HMCs, human mesangial
cells; RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; ov, overexpression vector; si, small interfering RNA; NC,
negative control; OD, optical density; TNF, tumour necrosis factor;
IL, interleukin; Col. IV, type IV collagen; FN, fibronectin; PAI-1,
plasminogen activator inhibitor 1. |
Discussion
Increasing evidence has indicated that
hyperglycaemia serves a major role in DN (32). Inflammatory and fibrotic reactions
in diabetic patients are mainly caused by hyperglycaemia and
ultimately accelerate the development of DN (33,34).
Recent studies have demonstrated that lncRNAs serve a critical role
in DN (25-27).
A previous study reported upregulation of lncRNA antisense
non-coding mitochondrial RNA-2 in DN tissues and HG-treated MCs
(35). Another study observed
significantly upregulated expression of lncRNA distal-less homeobox
6, opposite strand 1 (Dlx6os1) in MCs under HG conditions compared
with NG conditions (36).
Similarly, the present study identified that lncRNA NORAD was
highly expressed in HG-stimulated HMCs and DN tissues. Therefore,
NORAD may be a pathogenic factor and may serve as a biomarker for
the prognosis of DN.
In the last decade, lncRNAs have been identified as
important regulators of cell proliferation, inflammation and
fibrosis in DN (13,27,28).
Hyperglycaemia is widely proposed to affect different types of
nephrocytes (32). Furthermore, MC
proliferation, inflammation and fibrosis are the three major
features of DN (13,27,28).
Ma et al (37) reported that
downregulation of lncRNA NEAT1 inhibited the proliferation,
fibrosis and inflammation of mouse MCs in DN. Feng et al
(7) observed that lncRNA brown fat
lncRNA 1 interference attenuated renal inflammation fibrosis in DN.
Furthermore, another study demonstrated that inhibition of lncRNA
Dlx6os1 decreased cell proliferation and fibrosis in DN (36). In the present study, NORAD knockdown
suppressed HG-stimulated HMC proliferation, inflammation and
fibrosis. Similar to the results of the present study, a recent
study also demonstrated that knockdown of NORAD decreased cell
viability in mouse glomerular mesangial cells in DN (17). However, the previous study only
investigated the mechanism of NORAD on cell proliferation. The
results of the present study further revealed the involvement of
lncRNA NORAD in regulating HMC proliferation, inflammation and
fibrosis in DN.
Recent studies have reported that miRNAs act as
regulatory factors in various cellular processes. For example,
miRNAs affect cell proliferation, apoptosis, stress resistance and
angiogenesis (38). Yao et
al (23) reported that miR-874
alleviates renal injury and inflammatory response in DN. Jiang
et al (20) observed that
miR-342-3p-overexpression suppressed renal interstitial fibrosis in
DN. The present study revealed decreased miR-485 expression in
HG-stimulated HMCs and DN tissues. Furthermore, miR-485 was the
direct target of lncRNA NORAD and inhibited HG-stimulated HMC
proliferation, inflammation and fibrosis. Similar to the results of
the present study, Wu et al (25) also demonstrated that miR-485
overexpression suppressed HG-induced HMC proliferation. The results
demonstrated that miR-485 may inhibit DN progression. The present
study also demonstrated that NORAD overexpression attenuated the
inhibitory effects of miR-485 on HG-induced HMC proliferation,
inflammation and fibrosis. These results suggested that NORAD may
affect DN progression by regulating miR-485 expression.
Emerging evidence has suggested that NRF1 is an
important regulatory factor in apoptosis (39). Zhang et al (39) reported that NRF1 overexpression
inhibited the apoptosis of palmitate-stimulated human cardiac
myocytes. Zhang et al (40)
also indicated that NRF1 acts as a key regulator of chondrocyte
apoptosis in osteoarthritis. The present study demonstrated
significantly increased NRF1 expression in DN tissues and showed
that transfection of miR-485 mimics into HG-stimulated HMCs
inhibited NRF1 expression. Therefore, NRF1 was determined to be the
target gene of miR-485. Furthermore, it was demonstrated that NRF1
expression was negatively regulated by miR-485, and that
downregulation of miR-485 and upregulation of NRF1 reversed the
effects of NORAD-knockdown on HG-induced HMC proliferation,
inflammation and fibrosis. The results indicated that NORAD
knockdown inhibited HG-induced HMC proliferation, inflammation and
fibrosis by regulating miR-485 and NRF1 expression.
In conclusion, the results of the present study
revealed that NORAD knockdown inhibited the proliferation,
inflammation and fibrosis of HG-induced HMCs by regulating the
in vitro expression of miR-485 and NRF1. However, the
difference between in vitro and in vivo conditions is
a limitation of the present study. Further studies are warranted to
elucidate these issues. Despite this limitation, these findings
suggest the potential of a novel strategy for treating DN.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LW and HG were responsible for the conception and
design of the study, and project supervision and management. XY and
HZ performed data acquisition and analysis. LL and MZ were involved
in data analysis and visualization. All authors confirmed the
authenticity of all the raw data, gave final approval of the
version to be published, and agreed to be accountable for all
aspects of the work. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from each
patient. The present study was approved by the Ethics Committee of
Shengli Oilfield Central Hospital (approval no.
Q/ZXYY-ZY-YWB-LL202037).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fox CS, Matsushita K, Woodward M, Bilo HJ,
Chalmers J, Heerspink HJ, Lee BJ, Perkins RM, Rossing P, Sairenchi
T, et al: Associations of kidney disease measures with mortality
and end-stage renal disease in individuals with and without
diabetes: A meta-analysis. Lancet. 380:1662–1673. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhang L, Zhou Y, Zhou F, Yu X, Liu J, Liu
Y, Zhu Y, Wang W and Chen N: Altered expression of long noncoding
and messenger RNAs in diabetic nephropathy following treatment with
rosiglitazone. Biomed Res Int. 2020(1360843)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang ZS, Qiu T, Liu XH, Zhou JQ, Chen ZB,
Wang L and Zhang L, Shen Y and Zhang L: Tripterysium glycosides
preconditioning attenuates renal ischemia/reperfusion injury in a
rat model. Int Urol Nephrol. 48:213–221. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Xu X, Pan X and Li S: Prospective analysis
of the efficacy of beraprost sodium combined with alprostadil on
diabetic nephropathy and influence on rennin-angiotensin system and
TNF-α. Exp Ther Med. 19:639–645. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yang XM, Yang KN, Gao W and Cao XJ:
Analysis of effect and adverse reactions of enalapril on diabetic
nephropathy. J Clin Med Pract. (3)2012.
|
|
6
|
Cao Y, Yun N and Zou A: Meta-analysis of
ADR induced by tripterysium glycosides tablet. China Pharmacy.
29:125–130. 2018.
|
|
7
|
Feng X, Zhao J, Ding J, Shen X, Zhou J and
Xu Z: lncRNA Blnc1 expression and its effect on renal fibrosis in
diabetic nephropathy. Am J Transl Res. 11:5664–5672.
2019.PubMed/NCBI
|
|
8
|
Gao J, Wang W, Wang F and Guo C:
lncRNA-NR_033515 promotes proliferation, fibrogenesis and
epithelial-to-mesenchymal transition by targeting miR-743b-5p in
diabetic nephropathy. Biomed Pharmacother. 106:543–552.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Alvarez ML and DiStefano JK: Functional
characterization of the plasmacytoma variant translocation 1 gene
(PVT1) in diabetic nephropathy. PLoS One. 6(e18671)2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Alvarez ML, Khosroheidari M, Eddy E and
Kiefer J: Role of microRNA 1207-5P and its host gene, the long
non-coding RNA Pvt1, as mediators of extracellular matrix
accumulation in the kidney: Implications for diabetic nephropathy.
PLoS One. 8(e77468)2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li N, Jia T and Li Y: lncRNA NEAT1
accelerates the occurrence and development of diabetic nephropathy
by sponging miR-23c. Eur Rev Med Pharmacol Sci. 24:1325–1337.
2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lee S, Kopp F, Chang TC, Sataluri A, Chen
B, Sivakumar S, Yu H, Xie Y and Mendell JT: Noncoding RNA NORAD
regulates genomic stability by sequestering PUMILIO proteins. Cell.
164:69–80. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang H and Guo H: Long non-coding RNA
NORAD induces cell proliferation and migration in prostate cancer.
J Int Med Res. 47:3898–3904. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tong L, Ao Y, Zhang H, Wang K, Wang Y and
Ma Q: Long noncoding RNA NORAD is upregulated in epithelial ovarian
cancer and its downregulation suppressed cancer cell functions by
competing with miR-155-5p. Cancer Med. 8:4782–4791. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li J, Xu X, Wei C, Liu L and Wang T: Long
noncoding RNA NORAD regulates lung cancer cell proliferation,
apoptosis, migration, and invasion by the miR-30a-5p/ADAM19 axis.
Int J Clin Exp Pathol. 13:1–13. 2020.PubMed/NCBI
|
|
16
|
Tao W, Li Y, Zhu M, Li C and Li P: lncRNA
NORAD promotes proliferation and inhibits apoptosis of gastric
cancer by regulating miR-214/Akt/mTOR axis. Onco Targets Ther.
12:8841–8851. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Qi H, Yao L and Liu Q: NORAD affects the
progression of diabetic nephropathy through targeting miR-520h to
upregulate TLR4. Biochem Biophys Res Commun. 521:190–195.
2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang G, Sun H, Zhang Y, Zhao H, Fan W, Li
J, Lv Y, Song Q, Li J, Zhang M and Shi H: Characterization of
dysregulated lncRNA-mRNA network based on ceRNA hypothesis to
reveal the occurrence and recurrence of myocardial infarction. Cell
Death Discov. 4(35)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen L, Nan A, Zhang N, Jia Y, Li X, Ling
Y, Dai J, Zhang S, Yang Q, Yi Y and Jiang Y: Circular RNA 100146
functions as an oncogene through direct binding to miR-361-3p and
miR-615-5p in non-small cell lung cancer. Mol Cancer.
18(13)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Jiang ZH, Tang YZ, Song HN, Yang M, Li B
and Ni CL: miRNA-342 suppresses renal interstitial fibrosis in
diabetic nephropathy by targeting SOX6. Int J Mol Med. 45:45–52.
2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sun T, Liu Y, Liu L and Ma F: MicroRNA-544
attenuates diabetic renal injury via suppressing glomerulosclerosis
and inflammation by targeting FASN. Gene.
723(143986)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
He M, Wang J, Yin Z, Zhao Y, Hou H, Fan J,
Li H, Wen Z, Tang J, Wang Y, et al: MiR-320a induces diabetic
nephropathy via inhibiting MafB. Aging (Albany NY). 11:3055–3079.
2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yao T, Zha D, Gao P, Shui H and Wu X:
MiR-874 alleviates renal injury and inflammatory response in
diabetic nephropathy through targeting toll-like receptor-4. J Cell
Physiol. 234:871–879. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen HO, Zhang L, Tang ZY and Gong ZM:
MiR-485-5p promotes the development of osteoarthritis by inhibiting
cartilage differentiation in BMSCs. Eur Rev Med Pharmacol Sci.
22:3294–3302. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wu J, Lu K, Zhu M, Xie X, Ding Y, Shao X,
Chen Y, Liu J, Xu M, Xu Y, et al: miR-485 suppresses inflammation
and proliferation of mesangial cells in an in vitro model of
diabetic nephropathy by targeting NOX5. Biochem Biophys Res Commun.
521:984–990. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yang XL, Hao YJ, Wang B, Gu XL, Wang XX
and Sun JF: Long noncoding RNA NORAD promotes the progression of
retinoblastoma by sponging miR 136-5p/PBX3 axis. Eur Rev Med
Pharmacol Sci. 24(4055)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tian Q, Yan X, Yang L, Liu Z, Yuan Z, Shen
Z and Zhang Y: lncRNA NORAD promotes hepatocellular carcinoma
progression via regulating miR-144-3p/SEPT2. Am J Transl Res.
12:2257–2266. 2020.PubMed/NCBI
|
|
28
|
Wan Y, Yao Z, Chen W and Li D: The lncRNA
NORAD/miR-520a-3p facilitates malignancy in non-small cell lung
cancer via PI3k/Akt/mTOR signaling pathway. Onco Targets Ther.
13:1533–1544. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Huang HQ, Ni HF, Ma KL and Zou JH: ANGPTL2
regulates autophagy through the MEK/ERK/Nrf-1 pathway and affects
the progression of renal fibrosis in diabetic nephropathy. Am J
Transl Res. 11:5472–5486. 2019.PubMed/NCBI
|
|
31
|
Hsieh PF, Liu SF, Hung TJ, Hung CY, Liu
GZ, Chuang LY, Chen MF, Wang JL, Shi MD, Hsu CH, et al: Elucidation
of the therapeutic role of mitochondrial biogenesis transducers
NRF-1 in the regulation of renal fibrosis. Exp Cell Res. 349:23–31.
2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kaur H, Chien A and Jialal I:
Hyperglycemia induces Toll like receptor 4 expression and activity
in mouse mesangial cells: Relevance to diabetic nephropathy. Am J
Physiol Renal Physiol. 303:F1145–F1150. 2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sortica DA, Crispim D, Zaffari GP,
Friedman R and Canani LH: The role of ecto-nucleotide
pyrophosphatase/phosphodiesterase 1 in diabetic nephropathy. Arq
Bras Endocrinol Metabol. 55:677–685. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Burcelin R, Serino M, Chabo C,
Blasco-Baque V and Amar J: Gut microbiota and diabetes: From
pathogenesis to therapeutic perspective. Acta Diabetol. 48:257–273.
2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gao Y, Chen ZY, Wang Y, Liu Y, Ma JX and
Li YK: Long non-coding RNA ASncmtRNA-2 is upregulated in diabetic
kidneys and high glucose-treated mesangial cells. Exp Ther Med.
13:581–587. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cheng J, Cheng L, Tang Y, Li H, Peng W and
Huang S: Inhibition of lncRNA Dlx6os1 decreases cell proliferation
and fibrosis and increases cell apoptosis in diabetic nephropathy.
Int J Clin Exp Pathol. 11:3302–3309. 2018.PubMed/NCBI
|
|
37
|
Ma J, Zhao N, Du L and Wang Y:
Downregulation of lncRNA NEAT1 inhibits mouse mesangial cell
proliferation, fibrosis, and inflammation but promotes apoptosis in
diabetic nephropathy. Int J Clin Exp Pathol. 12:1174–1183.
2019.PubMed/NCBI
|
|
38
|
Liang YZ, Dong J, Zhang J, Wang S, He Y
and Yan YX: Identification of neuroendocrine stress
response-related circulating microRNAs as biomarkers for type 2
diabetes mellitus and insulin resistance. Front Endocrinol
(Lausanne). 9(132)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhang J, Gu JY, Chen ZS, Xing KC and Sun
B: Astragalus polysaccharide suppresses palmitate-induced apoptosis
in human cardiac myocytes: The role of Nrf1 and antioxidant
response. Int J Clin Exp Pathol. 8:2515–2524. 2015.PubMed/NCBI
|
|
40
|
Zhang M, Wang Z, Li B, Sun F, Chen A and
Gong M: Identification of microRNA-363-3p as an essential regulator
of chondrocyte apoptosis in osteoarthritis by targeting NRF1
through the p53-signaling pathway. Mol Med Rep. 21:1077–1088.
2020.PubMed/NCBI View Article : Google Scholar
|