MicroRNAs (miRNAs/miRs) are an evolutionarily
conserved class of small non-coding RNAs of 18-25 nucleotides in
length, which are considered to be involved in the majority of
known physiological and pathological processes, including
oncological and non-oncological diseases (1). miRNAs function as target gene
expression repressors through base-pairing with the 3'-untranslated
region (UTR) of endogenous mRNAs. The miR-129 family consists of
two members: miR-129-1 and miR-129-2. miR-129-1 (gene ID, 406917)
is located on chromosome 7q32.1, a region that is frequently
deleted in multiple tumors. miR-129-2 (gene ID, 406918) is located
on chromosome 11p11.2. Each of these miRNAs produces two mature
miRNA strands that are denoted with a -3p or -5p suffix (2). The biogenesis of mature miR-129
undergoes the following stages: Pri-miR-129 (the non-coding primary
transcript of the miR-129 gene transcribed by RNA polymerase II),
pre-miR-129 (the 70-nucleotide stem-loop precursor miR-129 obtained
by cleaving pri-miR-129 through Drosha ribonuclease III enzyme),
duplex (the complex composed of mature miRNA and antisense miRNA,
and obtained by cleaving pre-miR-129 by the cytoplasmic Dicer
ribonuclease) and the mature miR-129 strand (one of the strands
from the duplex that will be incorporated into Argonaute and then
into the RNA-induced silencing complex) (3). It has been reported that the
expression of the miR-129 family is aberrant in several types of
oncological and non-oncological diseases; it is downregulated in
prostate cancer (miR-129-5p and miR-129-3p) (4), osteosarcoma (miR-129-5p) (5), lung cancer (miR-129-5p) (6), breast cancer (miR-129-5p) (7), nasopharyngeal carcinoma (miR-129-5p)
(8), ovarian cancer (miR-129-5p and
miR-129-3p) (9), colon cancer
(miR-129-5p and miR-129-3p) (10),
heart failure (HF; miR-129-5p) (11), epilepsy (miR-129-5p) (12), intervertebral disc degeneration
(IVDD; miR-129-5p) (13) and
Alzheimer's disease (AD; miR-129-5p) (14), but upregulated in obesity
(miR-129-5p and miR-129-3p) (15)
and diabetes (miR-129-5p and miR-129-3p) (16). In different diseases, the miR-129
family has different targets for modulating various biological
processes (Tables I and II) and involves multiple signaling
pathways (Figs. 1 and 2), thus playing an important role in
promoting or blocking disease progression (Tables III and IV). Members of the miR-129 family have
been reported as potentially promising biomarkers in certain
diseases, such as diabetes and epilepsy (12,16).
Therefore, the aim of the present review was to summarize the
functions of the miR-129 family in the development of oncological
and non-oncological diseases, focusing on the role of miR-129 as a
potential therapeutic target and biomarker in future clinical
applications.
Prostate cancer is one of the most commonly
diagnosed diseases among elderly men (4). The Wnt/β-catenin signaling pathway is
one of the major signaling pathways of epithelial-mesenchymal
transition (EMT), and its target genes mediate cancer initiation
and progression by regulating cell proliferation and apoptosis
(17). In prostate cancer cells,
the Wnt protein can enhance resistance to treatment (18) and the EMT (4). miR-129-5p has a decreased expression
level in prostate cancer tissues (19). In a previous study, the
overexpression of miR-129 decreased Zic family member 2 (ZIC2)
expression and thus inhibited Wnt/β-catenin signaling pathway, that
was the expression of phosphorylated Wnt, β-catenin, N-cadherin and
vimentin decreased, but that of E-cadherin increased, demonstrating
that the overexpression of miR-129-5p might hinder the activation
of the Wnt/β-catenin signaling pathway by targeting (ZIC2 to
inhibit EMT, angiogenesis, tumorigenesis, migration and apoptosis
in prostate cancer (4). ETS variant
transcription factor 1 (ETV1) encodes a number of the E-twenty-six
family of transcription factors and accelerates prostate cancer
cell proliferation through the transcriptional activation of
yes-associated protein (YAP) (20).
One study has identified a negative targeting association between
the ETV1 gene and miR-129-5p, based on reverse transcription
(RT)-qPCR, bioinformatics and dual-luciferase reporter assay
experiments (20). In addition,
Hippo/YAP signaling has been shown to be suppressed through
ETV1(20). Therefore, when
miR-129-5p is overexpressed, the proliferation of prostate cancer
cells is suppressed through the ETV1/Hippo/YAP axis (20). SMAD family member 3 (SMAD3) is a
protein-coding gene; the SMADs protein family plays a key role in
transforming growth factor-β (TGF-β) signaling from cell surface
receptors to the nucleus, and different SMADs mediate the signal
transduction of different TGF-β family members (19). A negative correlation has also been
observed between miR-129-3p and its target gene SMAD3, which
inhibits the proliferation and invasion of prostate cancer cells by
decreasing the expression of anti-apoptosis protein Bcl-2 and
increasing that of the pro-apoptotic protein BAX to promote
apoptosis (19,21). The level of miR-129 in patients with
prostate cancer before and after chemotherapy increase following
treatment, indicating that a high miR-129 expression is beneficial
to patient survival (22).
Therefore, the increased expression of miR-129 plays an important
role in inhibiting prostate cancer cells.
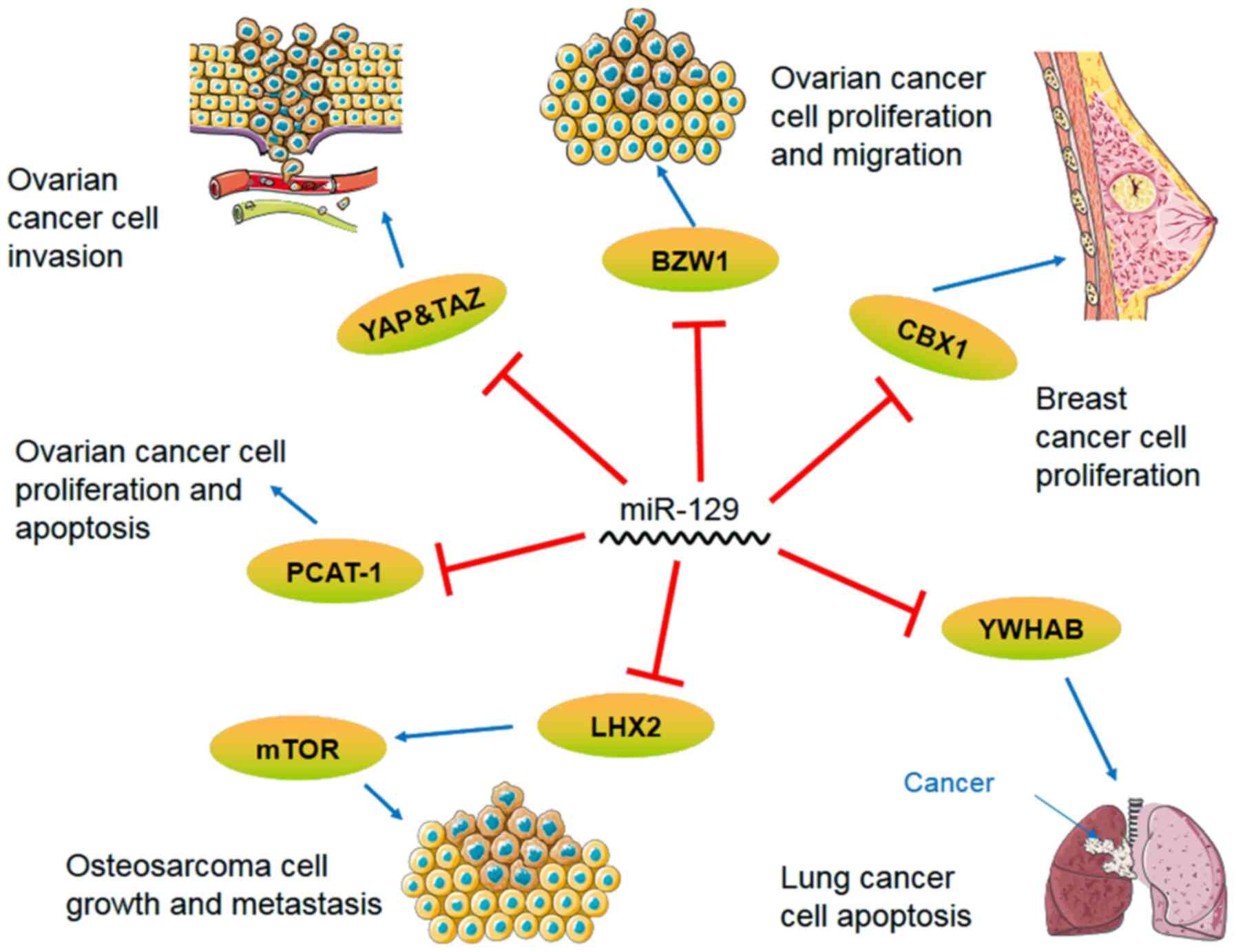
Osteosarcoma is the most common primary bone tumor
and it occurs most frequently in adolescents (23). HIF1A antisense RNA 2 (HIF1A-AS2) is
an RNA gene affiliated with the long non-coding RNA (lncRNAs) class
of RNAs. HIF1A-AS2 lncRNA has been shown to play a vital role in
bladder cancer, colorectal cancer and osteosarcoma (24-26),
among others. The expression of miR-129 has also been found to be
altered in osteosarcoma. Certain studies detected the expression of
HIF1A-AS2 and miR-129-5p in osteosarcoma cells, and used
dual-luciferase reporter assays and other methods to determine the
interaction between the two (24-26);
the results showed that the increased expression of HIF1A-AS2
inhibited osteosarcoma cell proliferation and cancer cell invasion
by altering miR-129-5p expression (5). LIM homeobox 2 (LHX2) is a
protein-coding gene; its protein, consisting of two zinc finger
domains, participates in cell differentiation and embryonic
development (27,28). LHX2 has been shown to play a vital
role in nasopharyngeal carcinoma by regulating Wnt signaling in
breast and lung cancer (29-31).
A previous study showed that in osteosarcoma, LHX2 silencing did
not alter the total amount of Akt or mechanistic target of
rapamycin (mTOR), but decreased the level of phosphorylated-Akt and
-mTOR, and that miR-129-5p knockdown in osteosarcoma promoted
tumorigenesis and inhibited autophagy through the transcription
factor LHX2/Akt/mTOR signaling axis (32). Therefore, miR-129 is associated with
osteosarcoma cell proliferation, cancer cell invasion and
autophagy.
Lung cancer is the second leading cause of mortality
in the rural population of China and is known to cause severe
health problems worldwide (33).
Tyrosine 3-plus monooxygenase/tryptophan 5-plus monooxygenase
activation protein β (YWHAB) interacts with a variety of proteins
and regulates various functions of the human body through both
dependent or independent serine phosphorylation (6). For example, YWHAB is involved in
signal transduction, proliferation and apoptosis, and is crucial to
the occurrence and development of tumors (34). Research has shown that miR-129-5p
overexpression could increase the apoptosis of lung cancer cells
following treatment with miR-129-5p mimics in etoposide-induced
lung cancer cells. Using the enzyme reporting method, western blot
analysis and RT-qPCR, miR-129-5p was proven to be negatively
correlated with YWHAB, and miR-129-5p regulated YWHAB in lung
cancer tissues (35). Zinc finger
E-box binding homeobox 2 (ZEB2) is a member of the ZEB family of
transcription factors and is known to help mediate EMT (36). In non-small cell lung cancer
(NSCLC), miR-129 was found to directly target ZEB2 to inhibit NSCLC
cell proliferation (37). miR-129
also regulates the Wnt/β-catenin signaling pathway by decreasing
the expression of activated β-catenin, c-Myc and cyclin D1.
Therefore, these studies have concluded that miR-129 inhibits NSCLC
cell proliferation, migration and invasion through the ZEB2 axis,
EMT and the Wnt/β-catenin axis. miR-129 has an important regulatory
role in the occurrence and development of lung cancer through
different signaling pathways.
The deregulation of miRNA is closely linked to
cancer development and progression, and miR-129 plays an important
role in breast cancer through different signaling pathways
(7). A previous study confirmed
that miR-129-5p could inhibit proliferation and metastasis in
breast cancer (38). Chromobox 4
(CBX4), also known as polycomb 2, is a special chromobox protein
that it is not only a transcriptional repressor, but also a small
ubiquitin-like modifier E3 ligase, which has been identified as an
oncogene (38). The expression of
CBX4 in MCF7, T47D, MDA-MB-468, MDA-MB-231, SKBR3 and normal MCF10A
breast cancer cells was detected by RT-qPCR and western blot
analysis, and it was found that the overexpression of CBX4 could
promote cell proliferation, while the depletion of CBX4 could
arrest cells in the G2/M phase. The results demonstrated
the regulatory relationship between the CBX4 gene and miR-129-5p by
showing that the increase of miR-129-5p depleted CBX4, which
inhibited cancer cell proliferation. This correlation may provide
new targets and ideas for the treatment of breast cancer (7).
Triple-negative breast cancer has high invasive
potential and a high recurrence rate, and its successful treatment
remains a challenge (39).
Silencing lncRNA metastasis-associated lung adenocarcinoma
transcript 1 (MALAT1) was shown to halt triple-negative breast
cancer cells at the G0/G1 stage through an
miR-129-5p-mediated mechanism, and could also markedly decrease
cell proliferation, migration and invasion (40). The continuous improvement of the
diagnosis and treatment of breast cancer, and the development of
novel treatments, is accompanied by drug resistance. Trastuzumab is
a drug specifically targeted to human epidermal growth factor
receptor 2 (Her-2)-positive breast cancer; it prevents the
combination of Her-2 and human epidermal growth factor, thus
leading to a therapeutic effect (41). In trastuzumab-resistant
Her-2-positive breast cancer cells, miR-129-5p was shown to be
downregulated, and a dual-luciferase reporter assay experiment
showed that miR-129-5p enhanced breast cancer sensitivity to
trastuzumab by targeting ribosomal protein S, which is crucial to
combating Her-2 trastuzumab resistance (42). The response to hormonal therapy is
affected by the progesterone receptor (PR)-status of patients with
breast cancer. The overexpression of miR-129-2 stimulated the
effect of progesterone treatment by downregulating PR, and the
inhibition of miR-129-2 repealed its interaction with PR in breast
cancer cells (43). To the best of
our knowledge, there have been no specific experimental studies on
luminal A and B breast cancer; however, clinically, the expression
of miR-129 in the serum of patients with breast cancer is lower
than that of healthy individuals (44,45).
The etiology of nasopharyngeal carcinoma involves
several factors and lacks obvious characteristics (8). ZIC2 is a transcription factor encoded
by the ZIC2 gene, which has been proven to play a role in a variety
of cancer types (46). A direct
targeting relationship between miR-129-5p and ZIC2 has been
confirmed, which eliminates the activation of the Hedgehog
signaling pathway, thereby inhibiting lymphangiogenesis and lymph
node metastasis in nasopharyngeal carcinoma. It has also been shown
that miR-129-5p can decrease the expression of associated genes
smoothened, frizzled class receptor and GLI family zinc finger 1 in
the Hedgehog signaling pathway, thus inhibiting the apoptosis of
nucleus pulposus cells (NPCs) (8).
In addition, WEE1 G2 checkpoint kinase (WEE1; molecular weight, 96
kDa) is a member of the serine/threonine-protein kinase family,
which can phosphorylate threonine 14 and tyrosine 15 of
cyclin-dependent kinase 1 (CDK1) to inhibit CDK1 kinase activity,
thereby inhibiting cell mitosis (47). The expression level of miR-129-3p in
cisplatin-resistant nasopharyngeal carcinoma cells was found to be
significantly lower than that in non-resistant nasopharyngeal
carcinoma cells, and a negative targeting relationship was
identified between miR-129-1-3p and WEE1. miR-129-1-3p was found to
decrease cisplatin resistance by inhibiting WEE1 in
cisplatin-resistant cells (48). In
conclusion, miR-129 plays an important role in the apoptosis and
drug resistance of nasopharyngeal carcinoma cells.
Ovarian cancer is one of the most common types of
cancer and the leading cause of cancer-associated mortality in
women, according to global statistics in 2019(49). Basic leucine zipper and W2 domains 1
(BZW1) is a member of the bZIP superfamily of transcription factors
(50). The overexpression of
miR-129-3p targets the cell cycle regulator BZW1 in SKOV3 cells,
which arrests most cells in the G0/G1 phase
and only permits a few cells to aggregate in the S phase,
suggesting that miR-129-3p inhibits ovarian cancer cell
proliferation (50). In addition,
studies have confirmed that miR-129-3p could inhibit ovarian cancer
cell migration, indicating that miR-129-3p also plays an important
role in the development of ovarian cancer (4,9,50).
Small nucleolus RNA host gene 12 (SNHG12) is an RNA gene associated
with the lncRNA class of RNAs. SNHG12 lncRNA has been shown to play
a vital role in gastric and colorectal cancer (51,52).
miR-129 expression was downregulated by SNHG12 overexpression in
ovarian cancer cells, and it could regulate ovarian cancer
development by promoting proliferation and enhancing the migratory
ability of cells through the downregulation of SRY-box
transcription factor 4 (SOX4) (53). The biological functions of prostate
cancer-associated transcript 1 (PCAT1), which was initially shown
to play oncogenic roles in prostate cancer (54), have been documented in multiple
types of human cancer, including gastric cancer (55), hepatocellular carcinoma and
colorectal cancer (56,57). PCAT-1 expression was also increased
in ovarian cancer and could promote cancer cell proliferation and
inhibit cancer cell apoptosis, the opposite trend of that observed
for miR-129 expression and the phenotype in ovarian cancer cells.
Bioinformatics predictions and dual-luciferase reporter assay
results showed that PCAT-1 and miR-129 were negatively correlated,
and that the overexpression of PCAT-1 inhibited miR-129, promoting
ovarian cancer development (49).
lncRNA overexpression of urothelial
carcinoma-associated protein 1 (UCA1) is associated with resistance
to chemotherapeutics and plays a key role in anticancer drug
resistance (58). A previous study
showed that lncRNA UCA1 was highly expressed in paclitaxel
(PTX)-resistant ovarian cancer, while its knockdown weakened that
resistance (9). It was further
demonstrated that UCA1 competitively bound to miR-129, which
triggered the release of ATP-binding cassette subfamily B member 1
(ABCB1) mRNA and increased the resistance of ovarian cancer cells
to PTX. Briefly, ABCB1 upregulation via the UCA1/miR-129 axis
promoted PTX resistance in ovarian cancer (9). YAP and tafazzin (TAZ) transcription
coactivators are key oncogenes in mammalian cells. The levels of
these two transcription coactivators were decreased in SKOV3 cells
transfected with miR-129-5p. Low miR-129-5p expression was shown to
increase YAP and TAZ expression, thus increasing the aggressiveness
of ovarian cancer and leading to a poor prognosis (59). Among all the changes in the
expression of breast- and ovarian-related genes and their
regulatory miRNA expression and the methylation of promoter CpG
islands, the methylation of miR-129-3p and SEMA3B has been found at
a high frequency in ovarian cancer. In addition, a correlation
between SEMA3B promoter hypermethylation and miR-129 gene
downregulation was identified in ovarian cancer and breast cancer
(60). In conclusion, miR-129 plays
a significant role in ovarian cancer cell proliferation and
migration.
MALAT1 has been proven to be an important regulator
of miR-129 in breast cancer (10),
and high-mobility group box-1 (HMGB1) is a vital member of the HMG
family, which has been shown to play a crucial role in pancreatic
(61), breast (62) and prostate (63) cancer. In colon cancer cells,
miR-129-5p was downregulated, and the expression of HMGB1 and
MALAT1 was increased. The MALAT1/miR-129-5p/HMGB1 axis was also
found to be an important target axis for regulating the progression
of colon cancer (10). p53 is a
tumor suppressor protein and transcription factor that can regulate
cell division, prevent DNA mutations or division of damaged cells,
and transmit apoptotic signals to those cells through
transcriptional regulation, thereby preventing tumor formation. The
active ingredient, Avenanthramide A, which is present in oats, was
found to exert a protective mechanism against colon cancer by
elevating miR-129-3p to inhibit ring finger and CHY zinc finger
domain containing 1 (Pirh2), resulting in a significant increase in
the protein levels of p53 and its downstream target p21, ultimately
leading to a significant increase in senescent cells (64). In other words, Avenanthramide A
induces cell senescence through the miR-129-3p/Pirh2/p53 axis
(64). The occurrence and
development of colon cancer are closely linked to the
downregulation of miR-129. Experiments using a new miR-129 mimic
(mimic-1) found that, compared with natural miR-129, mimic-1 had a
stronger inhibitory effect on colon cancer and induced colon cancer
cell apoptosis. Furthermore, the half-life of mimic-1 was longer
than that of natural miR-129, and so has a more profound impact as
a therapeutic candidate in colon cancer treatment (65).
Obesity is a metabolic disease and an important
inducing factor of metabolic disorders (70,71),
such as type II diabetes and cardiovascular disease (15).
An eicosapentaenoic acid-regulating differentially
expressed gene analysis of miRNAs in brown adipose tissue (BAT)
from diet-induced obese mice indicated that miR-129-5p and
miR-129-3p were significantly upregulated, which was subsequently
validated by RT-qPCR (15). Obesity
is defined as redundant fat accumulation in adipose tissue without
a clearly known pathogenesis (72).
Another analysis of autophagy-related genes showed that key
adipocyte differentiation pathways, including mTOR and insulin, and
adipocytokine signaling pathways may be upregulated by
autophagy-related gene 7 (ATG7)-mediated autophagy (73,74).
Autophagy is a cellular recycling pathway that delivers cytoplasmic
cargo into lysosomes for the maintenance of cellular quality
control (75), which may be
associated with obesity (76). ATG7
has been identified as one of the principal molecular executors of
autophagy (77). By removing
ubiquitinated AMP-activated protein kinase and thus stimulating
mTOR signaling pathways (78), the
activation of ATG7-mediated autophagy, which has been confirmed to
lead to increased white adipose tissue (WAT), is vital for
adipocyte differentiation. miR-129-5p may directly target ATG7 to
downregulate autophagy pathways, resulting in the inhibition of
white adipogenesis (79).
Consistently, when transfecting miR-129-5p mimics in the
subcutaneous fat tissues of mice, the expression of key effectors
in adipocyte differentiation, fatty acid-binding protein 4,
CCAAT/enhancer-binding protein and peroxisome
proliferator-activated receptor γ, were decreased (79).
In addition, the overexpression of miR-129-5p was
found to lead to the inhibition of brown adipogenesis, which may be
associated with the decrease protein expression of adipogenic genes
and specific markers of brown mature adipocytes, such as uncoupling
protein 1, PR/SET domain 16 and cell death-inducing DFFA-like
effector-a (79). The use of
miR-129-5p inhibitors led to an increase in the number of mature
brown adipocytes compared with the use of negative control
inhibitors (79). However, the
direct target of miR-129-5p in BAT remains unknown. Considering
that BAT activation abnormality may contribute to obesity,
exploring the regulatory mechanism of miR-129-5p in brown
adipogenesis is worthy of consideration.
An excessive body intake of triacylglycerols was
shown to lead to adipocyte hyperplasia and subsequent accumulation
of lipids in WAT, consequently promoting the development of obesity
(80). A series of predicted target
genes of miR-129 have been found to be associated with
triacylglycerol metabolism and thus, miR-129 may regulate the
progression of obesity (81).
However, the exact mechanism of the miR-129-mediated alteration of
triacylglycerol metabolism in obesity requires further study.
The miR-129 family negatively regulates the
formation of obesity using at least three different approaches,
involving white and brown adipogenesis and triacylglycerol
metabolism. To the best of our knowledge, miR-129 can directly
target and inhibit ATG7 to decrease the accumulation of WAT, while
its other targets remain uncharted or unsubstantiated in brown
adipogenesis and triacylglycerol metabolism.
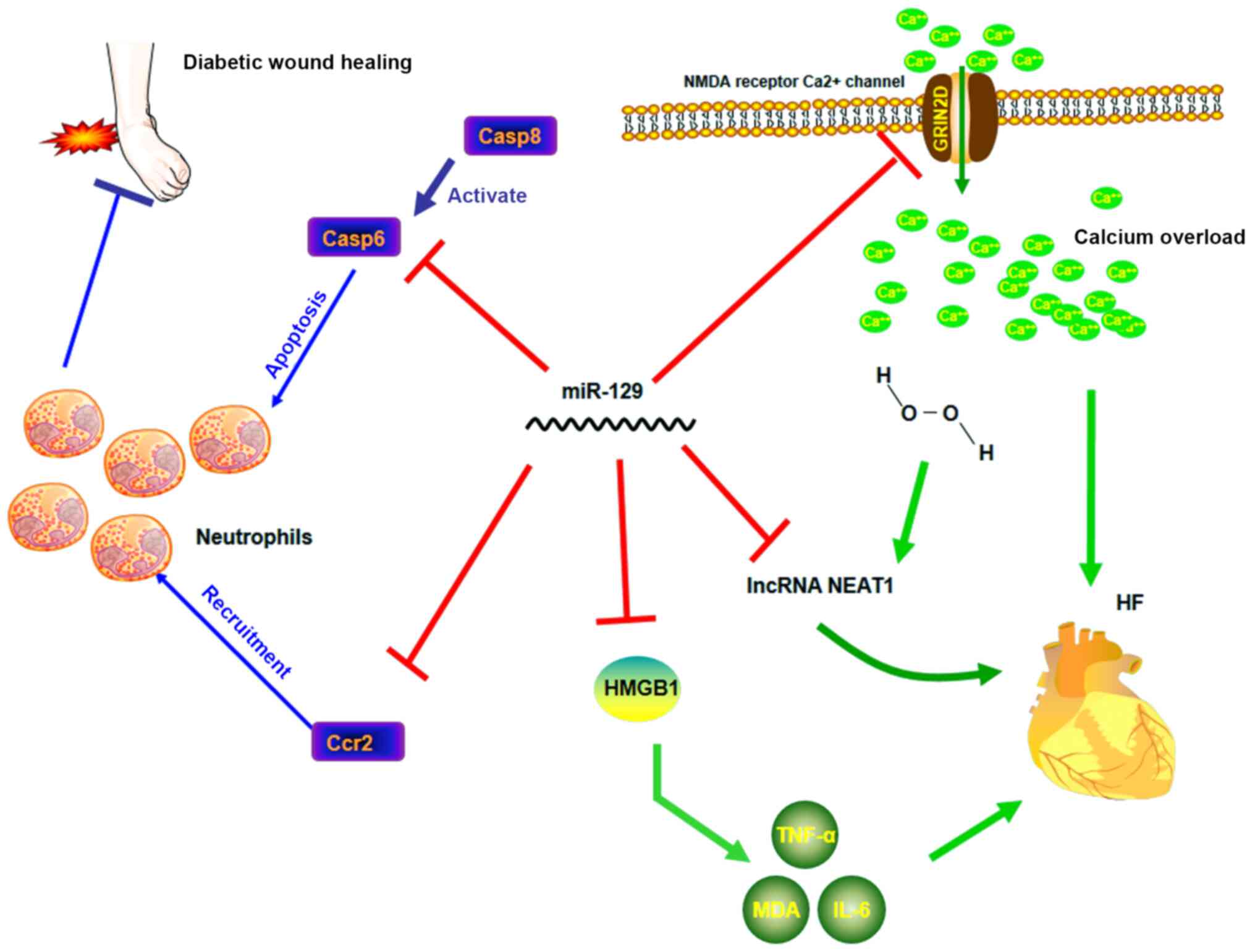
HF occurs in ~1/5 individuals during their lifetime,
particularly individuals aged >40 years, decreasing their
quality of life (82). As shown by
a longitudinal study, HF is associated with inflammatory reactions
(83). HMGB1 is an omnipresent
nuclear protein that is commonly discharged by dead or dying cells
(84). A previous study showed that
HMGB1 was an effective biomarker for predicting heart
transplantation in patients with HF (85). The persistence of HMGB1 in the
microenvironment is one of the main contributing factors to the
increase in inflammation and more severe symptoms (86). HMGB1 inhibition can alleviate
inflammation, thus affecting the pathogenesis and progression of HF
(11). miR-129-5p was found to
complementarily bind to the 3'-UTR of HMGB1 mRNA with its seed
region, which led to a decrease in the protein levels of HMGB1,
followed by a decrease in the levels of TNF-α and IL-6 to
ameliorate HF (11).
Anthracyclines are highly effective in the treatment
of a large number of malignancies (87). However, HF, a well-known side effect
of these drugs, limits its use in chemotherapy (88-90).
As shown by Kyoto Encyclopedia of Genes and Genomes pathway
enrichment analysis, Ca2+ pathway dysregulation is an
important cause of anthracycline-induced HF (91,92).
Ca2+ can trigger mitochondrial swelling in
anthracycline-damaged cardiac tissues (93) and Ca2+ overload may
result in the degradation of the myofilament protein titin, causing
sarcomere disruption and cell necrosis (91). Glutamate ionotropic receptor
N-methyl-D-aspartate (NMDA) type subunit 2D (GRIN2D) is an
essential subunit of the NMDA receptor Ca2+ channel,
which involves the formation of ligand-gated ion channels with high
Ca2+ permeability (94).
Since GRIN2D is considered a key regulator of Ca2+
influx, its expression is closely linked to cytoplasmic
Ca2+ content (10).
Calmodulin 1 (CALM1) and Ca2+/calmodulin-dependent
protein kinase II δ (CaMKIIδ), the key downstream activators of the
Ca2+ pathway in cardiomyocytes, have been linked to
myocardial cell death, cardiomyopathy and HF (95,96).
Therefore, GRIN2D overexpression caused Ca2+ overload,
leading to excessive activation of CALM1 and CaMKIIδ, and
eventually deteriorating HF (92).
By targeting GRIN2D and thus alleviating the GRIN2D-mediated
Ca2+ signaling pathway activity (92), miR-129-3p can downregulate
Ca2+ overload to prevent pirarubicin-induced
cardiomyocyte death and HF (92).
In addition, a study reported that the clearly
increased serum levels of lncRNA nuclear paraspeckle assembly
transcript 1 (NEAT1) in patients with HF were negatively correlated
with miR-129-5p levels (97).
Further research showed that the improvement in
H2O2-induced apoptosis mediated by a lncRNA
NEAT1 mimic in H2C9 immortalized cardiomyocytes could be strongly
attenuated by a miR-129-5p mimic, which indicated that the
interaction between lncRNA NEAT1 and miR-129-5p in myocardiocytes
may have implications in HF (97).
In conclusion, the miR-129 family, including miR-129-5p and
miR-129-3p, modulates the progression of HF through various
mechanisms, and current evidence suggests it has a favorable effect
on HF.
Epilepsy is a neurodegenerative disease that affects
>50 million individuals worldwide (98); its long-term predisposition to
seizures is caused by the hyperexcitability and hypersynchrony of
brain neurons (99,100). To date, there is a lack of
effective therapeutic targets or specific biomarkers for
epileptogenesis (101,102). Mitogen-activated protein kinases
(MAPKs) are serine-threonine kinases that regulate various cellular
activities, including proliferation, differentiation, survival and
death, through intracellular signaling (103). Therefore, MAPK pathway abnormality
is closely correlated with multiple types of cancer and
neurodegenerative diseases, including Parkinson's disease, AD and
epilepsy. At present, MAPK is considered an important regulator of
synaptic excitability, whose overactivation gives rise to epileptic
seizures (104). c-Fos is
considered a molecular sensor for MAPK signaling duration (105). Inhibiting c-Fos can block MAPK
signaling transduction. A previous study reported that miR-129-5p
could negatively target c-Fos (105), suggesting that miR-129-5p may
repress epilepsy seizures (103).
Therefore, miR-129-5p targets the sensor c-Fos to inhibit MAPK
signaling and then apoptosis, and promotes the proliferation of
hippocampal neurons, thereby suppressing the occurrence and
development of epilepsy (105).
However, the protective role of miR-129-5p is controversial due to
its synaptic downscaling function. Synaptic downscaling is a
homeostatic mechanism that decreases the firing rates of neurons
during chronically elevated network activity (106,107). Although synaptic downscaling is
crucial in epilepsy, the related pathways are poorly understood
(106,107). Following a small RNA profiling of
a synaptic downscaling model of picrotoxin-induced hippocampal
neurons, miR-129-5p was found to be increased in response to
picrotoxin (106,107). miR-129-5p inhibition led to the
inhibition of synaptic downscaling in vitro and decreased
epileptic seizure severity in vivo, indicating the potential
malignant influence of miR-129-5p on epilepsy (106,107).
By contrast, a different study reported the opposite
conclusion, supporting the beneficial function of miR-129-5p. The
study found that a high concentration of miR-129-5p significantly
enhanced the expression of pro-inflammatory factors such as IL-6,
cyclooxygenase-2 and TNF-α in epilepsy cell models, which increased
the frequency of epileptic seizures (108). The explanation of these two
opposite conclusions may be explained by the fact that miR-129-5p
plays different roles in each stage of epilepsy development;
therefore, its underlying mechanism requires further study.
Although diabetes has various symptoms, different
subtypes of diabetes share the same pathophysiological abnormality,
that is, damage to the islets (109). The timely detection of islet
destruction may benefit the protection of islet β cell function
(16). Exosomes, as newly
discovered essential conveyers of cellular information, have
attracted considerable attention as disease biomarkers in recent
years (110,111). Exosomes are extracellular small
membrane vesicles (30-100 nm) that contain proteins and nucleic
acids, including miRNA (111);
they can be secreted by various types of cells into the blood
circulation and then seized into recipient cells (110), which renders them efficient
intercellular information transmitters. Information in these
vesicles varies based on the status of the cells (110). As a consequence, the detection of
changes in functional biomolecules in exosomes may reveal cell
characteristics. miRNA profiles of exosomes derived from
streptozotocin- or mixed cytokine [TNF-α, IL-1β and interferon
(IFN-γ)]-induced injured islets demonstrated that six miRNAs,
including miR-129-5p, were differentially expressed at the
intersection between two injured conditions (112). Although debates on the mechanisms
of metformin in different tissues continue to this day (16), metformin has become a commonly used
drug in the treatment of type 2 diabetes since French scientist
Jean Sterne first produced it in 1957(113). One previous study reported that 3
months of metformin treatment for type 2 diabetes significantly
attenuated miR-129-3p expression, which may be a result of improved
islet function (16). This finding
showed that the differential expression of miR-129-5p and five
other miRNAs in islet-derived exosomes specifically indicated islet
β cell injury, regardless of the cause of injury.
Continuous accumulation of neutrophils and
macrophages is commonly detected in the wound sites of diabetic
patients (114), which can delay
wound healing and complications, such as foot ulcers (114,115). Previous studies have also
suggested that the caspase family is the main contributor of both
spontaneous and Fas receptor-mediated apoptosis in neutrophils
(116,117). Fas ligand and its death receptors
are the activators of caspase 8 (116,118), which mainly contribute to the
activation of death-inducing signaling complex by activating its
downstream caspases and can stimulate neutrophil apoptosis
(116). C-C chemokine receptor
type 2 (CCR2) is a chemokine receptor commonly expressed in
lymphocytes and monocytes, but not in neutrophils (88). However, specific inflammatory
stimuli in wounds have been confirmed to elevate the expression of
CCR2 in neutrophils (119,120). In diabetic wound sites, CCR2
expression was increased, which promoted wound recruitment in
neutrophils (120); accordingly,
CCR2 expression may play a key role in chronic inflammation in
diabetic patients (121).
miR-129-3p was previously found to directly regulate the
translation of caspase 6, the downstream activator of caspase 8,
and CCR2 to decrease apoptosis and neutrophil recruitment and
consequently accelerate diabetic wound healing (121). Similar to miR-129-3p, miR-129-5p
plays a beneficial role in diabetic wound healing (122).
Matrix metalloproteinases (MMPs) are widely known to
inhibit tissue remodeling by degrading extracellular matrix (ECM)
components (123). A high level of
MMP-9 expression causes excessive degradation of ECM components,
contributing to the imbalance between ECM synthesis and degradation
(120). The high level of MMP-9 is
therefore usually indicative of poor wound healing, as well as the
existence of diabetic wounds (120). MMP expression and activity can be
tightly regulated by methylation at specific DNA loci in the MMP-9
promoter region (124). Thus,
specificity protein-1 (Sp1), a transcription factor found to bind
to the MMP-9 promoter to initiate MMP-9 gene expression, can be an
important regulatory target for MMP-9(122). Through dual-luciferase reporter
assays, miR-129-5p was confirmed to directly regulate Sp1 protein
levels and significantly decrease the expression of MMP-9.
miR-129-5p thus modulates the SP1/MMP-9 axis and removes an
important obstacle in diabetic wound healing (122).
IVDD is a cause of lower back pain, which affects
>50% of the world's population during their lifetime, and is a
severe social burden (125-127).
Accumulating evidence shows that the apoptosis and proliferation of
fibrocartilaginous tissues on the intervertebral disc are
implicated in the progression of IVDD (128). NPCs, a type of chondroid cells
that exist in the intervertebral disc, lead the deposition of
fibrocartilage lamellas and fibers in a centripetal direction, thus
converting notochordal nucleus pulposus into fibrocartilaginous
nucleus pulposus (129). It has
been demonstrated that one of the main characteristics of IVDD is
the decrease in the number of NPCs, which leads to severe disc
deformation and dysregulation of its function (130).
Bone morphogenetic protein 2 (BMP2), first reported
as the anabolic growth factor regulating proteoglycan release
(131), was expressed at low
levels in NPCs collected from patients with IVDD, and its low
expression was accompanied by increased miR-129-5p expression
(13). Previous reports confirmed
that BMP2 was involved in the mediation of apoptosis, with its role
being dependent on cellular context and cell type (13). Yang and Sun (13) validated BMP2 as a promoter of
apoptosis and an inhibitor of NPC proliferation in IVDD, although
it was also found to stimulate proteoglycan secretion to prevent
the degradation of the ECM, which may help delay IVDD progression
(13). Fas-associated death domain
(FADD), an important agent for the triggering of cell death without
the enrollment of death receptors, was also demonstrated to
facilitate NPC apoptosis (132,133). Both BMP2 and FADD were confirmed
to be targeted by miR-129-5p, and the overexpression of miR-129-5p
was found to significantly facilitate the proliferation and
suppress the apoptosis of NPCs (13). Furthermore, high miR-129-5p
expression alleviated disc inflammation by promoting the apoptosis
of macrophages and CD8+ cells. Collectively, miR-129-5p
exerts effects not only on NPCs, but also on inflammatory cells,
although its targets in the latter have not yet been verified
(13,134). Contrary to the aforementioned
results, one study reported that miR-129-5p increased NPC apoptosis
by suppressing autophagy (135).
Specifically, autophagy progresses through the Beclin-1-dependent
pathway; therefore, Beclin-1 is essential for autophagy (136). Beclin-1 may promote autophagy,
while autophagy can decrease cathepsin B release into the cytoplasm
(135). In light of the positive
correlation between the cytoplasmic level of cathepsin B and
apoptosis, the Beclin-1-targeting capability of miR-129-5p
increased cytoplasmic release of cathepsin B from lysosomes and
resulted in the inhibition of autophagy and promotion of apoptosis.
This death-inducing effect to NPCs rendered miR-129-5p harmful to
the human intervertebral disc in that study (135). In conclusion, miR-129-5p appears
to modulate apoptosis in NPCs by targeting different factors, but
its implications in IVDD require further study.
As a significant neurodegenerative disease, AD is
characterized by the accumulation of β-amyloid plaques and the
dysfunction of τ-protein in the brain (137). In a longitudinal cohort study of
aging (based on 700 autopsy samples), miR-129-5p was shown to be
downregulated in the human dorsolateral prefrontal cortex of
patients with AD (14), and its
expression was found to be associated with the two characteristics
of AD, as shown by miRNA expression profiling (14,138).
In the study, miR-129-5p appeared to be involved in a variety of
pathways, the most significant of which was transcription
regulation (14). Notably,
miR-129-5p coordinated with another miRNA, miR-132, to target E1A
binding protein P300 (EP300), the encoding gene of the histone
acetyltransferase protein E1A-associated cellular p300
transcriptional co-activator, and each miRNA was responsible for
part of the variation in EP300 expression (14). A reasonable speculation is that
miR-129-5p influences histone acetylation and chromatin remodeling
through the regulation of EP300 to alter the expression of
β-amyloid and τ-protein in patients with AD.
The SOX gene family contains transcription
factor-coding genes that possess highly conserved HMG-box sequences
(138). A principal function of
SOX transcription factors is to regulate neurogenesis in the
embryonic and adult nervous system (139). It has been reported that the
expression of SOXB transcription factors may be involved in the
early disability of hippocampal neurogenesis in 5xFAD mouse models
of AD (139). A previous study
demonstrated that miR-129-5p overexpression decreased the
expression of its target SOX6(140), leading to the decrease of
apoptosis, the suppression of inflammatory reactions and the
increased proliferation of rat hippocampal neurons (140).
Certain studies have reported the role of the
miR-129 family in other diseases. For example, miR-129 was found to
target beclin-1 in atherosclerosis (141), HMGB1 in spinal cord injury
(142), spleen-associated tyrosine
kinase in ischemic stroke (143),
fibroblast growth factor 23 in chronic kidney disease (144) and NF-κB in hepatic fibrosis
(145).
Increasing evidence indicates the important
functions of the miR-129 family in various diseases, highlighting
its potential as a therapeutic candidate or prognostic biomarker.
In the present review, it was concluded that the dysregulated
expression of miR-129 is a common feature in several types of
cancer. In addition, it was shown that miR-129 plays different
roles in various types of cancer, depending on the disease type. In
most types, miR-129 regulates genes involved in a variety of
biological processes, including cell proliferation, migration,
invasion, cell apoptosis induction and drug resistance. In
conclusion, miR-129 family can be potentially used as a drug-tool
in the treatment for some of the aforementioned diseases with
little if any harm to the normal tissue. With regards to
non-cancerous diseases, previous research has supported the roles
of miR-129 in the treatment of HF and AD, and confirmed that
miR-129 has the potential to be a biomarker in diabetes and
epilepsy. It is worth noting that the suppressive effect of miR-129
on apoptosis has been consistently reported by multiple research
groups, providing a direction for future research. However, since
miR-129 could induce or suppress NPC apoptosis through different
signaling axes, its role in IVDD remains controversial and requires
further study. The present review systematically summarized the
role of the miR-129 family in oncological and non-oncological
diseases. To the best of our knowledge, this is the first review to
summarize the functions of miR-129 in non-cancerous diseases,
including obesity, HF, epilepsy, diabetes, IVDD and AD.
Not applicable.
Funding: Funding was provided by the Natural Science Foundation
of Hunan Province, China (grant no. 2019JJ40392) and the
Undergraduate Innovation and Entrepreneurship Training Program
Supportive Project (grant no. 2020105330143).
Not applicable.
BD and XT wrote the paper, and YW revised the
manuscript. All authors have read and approved the manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Zhang HD, Jiang LH, Sun DW, Li J and Ji
ZL: The role of miR-130a in cancer. Breast Cancer. 24:521–527.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gao Y, Feng B, Han S, Lu L, Chen Y, Chu X,
Wang R and Chen L: MicroRNA-129 in human cancers: From
tumorigenesis to clinical treatment. Cell Physiol Biochem.
39:2186–2202. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cai X, Hagedorn CH and Cullen BR: Human
microRNAs are processed from capped, polyadenylated transcripts
that can also function as mRNAs. RNA. 10:1957–1966. 2004.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jiang Z, Zhang Y, Chen X, Wu P and Chen D:
Inactivation of the Wnt/β-catenin signaling pathway underlies
inhibitory role of microRNA-129-5p in epithelial-mesenchymal
transition and angiogenesis of prostate cancer by targeting ZIC2.
Cancer Cell Int. 19(271)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang X, Peng L, Gong X, Zhang X and Sun R:
LncRNA HIF1A-AS2 promotes osteosarcoma progression by acting as a
sponge of miR-129-5p. Aging (Albany NY). 11:11803–11813.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Obsil T: 14-3-3 proteins-a family of
universal scaffolds and regulators. Semin Cell Dev Biol.
22:661–662. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Meng R, Fang J, Yu Y, Hou LK, Chi JR, Chen
AX, Zhao Y and Cao XC: miR-129-5p suppresses breast cancer
proliferation by targeting CBX4. Neoplasma. 65:572–578.
2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yu D, Han GH, Zhao X, Liu X, Xue K, Wang D
and Xu CB: MicroRNA-129-5p suppresses nasopharyngeal carcinoma
lymphangiogenesis and lymph node metastasis by targeting ZIC2. Cell
Oncol (Dordr). 43:249–261. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang J, Ye C, Liu J and Hu Y: UCA1 confers
paclitaxel resistance to ovarian cancer through miR-129/ABCB1 axis.
Biochem Biophys Res Commun. 501:1034–1040. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wu Q, Meng WY, Jie Y and Zhao H: LncRNA
MALAT1 induces colon cancer development by regulating
miR-129-5p/HMGB1 axis. J Cell Physiol. 233:6750–6757.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Xiao N, Zhang J, Chen C, Wan Y, Wang N and
Yang J: miR-129-5p improves cardiac function in rats with chronic
heart failure through targeting HMGB1. Mamm Genome. 30:276–288.
2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hübner A, Jaeschke A and Davis RJ:
Oncogene addiction: Role of signal attenuation. Dev Cell.
11:752–754. 2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yang W and Sun P: Downregulation of
microRNA-129-5p increases the risk of intervertebral disc
degeneration by promoting the apoptosis of nucleus pulposus cells
via targeting BMP2. J Cell Biochem. 120:19684–19690.
2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Patrick E, Rajagopal S, Wong HA, McCabe C,
Xu J, Tang A, Imboywa SH, Schneider JA, Pochet N, Krichevsky AM, et
al: Dissecting the role of non-coding RNAs in the accumulation of
amyloid and tau neuropathologies in Alzheimer's disease. Mol
Neurodegener. 12(51)2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Pahlavani M, Wijayatunga NN, Kalupahana
NS, Ramalingam L, Gunaratne PH, Coarfa C, Rajapakshe K, Kottapalli
P and Moustaid-Moussa N: Transcriptomic and microRNA analyses of
gene networks regulated by eicosapentaenoic acid in brown adipose
tissue of diet-induced obese mice. Biochim Biophys Acta Mol Cell
Biol Lipids. 1863:1523–1531. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Demirsoy IH, Ertural DY, Balci Ş, Çınkır
Ü, Sezer K, Tamer L and Aras N: Profiles of circulating MiRNAs
following metformin treatment in patients with type 2 diabetes. J
Med Biochem. 37:499–506. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Arend RC, Londoño-Joshi AI, Straughn JM Jr
and Buchsbaum DJ: The Wnt/β-catenin pathway in ovarian cancer: A
review. Gynecol Oncol. 131:772–779. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Murillo-Garzón V and Kypta R: WNT
signalling in prostate cancer. Nature reviews. Urology. 14:683–696.
2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tang PM, Zhou S, Meng XM, Wang QM, Li CJ,
Lian GY, Huang XR, Tang YJ, Guan XY, Yan BP, et al: Smad3 promotes
cancer progression by inhibiting E4BP4-mediated NK cell
development. Nat Commun. 8(14677)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Gao G, Xiu D, Yang B, Sun D, Wei X, Ding
Y, Ma Y and Wang Z: miR-129-5p inhibits prostate cancer
proliferation via targeting ETV1. OncoTargets and Ther.
12:3531–3544. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jia Y, Gao Y and Dou J: Effects of
miR-129-3p on biological functions of prostate cancer cells through
targeted regulation of Smad3. Oncol Lett. 19:1195–1202.
2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hu Z, Guo J, Zhao M, Jiang T and Yang X:
Predictive values of miR-129 and miR-139 for efficacy on patients
with prostate cancer after chemotherapy and prognostic correlation.
Oncol Lett. 18:6187–6195. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang RM, Tang T, Yu HM and Yao XD: LncRNA
DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma
through Wnt signaling. Biochem Biophys Res Commun. 507:260–266.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen X, Liu M, Meng F, Sun B, Jin X and
Jia C: The long noncoding RNA HIF1A-AS2 facilitates cisplatin
resistance in bladder cancer. J Cell Biochem. 120:243–252.
2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lin J, Shi Z, Yu Z and He Z: LncRNA
HIF1A-AS2 positively affects the progression and EMT formation of
colorectal cancer through regulating miR-129-5p and DNMT3A. Biomed
Pharmacother. 98:433–439. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lin H, Zhao Z, Hao Y and He J and He J:
Long noncoding RNA HIF1A-AS2 facilitates cell survival and
migration by sponging miR-33b-5p to modulate SIRT6 expression in
osteosarcoma. Biochem Cell Biol. 98:284–292. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hou PS, Chuang CY, Kao CF, Chou SJ, Stone
L, Ho HN, Chien CL and Kuo HC: LHX2 regulates the neural
differentiation of human embryonic stem cells via transcriptional
modulation of PAX6 and CER1. Nucleic Acids Res. 41:7753–7770.
2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tomann P, Paus R, Millar SE, Scheidereit C
and Schmidt-Ullrich R: Lhx2 is a direct NF-κB target gene that
promotes primary hair follicle placode down-growth. Development.
143:1512–1522. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liang TS, Zheng YJ, Wang J, Zhao JY, Yang
DK and Liu ZS: MicroRNA-506 inhibits tumor growth and metastasis in
nasopharyngeal carcinoma through the inactivation of the
Wnt/β-catenin signaling pathway by down-regulating LHX2. J Exp Clin
Cancer Res. 38(97)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kuzmanov A, Hopfer U, Marti P,
Meyer-Schaller N, Yilmaz M and Christofori G: LIM-homeobox gene 2
promotes tumor growth and metastasis by inducing autocrine and
paracrine PDGF-B signaling. Mol Oncol. 8:401–416. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhou F, Gou S, Xiong J, Wu H, Wang C and
Liu T: Oncogenicity of LHX2 in pancreatic ductal adenocarcinoma.
Mol Biol Rep. 41:8163–8167. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Song H, Liu J, Wu X, Zhou Y, Chen X, Chen
J, Deng K, Mao C, Huang S and Liu Z: LHX2 promotes malignancy and
inhibits autophagy via mTOR in osteosarcoma and is negatively
regulated by miR-129-5p. Aging (Albany NY). 11:9794–9810.
2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li G, Xie J and Wang J: Tumor suppressor
function of miR-129-5p in lung cancer. Oncol Lett. 17:5777–5783.
2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jeong BH, Jin HT, Choi EK, Carp RI and Kim
YS: Lack of association between 14-3-3 beta gene (YWHAB)
polymorphisms and sporadic Creutzfeldt-Jakob disease (CJD). Mol
Biol Rep. 39:10647–10653. 2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Xu C, Du Z, Ren S, Liang X and Li H:
MiR-129-5p sensitization of lung cancer cells to etoposide-induced
apoptosis by reducing YWHAB. J Cancer. 11:858–866. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Jacob S, Nayak S, Fernandes G, Barai RS,
Menon S, Chaudhari UK, Kholkute SD and Sachdeva G: Androgen
receptor as a regulator of ZEB2 expression and its implications in
epithelial-to-mesenchymal transition in prostate cancer. Endocr
Relat Cancer. 21:473–486. 2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li X, Li C, Bi H, Bai S, Zhao L, Zhang J
and Qi C: Targeting ZEB2 By microRNA-129 in non-small cell lung
cancer suppresses cell proliferation, invasion and migration via
regulating Wnt/beta-catenin signaling pathway and
epithelial-mesenchymal transition. Onco Targets Ther. 12:9165–9175.
2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang X, Li L, Wu Y, Zhang R, Zhang M, Liao
D, Wang G, Qin G, Xu RH and Kang T: CBX4 suppresses metastasis via
recruitment of HDAC3 to the Runx2 promoter in colorectal carcinoma.
Cancer Res. 76:7277–7289. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Chaudhary LN, Wilkinson KH and Kong A:
Triple-negative breast cancer: Who should receive neoadjuvant
chemotherapy? Surg Oncol Clin N Am. 27:141–153. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zuo Y, Li Y, Zhou Z, Ma M and Fu K: Long
non-coding RNA MALAT1 promotes proliferation and invasion via
targeting miR-129-5p in triple-negative breast cancer. Biomed
Pharmacother. 95:922–928. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Del Mastro L and De Laurentiis M: Clinical
applications of trastuzumab in the management of HER-2-positive
breast cancer. Recenti Prog Med. 110:594–603. 2019.PubMed/NCBI View Article : Google Scholar : (In Italian).
|
|
42
|
Lu X, Ma J, Chu J, Shao Q, Zhang Y, Lu G,
Li J, Huang X, Li W, Li Y, et al: MiR-129-5p sensitizes the
response of Her-2 positive breast cancer to trastuzumab by reducing
Rps6. Cell Physiol Biochem. 44:2346–2356. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Godbole M, Chandrani P, Gardi N, Dhamne H,
Patel K, Yadav N, Gupta S, Badwe R and Dutt A: miR-129-2 mediates
down-regulation of progesterone receptor in response to
progesterone in breast cancer cells. Cancer Biol Ther. 18:801–805.
2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Xue J, Zhu X, Huang P, He Y, Xiao Y, Liu R
and Zhao M: Expression of miR-129-5p and miR-433 in the serum of
breast cancer patients and their relationship with
clinicopathological features. Oncol Lett. 20:2771–2778.
2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zeng H, Wang L, Wang J, Chen T, Li H,
Zhang K, Chen J, Zhen S, Tuluhong D, Li J and Wang S:
microRNA-129-5p suppresses Adriamycin resistance in breast cancer
by targeting SOX2. Arch Biochem Biophys. 651:52–60. 2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yi W, Wang J, Yao Z, Kong Q, Zhang N, Mo
W, Xu L and Li X: The expression status of ZIC2 as a prognostic
marker for nasopharyngeal carcinoma. Int J Clin Exp Pathol.
11:4446–4460. 2018.PubMed/NCBI
|
|
47
|
Matheson CJ, Backos DS and Reigan P:
Targeting WEE1 kinase in cancer. Trends Pharmacol Sci. 37:872–881.
2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhang H, Lu J, Jiang C and Lu W:
miR-129-1-3p reverses cisplatin resistance of HNE1/CDDP human
nasopharyngeal carcinoma cells by targeting inhibition of WEE1
kinase. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 35:1014–1019.
2019.PubMed/NCBI(In Chinese).
|
|
49
|
Gu LP, Jin S, Xu RC, Zhang J, Geng YC,
Shao XY and Qin LB: Long non-coding RNA PCAT-1 promotes tumor
progression by inhibiting miR-129-5p in human ovarian cancer. Arch
Med Sci. 15:513–521. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Liu F, Zhao H, Gong L, Yao L, Li Y and
Zhang W: MicroRNA-129-3p functions as a tumor suppressor in serous
ovarian cancer by targeting BZW1. Int J Clin Exp Pathol.
11:5901–5908. 2018.PubMed/NCBI
|
|
51
|
Zhang H and Lu W: LncRNA SNHG12 regulates
gastric cancer progression by acting as a molecular sponge of
miR-320. Mol Med Rep. 17:2743–2749. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wang JZ, Xu CL, Wu H and Shen SJ: LncRNA
SNHG12 promotes cell growth and inhibits cell apoptosis in
colorectal cancer cells. Braz J Med Biol Res.
50(e6079)2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Sun D and Fan XH: LncRNA SNHG12
accelerates the progression of ovarian cancer via absorbing
miRNA-129 to upregulate SOX4. Eur Rev Med Pharmacol Sci.
23:2345–2352. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Bi M, Yu H, Huang B and Tang C: Long
non-coding RNA PCAT-1 over-expression promotes proliferation and
metastasis in gastric cancer cells through regulating CDKN1A. Gene.
626:337–343. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Wen J, Xu J, Sun Q, Xing C and Yin W:
Upregulation of long non coding RNA PCAT-1 contributes to cell
proliferation, migration and apoptosis in hepatocellular carcinoma.
Mol Med Rep. 13:4481–4486. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Qiao L, Liu X, Tang Y, Zhao Z, Zhang J and
Feng Y: Down regulation of the long non-coding RNA PCAT-1 induced
growth arrest and apoptosis of colorectal cancer cells. Life Sci.
188:37–44. 2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Wang H, Guan Z, He K, Qian J, Cao J and
Teng L: LncRNA UCA1 in anti-cancer drug resistance. Oncotarget.
8:64638–64650. 2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Tan G, Cao X, Dai Q, Zhang B, Huang J,
Xiong S, Zhang Yy, Chen W, Yang J and Li H: A novel role for
microRNA-129-5p in inhibiting ovarian cancer cell proliferation and
survival via direct suppression of transcriptional co-activators
YAP and TAZ. Oncotarget. 6:8676–8686. 2015.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Pronina IV, Loginov VI, Burdennyy AM,
Fridman MV, Kazubskaya TP, Dmitriev AA and Braga EA: Expression and
DNA methylation alterations of seven cancer-associated 3p genes and
their predicted regulator miRNAs (miR-129-2, miR-9-1) in breast and
ovarian cancers. Gene. 576:483–491. 2016.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Kang R, Xie Y, Zhang Q, Hou W, Jiang Q,
Zhu S, Liu J, Zeng D, Wang H, Bartlett DL, et al: Intracellular
HMGB1 as a novel tumor suppressor of pancreatic cancer. Cell Res.
27:916–932. 2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Amornsupak K, Jamjuntra P, Warnnissorn M,
O-Charoenrat P, Sa-Nguanraksa D, Thuwajit P, Eccles SA and Thuwajit
C: High ASMA+ fibroblasts and low cytoplasmic
HMGB1+ breast cancer cells predict poor prognosis. Clin
Breast Cancer. 17:441–452.e2. 2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Chou YE, Yang PJ, Lin CY, Chen YY, Chiang
WL, Lin PX, Huang ZY, Huang M, Ho YC and Yang SF: The impact of
HMGB1 polymorphisms on prostate cancer progression and
clinicopathological characteristics. Int J Environ Res Public
Health. 17(7247)2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Fu R, Yang P, Sajid A and Li Z:
Avenanthramide A induces cellular senescence via
miR-129-3p/Pirh2/p53 signaling pathway to suppress colon cancer
growth. J Agric Food Chem. 67:4808–4816. 2019.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Wu N, Fesler A, Liu H and Ju J:
Development of novel miR-129 mimics with enhanced efficacy to
eliminate chemoresistant colon cancer stem cells. Oncotarget.
9:8887–8897. 2017.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Kang M, Li Y, Liu W, Wang R, Tang A, Hao
H, Liu Z and Ou H: miR-129-2 suppresses proliferation and migration
of esophageal carcinoma cells through downregulation of SOX4
expression. Int J Mol Med. 32:51–58. 2013.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Li Z, Lu J, Zeng G, Pang J, Zheng X, Feng
J and Zhang J: MiR-129-5p inhibits liver cancer growth by targeting
calcium calmodulin-dependent protein kinase IV (CAMK4). Cell Death
Dis. 10(789)2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Lu JL, Zhao L, Han SC, Bi JL, Liu HX, Yue
C and Lin L: MiR-129 is involved in the occurrence of uterine
fibroid through inhibiting TET1. Eur Rev Med Pharmacol Sci.
22:4419–4426. 2018.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Wang YF, Yang HY, Shi XQ and Wang Y:
Upregulation of microRNA-129-5p inhibits cell invasion, migration
and tumor angiogenesis by inhibiting ZIC2 via downregulation of the
Hedgehog signaling pathway in cervical cancer. Cancer Biol Ther.
19:1162–1173. 2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Lv X, Zhou W, Sun J, Lin R, Ding L, Xu M,
Xu Y, Zhao Z, Chen Y, Bi Y, et al: Visceral adiposity is
significantly associated with type 2 diabetes in middle-aged and
elderly Chinese women: A cross-sectional study. J Diabetes.
9:920–928. 2017.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Hruby A and Hu FB: The epidemiology of
obesity: A big picture. Pharmacoeconomics. 33:673–689.
2015.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Yu YH: Making sense of metabolic obesity
and hedonic obesity. J Diabetes. 9:656–666. 2017.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Ahmed M, Nguyen HQ, Hwang JS, Zada S, Lai
TH, Kang SS and Kim DR: Systematic characterization of
autophagy-related genes during the adipocyte differentiation using
public-access data. Oncotarget. 9:15526–15541. 2018.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Zhang HY, Wang YH, Wang Y, Qu YN, Huang
XH, Yang HX, Zhao CM, He Y, Li SW, Zhou J, et al: miR-129-5p
regulates the immunomodulatory functions of adipose-derived stem
cells via targeting Stat1 signaling. Stem Cells Int.
2019(2631024)2019.PubMed/NCBI View Article : Google Scholar
|
|
75
|
He C and Klionsky DJ: Regulation
mechanisms and signaling pathways of autophagy. Annu Rev Genet.
43:67–93. 2009.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Suryawan A and Hu CY: Effect of serum on
differentiation of porcine adipose stromal-vascular cells in
primary culture. Comp Biochem Physiol Comp Physiol. 105:485–492.
1993.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Harding TM, Hefner-Gravink A, Thumm M and
Klionsky DJ: Genetic and phenotypic overlap between autophagy and
the cytoplasm to vacuole protein targeting pathway. J Biol Chem.
271:17621–17624. 1996.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Ahmed M, Hwang JS, Lai TH, Zada S, Nguyen
HQ, Pham TM, Yun M and Kim DR: Co-expression network analysis of
AMPK and autophagy gene products during adipocyte differentiation.
Int J Mol Sci. 19(1808)2018.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Fu X, Jin L, Han L, Yuan Y, Mu Q, Wang H,
Yang J, Ning G, Zhou D and Zhang Z: miR-129-5p inhibits
adipogenesis through autophagy and may be a potential biomarker for
obesity. Int J Endocrinol. 2019(5069578)2019.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Bielawiec P, Harasim-Symbor E and
Chabowski A: Phytocannabinoids: Useful drugs for the treatment of
obesity? Special focus on cannabidiol. Front Endocrinol (Lausanne).
11(114)2020.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Gracia A, Miranda J, Fernández-Quintela A,
Eseberri I, Garcia-Lacarte M, Milagro FI, Martínez JA, Aguirre L
and Portillo MP: Involvement of miR-539-5p in the inhibition of de
novo lipogenesis induced by resveratrol in white adipose tissue.
Food Funct. 7:1680–1688. 2016.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Dinatolo E, Sciatti E, Anker MS, Lombardi
C, Dasseni N and Metra M: Updates in heart failure: What last year
brought to us. ESC Heart Fail. 5:989–1007. 2018.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Murphy SP, Kakkar R, McCarthy CP and
Januzzi JL Jr: Inflammation in heart failure: JACC state-of-the-art
review. J Am Coll Cardiol. 75:1324–1340. 2020.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Martinotti S, Patrone M and Ranzato E:
Emerging roles for HMGB1 protein in immunity, inflammation, and
cancer. Immunotargets Ther. 4:101–109. 2015.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Volz HC, Laohachewin D, Schellberg D,
Wienbrandt AR, Nelles M, Zugck C, Kaya Z, Katus HA and Andrassy M:
HMGB1 is an independent predictor of death and heart
transplantation in heart failure. Clin Res Cardiol. 101:427–435.
2012.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Gorgulho CM, Romagnoli GG, Bharthi R and
Lotze MT: Johnny on the Spot-chronic inflammation is driven by
HMGB1. Front Immunol. 10(1561)2019.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Piorecka K, Smith D, Kurjata J, Stanczyk M
and Stanczyk WA: Synthetic routes to nanoconjugates of
anthracyclines. Bioorg Chem. 96(103617)2020.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Anakwue R: Cytotoxic-induced heart failure
among breast cancer patients in Nigeria: A call to prevent today's
cancer patients from being tomorrow's cardiac patients. Ann Afr
Med. 19:1–7. 2020.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Bansal N, Adams MJ, Ganatra S, Colan SD,
Aggarwal S, Steiner R, Amdani S, Lipshultz ER and Lipshultz SE:
Strategies to prevent anthracycline-induced cardiotoxicity in
cancer survivors. Cardiooncology. 5(18)2019.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Cai F, Luis MAF, Lin X, Wang M, Cai L, Cen
C and Biskup E: Anthracycline-induced cardiotoxicity in the
chemotherapy treatment of breast cancer: Preventive strategies and
treatment. Mol Clin Oncol. 11:15–23. 2019.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Lim CC, Zuppinger C, Guo X, Kuster GM,
Helmes M, Eppenberger HM, Suter TM, Liao R and Sawyer DB:
Anthracyclines induce calpain-dependent titin proteolysis and
necrosis in cardiomyocytes. J Biol Chem. 279:8290–8299.
2004.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Li Q, Qin M, Tan Q, Li T, Gu Z, Huang P
and Ren L: MicroRNA-129-1-3p protects cardiomyocytes from
pirarubicin-induced apoptosis by down-regulating the
GRIN2D-mediated Ca2+ signalling pathway. J Cell Mol Med.
24:2260–2271. 2020.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Javadov S, Hunter JC, Barreto-Torres G and
Parodi-Rullan R: Targeting the mitochondrial permeability
transition: Cardiac ischemia-reperfusion versus carcinogenesis.
Cell Physiol Biochem. 27:179–190. 2011.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Hajdú T, Juhász T, Szűcs-Somogyi C, Rácz K
and Zákány R: NR1 and NR3B Composed intranuclear
N-methyl-d-aspartate receptor complexes in human melanoma cells.
Int J Mol Sci. 19(1929)2018.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Liu XR, Rempel DL and Gross ML: Composite
conformational changes of signaling proteins upon ligand binding
revealed by a single approach: Calcium-calmodulin study. Anal Chem.
91:12560–12567. 2019.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Hwang HS, Baldo MP, Rodriguez JP, Faggioni
M and Knollmann BC: Efficacy of flecainide in catecholaminergic
polymorphic ventricular tachycardia is mutation-independent but
reduced by calcium overload. Front Physiol. 10(992)2019.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Wei Q, Zhou HY, Shi XD, Cao HY and Qin L:
Long noncoding RNA NEAT1 promotes myocardiocyte apoptosis and
suppresses proliferation through regulation of miR-129-5p. J
Cardiovasc Pharmacol. 74:535–541. 2019.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Camfield P and Camfield C: Regression in
children with epilepsy. Neurosci Biobehav Rev. 96:210–218.
2019.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Devinsky O, Vezzani A, Najjar S, De
Lanerolle NC and Rogawski MA: Glia and epilepsy: Excitability and
inflammation. Trends Neurosci. 36:174–184. 2013.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Fisher RS, Acevedo C, Arzimanoglou A,
Bogacz A, Cross JH, Elger CE, Engel J Jr, Forsgren L, French JA,
Glynn M, et al: ILAE official report: A practical clinical
definition of epilepsy. Epilepsia. 55:475–482. 2014.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Hunsberger JG, Bennett AH, Selvanayagam E,
Duman RS and Newton SS: Gene profiling the response to kainic acid
induced seizures. Brain research. Mol Brain Res. 141:95–112.
2005.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Sharma AK, Searfoss GH, Reams RY, Jordan
WH, Snyder PW, Chiang AY, Jolly RA and Ryan TP: Kainic acid-induced
F-344 rat model of mesial temporal lobe epilepsy: Gene expression
and canonical pathways. Toxicol Pathol. 37:776–789. 2009.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Gorter JA, Iyer A, White I, Colzi A, van
Vliet EA, Sisodiya S and Aronica E: Hippocampal subregion-specific
microRNA expression during epileptogenesis in experimental temporal
lobe epilepsy. Neurobiol Dis. 62:508–520. 2014.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Pernice HF, Schieweck R, Kiebler MA and
Popper B: mTOR and MAPK: From localized translation control to
epilepsy. BMC Neurosci. 17(73)2016.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Wu DM, Zhang YT, Lu J and Zheng YL:
Effects of microRNA-129 and its target gene c-Fos on proliferation
and apoptosis of hippocampal neurons in rats with epilepsy via the
MAPK signaling pathway. J Cell Physiol. 233:6632–6643.
2018.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Rajman M, Metge F, Fiore R, Khudayberdiev
S, Aksoy-Aksel A, Bicker S, Ruedell Reschke C, Raoof R, Brennan GP,
Delanty N, et al: A microRNA-129-5p/Rbfox crosstalk coordinates
homeostatic downscaling of excitatory synapses. EMBO J.
36:1770–1787. 2017.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Siddoway B, Hou H and Xia H: Molecular
mechanisms of homeostatic synaptic downscaling. Neuropharmacology.
78:38–44. 2014.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Wan Y and Yang ZQ: LncRNA NEAT1 affects
inflammatory response by targeting miR-129-5p and regulating Notch
signaling pathway in epilepsy. Cell Cycle. 19:419–431.
2020.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Tuomi T, Santoro N, Caprio S, Cai M, Weng
J and Groop L: The many faces of diabetes: A disease with
increasing heterogeneity. Lancet. 383:1084–1094. 2014.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Hoshino A, Costa-Silva B, Shen TL,
Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di
Giannatale A, Ceder S, et al: Tumour exosome integrins determine
organotropic metastasis. Nature. 527:329–335. 2015.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Melo SA, Luecke LB, Kahlert C, Fernandez
AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari
N, et al: Glypican-1 identifies cancer exosomes and detects early
pancreatic cancer. Nature. 523:177–182. 2015.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Fu Q, Jiang H, Wang Z, Wang X, Chen H,
Shen Z, Xiao L, Guo X and Yang T: Injury factors alter miRNAs
profiles of exosomes derived from islets and circulation. Aging
(Albany NY). 10:3986–3999. 2018.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Collinson P: Laboratory medicine is faced
with the evolution of medical practice. J Med Biochem. 36:211–215.
2017.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Wicks K, Torbica T and Mace KA: Myeloid
cell dysfunction and the pathogenesis of the diabetic chronic
wound. Semin Immunol. 26:341–353. 2014.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Williams MD and Nadler JL: Inflammatory
mechanisms of diabetic complications. Curr Diabet Rep. 7:242–248.
2007.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Daigle I and Simon HU: Critical role for
caspases 3 and 8 in neutrophil but not eosinophil apoptosis. Int
Arch Allergy Immuno. 126:147–156. 2001.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Pongracz J, Webb P, Wang K, Deacon E, Lunn
OJ and Lord JM: Spontaneous neutrophil apoptosis involves caspase
3-mediated activation of protein kinase C-delta. J Biol Chem.
274:37329–37334. 1999.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Zhao R, Guan DW, Zhang W, Du Y, Xiong CY,
Zhu BL and Zhang JJ: Increased expressions and activations of
apoptosis-related factors in cell signaling during incised skin
wound healing in mice: A preliminary study for forensic wound age
estimation. Leg Med (Tokyo, Japan). 11 (Suppl 1):S155–S160.
2009.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Willenborg S, Lucas T, van Loo G, Knipper
JA, Krieg T, Haase I, Brachvogel B, Hammerschmidt M, Nagy A,
Ferrara N, et al: CCR2 recruits an inflammatory macrophage
subpopulation critical for angiogenesis in tissue repair. Blood.
120:613–625. 2012.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Devalaraja RM, Nanney LB, Du J, Qian Q, Yu
Y, Devalaraja MN and Richmond A: Delayed wound healing in CXCR2
knockout mice. J Invest Dermatol. 115:234–244. 2000.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Umehara T, Mori R, Mace KA, Murase T, Abe
Y, Yamamoto T and Ikematsu K: Identification of specific miRNAs in
neutrophils of type 2 diabetic mice: Overexpression of
miRNA-129-2-3p accelerates diabetic wound healing. Diabetes.
68:617–630. 2019.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Wang W, Yang C, Wang XY, Zhou LY, Lao GJ,
Liu D, Wang C, Hu MD, Zeng TT, Yan L and Ren M: MicroRNA-129 and
-335 promote diabetic wound healing by inhibiting Sp1-mediated
MMP-9 expression. Diabetes. 67:1627–1638. 2018.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Jobin PG, Butler GS and Overall CM: New
intracellular activities of matrix metalloproteinases shine in the
moonlight. Biochim Biophys Acta Mol Cell Res. 1864:2043–2055.
2017.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Overall CM, Wrana JL and Sodek J:
Transcriptional and post-transcriptional regulation of 72-kDa
gelatinase/type IV collagenase by transforming growth factor-beta 1
in human fibroblasts. Comparisons with collagenase and tissue
inhibitor of matrix metalloproteinase gene expression. J Biol Chem.
266:14064–14071. 1991.PubMed/NCBI
|
|
125
|
McBeth J and Jones K: Epidemiology of
chronic musculoskeletal pain. Best Pract Res Clin Rheumatol.
21:403–425. 2007.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Wieser S, Horisberger B, Schmidhauser S,
Eisenring C, Brügger U, Ruckstuhl A, Dietrich J, Mannion AF,
Elfering A, Tamcan O and Müller U: Cost of low back pain in
Switzerland in 2005. Eur J Health Econ. 12:455–467. 2011.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Dario AB, Ferreira ML, Refshauge KM, Lima
TS, Ordoñana JR and Ferreira PH: The relationship between obesity,
low back pain, and lumbar disc degeneration when genetics and the
environment are considered: A systematic review of twin studies.
Spine J. 15:1106–1117. 2015.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Johnson WE, Eisenstein SM and Roberts S:
Cell cluster formation in degenerate lumbar intervertebral discs is
associated with increased disc cell proliferation. Connect Tissue
Res. 42:197–207. 2001.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Kim KW, Lim TH, Kim JG, Jeong ST, Masuda K
and An HS: The origin of chondrocytes in the nucleus pulposus and
histologic findings associated with the transition of a notochordal
nucleus pulposus to a fibrocartilaginous nucleus pulposus in intact
rabbit intervertebral discs. Spine (Phila Pa 1976). 28:982–990.
2003.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Patterson JE: The pre-travel medical
evaluation: The traveler with chronic illness and the geriatric
traveler. Yale J Biol Med. 65:317–327. 1992.PubMed/NCBI
|
|
131
|
Li J, Yoon ST and Hutton WC: Effect of
bone morphogenetic protein-2 (BMP-2) on matrix production, other
BMPs, and BMP receptors in rat intervertebral disc cells. J Spinal
Disord Tech. 17:423–428. 2004.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Antunovic M, Matic I, Nagy B, Caput
Mihalic K, Skelin J, Stambuk J, Josipovic P, Dzinic T, Paradzik M
and Marijanovic I: FADD-deficient mouse embryonic fibroblasts
undergo RIPK1-dependent apoptosis and autophagy after NB-UVB
irradiation. J Photochem Photobiol B. 194:32–45. 2019.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis
D, Jia LT, Wu SX, Huang J, Chen J and Luo ZJ: Deregulated miR-155
promotes Fas-mediated apoptosis in human intervertebral disc
degeneration by targeting FADD and caspase-3. J Pathol.
225:232–242. 2011.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Li N, Gao Q, Zhou W, Lv X, Yang X and Liu
X: MicroRNA-129-5p affects immune privilege and apoptosis of
nucleus pulposus cells via regulating FADD in intervertebral disc
degeneration. Cell Cycle. 19:933–948. 2020.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Zhao K, Zhang Y, Kang L, Song Y, Wang K,
Li S, Wu X, Hua W, Shao Z, Yang S and Yang C: Methylation of
microRNA-129-5P modulates nucleus pulposus cell autophagy by
targeting Beclin-1 in intervertebral disc degeneration. Oncotarget.
8:86264–86276. 2017.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Maiuri MC, Criollo A, Tasdemir E, Vicencio
JM, Tajeddine N, Hickman JA, Geneste O and Kroemer G: BH3-only
proteins and BH3 mimetics induce autophagy by competitively
disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L).
Autophagy. 3:374–376. 2007.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Pluta R, Ulamek-Koziol M, Januszewski S
and Czuczwar SJ: Gut microbiota and pro/prebiotics in Alzheimer's
disease. Aging (Albany NY). 12:5539–5550. 2020.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Shi Y, Han Y, Niu L, Li J and Chen Y:
MiR-499 inhibited hypoxia/reoxygenation induced cardiomyocytes
injury by targeting SOX6. Biotechnol Lett. 41:837–847.
2019.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Zaletel I, Schwirtlich M, Perović M,
Jovanović M, Stevanović M, Kanazir S and Puškaš N: Early
impairments of hippocampal neurogenesis in 5xFAD mouse model of
Alzheimer's disease are associated with altered expression of SOXB
transcription factors. J Alzheimer's Dis. 65:963–976.
2018.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Zeng Z, Liu Y, Zheng W, Liu L, Yin H,
Zhang S, Bai H, Hua L, Wang S, Wang Z, et al: MicroRNA-129-5p
alleviates nerve injury and inflammatory response of Alzheimer's
disease via downregulating SOX6. Cell Cycle. 18:3095–3110.
2019.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Geng Z, Xu F and Zhang Y:
MiR-129-5p-mediated Beclin-1 suppression inhibits endothelial cell
autophagy in atherosclerosis. Am J Transl Res. 8:1886–1894.
2016.PubMed/NCBI
|
|
142
|
Wan G, An Y, Tao J, Wang Y, Zhou Q, Yang R
and Liang Q: MicroRNA-129-5p alleviates spinal cord injury in mice
via suppressing the apoptosis and inflammatory response through
HMGB1/TLR4/NF-κB pathway. Biosci Rep.
40(BSR20193315)2020.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Huang S, Lv Z, Wen Y, Wei Y, Zhou L, Ke Y,
Zhang Y, Xu Q, Li L, Guo Y, et al: miR-129-2-3p directly targets
SYK gene and associates with the risk of ischaemic stroke in a
Chinese population. J Cell Mol Med. 23:167–176. 2019.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Xu M, Li H, Bai Y, He J, Chen R, An N, Li
Y and Dong Y: miR-129 blocks secondary hyperparathyroidism-inducing
Fgf23/αKlotho signaling in mice with chronic kidney disease. Am J
Med Sci. 361:624–634. 2021.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Zhu Y, Hu Y, Cheng X, Li Q and Niu Q:
Elevated miR-129-5p attenuates hepatic fibrosis through the NF-κB
signaling pathway via PEG3 in a carbon CCl4 rat model. J
Mol Histol. 52:491–501. 2021.PubMed/NCBI View Article : Google Scholar
|