Introduction
Osteoarthritis (OA) is the most common type of
arthritis, and it affects >10% of the adult population (1). Physicians consider the disease to be a
degenerative disease that involves all the tissues of the joint. In
OA, cartilage within a joint begins to break down, and the
underlying bone begins to change (2). OA can induce substantial limb pain,
stiffness, disability, and even loss of whole-body mobility. OA
occurs most frequently in the hands, hips and knees, leading to
difficulties in patients' daily activities (3). Therefore, numerous management
strategies to prevent cartilage degeneration, such as microfracture
surgery to stimulate a healing response, stem cell therapy to
repair damaged articular cartilage, cartilage transplantation, and
complementary treatments, have been developed (4-6).
Long noncoding RNAs (lncRNAs) are transcripts longer
than 200 nucleotides and are noncoding RNAs. Aberrant expression of
lncRNAs participate in numerous biological processes and various
diseases by posttranscriptional or posttranslational regulation
(7,8). Jiang et al (9) and Marques-Rocha et al (10) reported that patients with OA have
aberrant expression of certain lncRNAs, which indicates that
lncRNAs may play a key role in articular cartilage degeneration.
Recently, the novel lncRNA HOX antisense intergenic RNA myeloid 1
(HOTAIRM1) was found to be expressed in cells of the myeloid
lineage (11,12). Additional evidence has confirmed
that HOTAIRM1 plays a crucial role in the pathological progression
of colorectal cancer and several other diseases, such as acute
myeloid leukemia (13-15).
Nonetheless, the potential function of HOTAIRM1 in the development
of osteoarthritis remains unknown.
HOTAIRM1 is located between the HOXA1 and HOXA2 loci
and has been identified as a myeloid-specific regulator in the HOXA
gene family. HOTAIRM1 targets gene transcription during the
chromosome remodeling that occurs when myeloid cells are induced to
undergo chondrogenic differentiation (16). Furthermore, previous studies in
mesenchymal stem cells showed that miR-125b is a miRNA target of
HOTAIRM1 during osteoblastic differentiation (16,17).
In the present study, the hypothesis that HOTAIRM1-1 and miR-125b
have a functional association and that miR-125b is a potential
therapeutic target of articular cartilage denegation was
tested.
Materials and methods
Tissue collection
The present study was approved by the human ethics
committee of Tianjin Hospital. All the patients provided written
informed consent and participated in the study according to their
own will. A total of 15 articular cartilage samples were obtained
from patients with OA undergoing total knee arthroplasty (n=15;
mean age, 65.8 ± 6.2 years; 5 males and 10 females). Healthy
control articular cartilage samples were collected from trauma
patients who underwent amputation and did not have a history of
rheumatoid arthritis or OA (n=8; mean age, 41.2 ± 9.1 years; 5 male
and 3 female). There was no significant difference between the OA
group and the control group in terms of sex or age.
Cell culture
Chondrocytes from the cartilage of patients with OA
were cultured following a previously published protocol (18). In brief, small sections of OA
cartilage tissues were digested with trypsin (0.25%). Then, type II
collagenase (0.2%; Gibco; Thermo Fisher Scientific, Inc.) was
incubated with the samples. The chondrocytes were cultured in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS and maintained in a cell
incubator containing 5% CO2 at 37˚C.
Cell transfection
The plasmids containing small interfering (si)RNA
targeting HOTAIRM1-1 (si-HOTAIRM1-1,
5'-CACCGGAGACTGGTAGCTTATTATTCAAGAGATAATAAGCTACCAGTCTCCTTTTTTG-3')
or a si-HOTAIRM1-1 negative control sequence (si-NC,
5'-CACCGTTCTCCGAACGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATTTTTT-3')
were purchased from Gene-Pharma (Shanghai GenePharma Co., Ltd.).
Briefly, siRNA was transfected into cultured chondrocytes by using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) at
a concentration of 50 nM, following the manufacturer's
instructions, which were plated into 6-well plates the day before
transfection to achieve 60-70% confluency. After 48 h, the
transfected cells were used for further analysis.
Synthetic miRNA mimics miR-125b, miR-125b inhibitor
and their negative control (custom synthesized by Shanghai
GenePharma Co., Ltd.) were transfected into cells following the
Lipofectamine® RNAiMAX Reagent (Thermo Fisher
Scientific, Inc.) transfection protocol. After 48 h, the
transfected cells were used for further analysis.
Reverse transcription-quantitative
PCR
Total RNA was extracted from cultured cells by using
the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
Next, 1 mg RNA was reverse transcribed into complementary DNA using
the PrimeScript RT reagent kit (Takara Bio, Inc.) at 23˚C for 30
min. Quantification of mRNA was determined by qPCR using a
SYBRGreen supermix (Bio-Rad Laboratories, Inc.). The primers of
HOTAIRM1-1, miR-125b, IL-10 and MMP-13 were designed and
synthesized by Sangon Biotech Co., Ltd; The primer sequences are
shown in Table I. Amplification
conditions were 95˚C for 30 sec, followed by 35 cycles of 95˚C for
5 sec, and 60˚C for 30 sec. Moreover, to confirm that only one
product was amplified, melting curve analysis was set at from 58 to
95˚C with stepwise fluorescence acquisition at every 1˚C/sec.
Relative quantification was performed using the 2-ΔΔCq
method (19). U6 was used as the
endogenous reference gene for all experiments with ncRNA. GAPDH was
used as normalization for matrix metalloprotease (MMP)13 and
interleukin (IL)-10.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Gene | F, 5'-3' | R, 5'-3' |
|---|
| HOTAIRM1-1 |
AAACGAGGGATGGAAGGGAGCG |
CCAGGCATTCGGCAATGTG |
| miR-125b |
GCTTTGCTGCGTACTTCCA |
GTCCACACGGGTTCCAGA |
| MMP-13 |
GGCTTCGACACCCGTGTAA |
CGTCAAACCTCTTGTCATCCA |
| IL-10 |
GTAGAGGACACGGGCAAGAT |
TTCACGAACTGTCAACTGCAC |
| U6 |
CTCGGCTTCGGCAGCACA |
AACGCTTCACGAATTGCGT |
| GAPDH |
AGAAGGCTGGGGCATTTG |
AGGGGCCATCCACAGTCTTC |
Cell proliferation
To analyze cell proliferation, Cell Counting Kit-8
(Dojindo Molecular Technologies, Inc.) was used. The cells were
seeded into 96-well plates at ~1x10 cells in each well. Cell
proliferation was determined at 0, 24, 48 72 and 96 h. The 96-well
plates were analyzed on a microplate reader at OD 450 nm.
Luciferase reporter gene assay
To construct 3'-untranslated region (UTR) Green
Renilla Luciferase reporters (Thermo Fisher Scientific,
Inc.), mutant and wild-type sequences were simultaneously
constructed. Chondrocytes were cultured in 24-well plates, and
then, the miR-125b mimics, miR-NC and mutant or wild-type
HOTAIRM1-1 were transfected into the chondrocytes using a
transfection kit (Lipofectamine 2000®; Thermo Fisher
Scientific, Inc.) at 37˚C for 4 h. After 48 h, luciferase reporter
gene assays were performed using a pierce renilla luciferase glow
assay kit (Thermo Fisher Scientific, Inc.) to measure the
luciferase activity. The results are shown as the
firefly/Renilla ratio normalized to the Renilla
luciferase activity.
Western blotting
Proteins were obtained from cultured cells with
radioimmunoprecipitation assay (RIPA) buffer (Pierce; Thermo Fisher
Scientific, Inc.) on ice for 30 min. The protein concentration of
each cell lysate was determined by a BCA Assay kit (Thermo Fisher
Scientific, Inc.). The protein samples (50 µg) were separated on 8%
polyacrylamide gels by electrophoresis for 5 min at 50 V, and then
the voltage was increased to 100 V for 1 h. Then, the proteins were
transferred from the gels onto polyvinylidene difluoride (PVDF)
membranes (EMD Millipore) followed by blocking with 5% non-fat milk
in buffer [10 mM Tris-HCl (pH 7.6), 100 mM NaCl and 0.1% Tween-20]
at 23˚C for 1 h. The membranes were incubated with anti-collagen II
(cat. no. ab185430; 1:500 dilution; Abcam) and anti-aggrecan (cat.
no. ab36861, 1:1,000 dilution; Abcam) primary antibodies for 4 h at
room temperature. The membrane was washed twice and then incubated
with Goat Anti-Rabbit IgG H&L secondary antibodies (cat. no.
ab205718; 1:1,000; Abcam) for 60 min at room temperature. A
chemiluminescent substrate was applied to the blots with an ECL
detection kit (Thermo Fisher Scientific, Inc.). The
chemiluminescence signals were captured using a Chemi-Doc XRS
camera-based imager (Bio-Rad Laboratories, Inc.). Image analysis
software was used to analyze the bands of the targeting proteins
with Empiria Studio® Software (9141-500E; LI-COR
Biosciences), and β-actin was used as the internal control to
normalize the band intensity.
Terminal deoxynucleotidyl transferase
dUTP nick end labeling (TUNEL)
Cells were washed twice with cold PBS, after which
samples were fixed with 4% paraformaldehyde for 2 min at 23˚C and
washed twice with PBS. Terminal deoxynucleotidyl transferase dUTP
nick end labeling (TUNEL) staining was performed on cells with an
In-Situ Cell Death Detection kit (Roche Diagnostics). A total of 50
µl reaction mixture (terminal deoxynucleotidyl transferase from
calf thymus recombinant in E. coli. Buffer as enzyme
solution and nucleotide; provided in the kit) was incubated for 1 h
at 37˚C. After staining, samples were mounted with
4',6-diamidino-2-phenylindole (DAPI) mounting medium (Vector
Laboratories, Inc.). Nuclear staining was performed at the same
time as mounting, at room temperature for 10 min. A microscope was
used to visualize cell apoptosis under 10 fields of view per sample
(magnification, x200).
Identification of miRNA-mRNA and
miRNA-ncRNA interactions
To inspect genome-wide interactions between miRNAs
and their target genes, the conserved miRNA target sites were
predicted using algorithms from a public database (http://starbase.sysu.edu.cn/index.php).
starBase was intersected with the aforementioned Ago CLIP clusters
to gain CLIP-supported sites.
Statistical analyses
The sample size for each experiment was determined
based on power analysis calculations. All the data are shown as the
mean ± SD (standard deviation). Statistical analyses and graph
generation were completed by Prism GraphPad (GraphPad Software,
Inc.). Student's t test was used for the analysis of two groups.
One-way ANOVA followed by the Tukey-Kramer test was used to compare
multiple groups. Two-way ANOVA was used to determine the effect of
time, the effect of treatment, and their interaction. The Pearson's
correlation analysis test was used to evaluate the strength of the
association between two quantitative variables. P<0.05 was
considered to indicate a statistically significant in all the
tests.
Results
Expression of HOTAIRM1-1 and miR-125b
in chondrocytes is inversely affected by IL-6 and IL-1β
treatment
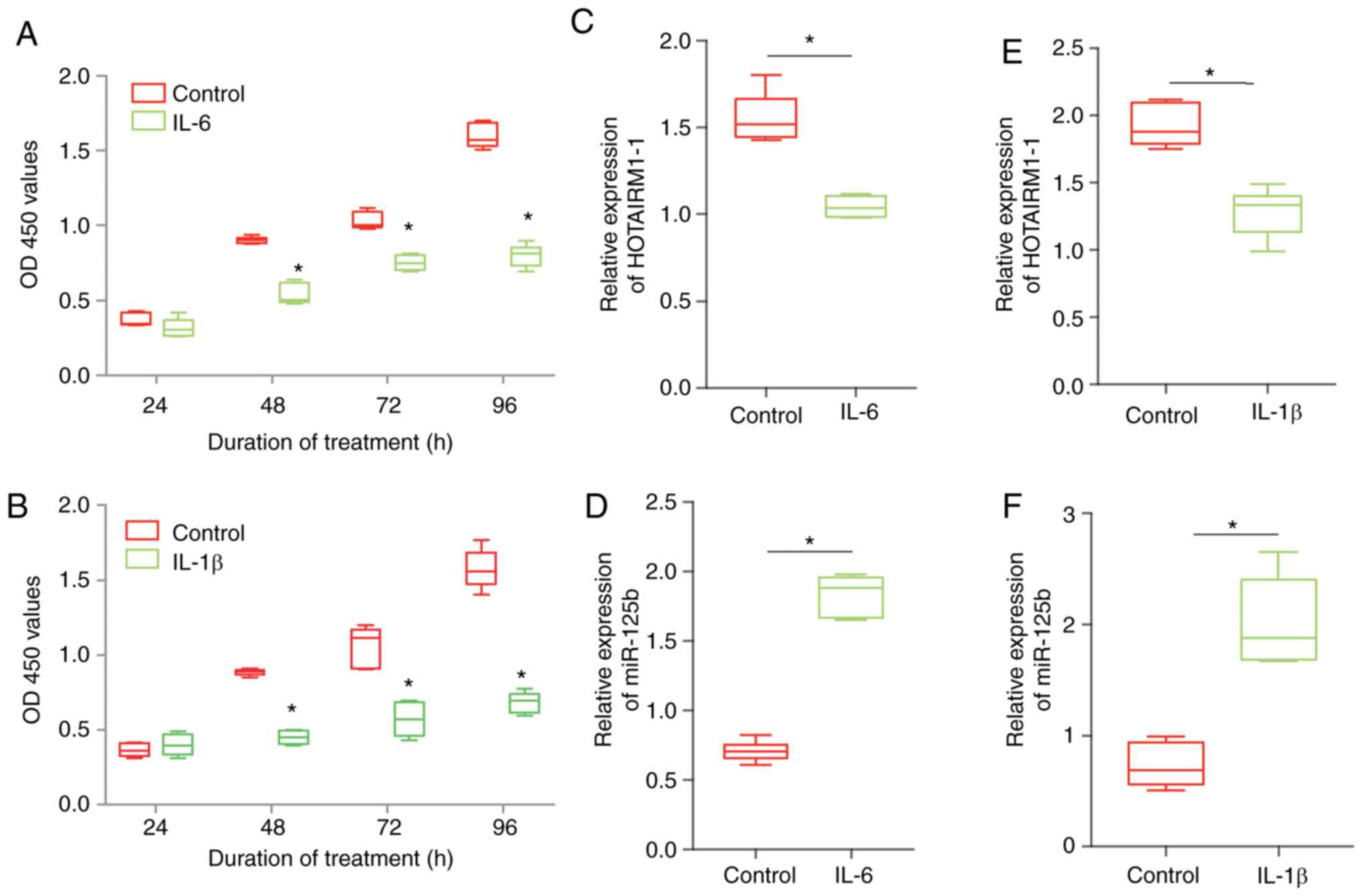
To mimic OA in vitro, the effect of IL-6 or
IL-1β on HOTAIRM1-1 and miR-125b expression was first tested in
chondrocytes. Chondrocytes were treated with IL-6 or IL-1β for 96
h. The data confirmed that IL-6 could significantly decreased
chondrocyte proliferation (Fig.
1A). Using the same method, IL-1β had a similar effect and
decreased chondrocyte proliferation (Fig. 1B). It was observed that the
expression of HOTAIRM1-1 was decreased in chondrocytes after IL-6
treatment, but the expression of miR-125b increased in chondrocytes
(Fig. 1C and D). Additionally, the results demonstrated
similar trends in the expression of HOTAIRM1-1 and miR-125b in the
group of chondrocytes treated with IL-1β (Fig. 1E and F).
Expression of HOTAIRM1-1 is
downregulated and the expression of miR-125b is upregulated in OA
tissues
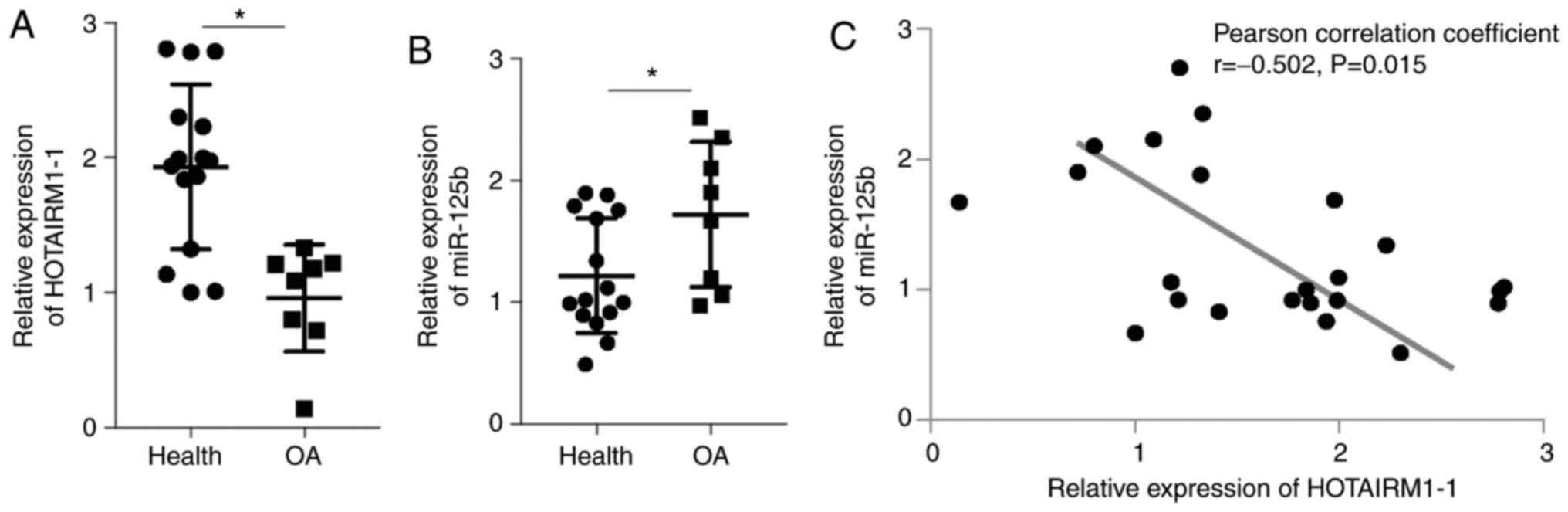
Moreover, the hypothesis that the expression of
HOTAIRM1-1 is downregulated in OA tissue was tested. The data
demonstrated that the expression of HOTAIRM1-1 was significantly
lower in the OA tissue samples compared with the healthy control
samples (Fig. 2A; P<0.05). In
addition, miR-125b expression was higher in the OA tissue samples
compared with the healthy control tissue samples (Fig. 2B; P<0.01). The correlation test
showed that the expression level of miR-125b was negatively
correlated with the expression level of HOTAIRM1-1 (Fig. 2C).
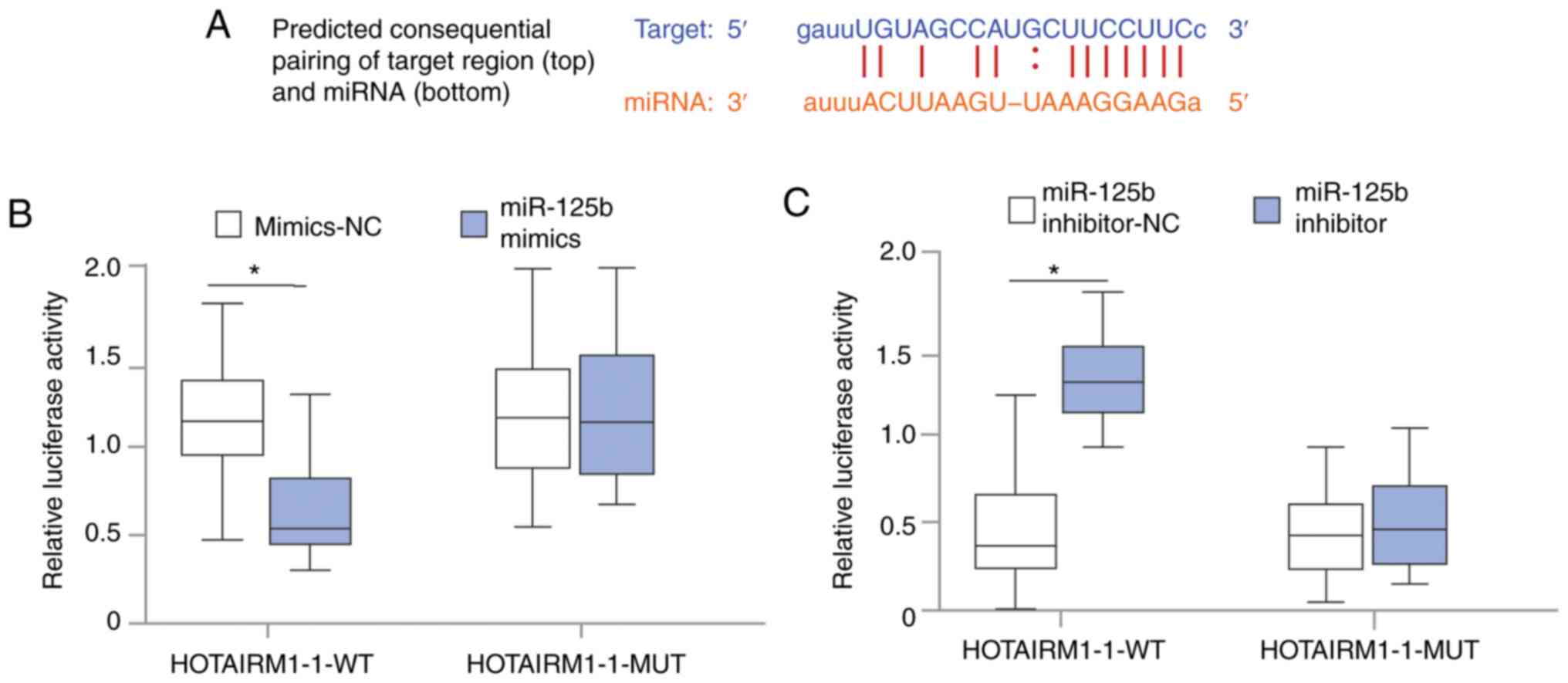
HOTAIRM1-1 directly interacts with
miR-125b in chondrocytes
Studies have reported that miR-125b is a directly
competitive endogenous RNA (ceRNA) that affects the function of
HOTAIRM1. However, whether HOTAIRM1-1 has a potential role in OA
progression remains unknown. To address this question, potential
miRNA binding sites that correlated with HOTAIRM1-1 were searched
for with the starBase online tool (http://starbase.sysu.edu.cn/index.php; Fig. 3A). Furthermore, the luciferase assay
results indicated that HOTAIRM1-1 directly targeted miR-125b, which
confirms a direct association between HOTAIRM1-1 and miR-125b
(Fig. 3B and C).
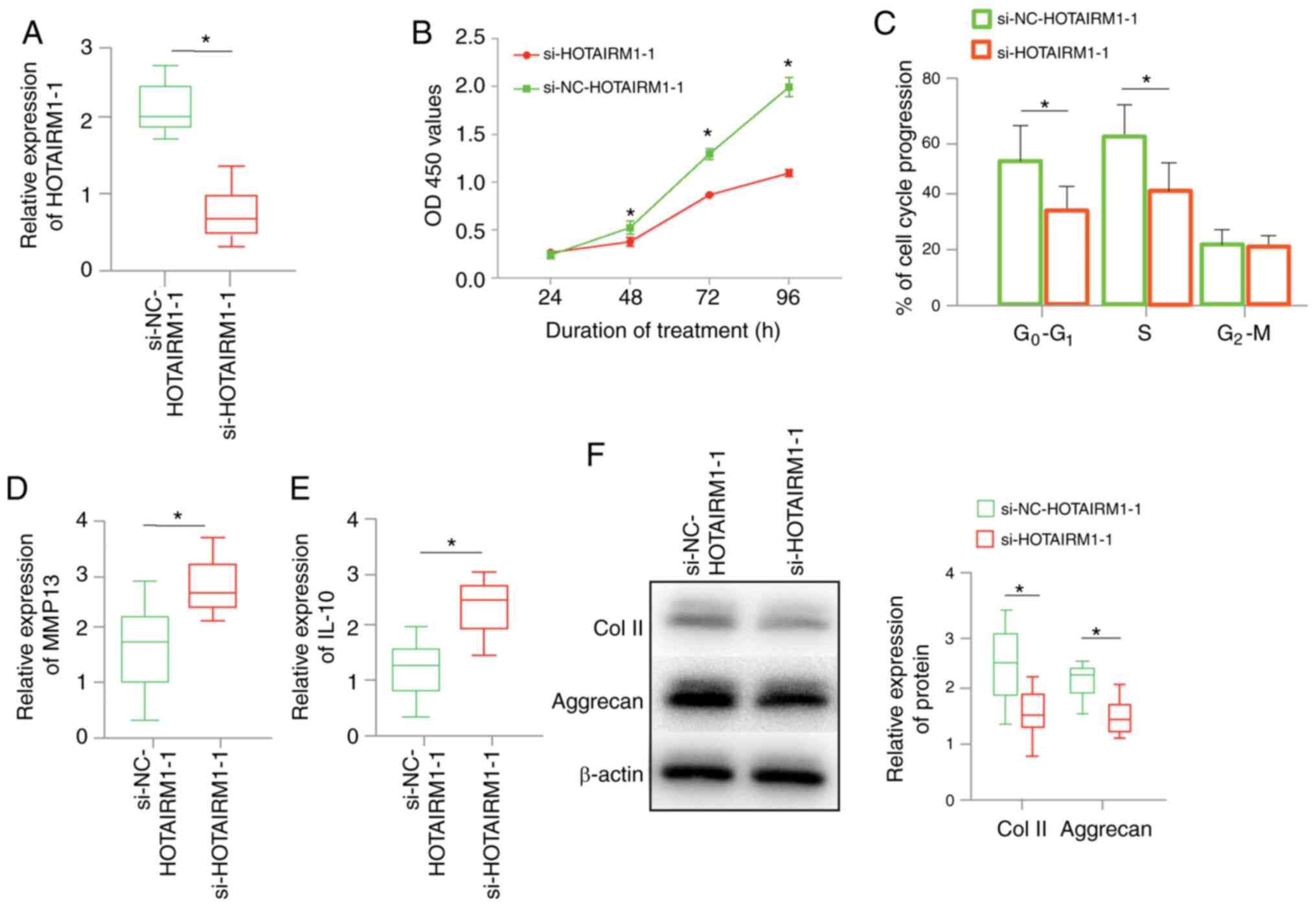
HOTAIRM1-1 knockdown is associated
with chondrocyte proliferation and extracellular matrix
degradation
Next, the effect of HOTAIRM1-1 knockdown on
chondrocytes treated with IL-6 for 48 h before transfection was
determined. Chondrocytes were transfected with an siRNA targeting
HOTAIRM1-1. The expression of HOTAIRM1-1 was significantly
downregulated in the si-HOTAIRM1-1 group compared with the
si-NC-HOTAIRM1-1 group (Fig. 4A).
The proliferation results demonstrated that HOTAIRM1-1 knockdown
significantly decreased cell proliferation (Fig. 4B) and decreased cell cycle
progression in the chondrocytes (Fig.
4C). MMP-13 and IL-10 were identified as the major
cartilage-degrading enzyme markers in chondrocytes. Collagen II and
aggrecan are considered to be fundamental extracellular matrix
(ECM) proteins in chondrocytes. The results revealed that MMP-13
and IL-10 expression was increased in the si-HOTAIRM1-1 group
(Fig. 4D and E). Conversely, ECM degeneration marker
expression was significantly lower in the si-HOTAIRM1-1 group
compared with the control group (Fig.
4F). In conclusion, cartilage-degrading enzymes may be
upregulated by HOTAIRM1-1 knockdown, which promotes ECM
degradation.
miR-125b reverses the effects of
HOTAIRM1-1 on proliferation and apoptosis in IL-6-treated
chondrocytes
To further explore whether HOTAIRM1-1 regulates
chondrocyte proliferation and apoptosis via miR-125b in OA a rescue
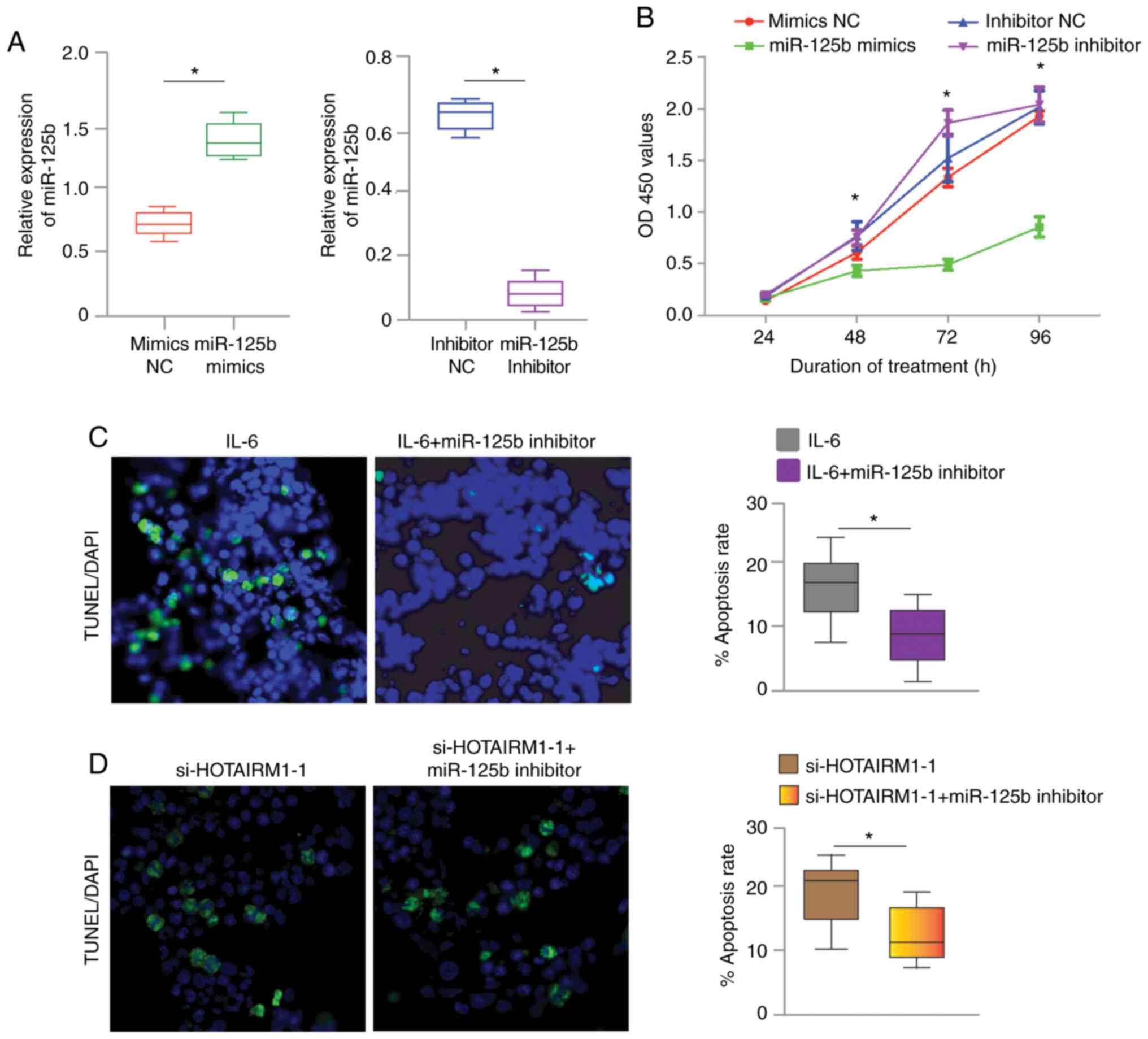
experiment was performed. As shown in Fig. 5A, miR-125b mimics transfection
effectively increased the expression of miR-125b compared with
mimics control transfection. The miR-125b inhibitor effectively
decreased the expression of miR-125b compared with inhibitor
negative control group. A CCK-8 assay was conducted to determine
the proliferation ability of the chondrocytes, as shown in Fig. 5B. Downregulation of miR-125b
strikingly promoted chondrocyte proliferation. However, miR-125b
inhibition dramatically blocked this effect. TUNEL staining is
shown in Fig. 5C. The miR-125b
inhibitor significantly blocked chondrocyte apoptosis when the
cells were treated with IL-6. Conversely, HOTAIRM1-1 knockdown
increased cell apoptosis, while the miR-125b inhibitor obviously
reversed this effect (Fig. 5d). It
is postulated that miR-125b competitively binds to HOTAIRM1-1 to
promote chondrocyte apoptosis.
Discussion
Understanding the association of key lncRNAs with
disease pathogenesis would help with disease diagnosis and
prognosis. Emerging reports have demonstrated that the lncRNA
HOTAIRM1-1 is an important lncRNA in several tissues and diseases,
such as colorectal cancer and acute myeloid leukemia (20,21).
Additionally, HOTAIRM1-1 is involved in the biological processes
associated with OA (22).
Presently, many studies on OA focus on cartilaginous tissues, since
cartilage is a key component of joints. Therefore, cartilage damage
is thought to be a landmark event during OA pathology progression
(23,24). Some studies have reported that the
proinflammatory cytokines IL-6 and IL-1β may play a significant
role in the pathology of OA (25).
IL-6 is a compound that is characterized by omnidirectional
interactions in the processes that occur in the human body. IL-6 is
considered a cytokine that strongly activates the immune system and
enhances the inflammatory response, although considering some of
its effects, IL-6 may be classified as an anti-inflammatory
cytokine (26). The production of
IL-6 in tissues of joints affected by OA usually occurs in response
to IL-1β and tumor necrosis factor (TNF)α and IL-6 and is mainly
produced by chondrocytes (27). A
study by Zanotti and Canalis (27)
showed that IL-6 mediates the induction of MMP13 expression through
Notch and contributes to the inhibitory effect of Notch on the mRNA
levels in cells of the chondrocyte lineage. Simsa-Maziel and
Monsonego-Ornan (28) suggest that
interleukin-1β promotes the proliferation and inhibits the
differentiation of chondrocytes through a mechanism involving the
downregulation of fibroblast growth factor receptor-3 and p21.
In the present study, an in vitro model was
established to mimic OA pathogenesis in chondrocytes. The results
demonstrated that the inflammatory mediators IL-6 and IL-1β
suppressed HOTAIRM1-1 expression in chondrocytes. In addition, the
expression of miR-125b exhibited opposite changes, and its
expression was increased after treatment with the inflammatory
mediators (Fig. 1). Furthermore,
the expression of HOTAIRM1-1 and miR-125b was investigated in
cartilage samples from patients with OA and healthy subjects. It
was found that the expression of HOTAIRM1-1 (downregulated) and
miR-125b (upregulated) was inversely correlated in OA cartilage vs.
healthy control cartilage (Fig. 2).
The preliminary data suggest that miR-125b might stimulate the
proliferation and apoptosis of chondrocytes through the development
of osteoarthritis. In cells of the myeloid lineage, miR-125b
represses interferon regulatory factor 4 and regulates inflammation
by downregulating its direct target, TNFα (29). Nagpal et al (30) reported that miR-125b upregulation is
critical for the TGF-β-induced fibroblast-to-myofibroblast
transition, which implies a potential association between miR-125b
and the TGF-β pathway. Zhen et al (31) demonstrated that inhibition of the
TGF-β pathway in subchondral bone can attenuate OA progression.
Moreover, in a bioinformatics study, the TNF signaling pathway was
reported to be closely associated with HOX genes by interacting
with HOX proteins (32). It was
thus speculated that HOTAIRM1-1 and miR-125b physically interact
with each other. The present study was performed to show an
interaction between miR-125b and HOTAIRM1-1, and the results proved
that HOTAIRM1-1 was a direct target of miR-125b (Fig. 3). Thus, HOTAIRM1-1 probably plays
its role by directly targeting miR-125b, thus influencing
chondrocytes in OA. As with many of the inflammatory cytokines
whose levels increase with age, the precise consequences of
dysregulated miR-125b expression on the functions of monocytes and
naïve CD8 T cells, such as inflammatory states or altered
migration, remain to be elucidated in OA.
Consistent with previous studies (25,31),
HOTAIRM1-1 knockdown in chondrocytes promoted cell cycle-induced
proliferation and cell apoptosis. ECM degradation is the main
factor that changes during chondrocyte destruction in OA. MMP13 and
IL-10 both play important roles in cartilage degradation (25,33,34).
Aggrecan and type II collagen primarily contribute to the formation
of cartilage tissue in the development of (35,36);
the present study measured these markers when HOTAIRM1-1 was
knocked down. MMP-13 and IL-10 expression was increased by
downregulating HOTAIRM1-1 in chondrocytes. On the other hand,
collagen II and aggrecan protein expression was decreased (Fig. 4). Thus, it was predicted that
HOTAIRM1-1 may play a key role in the pathogenesis of OA. Recently,
miR-125b was found to be stimulated as a possible consequence of
the inhibitory role of the IL-6 receptor and activator of
transcription 3 feedback loop (37,38).
Rescue experiments demonstrated that the miR-125b inhibitor
functionally reversed the impacts of HOTAIRM1-1 on cell
proliferation and apoptosis in chondrocytes (Fig. 5). Furthermore, these results suggest
that HOTAIRM1-1 downregulation may be required for cell
proliferation and cell apoptosis in OA.
In conclusion, the present study indicates that the
loss of HOTAIRM1-1 function leads to aberrant increase in the
proliferation and apoptosis of chondrocytes and causes a
comprehensive OA phenotype. miR-125b may be a potential downstream
mechanism that regulates the function of HOTAIRM1-1, and this
finding may provide a therapeutic strategy for OA.
Acknowledgements
Not applicable.
Funding
Funding: The current study was supported the science and
technology plan foundation of Yantai city (grant no. 2018SFGY094)
and Shan Dong Natural Science Foundation of China (grant no.
ZR2017LH022).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WBL and QJF contributed to the conception of the
study and analyzed the data. GSL and PS performed the experiments
and wrote the manuscript. YNL, FJZ and WBL analyzed the data and
wrote the manuscript. YNL and WBL analyzed the data and provided
constructive criticism. FJZ and PS confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the human ethics
committee of Tianjin Hospital (approval no. 2019-Yilunli-146). All
the patients provided written informed consent and participated in
the study according to their own will.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Thomas AC, Hubbard-Turner T, Wikstrom EA
and Palmieri-Smith RM: Epidemiology of posttraumatic
osteoarthritis. J Athl Train. 52:491–496. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Vina ER and Kwoh CK: Epidemiology of
osteoarthritis: Literature update. Curr Opin Rheumatol. 30:160–167.
2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Okada A and Okada Y: Progress of research
in osteoarthritis. Metalloproteinases in osteoarthritis. Clin
Calcium. 19:1593–1601. 2009.PubMed/NCBI(In Japanese).
|
|
4
|
Yoon DS, Lee KM, Kim SH, Kim SH, Jung Y,
Kim SH, Park KH, Choi Y, Ryu HA, Choi WJ and Lee JW: Synergistic
action of IL-8 and bone marrow concentrate on cartilage
regeneration through upregulation of chondrogenic transcription
factors. Tissue Eng Part A. 22:363–374. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Richardson SM, Kalamegam G, Pushparaj PN,
Matta C, Memic A, Khademhosseini A, Mobasheri R, Poletti FL,
Hoyland JA and Mobasheri A: Mesenchymal stem cells in regenerative
medicine: Focus on articular cartilage and intervertebral disc
regeneration. Methods. 99:69–80. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mobasheri A, Kalamegam G, Musumeci G and
Batt ME: Chondrocyte and mesenchymal stem cell-based therapies for
cartilage repair in osteoarthritis and related orthopaedic
conditions. Maturitas. 78:188–198. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Pant T, Dhanasekaran A, Fang J, Bai X,
Bosnjak ZJ, Liang M and Ge ZD: Current status and strategies of
long noncoding RNA research for diabetic cardiomyopathy. BMC
Cardiovasc Disord. 18(197)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ji D, Zhong X, Jiang X, Leng K, Xu Y, Li
Z, Huang L, Li J and Cui Y: The role of long non-coding RNA
AFAP1-AS1 in human malignant tumors. Pathol Res Pract.
214:1524–1531. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Jiang SD, Lu J, Deng ZH, Li YS and Lei GH:
Long noncoding RNAs in osteoarthritis. Joint Bone Spine.
84:553–556. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Marques-Rocha JL, Samblas M, Milagro FI,
Bressan J, Martinez JA and Marti A: Noncoding RNAs, cytokines, and
inflammation-related diseases. FASEB J. 29:3595–3611.
2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen L, Hu N, Wang C and Zhao H: HOTAIRM1
knockdown enhances cytarabine-induced cytotoxicity by suppression
of glycolysis through the Wnt/β-catenin/PFKP pathway in acute
myeloid leukemia cells. Arch Biochem Biophys.
680(108244)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bhan A, Hussain I, Ansari KI, Kasiri S,
Bashyal A and Mandal SS: Antisense transcript long noncoding RNA
(lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol
Biol. 425:3707–3722. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang Y, Hardin H, Chu YH, Esbona K, Zhang
R and Lloyd RV: Long non-coding RNA expression in anaplastic
thyroid carcinomas. Endocr Pathol. 30:262–269. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xiao Y, Yan X, Yang Y and Ma X:
Downregulation of long noncoding RNA HOTAIRM1 variant 1 contributes
to osteoarthritis via regulating miR-125b/BMPR2 axis and activating
JNK/MAPK/ERK pathway. Biomed Pharmacother. 109:1569–1577.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wei S, Zhao M, Wang X, Li Y and Wang K:
PU.1 controls the expression of long noncoding RNA HOTAIRM1 during
granulocytic differentiation. J Hematol Oncol. 9(44)2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li Y, Li S, Luo Y, Liu Y and Yu N: LncRNA
PVT1 Regulates chondrocyte apoptosis in osteoarthritis by acting as
a sponge for miR-488-3p. DNA Cell Biol. 36:571–580. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Huang K, Fu J, Zhou W, Li W, Dong S, Yu S,
Hu Z, Wang H and Xie Z: MicroRNA-125b regulates osteogenic
differentiation of mesenchymal stem cells by targeting Cbfβ in
vitro. Biochimie. 102:47–55. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang HF, Li ZJ, Fu X, Ma JX and Ma XL:
Interactions of bone marrow stromal cells with native and RGD
surface modified acellular bone matrix: A biocompatibility study.
Arch Med Res. 44:69–74. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li L, Wang Y, Song G, Zhang X, Gao S and
Liu H: HOX cluster-embedded antisense long non-coding RNAs in lung
cancer. Cancer Lett. 450:14–21. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gulei D, Mehterov N, Ling H, Stanta G,
Braicu C and Berindan-Neagoe I: The ‘good-cop bad-cop’ TGF-β role
in breast cancer modulated by non-coding RNAs. Biochim Biophys Acta
Gen Subj. 1861:1661–1675. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Park S, Lee M, Chun CH and Jin EJ: The
lncRNA, nespas, is associated with osteoarthritis progression and
serves as a potential new prognostic biomarker. Cartilage.
10:148–156. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yang S, Kim J, Ryu JH, Oh H, Chun CH, Kim
BJ, Min BH and Chun JS: Hypoxia-inducible factor-2alpha is a
catabolic regulator of osteoarthritic cartilage destruction. Nat
Med. 16:687–693. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Miyaki S, Sato T, Inoue A, Otsuki S, Ito
Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, et al:
MicroRNA-140 plays dual roles in both cartilage development and
homeostasis. Genes Dev. 24:1173–1185. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wojdasiewicz P, Poniatowski LA and
Szukiewicz D: The role of inflammatory and anti-inflammatory
cytokines in the pathogenesis of osteoarthritis. Mediators Inflamm.
2014(561459)2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Pearson MJ, Herndler-Brandstetter D, Tariq
MA, Nicholson TA, Philp AM, Smith HL, Davis ET, Jones SW and Lord
JM: IL-6 secretion in osteoarthritis patients is mediated by
chondrocyte-synovial fibroblast cross-talk and is enhanced by
obesity. Sci Rep. 7(1)(3451)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zanotti S and Canalis E: Interleukin 6
mediates selected effects of notch in chondrocytes. Osteoarthritis
Cartilage. 21:1766–1773. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Simsa-Maziel S and Monsonego-Ornan E:
Interleukin-1β promotes proliferation and inhibits differentiation
of chondrocytes through a mechanism involving down-regulation of
FGFR-3 and p21. Endocrinology. 153:2296–2310. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cheng NL, Chen X, Kim J, Shi AH, Nguyen C,
Wersto R and Weng NP: MicroRNA-125b modulates inflammatory
chemokine CCL4 expression in immune cells and its reduction causes
CCL4 increase with age. Aging Cell. 14:200–208. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Nagpal V, Rai R, Place AT, Murphy SB,
Verma SK, Ghosh AK and Vaughan DE: MiR-125b is critical for
fibroblast-to-myofibroblast transition and cardiac fibrosis.
Circulation. 133:291–301. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhen G, Wen C, Jia X, Li Y, Crane JL,
Mears SC, Askin FB, Frassica FJ, Chang W, Yao J, et al: Inhibition
of TGF-β signaling in mesenchymal stem cells of subchondral bone
attenuates osteoarthritis. Nat Med. 19:704–712. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
32
|
Shi X, Shao X, Liu B, Lv M, Pandey P, Guo
C, Zhang R and Zhang Y: Genome-wide screening of functional long
noncoding RNAs in the epicardial adipose tissues of atrial
fibrillation. Biochim Biophys Acta Mol Basis Dis.
1866(165757)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Charlier E, Deroyer C, Ciregia F, Malaise
O, Neuville S, Plener Z, Malaise M and de Seny D: Chondrocyte
dedifferentiation and osteoarthritis (OA). Biochem Pharmacol.
165:49–65. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Silawal S, Willauschus M, Schulze-Tanzil
G, Gogele C, Gesslein M and Schwarz S: IL-10 could play a role in
the interrelation between diabetes mellitus and osteoarthritis. Int
J Mol Sci. 20(768)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hu J, Wang Z, Shan Y, Pan Y, Ma J and Jia
L: Long non-coding RNA HOTAIR promotes osteoarthritis progression
via miR-17-5p/FUT2/β-catenin axis. Cell Death Dis.
9(711)2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Matyas JR, Adams ME, Huang D and Sandell
LJ: Discoordinate gene expression of aggrecan and type II collagen
in experimental osteoarthritis. Arthritis Rheum. 38:420–425.
1995.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Misso G, Zarone MR, Lombardi A, Grimaldi
A, Cossu AM, Ferri C, Russo M, Vuoso DC, Luce A, Kawasaki H, et al:
miR-125b upregulates miR-34a and sequentially activates stress
adaption and cell death mechanisms in multiple myeloma. Mol Ther
Nucleic Acids. 16:391–406. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Cui F, Li X, Zhu X, Huang L, Huang Y, Mao
C, Yan Q, Zhu J, Zhao W and Shi H: MiR-125b inhibits tumor growth
and promotes apoptosis of cervical cancer cells by targeting
phosphoinositide 3-kinase catalytic subunit delta. Cell Physiol
Biochem. 30:1310–1318. 2012.PubMed/NCBI View Article : Google Scholar
|