Introduction
In 2019, prostate cancer (PCa) was one of the most
common malignancies among males worldwide (1). Patients with early stages of PCa can
be cured by radical surgery or radiotherapy, but the treatment of
advanced prostate cancer is still dependent on androgen deprivation
therapy (ADT) (2). Although ADT can
effectively inhibit the progression of PCa, resistance of PCa cells
to ADT can lead to tumor metastasis (3). Therefore, novel therapy strategies are
urgently needed in the treatment of PCa.
Curcumin is a polyphenolic compound derived from
Curcuma longa L., which is used as perfume in India and
herbal medicine in China (4,5).
Previous studies indicate that curcumin has anti-inflammatory,
anti-oxidative, anti-infective, anti-fibrotic and
anti-atherosclerotic effects, and that it exhibits low toxicity
(5,6). The anticancer properties of curcumin
have attracted increasing attention. Accumulating studies have
indicated that curcumin inhibits the occurrence, development,
chemoresistance and radioresistance of various malignancies
(7-13).
For example, curcumin inhibits the proliferation of colon cancer
cells by inhibiting the Wnt/β-catenin signaling pathway, and can
increase the radiosensitivity of PCa cells by inhibiting autophagy
(11,12). In addition, curcumin suppresses the
activity of JNK signaling in PCa cells, thereby impeding cell
proliferation and promoting apoptosis (13). Nevertheless, the mechanism of
curcumin in blocking PCa progression has not been clearly
clarified.
Certain studies have reported that curcumin can
inhibit cancer progression by regulating microRNA (miRNA/miR)
expression (7,14-15).
For example, in PCa, curcumin inhibits cancer cell proliferation
and migration by regulating the miR-143/phosphoglycerate kinase 1
axis (14); curcumin induces the
upregulation of miR-145, which in turn inhibits PCa cell
proliferation by repressing octamer-binding transcription factor 4
expression (15). Therefore, the
present study was designed to uncover the biological functions and
specific mechanisms of curcumin/miR-30a-5p/PCNA clamp associated
factor (PCLAF) axis in PCa via in vitro assays.
Materials and methods
Clinical specimens
PCa tissues and the adjacent benign tissues (at 5-cm
distance) of 35 patients with PCa who visited Zhongshan Hospital
Affiliated to Fudan University (Shanghai, China) from March 2017 to
March 2019 were collected. All of the patients were diagnosed via
biopsy and received radical prostatectomy. There were 21 male and
14 female patients. Their age range was 49-71 years (mean,
60.9±5.1). Among the patients enrolled, the Gleason score was ≥8
points in 11 cases and <8 points in 24 cases, and the serum
prostate-specific antigen levels were ≤10 ng/ml in 19 cases and
>10 ng/ml in 16 cases. All tissue specimens were surgically
removed and quickly frozen in liquid nitrogen. The study was
approved by The Ethics Review Board of Zhongshan Hospital
Affiliated to Fudan University, and all participants provided
written informed consent.
Cell culture, drug treatment and
transfection
Both PC-3 and DU145 cells were purchased from the
China Center for Type Culture Collection. The miR-30a-5p inhibitor
(5'-CUUCCAGUCGAGGAUGUUUACA-3') and non-targeting control
(5'-ACAUAGGGCCCAUGCUAACUGC-3') were procured from Guangzhou RiboBio
Co., Ltd. PC-3 and DU145 cells were cultured in RPMI-1640 medium
(Thermo Fisher Scientific, Inc.) containing 10% FBS (Thermo Fisher
Scientific, Inc), 100 µg/ml penicillin and 100 µg/ml streptomycin
(Sigma-Aldrich; Merck KGaA) at 37˚C in 5% CO2. When the
cells had reached 70-80% confluence, the miR-30a-5p inhibitor and
control were transfected into PC-3 and DU145 cells at 37˚C for 6 h
at a final concentration of 50 µM according to the manufacturer's
instructions of Lipofectamine® 2000 (Thermo Fisher
Science, Inc.). After 24 h, cells were treated with curcumin (cat.
no. 239802; Sigma-Aldrich; Merck KGaA) at different concentrations
(10, 20, 30, 40 or 50 µmol/l) at 37˚C for 12, 24 or 48 h. After
that, cells were harvested for subsequent experiments.
Cell Counting Kit-8 (CCK-8) assay
PC-3 and DU145 cells in the logarithmic growth phase
were seeded in a 96-well cell plate (2x103 cells/well).
Subsequently, the cells were randomly divided into six groups with
different concentrations of curcumin: i) Control group (0 µmol/l);
ii) 10 µmol/l curcumin group; iii) 20 µmol/l curcumin group; iv) 30
µmol/l curcumin group; v) 40 µmol/l curcumin group; and vi) 50
µmol/l curcumin group. CCK-8 solution (10 µl; Beyotime Institute of
Biotechnology) was supplemented to each well at 12, 24 and 48 h,
and then the cells were incubated at 37˚C for a further 4 h. After
that, the absorbance value of each well was detected by a
microplate reader (GENios FL Fluorescence Microplate Reader FI TRF;
Tecan Group, Ltd.) at a wavelength of 450 nm, and the average
absorbance value of three wells were used to evaluate the viability
of cells.
Flow cytometry analysis
The Annexin V-FITC/propidium iodide (PI) double
staining method was used to analyze the apoptosis of PCa cells in
this study. A total of 1x106 PC-3 or DU145 cells from
each group were harvested and resuspended using 100 µl 1X binding
buffer (BSL Bioservice), followed by the addition of 5 µl Annexin
V-FITC solution and 5 µl PI solution (both from Beyotime Institute
of Biotechnology), mixed thoroughly and incubated in the dark at
room temperature for 15 min. Subsequently, the apoptosis rate was
detected using a flow cytometer (BD Biosciences) within 1 h and the
data were analyzed using FlowJo v10.07 software (FlowJo, LLC).
Reverse transcription-quantitative PCR
(RT-qPCR)
PC-3 and DU145 cells treated with 30 µmol/l curcumin
for 24 h were collected. Total cellular RNA was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RNA concentration and purity were measured on the NanoDrop
2000 (NanoDrop Technologies; Thermo Fisher Scientific, Inc.) using
1 µl of RNA. After the concentration and purity of RNA were
determined, total RNA was reverse transcribed into cDNA using a
RevertAid First Strand cDNA Synthesis Kit (1 h at 37˚C; Thermo
Fisher Scientific, Inc.). RT-qPCR was conducted using a CFX96
quantitative PCR system (Bio-Rad Laboratories, Inc.) with an
SYBR® Green Premix Ex Taq II kit (Takara Biotechnology
Co., Ltd.) in accordance with the manufacturer's instructions. The
following temperature protocol was used for reverse transcription:
37˚C For 2 min, 23˚C for 10 min, 55˚C for 10 min and 85˚C for 10
min. U6 was identified as the internal reference for miR-30a-5p,
and β-actin was used as the internal reference for PCLAF. The
levels of miR-30a-5p and PCLAF mRNA expression were calculated
using the 2-ΔΔCq method (16). The primer sequences were shown in
Table I.
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Name | Primer sequences,
5'-3' |
|---|
| miR-30a-5p
forward |
GGGCCTGTAAACATCCTCG |
| miR-30a-5p
reverse |
GAATACCTCGGACCCTGC |
| PCLAF forward |
ATGGTGCGGACTAAAGCAGAC |
| PCLAF reverse |
CCTCGATGAAACTGATGTCGAAT |
| U6 forward |
GCTTCGGCAGCACATATACTAAAAT |
| U6 reverse |
CGCTTCACGAATTTGCGTGTCAT |
| β-actin
forward |
TCCCTCAAGATTGCTAGCAA |
| β-actin
reverse |
AGATCCACAACGGATACATT |
BrdU experiment
PC-3 and DU145 cells were prepared into a
single-cell suspension and inoculated in a 96-well plate
(1x104 cells/well). The cells were then divided into
control- and curcumin-treated groups. After being treated with 30
µmol/l curcumin for 24 h, BrdU solution (Beyotime Institute of
Biotechnology) was added at a final concentration of 30 µmol/l and
then the cells were incubated at 37˚C for 8 h. The cells were
washed with PBS three times. Subsequently, the cells were fixed
with 4% paraformaldehyde at room temperature for 30 min and rinsed
with PBS for three times again. A total of 2 mol/l HCl was added to
denature the DNA for 30 min at 37˚C and then cells were immersed in
PBS containing 0.1% Tween-20 for 30 min. After that, non-specific
antigens were blocked with 3% BSA (Sigma-Aldrich; Merck KGaA) for 1
h at room temperature. Following which, mouse anti-BrdU antibody
(1:1400; cat. no. 5292S; Cell Signaling Technology, Inc.) was
incubated with the cells for 2 h at room temperature. The cells
were rinsed for three times with PBS, and the secondary antibody
incubated with the cells for 1 h at room temperature. Subsequently,
DAPI staining solution was used to stain the nuclei at 37˚C for 30
min. The cells were observed and images captured under a
fluorescence microscope (magnification, x400; Olympus Corporation).
Overall, five visual fields were observed under fluorescence
microscope for each sample. Cell proliferation rate = the number of
BrdU staining positive cells/total DAPI staining positive cells
x100%.
Scratch assay
Wound healing was used to evaluate PCa cell
migration. PC-3 and DU145 cells were seeded into six-well plates at
a density of 5x105 cells/well. After the cells were
cultured to 100% confluence and serum starved for 24 h, a sterile
200-µl pipette tip was used to scratch the cells at the center of
the well. The cells were washed with PBS to remove the cell debris,
and the cells were cultured in serum-free medium. Closure of the
wound was observed under an inverted fluorescence microscope
(magnification, x400; Olympus Corporation) at 0 and 24 h after
scratching.
Transwell assay
Cells in each group were trypsinized, harvested and
resuspended in serum-free medium to adjust the cell concentration
to 5x105 cells/ml. To evaluate the migration of cells, a
Transwell system (8-µm pore-size; Corning, Inc.) was used. A total
of 200 µl cell suspension was added to the upper chamber, 600 µl
RPMI-1640 medium containing 10% FBS (both from Thermo Fisher
Scientific, Inc.) was added to the lower chamber and the cells were
cultured in an incubator for 24 h. Subsequently, the cells in the
upper chamber were carefully wiped off. The migrated cells were
fixed with formaldehyde at 37˚C for 15 min, and then stained with
crystal violet solution at 37˚C for 30 min. After being rinsed with
PBS, the cells were dried and observed under an inverted
fluorescence microscope (magnification, x400). Overall, five visual
fields were observed under fluorescence microscope for each sample.
To test cell invasion, the membrane of the Transwell system was
coated with Matrigel for 1 h at 37˚C (BD Biosciences) before the
experiment, and the other steps were as the same as the migration
assay.
Western blotting
PC-3 and DU145 cells were treated with 30 µmol/l
curcumin for 24 h, and then the protease inhibitor-containing RIPA
buffer (Beyotime Institute of Biotechnology) was added to lyse the
cells on ice for 40 min. Then, the lysate was centrifuged at 1,500
x g for 15 min to obtain the supernatant. Protein concentration
determination was performed using the BCA protein determination
assay and 30 µg of protein was loaded per lane. Thereafter, the
protein samples were loaded, 10% SDS-PAGE was performed and then
the proteins were electrically transferred onto the PVDF membrane
(EMD Millipore). After that, 5% skim-milk was used to block the
non-specific antigens at room temperature. Next, primary antibodies
were added to incubate the membrane at 4˚C overnight, and then the
membrane was subsequently incubated with horseradish
peroxidase-conjugated secondary antibody for 1 h at room
temperature. Then, ECL chemiluminescent solution (Beyotime
Institute of Biotechnology) was used to visualize the bands and the
bands were quantified using ImageJ software (v1.52; National
Institutes of Health). The antibodies used in this study were
obtained from Abcam: Anti-PCLAF Antibody (cat. no. ab226255;
1:1,000), anti-Bax antibody (cat. no. ab32503; 1:1,000), anti-Bcl-2
antibody (cat. no. ab185002; 1:1,000), anti-caspase-3 antibody
(cat. no. ab13847, 1:1,000), anti-cleaved caspase-3 antibody (cat.
no. ab2302; 1:500), anti-β-actin antibody (cat. no. ab8227; 1:500)
and horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody (cat. no. ab205718; 1:1,000).
Bioinformatics analysis
The Gene Expression Profiling Interactive Analysis
(GEPIA) database (http://gepia.cancer-pku.cn/) was used to analyze PCLAF
expression in PCa samples. The StarBase v3.0 database (http://starbase.sysu.edu.cn/) was used to analyze the
correlation between miR-30a-5p expression and PCLAF expression in
PCa samples. Cut-off values were as follows: |Log2FC| Cut-off =1,
P-value cut-off =0.01.
Statistical analysis
All data are expressed as mean ± standard deviation
(unless otherwise shown). SPSS v22.0 (IBM Corp.) and GraphPad Prism
8.0 (Version X; GraphPad Software, Inc.) were applied for
statistical analysis. All experiments were performed in triplicate.
Whether the data were normally distributed was examined using a
one-sample Kolmogorov-Smirnov test. For normally distributed data,
an independent sample t-test was used to make the comparison
between two groups. One-way ANOVA was used to compare three or more
groups. If there was a significant difference, Student-Neuman-Keuls
or Tukey's tests were used to determine significant differences
between two groups. Correlation was analyzed using Pearson
correlation coefficient. P<0.05 was considered to indicate a
statistically significant difference.
Results
Function of curcumin on the viability
and apoptosis of PCa cells
PCa cell lines PC-3 and DU145 cells were treated
with different concentrations of curcumin (0, 10, 20, 30, 40 and 50
µmol/l), and a CCK-8 assay was employed to detect cell viability at
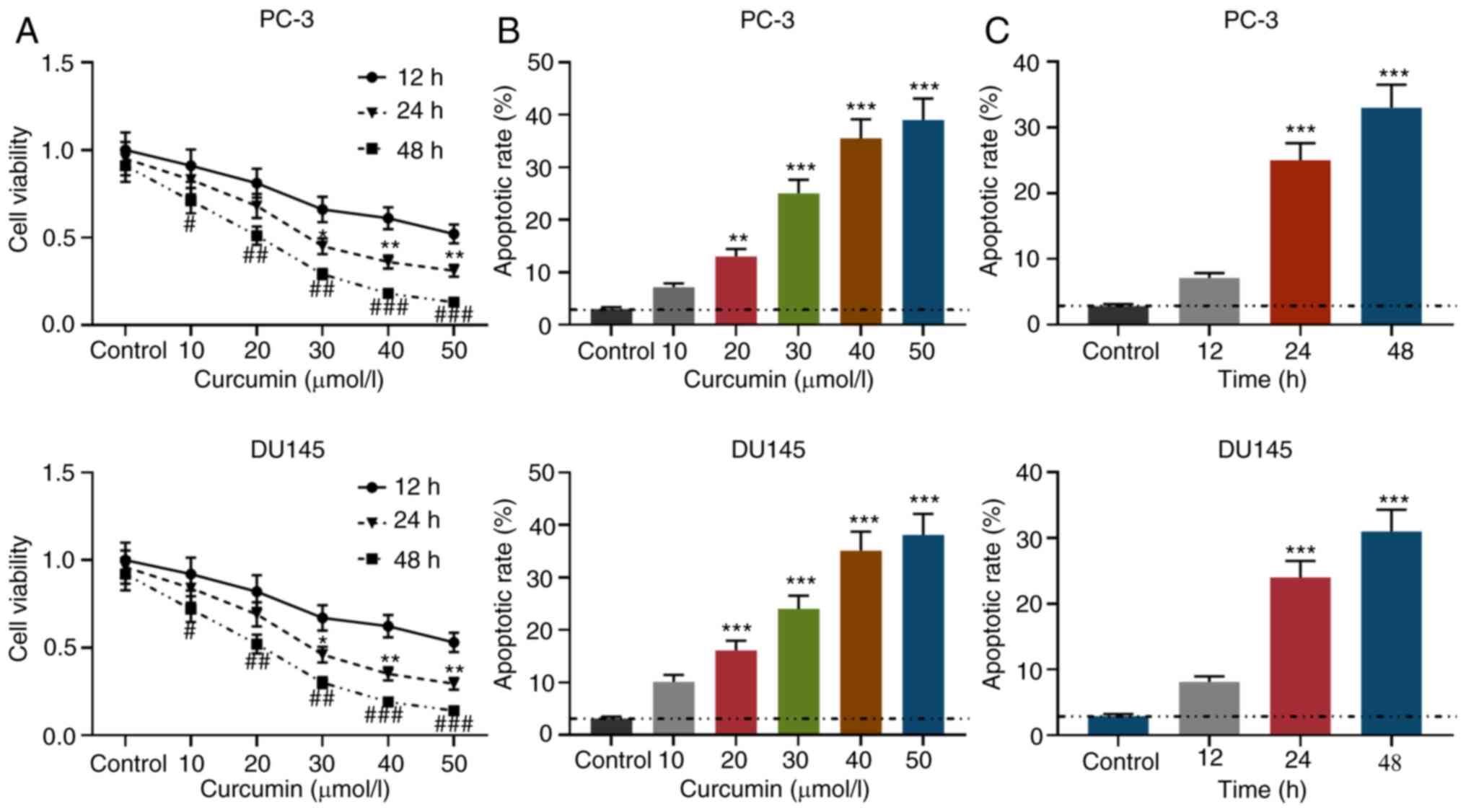
12, 24 and 48 h after treatment. As shown in Fig. 1A, compared with the untreated group,
curcumin inhibited the viability of PCa in concentration- and
time-dependent manners. As seen in Figs. 1B and S1, after being treated with curcumin for
24 h, the apoptosis rate of PCa cells gradually increased with
increasing concentrations of curcumin. Moreover, when PCa cells
were treated with 30 µmol/l curcumin, the apoptotic rate gradually
increased with the treatment time (Figs. 1C and S1). These observations suggested that
curcumin can inhibit cell viability and promote apoptosis.
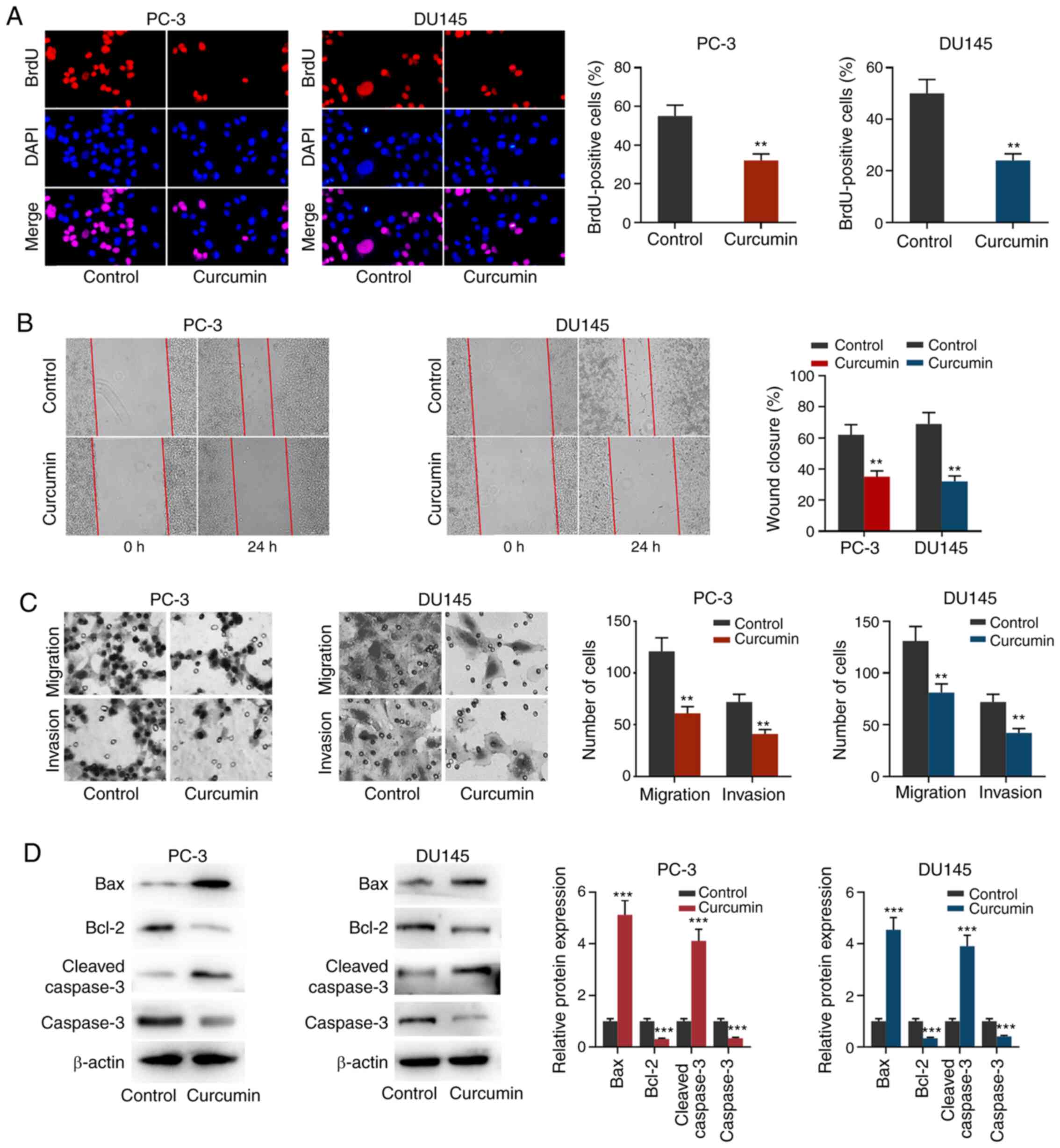
Curcumin inhibits the proliferation,
migration and invasion of PCa cells and promotes apoptosis
To further examine the role of curcumin on PCa cell
proliferation, migration and invasion, BrdU, wound healing and
Transwell assays were performed. As shown in Fig. 2A-C, after PC-3 and DU145 cells were
treated with 30 µmol/l curcumin for 24 h, cell proliferation,
migration and invasion were significantly reduced compared with the
control untreated group. Western blotting was used to detect the
expression of apoptosis-related proteins. As shown in Fig. 2D, after treatment with curcumin, the
expression levels of Bax and cleaved caspase-3 were increased in
both PC-3 cells and DU145 cells, while the expression levels of
Bcl-2 and caspase-3 were decreased. These results further indicated
that curcumin exhibited inhibitory effects on PCa cells.
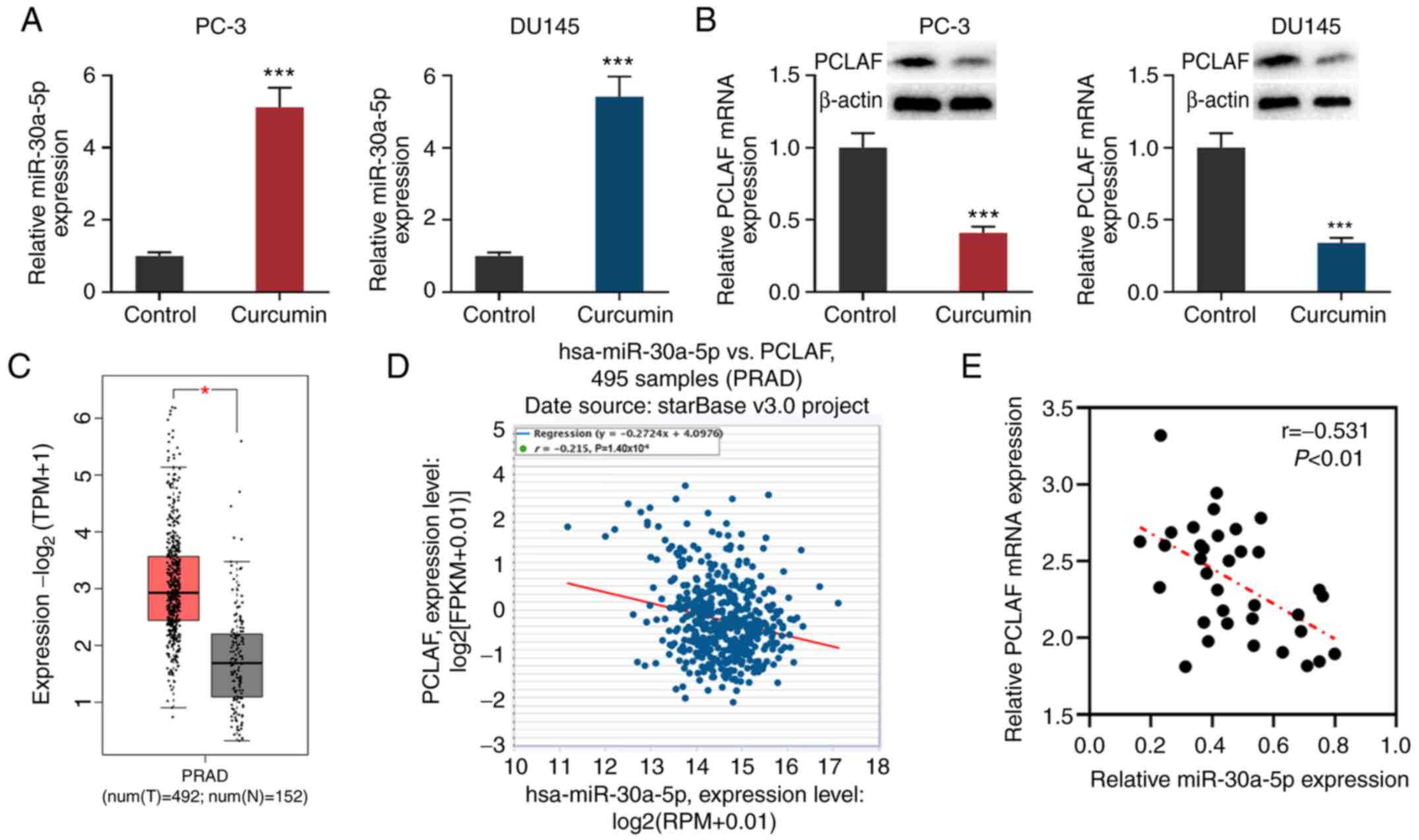
Curcumin suppresses PCLAF expression
by upregulating miR-30a-5p in PCa cells
miR-30a-5p exerts a tumor-suppressive effect in
colorectal cancer and melanoma by inhibiting cell proliferation,
metastasis, differentiation and the cell cycle (17-19).
A recent study reported that miR-30a-5p has low expression levels
in PCa, and downregulation of miR-30a-5p promotes the progression
of PCa by targeting PCLAF (20).
After PC-3 and DU145 cells were treated with 30 µmol/l curcumin for
24 h, the cells were collected and the levels of PCLAF and
miR-30a-5p expression in PC-3 and DU145 cells were detected via
RT-qPCR and western blotting. The results showed that in comparison
with the untreated group, miR-30a-5p expression was significantly
increased in PCa cells, while PCLAF expression was decreased
(Fig. 3A and B).
Through analyzing the data from GEPIA, it was
demonstrated that PCLAF expression was upregulated in PCa samples
compared with normal tissues adjacent to PCa cancer tissues
(Fig. 3C). Further analysis of the
data from the StarBase database found that in prostate
adenocarcinoma samples, miR-30a-5p expression and PCLAF expression
were significant (r=-0.215; Fig.
3D). Levels of miR-30a-5p and PCLAF mRNA expression were also
detected in the 35 pairs of PCa tissues/adjacent tissues collected
from patients; miR-30a-5p was downregulated in PCa tissues, while
PCLAF was upregulated (Fig. S2A
and B). Additionally, the
expression levels of miR-30a-5p and PCLAF were negatively
correlated in PCa tissues (Fig.
3E). Considering that PCLAF is a target gene of miR-30a-5p
(20), it was concluded that
curcumin could inhibit the expression of PCLAF by promoting the
expression of miR-30a-5p.
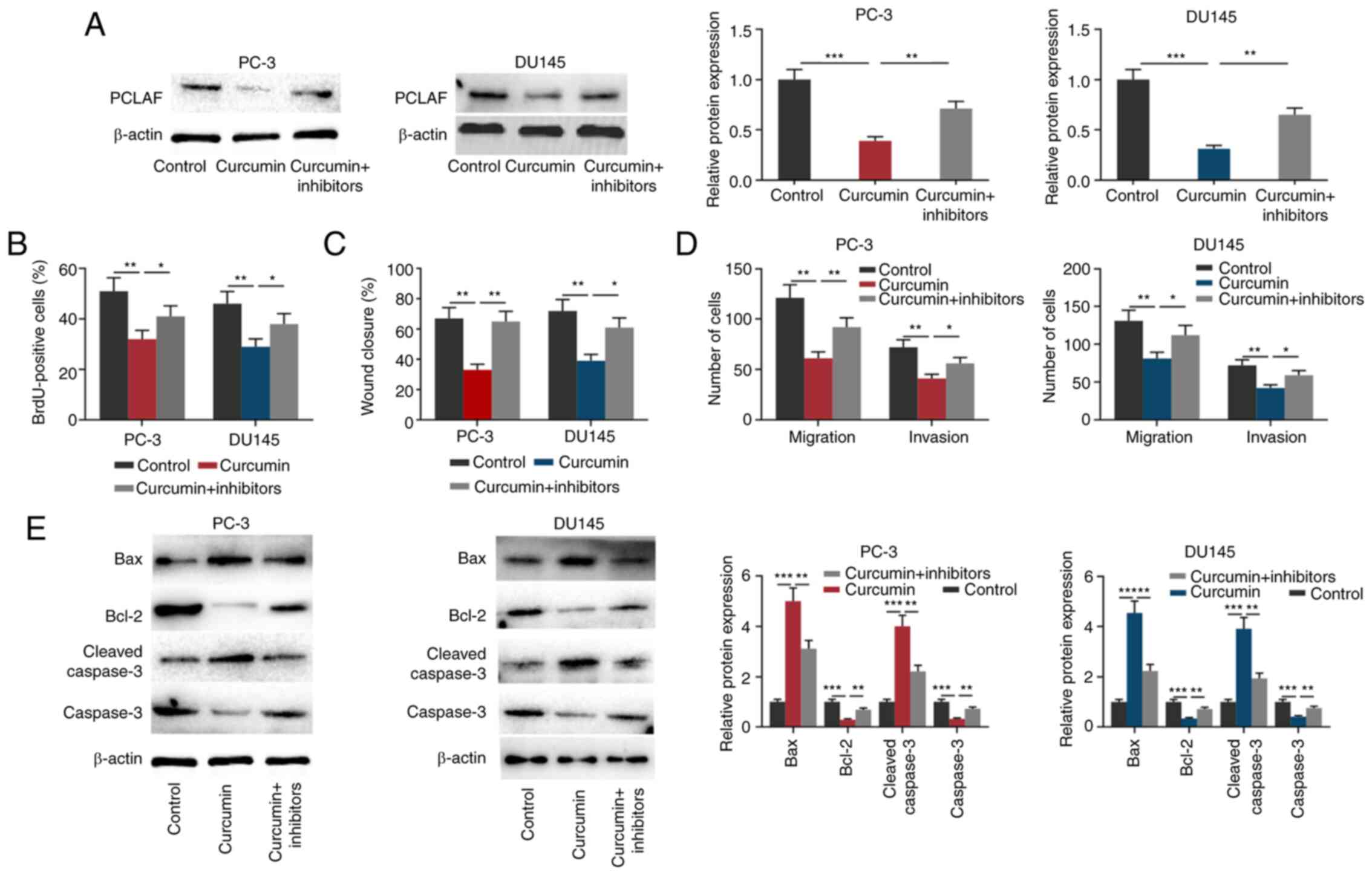
Curcumin inhibits the malignant
biological behaviors of PCa cells partly by regulating
miR-30a-5p
To investigate whether curcumin affected PCa cells
through regulating miR-30a-5p and PCLAF, before PCa cells were
treated with 30 µmol/l curcumin for 24 h, miR-30a-5p inhibitors
were transfected into PCa cells (Fig.
S2C). As shown in Fig. 4A, the
transfection of miR-30a-5p inhibitors partially reversed the
inhibitory effect of curcumin on the expression of PCLAF in both
PCa cell lines. Subsequently, through the BrdU, wound healing and
Transwell assays, it was found that curcumin significantly
suppressed PCa cell proliferation, migration and invasion, while
miR-30a-5p inhibitors partially reversed these effects (Figs. 4B-D and S1). Additionally, transfection of
miR-30a-5p inhibitors partially reversed the effects of curcumin on
the expression of Bax, Bcl2, cleaved caspase-3 and caspase-3
(Fig. 4E). These results further
validated that the miR-30a-5p/PCLAF axis was a significant
downstream effector of curcumin.
Discussion
A number of studies have shown that curcumin exerts
significant anticancer effects in diverse malignancies, inhibiting
cancer cell proliferation and metastasis, promoting apoptosis, and
increasing chemosensitivity and radiosensitivity (21-24).
Curcumin suppresses the proliferation, migration and invasion of
PCa cells by regulating multiple signaling pathways. For example,
it inhibits the expression of membrane type 1-MMP and MMP2 in PCa
cells, and suppresses the viability and metastasis of cancer cells
via the Notch-1 signaling pathway (21). A similar study reports that curcumin
induces PCa apoptosis and G0/G1 arrest by inhibiting Notch
signaling (22). In addition,
curcumin represses the monoamine oxidase A/mTOR/hypoxia inducible
factor-1α signaling pathway, impedes the production of reactive
oxygen species, inhibits the expression levels of C-X-C chemokine
receptor 4 and IL-6 receptors, and suppresses the
epithelial-mesenchymal transition of PCa cells mediated by
cancer-associated fibroblasts (23). Moreover, curcumin inhibits nuclear
β-catenin transcription activity in PCa cells by activating
polycystin-1, thereby suppressing the proliferation, colony
formation and motility of cancer cells (24). In the present study, it was observed
that curcumin could inhibit PCa cell proliferation, migration and
invasion, and promote apoptosis in a concentration- and time-
dependent manner. These experiments indicated that curcumin
exhibited tumor-suppressive properties in PCa, which is consistent
with previous reports.
An increasing number of studies have shown that
miRNAs have notable effects on cell proliferation, apoptosis and
differentiation (25-28).
A number of studies report that miRNAs are implicated in
carcinogenesis and cancer progression. For example, miR-9-5p can
suppress the proliferation of glioma cells by downregulating
forkhead box P2 expression (28).
In PCa, miR-381 suppresses the proliferation and invasion of PCa
cells by downregulating androgen receptor expression (29). Curcumin can exert tumor-suppressive
effects by regulating the expression of a diverse number of miRs
(12,30-33).
For example, in laryngeal squamous cell carcinoma, curcumin
inhibits the activity of the PI3K/Akt/mTOR signaling pathway by
upregulating miR-145 expression, thereby inhibiting the
proliferation, cell cycle procession and metastasis of cancer cells
(32). In nasopharyngeal carcinoma,
curcumin suppresses cancer cell proliferation and metastasis by
regulating the miR-7/Skp2/p21 axis (33). In PCa, curcumin represses autophagy
through the miR-143/ATG2B axis, thereby increasing the
radiosensitivity of cancer cells (12).
The present study observed that miR-30a-5p
expression was induced by curcumin treatment in both PC-3 and
DU-145 cells. Notably, after PCa cells were transfected with
miR-30a-5p inhibitors, the antitumor activity of curcumin was
attenuated. These results suggested that miR-30a-5p was a
downstream effector of curcumin, and the function of curcumin in
inhibiting PCa progression was partly dependent on miR-30a-5p.
miR-30a-5p is well known as a tumor suppressor
(34-36).
In breast cancer, miR-30a-5p targets the regulation of lactate
dehydrogenase A to inhibit the Warburg effect, and impedes the
proliferation and metastasis of cancer cells (37). In non-small cell lung cancer,
miR-30a-5p downregulates CD73 expression and inhibits
proliferation, metastasis and epithelial-mesenchymal transition by
regulating the EGF signaling pathway (38). In PCa, miR-30a-5p is also
significantly downregulated in cancer tissues, and it suppresses
the proliferation of PCa cells by inhibiting PCLAF (20). PCLAF, also known as KIAA0101, is an
upregulated oncogene in a variety of tumors (39). Previous studies have shown that
PCLAF plays a vital role in cell proliferation, cell survival, DNA
repair and tumorigenesis (40-42).
For example, PCLAF-knockdown suppresses cancer cell proliferation
and cell cycle progression by promoting the formation of p53/Sp1
complex in breast cancer (41).
miR-429 inhibits the progression of epithelial ovarian cancer by
targeting PCLAF (42). The present
study demonstrated that PCLAF was highly expressed in PCa samples
and that miR-30a-5p expression was negatively associated with PCLAF
expression in PCa samples. These data further evidenced the
regulatory effect of miR-30a-5p on PCLAF, which is consistent with
previous reports (20). Moreover,
curcumin treatment downregulated PCLAF expression in both PC-3 and
DU145 cell lines. Collectively, these data showed that curcumin
could inhibit PCLAF to block the progression of PCa by inducing the
expression of miR-30a-5p. However, the weak correlation between
miR-30a-5p and PCLAF predicted by bioinformatics in the present
study is limitations the conclusions of this study.
In summary, curcumin inhibited the proliferation and
metastasis, and promoted the apoptosis of PCa cells by regulating
the miR-30a-5p/PCLAF axis. The present study provides novel
insights into the mechanism of curcumin's tumor-suppressive
functions. Notably, that curcumin may repress the malignant
phenotypes of PCa cells by regulating other miRNAs and proteins,
which remains to be clarified in future studies.
Supplementary Material
Representative images of flow
cytometry, BrdU, wound healing and Transwell assays (magnification,
x400). PI, propidium iodide.
Transfection of miR inhibitors.
Expression levels of (A) miR-30a-5p and (B) PCLAF mRNA in PCa
tissues and adjacent prostate tissues were detected via RT-qPCR.
(C) RT-qPCR was used to detect the transfection efficacy of
miR-30a-5p inhibitors. ***P<0.001 vs. Control or as
indicated. PCa, prostate cancer; miR, microRNA; PCLAF, PCNA clamp
associated factor; RT-qPCR, reverse transcription-quantitative
PCR.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by a grant from The
Department of Urology, Xuhui Hospital, Zhongshan Hospital
Affiliated to Fudan University (grant no. 2017XHYY-05).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LP, JS and JG conceived and designed the
experiments. LP, WL and TB performed the experiments. YW and WL
performed the statistical analysis. LP, JS and TB drafted the
manuscript. LP, JS and JG confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics Review
Board of Zhongshan Hospital Affiliated to Fudan University and all
participants provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Guo Z, He C, Yang F, Qin L, Lu X and Wu J:
Long non-coding RNA-NEAT1, a sponge for miR-98-5p, promotes
expression of oncogene HMGA2 in prostate cancer. Biosci Rep: Sep
24, 2019 (Epub ahead of print). doi: 10.1042/BSR20190635.
|
|
2
|
Song Z, Zhuo Z, Ma Z, Hou C, Chen G and Xu
G: Hsa_Circ_0001206 is downregulated and inhibits cell
proliferation, migration and invasion in prostate cancer. Artif
Cells Nanomed Biotechnol. 47:2449–2464. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Dart DA, Koushyar S, Lanning BE and Jiang
W: MiR-221 is specifically elevated in PC3 cells and its deletion
reduces adhesion, motility and growth. Anticancer Res.
39:5311–5327. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Li H, Yue L, Xu H, Li N, Li J, Zhang Z and
Zhao RC: Curcumin suppresses osteogenesis by inducing miR-126a-3p
and subsequently suppressing the WNT/LRP6 pathway. Aging (Albany
NY). 11:6983–6998. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mou S, Zhou Z, He Y, Liu F and Gong L:
Curcumin inhibits cell proliferation and promotes apoptosis of
laryngeal cancer cells through Bcl-2 and PI3K/Akt, and by
upregulating miR-15a. Oncol Lett. 14:4937–4942. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhan JW, Jiao DM, Wang Y, Song J, Wu JH,
Wu LJ, Chen QY and Ma SL: Integrated microRNA and gene expression
profiling reveals the crucial miRNAs in curcumin anti-lung cancer
cell invasion. Thorac Cancer. 8:461–470. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sun C, Zhang S, Liu C and Liu X: Curcumin
promoted miR-34a expression and suppressed proliferation of gastric
cancer cells. Cancer Biother Radiopharm. 34:634–641.
2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Qian C, Wang B, Zou Y, Zhang Y, Hu X, Sun
W, Xiao H, Liu H and Shi L: MicroRNA 145 enhances chemosensitivity
of glioblastoma stem cells to demethoxycurcumin. Cancer Manag Res.
11:6829–6840. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sak K: Radiosensitizing potential of
curcumin in different cancer models. Nutr Cancer. 72:1276–1289.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Xu R, Li H, Wu S, Qu J, Yuan H, Zhou Y and
Lu Q: MicroRNA-1246 regulates the radio-sensitizing effect of
curcumin in bladder cancer cells via activating P53. Int Urol
Nephrol. 51:1771–1779. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dou H, Shen R, Tao J, Huang L, Shi H, Chen
H, Wang Y and Wang T: Curcumin suppresses the colon cancer
proliferation by inhibiting Wnt/β-catenin pathways via miR-130a.
Front Pharmacol. 8(877)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu J, Li M, Wang Y and Luo J: Curcumin
sensitizes prostate cancer cells to radiation partly via epigenetic
activation of miR-143 and miR-143 mediated autophagy inhibition. J
Drug Target. 25:645–652. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhao W, Zhou X, Qi G and Guo Y: Curcumin
suppressed the prostate cancer by inhibiting JNK pathways via
epigenetic regulation. J Biochem Mol Toxicol.
32(e22049)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cao H, Yu H, Feng Y, Chen L and Liang F:
Curcumin inhibits prostate cancer by targeting PGK1 in the
FOXD3/miR-143 axis. Cancer Chemother Pharmacol. 79:985–994.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Liu T, Chi H, Chen J, Chen C, Huang Y, Xi
H, Xue J and Si Y: Curcumin suppresses proliferation and in vitro
invasion of human prostate cancer stem cells by ceRNA effect of
miR-145 and lncRNA-ROR. Gene. 631:29–38. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wu X, Jia R, Wang M, Chen S, Liu M, Zhu D,
Zhao X, Yang Q, Wu Y, Yin Z, et al: Downregulation of
microRNA-30a-5p contributes to the replication of duck enteritis
virus by regulating Beclin-1-mediated autophagy. Virol J.
16(144)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li J, Zhao LM, Zhang C, Li M, Gao B, Hu
XH, Cao J and Wang GY: The lncRNA FEZF1-AS1 promotes the
progression of colorectal cancer through regulating OTX1 and
targeting miR-30a-5p. Oncol Res. 28:51–63. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Noori J, Haghjooy Javanmard S and Sharifi
M: The role of microRNA-30a and downstream snail1 on the growth and
metastasis of melanoma tumor. Iran J Basic Med Sci. 22:534–540.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhao H, Lai X, Zhang W, Zhu H, Zhang S, Wu
W, Wang S, Tang M, Deng Z and Tan J: MiR-30a-5p frequently
downregulated in prostate cancer inhibits cell proliferation via
targeting PCLAF. Artif Cells Nanomed Biotechnol. 47:278–289.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yang J, Wang C, Zhang Z, Chen X, Jia Y,
Wang B and Kong T: Curcumin inhibits the survival and metastasis of
prostate cancer cells via the Notch-1 signaling pathway. APMIS.
125:134–140. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sha J, Li J, Wang W, Pan L, Cheng J, Li L,
Zhao H and Lin W: Curcumin induces G0/G1 arrest and apoptosis in
hormone independent prostate cancer DU-145 cells by down regulating
Notch signaling. Biomed Pharmacother. 84:177–184. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Du Y, Long Q, Zhang L, Shi Y, Liu X, Li X,
Guan B, Tian Y, Wang X, Li L, et al: Curcumin inhibits
cancer-associated fibroblast-driven prostate cancer invasion
through MAOA/mTOR/HIF-1α signaling. Int J Oncol. 47:2064–2072.
2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sundram V, Chauhan SC, Ebeling M and Jaggi
M: Curcumin attenuates β-catenin signaling in prostate cancer cells
through activation of protein kinase D1. PLoS One.
7(e35368)2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wei Q, Tu Y, Zuo L, Zhao J, Chang Z, Zou Y
and Qiu J: MiR-345-3p attenuates apoptosis and inflammation caused
by oxidized low-density lipoprotein by targeting TRAF6 via
TAK1/p38/NF-kB signaling in endothelial cells. Life Sci.
241(117142)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wu Z, Chen D, Wang K, Cao C and Xu X: Long
non-coding RNA SNHG12 functions as a competing endogenous RNA to
regulate MDM4 expression by sponging miR-129-5p in clear cell renal
cell carcinoma. Front Oncol. 9(1260)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ye J, Lei J, Fang Q, Shen Y, Xia W, Hu X,
Xu Q, Yuan H, Huang J and Ni C: miR-4666-3p and miR-329
synergistically suppress the stemness of colorectal cancer cells
via targeting TGF-β/Smad pathway. Front Oncol.
9(1251)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang H, Li Y, Tan Y, Liu Q, Jiang S, Liu
D, Chen Q and Zhang S: MiR-9-5p inhibits glioblastoma cells
proliferation through directly targeting FOXP2 (Forkhead Box P2).
Front Oncol. 9(1176)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Rui X, Gu TT, Pan HF, Shao SL and Shao HX:
MicroRNA-381 suppresses proliferation and invasion of prostate
cancer cells through downregulation of the androgen receptor. Oncol
Lett. 18:2066–2072. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jiao DM, Yan L, Wang LS, Hu HZ, Tang XL,
Chen J, Wang J, Li Y and Chen QY: Exploration of inhibitory
mechanisms of curcumin in lung cancer metastasis using a miRNA-
transcription factor-target gene network. PLoS One.
12(e0172470)2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yin S, Du W, Wang F, Han B, Cui Y, Yang D,
Chen H, Liu D, Liu X, Zhai X, et al: MicroRNA-326 sensitizes human
glioblastoma cells to curcumin via the SHH/GLI1 signaling pathway.
Cancer Biol Ther. 19:260–270. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhu X and Zhu R: Curcumin suppresses the
progression of laryngeal squamous cell carcinoma through the
upregulation of miR-145 and inhibition of the PI3K/Akt/mTOR
pathway. OncoTargets Ther. 11:3521–3531. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Feng S, Wang Y, Zhang R, Yang G, Liang Z,
Wang Z and Zhang G: Curcumin exerts its antitumor activity through
regulation of miR-7/Skp2/p21 in nasopharyngeal carcinoma cells.
OncoTargets Ther. 10:2377–2388. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhang L, Zhang XW, Liu CH, Lu K, Huang YQ,
Wang YD, Xing L, Zhang LJ, Liu N, Jiang H, et al: miRNA-30a
functions as a tumor suppressor by downregulating cyclin E2
expression in castration-resistant prostate cancer. Mol Med Rep.
14:2077–2084. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang J, Shen C, Wang L, Ma Q, Xia P, Qi
M, Yang M and Han B: Metformin inhibits epithelial-mesenchymal
transition in prostate cancer cells: Involvement of the tumor
suppressor miR30a and its target gene SOX4. Biochem Biophys Res
Commun. 452:746–752. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Jiang LH, Zhang HD and Tang JH: MiR-30a: A
novel biomarker and potential therapeutic target for cancer. J
Oncol. 2018(5167829)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li L, Kang L, Zhao W, Feng Y, Liu W, Wang
T, Mai H, Huang J, Chen S, Liang Y, et al: miR-30a-5p suppresses
breast tumor growth and metastasis through inhibition of
LDHA-mediated Warburg effect. Cancer Lett. 400:89–98.
2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhu J, Zeng Y, Li W, Qin H, Lei Z, Shen D,
Gu D, Huang JA and Liu Z: CD73/NT5E is a target of miR-30a-5p and
plays an important role in the pathogenesis of non-small cell lung
cancer. Mol Cancer. 16(34)2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhang T, Guo J, Gu J, Chen K, Wang Z, Li
H, Wang G and Wang J: KIAA0101 is a novel transcriptional target of
FoxM1 and is involved in the regulation of hepatocellular carcinoma
microvascular invasion by regulating epithelial-mesenchymal
transition. J Cancer. 10:3501–3516. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Abdelgawad IA, Radwan NH and Hassanein HR:
KIAA0101 mRNA expression in the peripheral blood of hepatocellular
carcinoma patients: Association with some clinicopathological
features. Clin Biochem. 49:787–791. 2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Lv W, Su B, Li Y, Geng C and Chen N:
KIAA0101 inhibition suppresses cell proliferation and cell cycle
progression by promoting the interaction between p53 and Sp1 in
breast cancer. Biochem Biophys Res Commun. 503:600–606.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen H, Xia B, Liu T, Lin M and Lou G:
KIAA0101, a target gene of miR-429, enhances migration and
chemoresistance of epithelial ovarian cancer cells. Cancer Cell
Int. 16(74)2016.PubMed/NCBI View Article : Google Scholar
|