Introduction
Rheumatoid arthritis (RA) is an autoimmune disease
that causes chronic inflammation in synovial tissues. Hyperplasia
of synovial tissues leads to the formation of pannus, which invades
joint cartilage and bone, resulting in joint destruction. Previous
reports have indicated that a number of features of transformed
long-lived cells are observed in hyperplastic synovial tissues of
patients with RA, such as oncogene expression, resistance to
apoptosis, and the presence of somatic mutations (1-3).
Several explanations for resistance to apoptosis of rheumatoid
fibroblast-like synoviocytes (RA-FLS) have been proposed, including
deregulation of the Bcl-2 family of proteins critical to the
intrinsic apoptosis pathway, deregulation of NF-κB signaling, p53
mutations, and low expression of PUMA; these are all found in RA
synovium and FLS, which provides an explanation for the lack of
p53-induced FLS apoptosis (4). In
addition, it has been reported that hyperproliferation of RA
synovial cells involves the abnormal function of death receptors
such as Fas and death receptor 3 (5,6).
Fas ligand (FasL)/TNFSF6, a member of the tumor
necrosis factor (TNF) superfamily, is expressed by various cell
types in arthritic synovium, including T cells, synovial
fibroblasts, and macrophages (7),
and can promote apoptosis in activated primary B cells, T cells,
dendritic cells, and synovial fibroblasts through Fas (8,9).
Inhibition of the Fas/FasL pathway contributes to synovial
hyperplasia of RA (10-12).
Apoptosis through the Fas/FasL pathway in RA synovial cells is
inhibited by pro-inflammatory cytokines present within the synovium
(8). Meanwhile, the Fas/FasL system
may have a pro-inflammatory effect in RA (13,14).
Audo et al demonstrated that membrane-bound FasL induces
apoptosis as well as proliferation, whereas soluble FasL stimulates
only proliferation (13). Moreover,
soluble FasL activates several signaling pathways in RA-FLS, such
as extracellular signal-regulated kinase (ERK)-1/2,
phosphatidyl-inositol 3- kinase, caspase 8, and c-jun N-terminal
kinase (13). However, the
mechanisms and cell targets for these effects are still poorly
understood.
Decoy receptor 3 (DcR3)/TR6/M68/TNFRSF6b, a member
of the TNF receptor superfamily, binds to 3 ligands belonging to
the TNF superfamily: FasL, LIGHT, and TL1A (15). Overexpression of DcR3 may benefit
tumors by helping them avoid the cytotoxic and regulatory effects
of FasL (16,17), LIGHT (18), and TL1A (19). In our previous studies, we
demonstrated that DcR3 overexpressed in RA-FLS and stimulated by
TNFα protects cells from Fas-induced apoptosis (20). We previously also reported that DcR3
could play a role as a ligand by binding to membrane-bound TL1A in
the pathogenesis of RA (21-24).
Furthermore, the expression profiles of genes
regulated by DcR3 and TL1A in RA-FLS have been revealed by the use
of cDNA microarrays in our previous studies (25,26),
suggesting that signaling through DcR3 and its ligands is involved
in the pathogenesis of RA. However, the contribution of FasL,
another ligand of DcR3, to the pathogenesis of RA remains to be
fully elucidated.
In the current study, we searched for genes whose
expressions in RA-FLS were regulated by FasL using a cDNA
microarray. The gene expression profiles revealed a series of genes
that may play a significant role in the pathogenesis of RA via the
FasL-Fas signaling pathway. Further study is needed to reveal the
difference of the gene expression profiles among the ligands, which
might result in better understanding the role of the FasL/TL1A/DcR3
signaling system in the pathogenesis of RA.
Materials and methods
Isolation and culture of synovial
fibroblasts
RA-FLS were obtained from ten patients (samples
1-10) with RA who fulfilled the 1987 criteria of the American
College of Rheumatology (formerly, the American Rheumatism
Association) (27) during total
knee replacement surgery from September 2014 to April 2019.
Patients included one male and nine females aged 70.4±8.5 years
old. Their C-reactive protein levels and erythrocyte sedimentation
rates were 1.4±2.6 mg/dl and 25.6±14.0 mm/h, respectively. As for
the drug therapy for RA, five patients were administered oral
methotrexate (MTX) (average MTX dose, 8.8±4.6 mg/week), two were
administered tacrolimus (1.5±0.5 g/day), two were administered
salazosulfapyridine (1.0±0.0 g/day), and two were administered
bucillamine (150.0±50.0 mg/day). Prednisolone (PSL) was used to
treat three patients (average PSL dose, 4.7±0.6 mg/day). None of
the patients had been treated with biological disease-modifying
anti-rheumatic drugs (bioDMARDs) or Janus kinase inhibitors.
Synovial samples were collected from the patients,
all of whom provided informed written consent to participate in
this study in accordance with the World Medical Association
Declaration of Helsinki Ethical Principles for Medical Research
Involving Human Subjects. The protocol, including consent
procedures, was approved by the Kobe University Graduate School of
Health Sciences Ethics Committee (approval no. 308). Tissue
specimens were minced and digested in Dulbecco's modified Eagle's
medium (DMEM; Merck KGaA) containing 0.2% collagenase (Merck KGaA)
for 2 h at 37˚C with 5% CO2. Dissociated cells were
cultured in DMEM supplemented with 10% fetal bovine serum (Merck
KGaA) and 100 U/ml of penicillin/streptomycin (Meiji Seika Pharma
Co., Ltd.). Following incubation overnight and the removal of
non-adherent cells, adherent cells were further incubated in fresh
medium. All experiments were conducted using cells from passages 3
to 4(20).
Gene expression profiling
Four individual cell lines (samples 1-4) of primary
cultured RA-FLS (2x106 cells/well) were incubated with
1,000 ng/ml of recombinant human FasL protein (R&D Systems) or
were left untreated with OPTI-MEM medium (Thermo Fisher Scientific,
Inc.) as control for 12 h at 37˚C with 5% CO2. The
concentration of FasL was determined by a preliminary experiment
based of the previous reports using FasL (28,29).
After incubation, RNA was extracted with QIAshredder (Qiagen GmbH)
and an RNeasy Mini kit (Qiagen GmbH) according to the
manufacturer's protocol. Extraction of total RNA was performed for
each sample separately.
Gene expressions were detected by a microarray assay
(Human Genome U133 Plus 2.0, GeneChip® 3' Expression
Array; Thermo Fisher Scientific, Inc.). The labeling of RNA probes,
hybridization, and washing were carried out according to the
manufacturer's protocol.
RT-qPCR analysis for mRNA expression
of genes regulated by FasL
RA-FLS (samples 5-10) were cultured in six-well
plates at a density of 2x106 cells/well with 1,000 ng/ml
of FasL or serum-free medium only as a control. RNA was extracted
using the QIAshredder and RNeasy mini kits according to the
manufacturer's protocols. Oligo (dT)-primed first-strand
complementary DNA (cDNA) was synthesized (2 µg total RNA) using a
High Capacity cDNA Transcription kit (Applied Biosystems; Thermo
Fisher Scientific). Relative expression levels of mRNA encoding
DUSP6, EREG, IL-11, ANGPTL7, PIAS2, and GDF5 were compared using
TaqMan® real-time PCR on a StepOne™ real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Pre-designed
primers and probes for DUSP6 (Hs04329643_s1), EREG (Hs00914313_m1),
IL-11 (Hs01055413_g1), ANGPTL7 (Hs00221727_m1), PIAS2
(Hs00915227_m1), GDF5 (Hs00167060_m1), and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Hs99999905_m1)
were obtained from Applied Biosystems (Thermo Fisher Scientific,
Inc.). Comparative analysis of each of these genes in individual
patients was performed using StepOne™ 2.1 software (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. All amplifications were conducted in
duplicate. The mRNA expression levels of each gene were calculated
using the comparative threshold cycle (ΔΔCq) method as previously
described (30).
Statistical analysis
Values are expressed as the mean ± standard
deviation unless otherwise indicated. As for the data analysis of
the microarray assay, Avadis 3.3 Prophetic software (Strand Life
Sciences) was used for statistical analysis (31). Differentially expressed genes were
extracted by a paired t-test, with P values <0.05 considered to
indicate statistical significance and fold-change >2.0, and
ordered into hierarchical clusters using the Euclidean algorithm as
the distance measure and the complete algorithm as the linkage
method.
The data analysis of the RT-qPCR assay was as
follows. The Wilcoxon signed ranked test was used to evaluate the
differences between mRNA expression levels of genes in the control
group and FasL-stimulated group. Statistical analyses were
conducted using Statcel (version 3; OMS Publishing, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Microarray analysis (gene expression
profiling of RA-FLS stimulated by FasL)
The microarray analysis used in the current
study (Human Genome U133 Plus 2.0, GeneChip® 3'
Expression Array) was able to detect the expression of 27,420
genes.
The microarray analysis revealed that FasL
upregulates or downregulates the expressions of various genes in
RA-FLS. We used the NCBI's UniGene database (https://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?ORG=Hs&CID=55682)
to identify the genes. Among the 1039 genes differentially
upregulated by FasL, 806 were annotated in the database. Twenty of
the 806 genes upregulated by FasL are shown in Table I. Gene annotations of 1190 among the
1518 genes differentially downregulated by FasL were also in the
database. Twenty of the 1190 downregulated genes by FasL are shown
in Table II. Hierarchical
clustering analysis was performed for genes for which expression
changes were detected in at least 2 of the 4 samples, which was 247
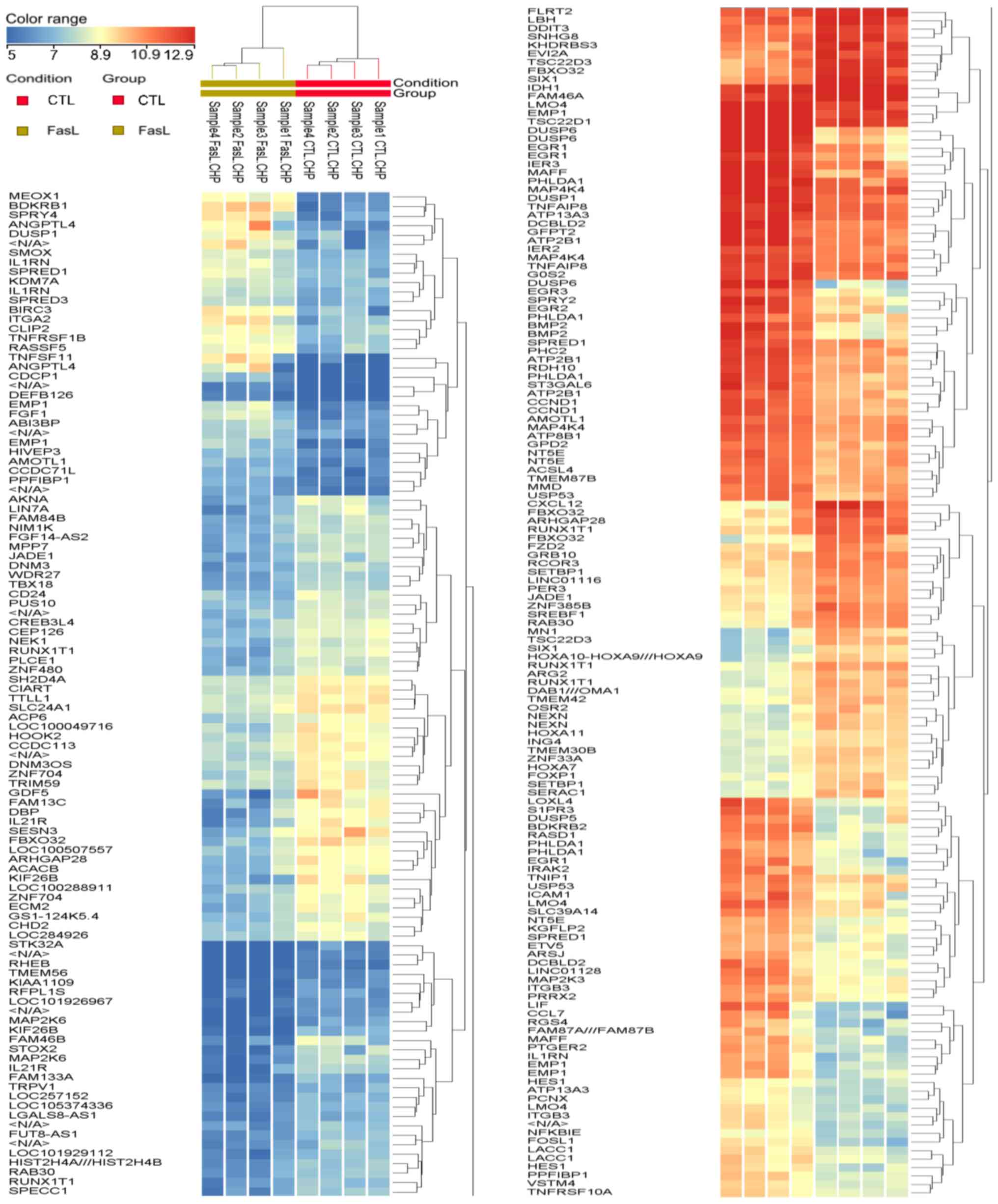
genes. The results of hierarchical clustering analysis for these
247 genes are illustrated in Fig.
1.
 | Table IThe 20 genes most upregulated by
FasL. P-values were detected by a paired t-test. |
Table I
The 20 genes most upregulated by
FasL. P-values were detected by a paired t-test.
| Gene
abbreviation | P-value | FC (abs) | Gene name |
|---|
| DUSP6 | 0.000018 | 34.61 | Dual specificity
phosphatase 6 |
| EREG | 0.019622 | 29.23 | Epiregulin |
| IL-11 | 0.007275 | 25.28 | Interleukin 11 |
| ANGPTL4 | 0.002853 | 23.50 | Angiopoietin-like
4 |
| SLCO4A1 | 0.002094 | 20.39 | Solute carrier
organic anion transporter family, member 4A1 |
| TNFSF11 | 0.006236 | 18.48 | Tumor necrosis
factor (ligand) superfamily, member 11 |
| BDKRB1 | 0.000004 | 14.39 | Bradykinin receptor
B1 |
|
OTTHUMG00000172357//RP11-475A13.2 | 0.000002 | 14.12 | NULL//NULL |
| AREG//AREGB | 0.030537 | 13.77 |
Amphiregulin//amphiregulin B |
| LIF | 0.000498 | 13.53 | Leukemia inhibitory
factor |
| IFNA8 | 0.000039 | 12.09 | Interferon, α
8 |
|
OTTHUMG00000175763//RP11-744D14.2 | 0.000716 | 11.73 | NULL//NULL |
| HBEGF | 0.014773 | 11.34 | Heparin-binding
EGF-like growth factor |
| PPP4R4 | 0.023739 | 11.03 | Protein phosphatase
4, regulatory subunit 4 |
| NDP | 0.008832 | 10.67 | Norrie disease
(pseudoglioma) |
| NR4A3 | 0.000432 | 10.59 | Nuclear receptor
subfamily 4, group A, member 3 |
| EGLN3 | 0.028458 | 9.95 | Egl nine homolog 3
(C. elegans) |
| BMP2 | 0.000082 | 9.92 | Bone morphogenetic
protein 2 |
| UBR2 | 0.007847 | 9.91 | Ubiquitin protein
ligase E3 component n-recognin 2 |
| SLC38A10 | 0.001400 | 9.34 | Solute carrier
family 38, member 10 |
 | Table IIThe 20 genes most downregulated by
FasL. P-values were detected by a paired t-test. |
Table II
The 20 genes most downregulated by
FasL. P-values were detected by a paired t-test.
| Gene
abbreviation | P-value | FC (abs) | Gene name |
|---|
| ANGPTL7 | 0.000283 | 11.61 | Angiopoietin-like
7 |
| PIAS2 | 0.001099 | 11.34 | Protein inhibitor
of activated STAT, 2 |
| LINC00310 | 0.000038 | 11.30 | Long intergenic
non-protein coding RNA 310 |
| GDF5 | 0.004260 | 11.12 | Growth
differentiation factor 5 |
| TBX22 | 0.000755 | 11.11 | T-box 22 |
| DCAF4L1 | 0.000734 | 10.64 | DDB1 and CUL4
associated factor 4-like 1 |
| KRT16 | 0.013331 | 10.62 | Keratin 16 |
|
OTTHUMG00000180314//RP1-193H18.2 | 0.003726 | 10.31 | NULL//NULL |
| TAS2R40 | 0.000202 | 10.14 | Taste receptor,
type 2, member 40 |
| HEPACAM2 | 0.001656 | 9.93 | HEPACAM family
member 2 |
| CSMD1 | 0.000096 | 9.87 | CUB and Sushi
multiple domains 1 |
| IQCA1 | 0.009692 | 9.63 | IQ motif containing
with AAA domain 1 |
|
LOC100996810//LOC283861 | 0.003182 | 9.26 | Uncharacterized
LOC100996810//uncharacterized LOC283861 |
| FGFR2 | 0.029699 | 9.25 | Fibroblast growth
factor receptor 2 |
| WDR65 | 0.000045 | 9.21 | WD repeat domain
65 |
| LOC253573 | 0.001556 | 9.18 | Uncharacterized
LOC253573 |
| PHLDB2 | 0.001030 | 9.06 | Pleckstrin
homology-like domain, family B, member 2 |
| PCDHAC2 | 0.017711 | 9.01 | Protocadherin alpha
subfamily C, 2 |
| LOC100506629 | 0.002936 | 8.72 | Uncharacterized
LOC100506629 |
| FAM66C | 0.003647 | 8.68 | Family with
sequence similarity 66, member C |
Functional annotation
The 246 genes regulated by FasL were classified into
categories registered in the David Bioinformatics Database
(https://david.ncifcrf.gov/) according to
their biological functions. The most significant 10 functional
categories were as follows: Transcriptional activator activity,
positive regulation of metabolic process, positive regulation of
cellular metabolic process, positive regulation of macromolecule
metabolic process, positive regulation of nitrogen compound
metabolic process, regulation of phosphorylation, positive
regulation of biological process, regulation of phosphate metabolic
process, regulation of MAPK cascade, regulation of multicellular
organismal process (Table
III).
 | Table IIIThe 10 most significant functional
categories of the 246 genes most differentially expressed by FasL
exposure in RA-FLS. P-values were detected by a paired t-test. |
Table III
The 10 most significant functional
categories of the 246 genes most differentially expressed by FasL
exposure in RA-FLS. P-values were detected by a paired t-test.
| GO Accession | GO Term | Corrected
P-value |
|---|
| GO:0001228 | Transcriptional
activator activity, RNA polymerase II transcription Regulatory
region sequence-specific DNA binding | 0.000028 |
| GO:0009893|
GO:0044253 | Positive regulation
of metabolic process | 0.000028 |
| GO:0031325 | Positive regulation
of cellular metabolic process | 0.000028 |
| GO:0010604 | Positive regulation
of macromolecule metabolic process | 0.000028 |
| GO:0051173 | Positive regulation
of nitrogen compound metabolic process | 0.000028 |
| GO:0042325 | Regulation of
phosphorylation | 0.000066 |
| GO:0048518|
GO:0043119 | Positive regulation
of biological process | 0.000066 |
| GO:0019220 | Regulation of
phosphate metabolic process | 0.000087 |
| GO:0043408 | Regulation of MAPK
cascade | 0.000087 |
| GO:0051239 | Regulation of
multicellular organismal process | 0.000087 |
mRNA expression detected by
RT-qPCR
Based on the microarray assay, we confirmed the mRNA
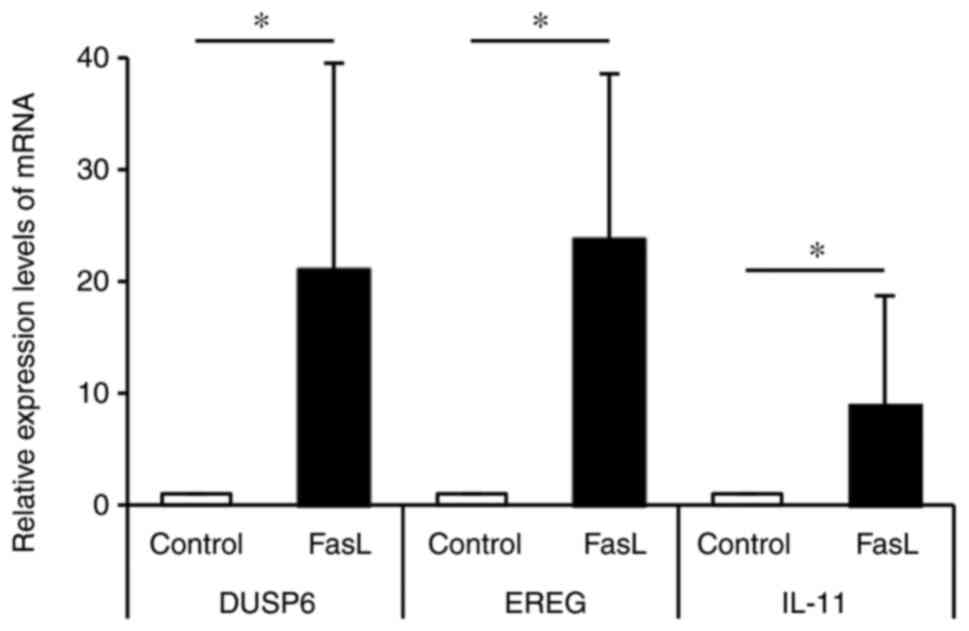
expressions of genes by real-time PCR. Fig. 2 shows the mRNA expression levels of
the 3 most upregulated genes. DUSP6 was upregulated 21 times by
FasL compared to the control, EREG was upregulated 24 times by FasL
compared to the control, and IL-11 was upregulated 9 times by FasL
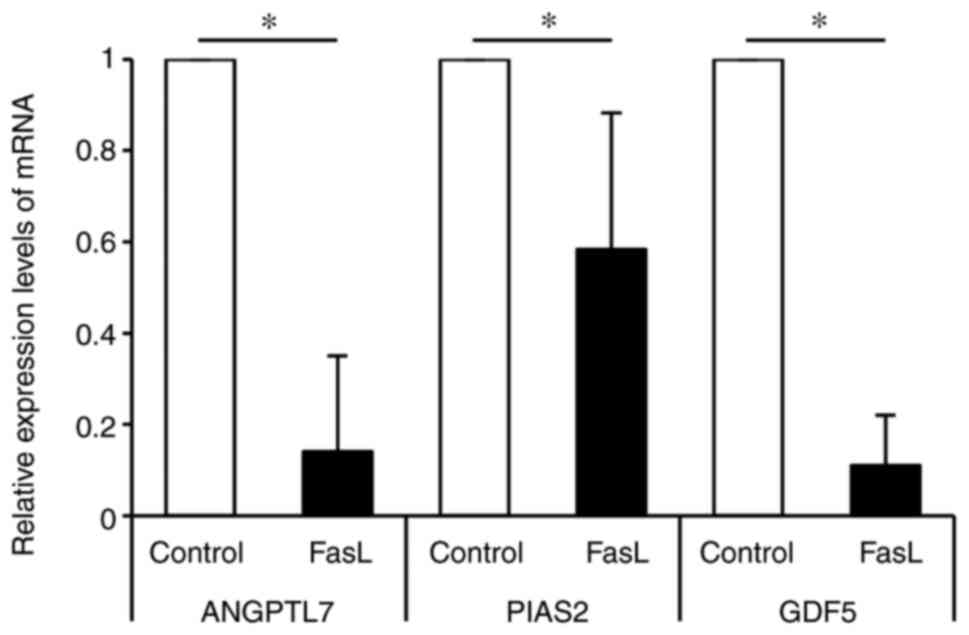
compared to the control. Fig. 3
shows the mRNA expression levels of the 3 most downregulated genes.
ANGPTL7 was downregulated 0.15 times by FasL compared to the
control, PIAS2 was downregulated 0.58 times by FasL compared to the
control, and GDF5 was downregulated 0.11 times by FasL compared to
the control.
Discussion
Genome-wide gene expression cDNA microarrays provide
a powerful technique to investigate the pathophysiology of a
variety of diseases, including tumors (32-34),
immune-mediated diseases (35,36),
and inflammatory diseases (37-39).
Using microarray assays, we previously revealed the expression
profiles of genes in RA-FLS regulated by DcR3(25) and TL1A (26). Subsequently, based on the profile
regulated by DcR3, we investigated the significance of IL-12B
p40(22), tryptophan hydroxylase
1(24), and centrosomal protein 70
kDa (23) as regulated by DcR3 in
RA-FLS in detail. The profile regulated by TL1A included the
following noteworthy genes: Spectrin repeat-containing nuclear
envelope 1, Fc receptor-like 2, PYD (pyrin domain)-containing 1,
cell division cycle 45 homolog, signal transducer and activator of
transcription 5B, and interferon regulatory factor 4(26).
To the best of our knowledge, this is the first
study to reveal the expression profiles of genes in RA-FLS
regulated by FasL. Among the genes in this profile, the following
genes were of note: Dual specificity phosphatase 6 (DUSP6),
epiregulin (EREG), interleukin 11 (IL-11), angiopoietin-like 7
(ANGPTL7), protein inhibitor of activated STAT 2 (PIAS2), and
growth differentiation factor 5 (GDF5); these genes were all highly
regulated by FasL.
DUSP6 regulates CD4+ T-cell activation
and differentiation by inhibiting T-cell receptor dependent ERK 1/2
activation (40). It has been
reported that DUSP6 promotes endothelial inflammation through the
inducible expression of TNF-α-induced intercellular adhesion
molecule-1 via nuclear factor-κB, which is independent of ERK
signaling (41).
Epiregulin is a growth regulator that belongs to the
epidermal growth factor (EGF) family and mediates the
dose-dependent increase in proliferation of primary mouse
keratinocytes (42). EREG is
increased in patients with RA and is associated with the
development of IL-6 amplifier activation (43). EREG triggers the temporal regulation
of growth factors such as amphiregulin, betacellulin, transforming
growth factor (TGF)-α, fibroblast growth factor 2, placental growth
factor 2, and tenascin C, contributing to the early phase of
inflammation; each growth factor reciprocally regulates EREG in
affected tissue during the late phase of inflammatory disease
development (44). Secretion of
vascular endothelial growth factor-A and EREG from RA-FLS was
inhibited upon treatment with the aryl hydrocarbon receptor
antagonist GNF351, resulting in attenuation of RA-FLS cell
migration, along with cytokine-induced RA-FLS cell proliferation
(45).
IL-11 signaling appears to be initiated by the
binding of IL-11 to IL-11 receptor α chain (IL-11Rα), which then
binds gp130, the signaling unit of the IL-6 cytokine family
(46). IL-11 attenuates the
inflammatory response through downregulation of proinflammatory
cytokine release and nitric oxide production (47,48).
IL-11 contributes to RA angiogenesis directly and indirectly. IL-11
promotes endothelial cell migration and tube formation mediated
through IL-11Rα ligation. Vascular endothelial growth factor and
IL-8 produced from IL-11-treated RA-FLS contribute to the indirect
effect of IL-11 on angiogenesis (49). In addition, IL-11 plays a key role
in osteoclast formation via the gp130/Jak signaling pathway
(50).
ANGPTL7 is a member of angiopoietin family that
exerts pro-angiogenic activities on endothelial cells. ANGPTL7
expression has been identified in some cancer cells induced by
hypoxia (51). ANGPTL7 induces
proinflammatory responses in macrophages, including the induction
of immune gene expression, the promotion of proinflammatory
cytokine secretion, enhanced phagocytosis, and antagonized
anti-inflammatory signaling through the P38 MAPK signaling pathway
(52). Down-regulation of a
positive regulator of inflammation might have a negative effect for
inflammation in patients with RA.
PIAS proteins inhibit activated STAT and play
important roles in regulating many important cellular events, such
as cell survival, migration, and signal transduction in many cell
types (53,54). PIAS proteins also modulate the
activity of several transcription factors and act as E3 ubiquitin
ligases in the sumoylation pathway (54-57).
In a similar fashion, down-regulation of a negative regulator of
inflammation might have a positive effect for inflammation in
patients with RA. Lao et al reported that PIAS3 regulates
migration and invasion through the Rac1/PAK1/JNK pathway in RA-FLSs
(53).
GDF5 is a member of the TGF-β superfamily and is
most closely related to the bone morphogenetic protein subfamily.
GDF5 increases glycosaminoglycan synthesis (58) and cartilage and bone formation
(59). GDF5 is present in the
synovium membrane and cartilage of patients with RA and is actively
involved in the regulation of cartilage maintenance and repair
(60). GDF5 is associated with
joint destruction in patients with osteoarthritis (61) and RA (62). GDF5 in RA-FLS was suppressed by
IL-1β and had a strong chondrogenic-promoting effect on
TGF-β3-induced chondrocyte differentiation in RA-FLS (60).
In the present study, at the first brush, we
exhaustively investigated and revealed the gene expression profiles
regulated by FasL in RA-FLS by the microarray assay. Secondly, we
confirmed the universality of the gene expression pattern by a
different method, RT-qPCR assay, using the different samples of
RA-FLS from those used for the microarray assay. In order to obtain
the pathological homogeneity among the samples as much as possible,
the patients who underwent similar clinical features were
recruited; who had been treated only with conventional DMARDs, not
with biological DMARDs or targeted synthetic DMARDs, and who had
their knee joint destructed severely resulting total knee
replacement surgery. Therefore, we considered that there were no
differences among the 10 samples. The samples 1-4 used for the
microarray and 5-10 for the RT-qPCR assay were randomly
selected.
The expression profiles of genes regulated by TL1A
were elucidated by use of a microarray assay in our previous report
(26). TL1A and FasL are bound and
inhibited by the common decoy receptor, DcR3. Therefore, clarifying
the relationship between the expression profiles of genes regulated
by FasL and those regulated by TL1A might help us to better
understand the role of the FasL/TL1A/DcR3 signaling system in the
pathogenesis of RA. Further study is needed to reveal the
relationship between these gene expression profiles.
The limitations of the current study include its
small sample size and that it presents gene expression data only.
The results of the current study revealed a series of genes whose
expression is regulated by FasL in RA-FLS with microarray analysis,
and the mRNA expression of some genes of note was confirmed by
RT-qPCR assay. However, in addition to the expression analysis of
each gene, how the genes regulated by FasL in RA-FLS are involved
in the pathogenesis of RA also requires further investigation. In
the current study, we aimed to analyze exhaustively the gene
expression pattern regulated by FasL in RA-FLS. Therefore, the
assay for expression of proteins coded by each gene should also be
performed in the further studies.
In conclusion, the current study is the first, to
the best of our knowledge, to report the expression profile of
genes in RA-FLS regulated by FasL. These data demonstrate that FasL
may regulate the gene expression of various key molecules in
RA-FLS, thus affecting the pathogenesis of RA, including apoptosis,
proliferation, cytokine production, cytokine-induced inflammation,
intracellular signaling, angiogenesis, bone destruction, and
chondrogenesis. FasL may have pleiotropic actions not only
protectively but also detrimentally for RA. Further investigation
of the genes detected in this profile may provide a deeper
understanding of the pathogenesis of RA and new targets for its
treatment.
Acknowledgements
The authors would like to thank Ms. Kyoko Tanaka,
Ms. Minako Nagata, and Ms. Maya Yasuda (all from Department of
Orthopaedic Surgery, Kobe University Graduate School of Medicine)
for their technical assistance.
Funding
Funding: The current study was supported by Grants-in-Aid for
Scientific Research (KAKENHI) (grant nos. 15K10473 and
18K09106).
Availability of data and materials
The datasets generated and analyzed during the
current study are available in the NCBI's Gene Expression Omnibus
(GEO) repository, GEO series accession no. GSE153378 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153378).
Authors' contributions
KF conceived and designed the current study, was
involved in data collection and analysis, confirmed the
authenticity of all the raw data, wrote and gave final approval of
the manuscript. YM conceived and designed the current study, was
involved in data collection and analysis, confirmed the
authenticity of all the raw data and gave final approval of the
manuscript. TosM and TomM collected the data and gave final
approval of the manuscript. SH conceived and designed the current
study, was involved in data collection and analysis and gave final
approval of the manuscript. RK conceived and designed the current
study and gave final approval of the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Kobe
University Graduate School of Health Sciences Ethics Committee
(approval no. 308). All the participants provided written informed
consent to participate in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chou CT, Yang JS and Lee MR: Apoptosis in
rheumatoid arthritis-expression of Fas, Fas-L, p53, and Bcl-2 in
rheumatoid synovial tissues. J Pathol. 193:110–116. 2001.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tak PP, Zvaifler NJ, Green DR and
Firestein GS: Rheumatoid arthritis and p53: How oxidative stress
might alter the course of inflammatory diseases. Immunol Today.
21:78–82. 2000.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yamanishi Y, Boyle DL, Rosengren S, Green
DR, Zvaifler NJ and Firestein GS: Regional analysis of p53
mutations in rheumatoid arthritis synovium. Proc Natl Acad Sci USA.
99:10025–10030. 2002.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bustamante MF, Garcia-Carbonell R,
Whisenant KD and Guma M: Fibroblast-like synoviocyte metabolism in
the pathogenesis of rheumatoid arthritis. Arthritis Res Ther.
19(110)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Baier A, Meineckel I, Gay S and Pap T:
Apoptosis in rheumatoid arthritis. Curr Opin Rheumatol. 15:274–279.
2003.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Takami N, Osawa K, Miura Y, Komai K,
Taniguchi M, Shiraishi M, Sato K, Iguchi T, Shiozawa K, Hashiramoto
A and Shiozawa S: Hypermethylated promoter region of DR3, the death
receptor 3 gene, in rheumatoid arthritis synovial cells. Arthritis
Rheum. 54:779–787. 2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Peng SL: Fas (CD95)-related apoptosis and
rheumatoid arthritis. Rheumatology (Oxford). 45:26–30.
2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wakisaka S, Suzuki N, Takeba Y, Shimoyama
Y, Nagafuchi H, Takeno M, Saito N, Yokoe T, Kaneko A, Asai T and
Sakane T: Modulation by proinflammatory cytokines of Fas/Fas
ligand-mediated apoptotic cell death of synovial cells in patients
with rheumatoid arthritis (RA). Clin Exp Immunol. 114:119–128.
1998.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Croft M and Siegel RM: Beyond TNF: TNF
superfamily cytokines as targets for the treatment of rheumatic
diseases. Nat Rev Rheumatol. 13:217–233. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Schedel J, Gay RE, Kuenzler P, Seemayer C,
Simmen B, Michel BA and Gay S: FLICE-inhibitory protein expression
in synovial fibroblasts and at sites of cartilage and bone erosion
in rheumatoid arthritis. Arthritis Rheum. 46:1512–1518.
2002.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kobayashi T, Okamoto K, Kobata T, Hasunuma
T, Kato T, Hamada H and Nishioka K: Differential regulation of
Fas-mediated apoptosis of rheumatoid synoviocytes by tumor necrosis
factor alpha and basic fibroblast growth factor is associated with
the expression of apoptosis-related molecules. Arthritis Rheum.
43:1106–1114. 2000.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Scaffidi C, Fulda S, Srinivasan A, Friesen
C, Li F, Tomaselli KJ, Debatin KM, Krammer PH and Peter ME: Two
CD95 (APO-1/Fas) signaling pathways. EMBO J. 17:1675–1687.
1998.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Audo R, Calmon-Hamaty F, Papon L, Combe B,
Morel J and Hahne M: Distinct effects of soluble and membrane-bound
fas ligand on fibroblast-like synoviocytes from rheumatoid
arthritis patients. Arthritis Rheumatol. 66:3289–3299.
2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Guegan JP and Legembre P: Nonapoptotic
functions of Fas/CD95 in the immune response. FEBS J. 285:809–827.
2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Shi G, Wu Y, Zhang J and Wu J: Death decoy
receptor TR6/DcR3 inhibits T cell chemotaxis in vitro and in vivo.
J Immunol. 171:3407–3414. 2003.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Pitti RM, Marsters SA, Lawrence DA, Roy M,
Kischkel FC, Dowd P, Huang A, Donahue CJ, Sherwood SW, Baldwin DT,
et al: Genomic amplification of a decoy receptor for Fas ligand in
lung and colon cancer. Nature. 396:699–703. 1998.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Tsuji S, Hosotani R, Yonehara S, Masui T,
Tulachan SS, Nakajima S, Kobayashi H, Koizumi M, Toyoda E, Ito D,
et al: Endogenous decoy receptor 3 blocks the growth inhibition
signals mediated by Fas ligand in human pancreatic adenocarcinoma.
Int J Cancer. 106:17–25. 2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yu KY, Kwon B, Ni J, Zhai Y, Ebner R and
Kwon BS: A newly identified member of tumor necrosis factor
receptor superfamily (TR6) suppresses LIGHT-mediated apoptosis. J
Biol Chem. 274:13733–13736. 1999.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Migone TS, Zhang J, Luo X, Zhuang L, Chen
C, Hu B, Hong JS, Perry JW, Chen SF, Zhou JX, et al: TL1A is a
TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell
costimulator. Immunity. 16:479–492. 2002.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hayashi S, Miura Y, Nishiyama T, Mitani M,
Tateishi K, Sakai Y, Hashiramoto A, Kurosaka M, Shiozawa S and
Doita M: Decoy receptor 3 expressed in rheumatoid synovial
fibroblasts protects the cells against Fas-induced apoptosis.
Arthritis Rheum. 56:1067–1075. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Takahashi M, Miura Y, Hayashi S, Tateishi
K, Fukuda K and Kurosaka M: DcR3-TL1A signalling inhibits
cytokine-induced proliferation of rheumatoid synovial fibroblasts.
Int J Mol Med. 28:423–427. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fukuda K, Miura Y, Maeda T, Hayashi S and
Kurosaka M: Interleukin12B is upregulated by decoy receptor 3 in
rheumatoid synovial fibroblasts. Mol Med Rep. 13:3647–3652.
2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fukuda K, Miura Y, Maeda T, Hayashi S and
Kuroda R: Decoy receptor 3 down-regulates centrosomal protein 70
kDa specifically in rheumatoid synovial fibroblasts. Mod Rheumatol.
28:287–292. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Maeda T, Miura Y, Fukuda K, Hayashi S and
Kurosaka M: Decoy receptor 3 regulates the expression of tryptophan
hydroxylase 1 in rheumatoid synovial fibroblasts. Mol Med Rep.
12:5191–5196. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fukuda K, Miura Y, Maeda T, Takahashi M,
Hayashi S and Kurosaka M: Decoy receptor 3 regulates the expression
of various genes in rheumatoid arthritis synovial fibroblasts. Int
J Mol Med. 32:910–916. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Fukuda K, Miura Y, Maeda T, Hayashi S and
Kuroda R: Expression profiling of genes in rheumatoid
fibroblast-like synoviocytes regulated by tumor necrosis
factor-like ligand 1A using cDNA microarray analysis. Biomed Rep.
1:1–5. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Arnett FC, Edworthy SM, Bloch DA, McShane
DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS,
et al: The American Rheumatism Association 1987 revised criteria
for the classification of rheumatoid arthritis. Arthritis Rheum.
31:315–324. 1988.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chang Q, Peter ME and Grassi MA: Fas
ligand-Fas signaling participates in light-induced apoptotic death
in photoreceptor cells. Invest Ophthalmol Vis Sci. 53:3703–3716.
2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Nitobe J, Yamaguchi S, Okuyama M, Nozaki
N, Sata M, Miyamoto T, Takeishi Y, Kubota I and Tomoike H: Reactive
oxygen species regulate FLICE inhibitory protein (FLIP) and
susceptibility to Fas-mediated apoptosis in cardiac myocytes.
Cardiovasc Res. 57:119–128. 2003.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Thiel CT, Kraus C, Rauch A, Ekici AB,
Rautenstrauss B and Reis A: A new quantitative PCR multiplex assay
for rapid analysis of chromosome 17p11.2-12 duplications and
deletions leading to HMSN/HNPP. Eur J Hum Genet. 11:170–178.
2003.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Choi YJ and Yun HK: Transcriptional
profiles of Rhizobium vitis-inoculated and salicylic acid-treated
‘Tamnara’ grapevines based on microarray analysis. J Plant
Biotechnol. 43:37–48. 2016.
|
|
32
|
Chang YC, Chen TC, Lee CT, Yang CY, Wang
HW, Wang CC and Hsieh SL: Epigenetic control of MHC class II
expression in tumor-associated macrophages by decoy receptor 3.
Blood. 111:5054–5063. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Espinosa I, Catasus L, Canet B, D'Angelo
E, Munoz J and Prat J: Gene expression analysis identifies two
groups of ovarian high-grade serous carcinomas with different
prognosis. Mod Pathol. 24:846–854. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Khan J, Simon R, Bittner M, Chen Y,
Leighton SB, Pohida T, Smith PD, Jiang Y, Gooden GC, Trent JM, et
al: Gene expression profiling of alveolar rhabdomyosarcoma with
cDNA microarrays. Cancer Res. 58:5009–5013. 1998.PubMed/NCBI
|
|
35
|
Whitney LW, Becker KG, Tresser NJ,
Caballero-Ramos CI, Munson PJ, Prabhu VV, Trent JM, McFarland HF
and Biddison WE: Analysis of gene expression in mutiple sclerosis
lesions using cDNA microarrays. Ann Neurol. 46:425–428.
1999.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Li J, Yang S, Lu S, Zhao H, Feng J, Li W,
Ma F, Ren Q, Liu B, Zhang L, et al: Differential gene expression
profile associated with the abnormality of bone marrow mesenchymal
stem cells in aplastic anemia. PLoS One. 7(e47764)2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
van der Pouw Kraan TC, van Gaalen FA,
Kasperkovitz PV, Verbeet NL, Smeets TJ, Kraan MC, Fero M, Tak PP,
Huizinga TW, Pieterman E, et al: Rheumatoid arthritis is a
heterogeneous disease: Evidence for differences in the activation
of the STAT-1 pathway between rheumatoid tissues. Arthritis Rheum.
48:2132–2145. 2003.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Lee SK, Jeon EK, Kim YJ, Seo SH, Kim CD,
Lim JS and Lee JH: A global gene expression analysis of the
peripheral blood mononuclear cells reveals the gene expression
signature in psoriasis. Ann Dermatol. 21:237–242. 2009.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Heller RA, Schena M, Chai A, Shalon D,
Bedilion T, Gilmore J, Woolley DE and Davis RW: Discovery and
analysis of inflammatory disease-related genes using cDNA
microarrays. Proc Natl Acad Sci USA. 94:2150–2155. 1997.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Bertin S, Lozano-Ruiz B, Bachiller V,
Garcia-Martinez I, Herdman S, Zapater P, Frances R, Such J, Lee J,
Raz E and González-Navajas JM: Dual-specificity phosphatase 6
regulates CD4+ T-cell functions and restrains spontaneous colitis
in IL-10-deficient mice. Mucosal Immunol. 8:505–515.
2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hsu SF, Lee YB, Lee YC, Chung AL, Apaya
MK, Shyur LF, Cheng CF, Ho FM and Meng TC: Dual specificity
phosphatase DUSP6 promotes endothelial inflammation through
inducible expression of ICAM-1. FEBS J. 285:1593–1610.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Patel RD, Kim DJ, Peters JM and Perdew GH:
The aryl hydrocarbon receptor directly regulates expression of the
potent mitogen epiregulin. Toxicol Sci. 89:75–82. 2006.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Murakami M, Harada M, Kamimura D, Ogura H,
Okuyama Y, Kumai N, Okuyama A, Singh R, Jiang JJ, Atsumi T, et al:
Disease-association analysis of an inflammation-related feedback
loop. Cell Rep. 3:946–959. 2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Harada M, Kamimura D, Arima Y, Kohsaka H,
Nakatsuji Y, Nishida M, Atsumi T, Meng J, Bando H, Singh R, et al:
Temporal expression of growth factors triggered by epiregulin
regulates inflammation development. J Immunol. 194:1039–1046.
2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lahoti TS, Hughes JM, Kusnadi A, John K,
Zhu B, Murray IA, Gowda K, Peters JM, Amin SG and Perdew GH: Aryl
hydrocarbon receptor antagonism attenuates growth factor
expression, proliferation, and migration in fibroblast-like
synoviocytes from patients with rheumatoid arthritis. J Pharmacol
Exp Ther. 348:236–245. 2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yin T, Taga T, Tsang ML, Yasukawa K,
Kishimoto T and Yang YC: Involvement of IL-6 signal transducer
gp130 in IL-11-mediated signal transduction. J Immunol.
151:2555–2561. 1993.PubMed/NCBI
|
|
47
|
Trepicchio WL, Bozza M, Pedneault G and
Dorner AJ: Recombinant human IL-11 attenuates the inflammatory
response through down-regulation of proinflammatory cytokine
release and nitric oxide production. J Immunol. 157:3627–3634.
1996.PubMed/NCBI
|
|
48
|
Hermann JA, Hall MA, Maini RN, Feldmann M
and Brennan FM: Important immunoregulatory role of interleukin-11
in the inflammatory process in rheumatoid arthritis. Arthritis
Rheum. 41:1388–1397. 1998.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Elshabrawy HA, Volin MV, Essani AB, Chen
Z, McInnes IB, Van Raemdonck K, Palasiewicz K, Arami S, Gonzalez M,
Ashour HM, et al: IL-11 facilitates a novel connection between RA
joint fibroblasts and endothelial cells. Angiogenesis. 21:215–228.
2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Murakami K, Kobayashi Y, Uehara S, Suzuki
T, Koide M, Yamashita T, Nakamura M, Takahashi N, Kato H, Udagawa N
and Nakamura Y: A Jak1/2 inhibitor, baricitinib, inhibits
osteoclastogenesis by suppressing RANKL expression in osteoblasts
in vitro. PLoS One. 12(e0181126)2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Parri M, Pietrovito L, Grandi A,
Campagnoli S, De Camilli E, Bianchini F, Marchio S, Bussolino F,
Jin B, Sarmientos P, et al: Angiopoietin-like 7, a novel
pro-angiogenetic factor over-expressed in cancer. Angiogenesis.
17:881–896. 2014.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Qian T, Wang K, Cui J, He Y and Yang Z:
Angiopoietin-Like Protein 7 promotes an inflammatory phenotype in
RAW264.7 macrophages through the P38 MAPK signaling pathway.
Inflammation. 39:974–985. 2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Lao M, Shi M, Zou Y, Huang M, Ye Y, Qiu Q,
Xiao Y, Zeng S, Liang L, Yang X and Xu H: Protein inhibitor of
activated STAT3 regulates migration, invasion, and activation of
Fibroblast-like synoviocytes in rheumatoid arthritis. J Immunol.
196:596–606. 2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Muller S, Ledl A and Schmidt D: SUMO: A
regulator of gene expression and genome integrity. Oncogene.
23:1998–2008. 2004.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Schmidt D and Muller S: PIAS/SUMO: New
partners in transcriptional regulation. Cell Mol Life Sci.
60:2561–2574. 2003.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Wu Y, Guo Z, Wu H, Wang X, Yang L, Shi X,
Du J, Tang B, Li W, Yang L and Zhang Y: SUMOylation represses Nanog
expression via modulating transcription factors Oct4 and Sox2. PLoS
One. 7(e39606)2012.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Chowdhury D, Singh A, Gupta A, Tulsawani
R, Meena RC and Chakrabarti A: p38 MAPK pathway-dependent
SUMOylation of Elk-1 and phosphorylation of PIAS2 correlate with
the downregulation of Elk-1 activity in heat-stressed HeLa cells.
Cell Stress Chaperones. 24:393–407. 2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Bobacz K, Gruber R, Soleiman A, Graninger
WB, Luyten FP and Erlacher L: Cartilage-derived morphogenetic
protein-1 and -2 are endogenously expressed in healthy and
osteoarthritic human articular chondrocytes and stimulate matrix
synthesis. Osteoarthritis Cartilage. 10:394–401. 2002.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Hotten GC, Matsumoto T, Kimura M, Bechtold
RF, Kron R, Ohara T, Tanaka H, Satoh Y, Okazaki M, Shirai T, et al:
Recombinant human growth/differentiation factor 5 stimulates
mesenchyme aggregation and chondrogenesis responsible for the
skeletal development of limbs. Growth Factors. 13:65–74.
1996.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Liu FL, Lin LH, Sytwu HK and Chang DM:
GDF-5 is suppressed by IL-1beta and enhances TGF-beta3-mediated
chondrogenic differentiation in human rheumatoid fibroblast-like
synoviocytes. Exp Mol Pathol. 88:163–170. 2010.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Miyamoto Y, Mabuchi A, Shi D, Kubo T,
Takatori Y, Saito S, Fujioka M, Sudo A, Uchida A, Yamamoto S, et
al: A functional polymorphism in the 5' UTR of GDF5 is associated
with susceptibility to osteoarthritis. Nat Genet. 39:529–533.
2007.PubMed/NCBI View
Article : Google Scholar
|
|
62
|
Martinez A, Varade J, Lamas JR,
Fernandez-Arquero M, Jover JA, de la Concha EG, Fernandez-Gutierrez
B and Urcelay E: GDF5 Polymorphism associated with osteoarthritis:
Risk for rheumatoid arthritis. Ann Rheum Dis. 67:1352–1353.
2008.PubMed/NCBI View Article : Google Scholar
|