Introduction
Breast cancer is the most common cancer and the
leading cause of cancer-associated mortality among females
worldwide (1,2). Triple-negative breast cancer (TNBC) is
characterized by a lack of expression of estrogen receptor (ER),
progesterone receptor (PR) and human epidermal growth factor
receptor 2 (HER2). Owing to the early metastasis and a lack of
available therapeutic targets, patients suffering from TNBC have
been reported to exhibit a poor prognosis and low survival rate
compared with most of other types breast cancer (3,4),
indicating a requirement to identify novel biomarkers for the early
diagnosis of TNBC, as well as potential molecular targets and novel
treatment strategies (5).
In TNBC, p53 is the most frequently mutated gene,
the mutation rate is as high as 80% (6). Mutant p53 lacks the wild-type
tumor-suppressor activity of p53 and acts as an oncogene via
gain-of-function activities, including induction of cell
proliferation, invasion, migration and inhibition of cell
apoptosis, which promote increased aggressiveness in cancer cells
(7-9).
MicroRNAs (miRNAs/miRs) are a class of small
non-coding RNAs with 22 nucleotides, which regulate gene expression
by interacting with a number of proteins to form an RNA-induced
silencing complex, which subsequently targets and binds to the
3'-untranslated region of target mRNAs to mediate mRNA degradation
or translational repression (10).
Previous studies have reported the dysregulation of miRNAs in
various cancer cells and demonstrated that this dysregulation may
be associated with tumorigenesis, cancer progression, migration and
invasion (11-14).
A previous study revealed that miR-449a was
downregulated in breast cancer tissues, especially in TNBC tissues
from patients carrying mutant p53(15). miR-449a has been indicated to reduce
the growth of cancer cells via targeting and inhibiting the
expression of histone deacetylase 1, which is involved in
maintaining the expression of mutant p53 (16-18).
Furthermore, miR-449a has been reported to act as a tumor
suppressor in colorectal cancer, hepatocellular carcinoma,
non-small cell lung cancer and gastric adenocarcinoma (19-22).
However, the role of miR-449a in breast cancer and its associated
mechanisms, especially in TNBC harboring mutant p53, are yet to be
elucidated.
In the present study, the role of miR-449a in the
proliferation, migration and apoptosis of MDA-MB-468 breast cancer
cells was investigated. The cellular signaling pathway of p53 and
the association of miR-449a with mutant p53 in MDA-MB-468 cells
were also examined.
Materials and methods
Cell culture, transfection and
infection
293T cells were purchased from ATCC, human normal
mammary epithelial cells MCF-10A and TNBC cell lines with p53
mutation [MDA-MB-468 (R273H) and MDA-MB-231 (R280K)] were obtained
from the Department of Laboratory Medicine, Chongqing Medical
University (Chongqing, China). All the cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.), at 37˚C with 5%
CO2. The retroviral pSuper-Retro-puro-short hairpin
(sh)-hp53 vector donated by the Hormel Institute of the University
of Minnesota was used to knock-down p53 expression.
2x106 293T cells were seeded into 6 cm dish,
pSuper-Retro-puro-sh-hp53 (3 µg) or pSuper-Retro-puro- sh-vector (3
µg) was co-transfected with the packaging plasmid pAmpho (1.5 µg)
into 293T cells using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), the retroviruses were
harvested from medium supernatant at 48 and 72 h. The MDA-MB-468
cells infected by retroviral supernatant (6x108 MOI/ml)
for 72 h, then treated by 1.5 µg/ml puromycin for 2 weeks to obtain
stably transfected cells. MDA-MB-468 cells were transfected with
miR-449a-mimic or miR-449a-NC (100 nM; designed and synthesized by
Guangzhou RiboBio Co., Ltd.) for 8 h using
Lipofectamine® 2000 reagent, and all the subsequent
experimentations were completed within 96 h.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from MDA-MB-468, MDA-MB-231
and MCF-10A cells using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. RNA (1.8 µg) and M-MLV reverse transcriptase (Promega
Corporation) were used to synthesize first-strand cDNA, according
to the manufacturer's protocol. For miRNA reverse transcription,
specific stem-loop primers were used. qPCR was carried out using
FastStart Essential DNA Green Master kit (Roche Diagnostics) by the
manufacturer's protocol. The thermocycling conditions for qPCR were
as follows: 10 min at 95˚C, and then 40 cycles of 30 sec at 95˚C,
30 sec at 60˚C (62˚C for p53 and miR-449a) and the
2-ΔΔCq (23) method was
used to calculate the relative expression level of miR-449a, p53,
E-cadherin, Vimentin, Twist and Snail. GAPDH and U6 were used as
control genes for mRNA and miR-449a expression detection,
respectively. The primers (Table I)
used for PCR were synthesized by Sangon Biotech Co., Ltd.
 | Table IPrimer sequences used in RT-PCR and
RT-qPCR. |
Table I
Primer sequences used in RT-PCR and
RT-qPCR.
| Reaction type | Gene | Forward/Reverse
primer | Sequence
(5'-3') | Product size
(bp) |
|---|
| RT-PCR | miR-449a | - |
5'-GTCGTATCCAGTGCAGGGTCCGAG | - |
| | | |
GTATTCGCACTGGATACGACACCAGC-3' | |
| | U6 | - |
5'-AACGCTTCACGAATTTGCGT-3' | - |
| RT-qPCR | miR-449a | Forward |
5'-TGCGGTGGCAGTGTATTGTTAGC-3' | 64 |
| | | Reverse |
5'-CCAGTGCAGGGTCCGAGGTA-3' | |
| | U6 | Forward |
5'-CTCGCTTCGGCAGCACA-3' | 94 |
| | | Reverse |
5'-AACGCTTCACGAATTTGCGT-3' | |
| | E-cadherin | Forward |
5'-AATCCCACCACGTACAAGGG-3' | 93 |
| | | Reverse |
5'-GGTATTGGGGGCATCATCAT-3' | |
| | Vimentin | Forward |
5'-TCCAGCAGCTTCCTGTAGGT-3' | 241 |
| | | Reverse |
5'-CCCTCACCTGTGAAGTGGAT-3' | |
| | Twist | Forward |
5'-GACAGTGATTCCCAGACGG-3' | 190 |
| | | Reverse |
5'-GTCCATAGTGATGCCTTTCCT-3' | |
| | Snail | Forward |
5'-TTTACCTTCCAGCAGCCCTA-3' | 108 |
| | | Reverse |
5'-GACAGAGTCCCAGATGAGCA-3' | |
| | p53 | Forward |
5'-CCAGGGCAGCTACGGTTTC-3' | 205 |
| | | Reverse |
5'-CTCCGTCATGTGCTGTGACTG-3' | |
| | GAPDH | Forward |
5'-ACAACTTTGGTATCGTGGAAGG-3' | 101 |
| | | Reverse |
5'-GCCATCACGCCACAGTTTC-3' | |
Western blot assay
MDA-MB-468, MAD-MB-231 and MCF-10A cells were lysed
with NP-40 lysis buffer (Beyotime Institute of Biotechnology) to
extract total proteins, and a BCA kit (Thermo Fisher Scientific,
Inc.) was used to determine the protein concentration. A total of
40 µg protein was separated by 10% SDS-PAGE (8% for mTOR and
p-mTOR) and transferred to a PVDF membrane (Bio-Rad Laboratories
Inc.). The membrane was blocked with 5% non-fat milk for 1 h at
room temperature. The primary antibodies were diluted to 1:1,000
and incubated with the PVDF membrane overnight at 4˚C. The
secondary antibody was diluted to 1:3,000 and incubated PVDF
membrane for 1 h at room temperature. The primary antibodies
specific to p53 (cat. no. 2527), AKT1 (cat. no. 2938),
phosphorylated (p)-AKT (cat. no. 4060), m-TOR (cat. no. 2983),
p-mTOR (cat. no. 2971), Bcl-2 (cat. no. 4223), caspase-3 (cat. no.
14220), cleaved caspase-3 (cat. no. 9661) and GAPDH (cat. no. 2118)
and the secondary antibody Anti-rabbit IgG (cat. no. 7074) were
purchased from Cell Signaling Technology, Inc. The proteins were
visualized using an ECL reagent kit (EMD Millipore) using Vilber
fusion FX7 spectra chemiluminescence apparatus (Vilber
Lourmat).
Cell migration assays
Transwell migration and wound healing assays were
carried out to assess the cell migratory ability. For the Transwell
migration assay, Transwell chambers (pore size, 6.5 mm) were put
into 24-well plates, a total of 1x105 MDA-MB-468 cells
resuspended in 200 µl serum-free DMEM were placed into the upper
Transwell chamber (Corning, Inc.), and 600 µl DMEM with 10% FBS was
added into the lower chamber. After 24 h, the cells in the upper
chamber were removed using a cotton swab, and the cells on the
surface of the lower chamber membrane were fixed by 4%
paraformaldehyde for 15 min and stained with 0.1% crystal violet
for 20 min at room temperature. Image acquiring and cells counting
were performed with BX51 light microscope (Olympus Corporation) at
magnification of x200 in eight randomly selected fields.
For the wound healing assay, 1.8x106
MDA-MB-468 cells resuspended in DMEM with 3% FBS were seeded into
6-well plates. The scratches were created using a 200 µl pipette
tip when cells reached 90% confluency. Then the medium was removed
from plate and the plate was washed using PBS to remove
non-attached cells, the attached cells were cultured in serum-free
DMEM. The width of the scratch was observed and images were
captured after 48 h using a BX51 light microscope at magnification
of x100.
Cell proliferation assay
A growth curve was used to assess cell
proliferation. 1x105 MDA-MB-468 cells were seeded into
6-well plates in DMEM supplemented with 10% FBS. Cell proliferation
was evaluated every 24 h for 96 h. The cells were fixed using 4%
paraformaldehyde for 15 min, then stained using 0.1% crystal violet
for 20 min, before crystal violet-stained cells were decolorized
using 10% acetic acid for 10 min. The OD values of the de-staining
solution was detected at 590 nm to generate a growth curve.
Statistical analysis
All data are presented as the mean ± SD from at
least three independent experiments with similar results.
Significant differences were analyzed by Student's t-test
(independent two-sample t-test) or one-way ANOVA (followed by
Fisher's Least Significant Difference post hoc test) using SPSS
v22.0 software (IBM Corp.). P<0.05 was considered to indicate a
statistically significant difference.
Results
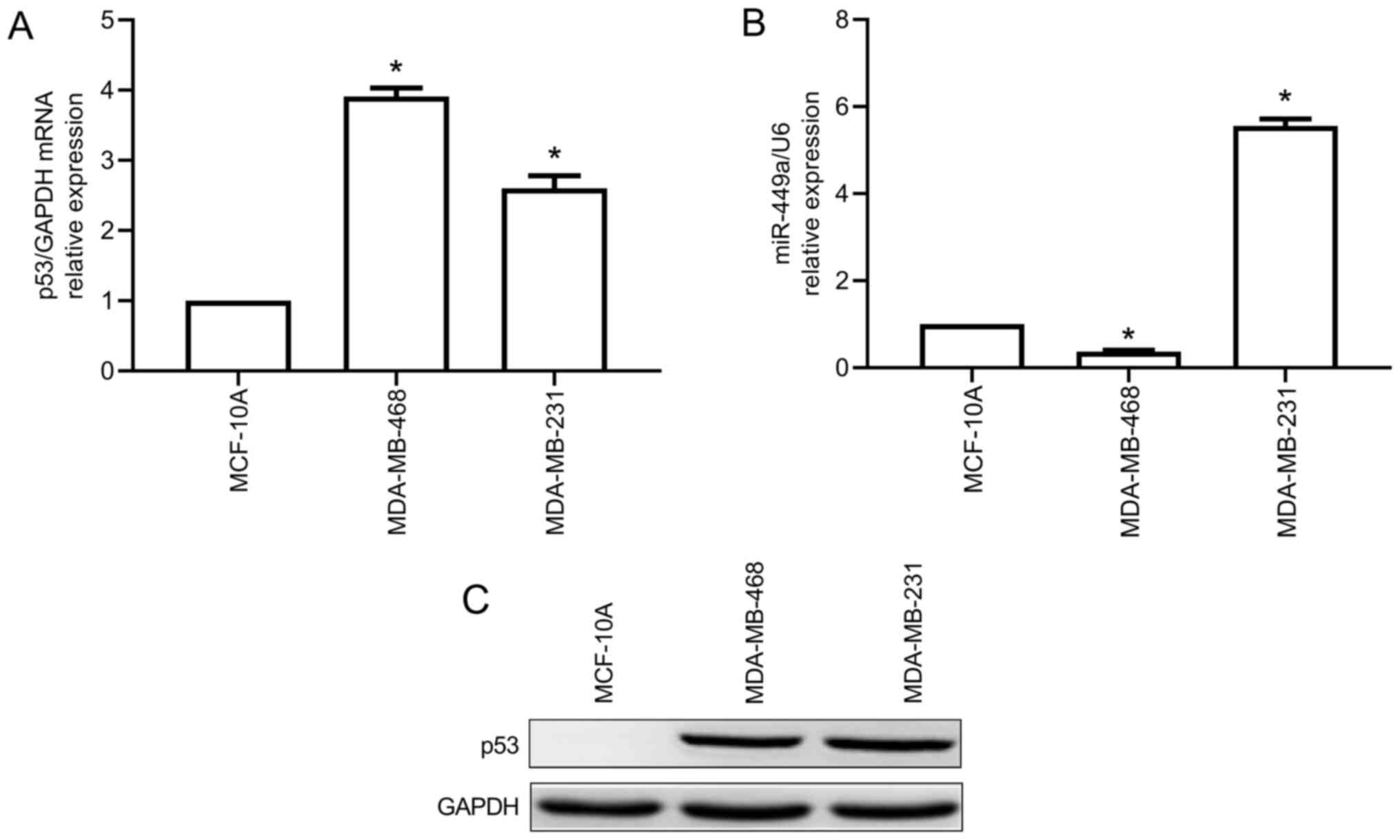
miR-449a expression level is decreased
in MDA-MB-468 cells overexpressing mutant p53
The expression levels of miR-449a and p53 were
detected via RT-qPCR. Mutant p53 protein level was determined via
western blotting. The results revealed that the expression level of
mutant p53 was significantly higher in MDA-MB-468 and MDA-MB-231
cells compared with that in MCF-10A cells (Fig. 1A and B); However, miR-449a expression level was
lower in MDA-MB-468 cells compared with that in MCF-10A cells, but
significant higher in MDA-MB-231 cells (Fig. 1C).
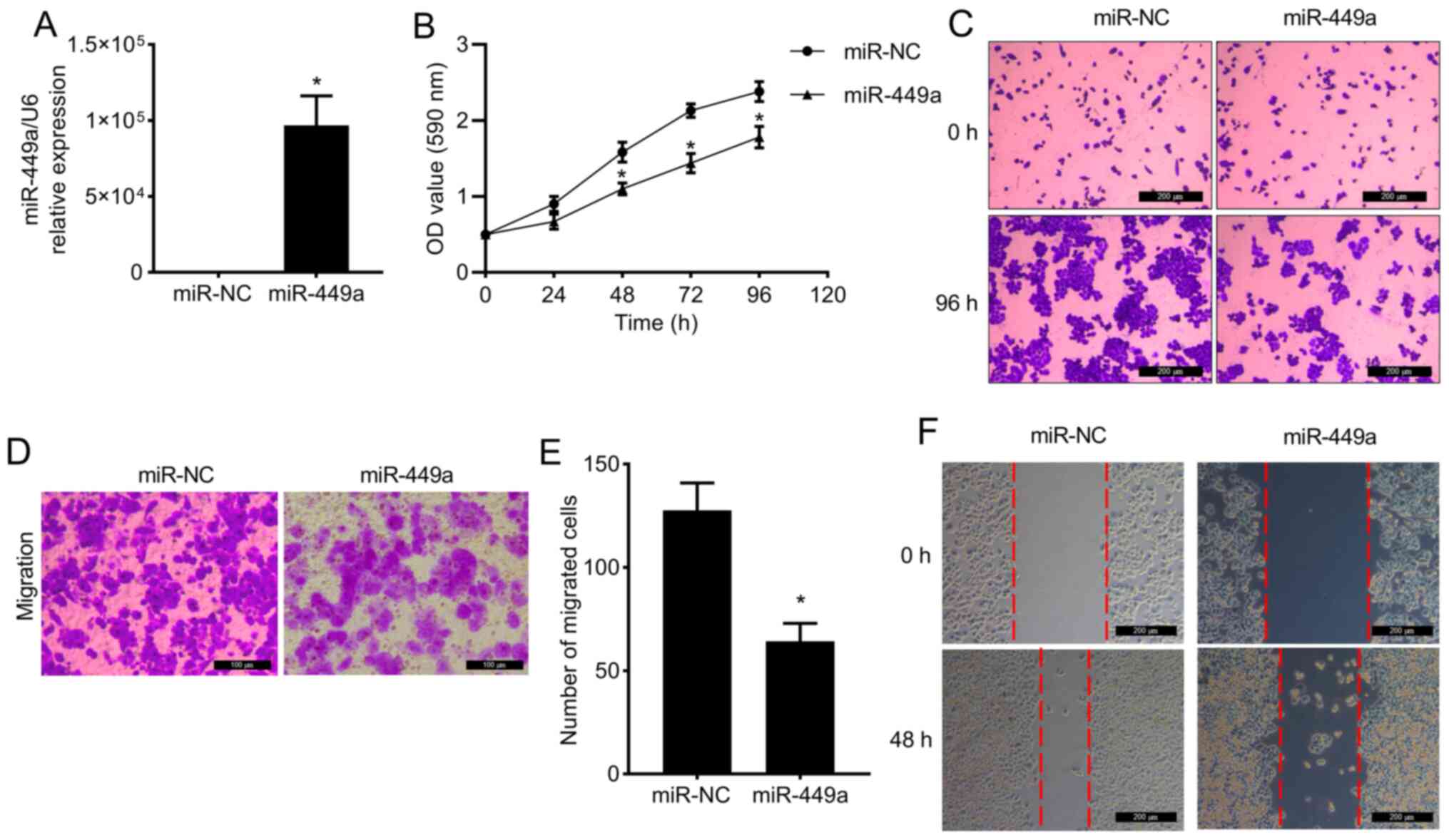
Overexpression of miR-449a suppresses
the proliferation and migration of MDA-MB-468 cells
miR-449a was indicated to be expressed at a low
level in MDA-MB-468 cells, as aforementioned. To examine the
physiological effects of miR-449a, miR-449a mimic was transfected
into MDA-MB-468 cells, and overexpression of miR-449a was validated
by RT-qPCR (Fig. 2A). As presented
in Fig. 2B and C, overexpression of miR-449a suppressed
MDA-MB-468 cell proliferation compared with the miR-NC group.
Furthermore, the migratory capacity of MDA-MB-468 cells was
inhibited by miR-449a overexpression, as indicated by the results
of the Transwell migration (Fig. 2D
and E) and wound healing assays
(Fig. 2F).
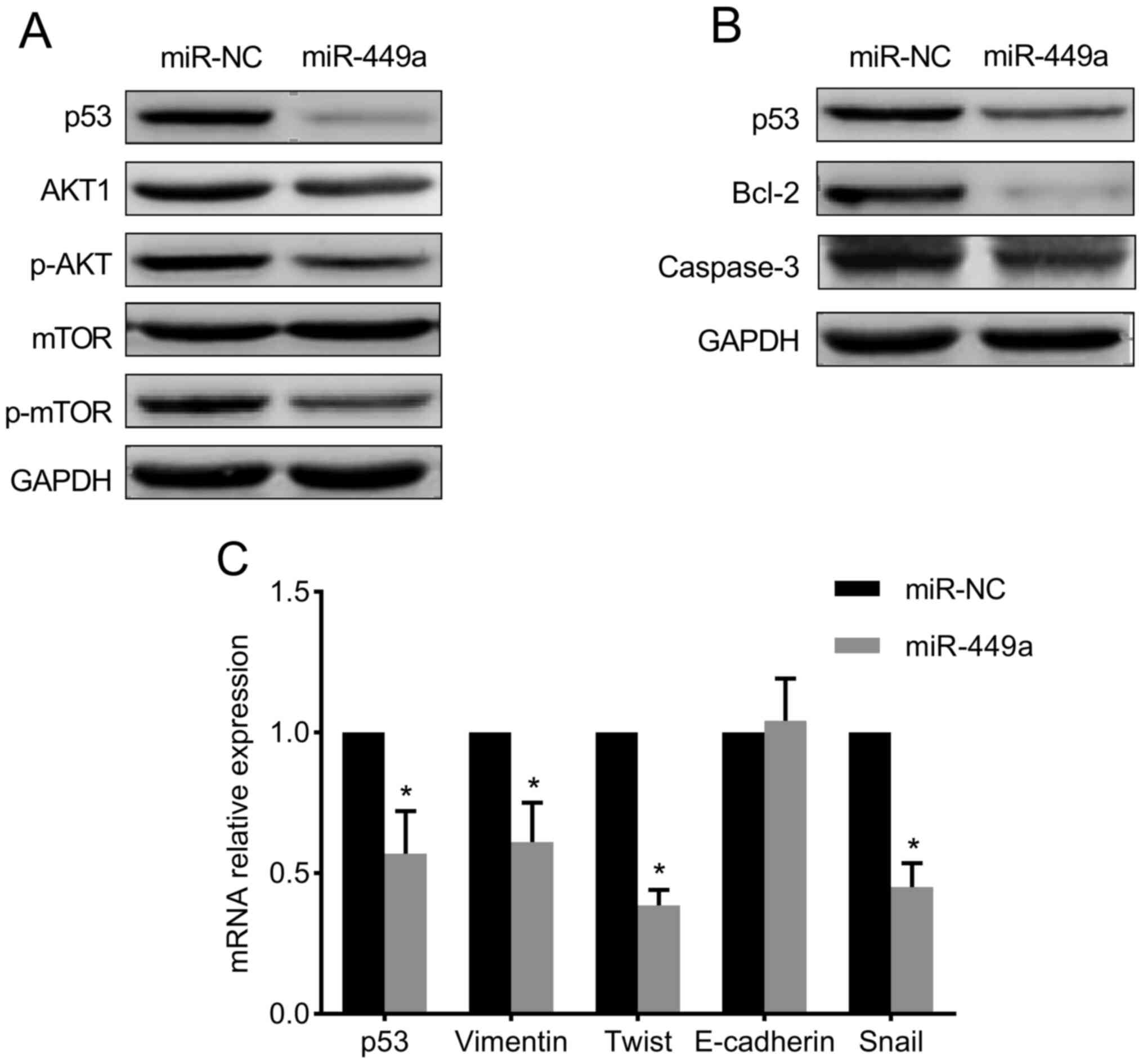
miR-449a reduces the expression of
mutant p53 and inhibits the PI3K/AKT/mTOR signaling pathway and
epithelial-mesenchymal transition (EMT) in MDA-MB-468 cells
To further explore the molecular mechanisms of
miR-449a-mediated biological activities, miR-449a mimic was
transfected into MDA-MB-468 cells. As illustrated in Fig. 3A, overexpression of miR-449a reduced
the p53 protein level in MDA-MB-468 cells, and the level of the
PI3K/AKT/mTOR pathway targets p-AKT and p-mTOR was markedly reduced
compared with the miR-NC group. Moreover, the expression level of
Bcl-2 and caspase-3 was decreased by miR-449a overexpression,
although cleaved caspase-3 expression was not detected (Fig. 3B).
Furthermore, the expression of EMT-related genes was
examined by RT-qPCR after miR-449a overexpression, and the results
indicated that the expression level of vimentin, Twist and Snail
was significantly reduced, while that of E-cadherin was slightly
increased compared with miR-NC-transfected cells (Fig. 3C).
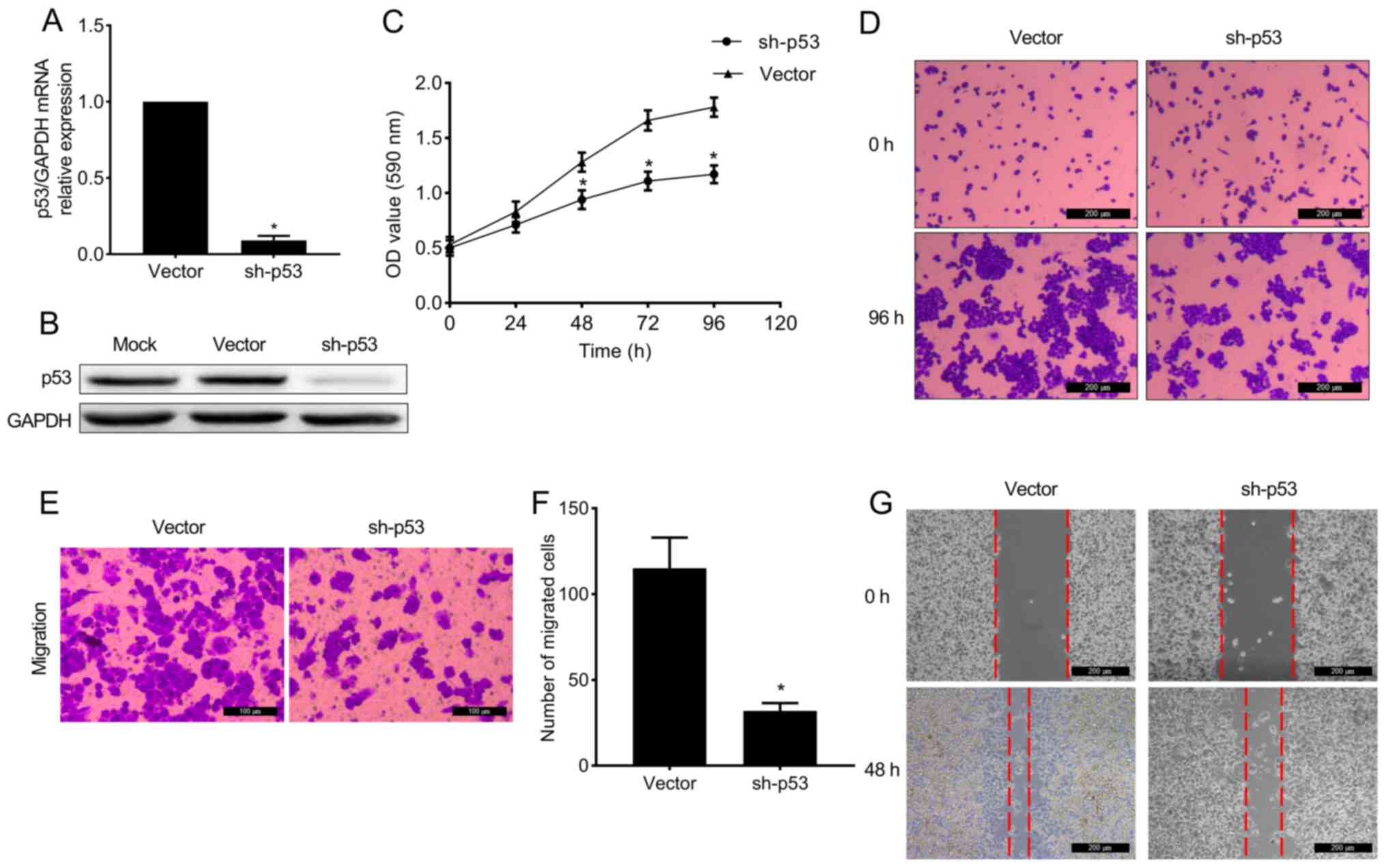
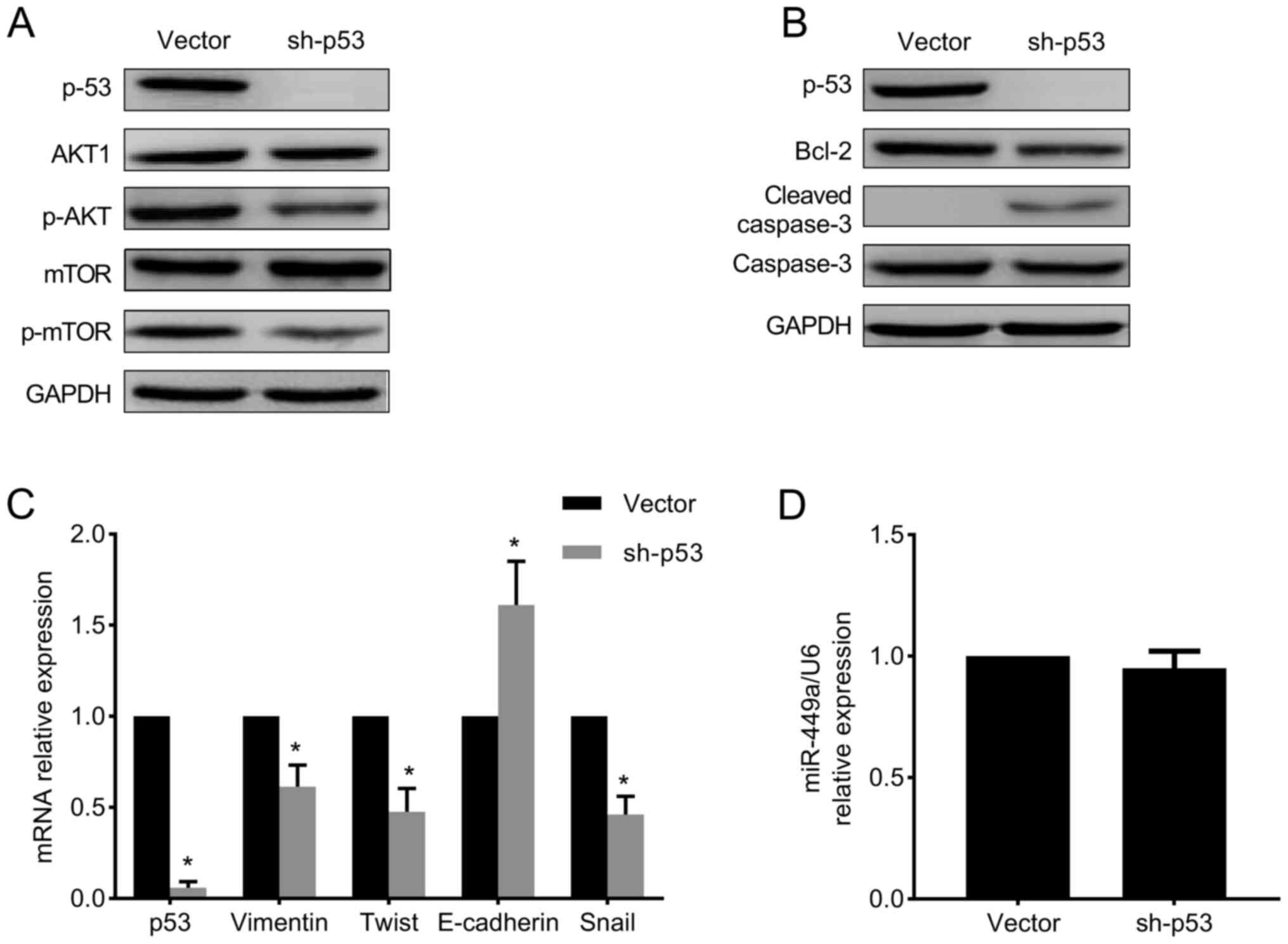
Knockdown of mutant p53 suppresses the
proliferation and migration of MDA-MB-468 cells
The p53 mRNA and protein were knockdown in
MDA-MB-468 cells by retroviral pSuper-Retro-puro- short hairpin
(sh)-hp53 vector (Fig. 4A and
B). It was revealed that the
proliferation of p53 knocked-down MDA-MB-468 cells was notably
inhibited compared with vector-transduced cells (Fig. 4C and D), and the migratory capacity exhibited
the same trend (Fig. 4E-G).
Mutant p53 regulates proliferation and
migration via the PI3K/AKT/mTOR signaling pathway and EMT in
MDA-MB-468 cells
The PI3K/AKT/mTOR pathway has been indicated to
regulate the survival, proliferation, invasion and migration of
cancer cells (24). In p53
knocked-down MDA-MB-468 cells the protein level of several members
of this pathway was detected, and it was observed that the
expression level of p-AKT and p-mTOR was remarkably decreased
(Fig. 5A), which indicated that the
PI3K/AKT/mTOR pathway was inhibited. In addition, apoptosis was
examined in p53-depleted cells, and the results indicated that
Bcl-2 expression level was decreased, while cleaved caspase-3
expression level was increased compared with vector-transduced
cells (Fig. 5B), suggesting that
the apoptotic pathway was activated.
In the present study, the expression levels of
E-cadherin, vimentin, Twist and Snail were evaluated by RT-qPCR.
The results demonstrated that in p53 knocked-down MDA-MB-468 cells,
the expression level of E-cadherin was increased, while the
expression levels of vimentin, Twist and Snail were markedly
reduced (Fig. 5C), indicating that
EMT was inhibited upon knockdown of mutant p53. On the other hand,
miR-449a level was detected in p53 knocked-down cells, while no
significant difference was observed in the miR-449a level compared
with vector-transduced cells (Fig.
5D). These data collectively suggested that the repressive
effects of miR-449a on cell proliferation and migration may be
partly facilitated by a decrease in mutant p53 in MDA-MB-468
cells.
Discussion
p53 is the most extensively studied tumor suppressor
gene, and mutant p53 proteins not only lose their tumor suppressive
abilities, but also gain additional oncogenic functions that
provide cells with growth and survival advantages (25). Previous studies have demonstrated
that restoration of wild-type p53 of colorectal cancer or removing
the mutant p53 from a mutant p53 oncogene addiction mice model were
help for cancer therapy (26,27).
In the current study, it was indicated that the
reduction of mutant p53 expression level effectively suppressed the
proliferation and migration of MDA-MB-468 cells. Subsequent
analysis revealed that the PI3K/AKT/mTOR signaling pathway was
notably downregulated by the knockdown of mutant p53. The
PI3K/AKT/mTOR signaling pathway is involved in cell proliferation,
survival, apoptosis and migration (24). Therefore, it can be concluded that
mutant p53 regulates the PI3K/AKT/mTOR signaling pathway in
MDA-MB-468 cells and decreasing mutant p53 expression level may
suppress cell proliferation and migration by inhibiting the
PI3K/AKT/mTOR signaling pathway.
EMT represents a process that results in a complete
loss of epithelial traits in epithelial cells accompanied by the
acquisition of mesenchymal traits. More specifically, epithelial
cell layers lose their polarity and the cell-cell contacts while
undergoing important cytoskeletal remodeling (28). A hallmark of the EMT process is the
acquisition of the ability to migrate and invade the extracellular
matrix as single cells (29). EMT
involves the loss of E-cadherin expression and the acquisition of
the expression of the mesenchymal marker vimentin (30). EMT enhances the mobility of cancer
cells and drives cancer metastasis (31). A number of studies have demonstrated
that mutant p53 can promote EMT (32-34),
and in the current study it was revealed that EMT was suppressed by
the knockdown of mutant p53, suggesting that downregulation of
mutant p53 expression may inhibit the migration of cancer cells by
suppressing EMT.
The protein expression levels of Bcl-2, caspase-3
and cleaved caspase-3, which are indicative of apoptosis, were also
detected. Bcl-2 belongs to the Bcl-2 family members that exert both
pro- and anti-apoptotic function, and the reduced expression level
of Bcl-2 can induce apoptosis in cancer cells (35). Cleaved caspase-3 is produced by the
activation of caspase-3 and induces cell apoptosis (36). In the present study, knockdown of
mutant p53 decreased the expression level of Bcl-2, while that of
cleaved caspase-3 was increased, suggesting that the apoptotic
pathway was activated.
In previous studies, miR-449a has been reported to
act as a tumor suppressor to inhibit cell migration, metastasis and
proliferation and induce cell senescence in NSCLC and prostatic
cancer (37-39).
However, the function of miR-449a in breast cancer is still
controversial (15,40), and the interaction between miR-449a
and mutant p53 remains elusive. In the current study, it was
demonstrated that overexpression of miR-449a suppressed
proliferation and migration of MDA-MB-468 cells, resulting in a
similar effect to the knockdown of mutant p53. Subsequent analysis
revealed that the expression of mutant p53 was markedly decreased
in MDA-MB-468 cells transfected with miR-449a mimic. Additionally,
the PI3K/AKT/mTOR pathway and EMT were suppressed following
miR-449a overexpression, suggesting that miR-449a may function as a
tumor suppressor to inhibit proliferation and migration of breast
cancer cells via downregulation of mutant p53. In addition, Bcl-2
expression level was reduced following overexpression of miR-449a,
which is consistent with Bcl-2 being previously reported as a
target of miR-449a (20). Knockdown
of p53 also decreased Bcl-2 expression level, indicating that Bcl-2
expression was regulated by both miR-449a and mutant p53. However,
the expression level of cleaved-caspase-3 was not detected in
miR-449a-overexpressing cells. Taken together, the aforementioned
results suggested that mutant p53 participates in the mechanism of
miR-449a function, affecting the proliferation and migration of
MDA-MB-468 cells.
To verify the results associated with miR-449a in
MDA-MB-468 cells, MDA-MB-231 cells overexpressing miR-449a and
mutant p53 (R280K) were used. However, the results obtained were
not in agreement with those observed in MDA-MB-468 cells. This
controversial outcome may be associated with the type of mutant p53
in each cell line.
The present study demonstrated that miR-449a
functions as a tumor suppressor to inhibit proliferation and
migration and induce apoptosis by downregulating the expression of
mutant p53 in MDA-MB-468 breast cancer cells. These results
indicated that miR-449a is a key component that targets the p53
pathway, highlighting that inhibiting mutant p53 expression via
miR-449a may be a potential therapeutic strategy for TNBC. However,
additional experiments are required in future studies. The protein
expression of EMT-associated genes should be investigated, the
effect of miR-449a on cell apoptosis should be detected via flow
cytometry and the level of additional apoptosis-related proteins
[cytochrome c, Bad, and poly (ADP-ribose) polymerase] should
be examined by western blotting.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported from Science and
Technology Support Program of Nanchong (grant no. 18SXHZ0487 and
18SXHZ0513) and Scientific Research Project of Sichuan Health
Commission (grant no. 19PJ200).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XLG and GCH designed the study. GCH and XWZ
performed the experiments and wrote the initial draft of the
manuscript. LHY, RS and JZ were involved in literature search and
statistical analysis. HBL, QM, LX and DSW were involved in
performing experiments. XLG reviewed and edited the manuscript. All
authors read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tharmapalan P, Mahendralingam M, Berman HK
and Khokha R: Mammary stem cells and progenitors: Targeting the
roots of breast cancer for prevention. EMBO J.
15(e100852)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Garrido-Castro AC, Lin NU and Polyak K:
Insights into molecular classifications of triple-negative breast
cancer: Improving patient selection for treatment. Cancer Discov.
9:176–198. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Nedeljković M and Damjanović A: Mechanisms
of chemotherapy resistance in triple-negative breast cancer-how we
can rise to the challenge. Cells. 8(957)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kandoth C, McLellan MD, Vandin F, Ye K,
Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al:
Mutational landscape and significance across 12 major cancer types.
Nature. 502:333–339. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Synnott NC, Murray A, McGowan PM, Kiely M,
Kiely PA, O'Donovan N, O'Connor DP, Gallagher WM, Crown J and Duffy
MJ: Mutant p53: A novel target for the treatment of patients with
triple-negative breast cancer? Int J Cancer. 140:234–246.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cordani M, Pacchiana R, Butera G, D'Orazi
G, Scarpa A and Donadelli M: Mutant p53 proteins alter cancer cell
secretome and tumour microenvironment: Involvement in cancer
invasion and metastasis. Cancer Lett. 376:303–309. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tan BS, Tiong KH, Choo HL, Chung FFL, Hii
LW, Tan SH, Yap IK, Pani S, Khor NT, Wong SF, et al: Mutant
p53-R273H mediates cancer cell survival and anoikis resistance
through AKT-dependent suppression of BCL2-modifying factor (BMF).
Cell Death Dis. 6(e1826)2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Muller PA and Vousden KH: Mutant p53 in
cancer: New functions and therapeutic opportunities. Cancer Cell.
25:304–317. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bertoli G, Cava C, Diceglie C, Martelli C,
Rizzo G, Piccotti F, Ottobrini L and Castiglioni I: MicroRNA-567
dysregulation contributes to carcinogenesis of breast cancer,
targeting tumor cell proliferation, and migration. Breast Cancer
Res Treat. 161:605–616. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ren Y, Chen Y, Liang X, Lu Y, Pan W and
Yang M: miRNA-638 promotes autophagy and malignant phenotypes of
cancer cells via directly suppressing DACT3. Cancer Lett.
390:126–136. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li Z, Meng Q, Pan A, Wu X, Cui J, Wang Y
and Li L: MicroRNA-455-3p promotes invasion and migration in triple
negative breast cancer by targeting tumor suppressor EI24.
Oncotarget. 8:19455–19466. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Humphries B and Yang C: The microRNA-200
family: Small molecules with novel roles in cancer development,
progression and therapy. Oncotarget. 6:6472–6498. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chikh A, Ferro R, Abbott JJ, Piñeiro R,
Buus R, Iezzi M, Ricci F, Bergamaschi D, Ostano P, Chiorino G, et
al: Class II phosphoinositide 3-kinase C2β regulates a novel
signaling pathway involved in breast cancer progression.
Oncotarget. 7:18325–18345. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Noonan EJ, Place RF, Pookot D, Basak S,
Whitson JM, Hirata H, Giardina C and Dahiya R: miR-449a targets
HDAC-1 and induces growth arrest in prostate cancer. Oncogene.
28:1714–1724. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Jeon HS, Lee SY, Lee EJ, Yun SC, Cha EJ,
Choi E, Na MJ, Park JY, Kang J and Son JW: Combining
microRNA-449a/b with a HDAC inhibitor has a synergistic effect on
growth arrest in lung cancer. Lung Cancer. 76:171–176.
2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Stojanovic N, Hassan Z, Wirth M, Wenzel P,
Beyer M, Schäfer C, Brand P, Kroemer A, Stauber RH, Schmid RM, et
al: HDAC1 and HDAC2 integrate the expression of p53 mutants in
pancreatic cancer. Oncogene. 36:1804–1815. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sun X, Liu S, Chen P, Fu D, Hou Y, Hu J,
Liu Z, Jiang Y, Cao X, Cheng C, et al: miR-449a inhibits colorectal
cancer progression by targeting SATB2. Oncotarget. 8:100975–100988.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chen SP, Liu BX, Xu J, Pei XF, Liao YJ,
Yuan F and Zheng F: miR-449a suppresses the epithelial-mesenchymal
transition and metastasis of hepatocellular carcinoma by multiple
targets. BMC Cancer. 15(706)2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Luo W, Huang B, Li Z, Li H, Sun L, Zhang
Q, Qiu X and Wang E: MicroRNA-449a is downregulated in non-small
cell lung cancer and inhibits migration and invasion by targeting
c-Met. PLoS One. 8(e64759)2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wei B, Song Y, Zhang Y and Hu M:
MicroRNA-449a functions as a tumor-suppressor in gastric
adenocarcinoma by targeting Bcl-2. Oncol Lett. 6:1713–1718.
2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Fruman DA and Rommel C: PI3K and cancer:
Lessons, challenges and opportunities. Nat Rev Drug Discov.
13:140–156. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
Subramanian M, Francis P, Bilke S, Li XL,
Hara T, Lu X, Jones MF, Walker RL, Zhu Y, Pineda M, et al: A mutant
p53/let-7i-axis-regulated gene network drives cell migration,
invasion and metastasis. Oncogene. 34:1094–1104. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang S, Zhou L, Hong B, van den Heuvel
AP, Prabhu VV, Warfel NA, Kline CL, Dicker DT, Kopelovich L and
El-Deiry WS: Small-molecule NSC59984 restores p53 pathway signaling
and antitumor effects against colorectal cancer via p73 activation
and degradation of mutant p53. Cancer Res. 75:3842–3852.
2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Jung CL, Mun H, Jo SY, Oh JH, Lee C, Choi
EK, Jang SJ and Suh YA: Suppression of gain-of-function mutant p53
with metabolic inhibitors reduces tumor growth in vivo. Oncotarget.
7:77664–77682. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen T, You Y, Jiang H and Wang ZZ:
Epithelial-mesenchymal transition (EMT): A biological process in
the development, stem cell differentiation, and tumorigenesis. J
Cell Physiol. 232:3261–3272. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tao Y, Han T, Zhang T, Ma C and Sun C:
LncRNA CHRF-induced miR-489 loss promotes metastasis of colorectal
cancer via TWIST1/EMT signaling pathway. Oncotarget. 8:36410–36422.
2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Heerboth S, Housman G, Leary M, Longacre
M, Byler S, Lapinska K, Willbanks A and Sarkar S: EMT and tumor
metastasis. Clin Transl Med. 4(6)2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hori S, Wadhwa K, Pisupati V, Zecchini V,
Ramos-Montoya A, Warren AY, Neal DE and Gnanapragasam VJ: Loss of
hSef promotes metastasis through upregulation of EMT in prostate
cancer. Int J Cancer. 140:1881–1887. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kim T, Veronese A, Pichiorri F, Lee TJ,
Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, et al:
p53 regulates epithelial-mesenchymal transition through microRNAs
targeting ZEB1 and ZEB2. J Exp Med. 208:875–883. 2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Dong P, Karaayvaz M, Jia N, Kaneuchi M,
Hamada J, Watari H, Sudo S, Ju J and Sakuragi N: Mutant p53
gain-of-function induces epithelial-mesenchymal transition through
modulation of the miR-130b-ZEB1 axis. Oncogene. 32:3286–3295.
2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hosain SB, Khiste SK, Uddin MB, Vorubindi
V, Ingram C, Zhang S, Hill RA, Gu X and Liu YY: Inhibition of
glucosylceramide synthase eliminates the oncogenic function of p53
R273H mutant in the epithelial-mesenchymal transition and induced
pluripotency of colon cancer cells. Oncotarget. 7:60575–60592.
2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Raha P, Thomas S, Thurn KT, Park J and
Munster PN: Combined histone deacetylase inhibition and tamoxifen
induces apoptosis in tamoxifen-resistant breast cancer models, by
reversing Bcl-2 overexpression. Breast Cancer Res.
17(26)2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shoja MH, Reddy ND, Nayak PG, Srinivasan
KK and Rao CM: Glycosmis pentaphylla (Retz.) DC arrests cell cycle
and induces apoptosis via caspase-3/7 activation in breast cancer
cells. J Ethnopharmacol. 168:50–60. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
You J, Zhang Y, Li Y, Fang N, Liu B, Zu L
and Zhou Q: miR-449a suppresses cell invasion by inhibiting MAP2K1
in non-small cell lung cancer. Am J Cancer Res. 5:2730–2744.
2015.PubMed/NCBI
|
|
38
|
Noonan EJ, Place RF, Basak S, Pookot D and
Li LC: miR-449a causes Rb-dependent cell cycle arrest and
senescence in prostate cancer cells. Oncotarget. 1:349–358.
2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kumar P, Sharad S, Petrovics G, Mohamed A,
Dobi A, Sreenath TL, Srivastava S and Biswas R: Loss of miR-449a in
ERG-associated prostate cancer promotes the invasive phenotype by
inducing SIRT1. Oncotarget. 7:22791–22806. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Shi W, Bruce J, Lee M, Yue S, Rowe M,
Pintilie M, Kogo R, Bissey PA, Fyles A, Yip KW and Liu FF: miR-449a
promotes breast cancer progression by targeting CRIP2. Oncotarget.
7:18906–18918. 2016.PubMed/NCBI View Article : Google Scholar
|