Introduction
Increasing interest has arisen for the study of
periodontitis and chronic apical periodontitis, which cause
irreversible destruction of periodontal support tissues, including
the alveolar bone, the periodontal ligament and the root cementum
(1,2). Several systemic diseases, such as
diabetes, endocrine disease and hypertension, are the main causes
of tooth loss (2). The combination
of membranes and fillers, demineralized freeze-dried bone
allografts, bovine-derived xenografts and barrier membranes have
been adopted in modern clinical practice to treat periodontitis and
chronic apical periodontitis; however, only a small number of these
therapies have been accepted as regenerative techniques, and most
have had limited success and generally unsatisfactory outcomes
(2). Therefore, it is of great
interest to find better treatments for periodontitis and chronic
apical periodontitis.
Human dental pulp stem cells (hDPSCs) and
appropriate growth factors, such as TGF-β1, are necessary for
functional periodontal tissue regeneration (3). hDPSCs are an undifferentiated
mesenchymal cell type in dental pulp that differentiate into a
variety of cells, such as those which form dentin (4). In recent years, the MAPK signaling
pathway has been widely investigated in the fields of cell
proliferation, differentiation and apoptosis (5). Concentrated growth factor (CGF) is
known to be enriched in growth factors and fibrin (5-7).
CGF is a gel-like substance that can be obtained by centrifugation
of venous blood (7). CGF combined
with bone graft material was indicated to promote immediate
periodontal tissue regeneration and osteogenic differentiation
(5-7).
CGF exudate (CGFe) is extracted from CGF and used in research
experiments to study the effects of CGF in vitro (1). Previous studies have indicated that
CGFe shortened the duration before osteogenesis onset in the
operational area of periodontal tissue and notably improved the
quality of bone formation (6-8).
For instance, Park et al (8)
reported that CGFe stimulated the proliferation of beagle
periodontal ligament stem cells in vitro.
TGF-β is a multifunctional cytokine involved in the
regulation of cell proliferation, migration, differentiation,
apoptosis and extracellular matrix formation (9,10). It
also plays an important role in bone repair, vascular regeneration
and immune system regulation (9,10).
TGF-β family includes three homologous isoforms, namely TGF-β1,
TGF-β2 and TGF-β3. Among these, TGF-β1 is the most abundant and
widely distributed (9,10). It also has been indicated to induce
mesenchymal stem cells to differentiate into osteoblasts by
recruiting osteoblasts and coupling them to promote bone tissue
reconstruction (11). The
combination of TGF-β1 and platelet-derived growth factor (PDGF) has
been demonstrated to induce osteogenesis in osteoblast-like cells,
and TGF-β1 at a concentration of 5 ng/ml promoted the formation of
mineralized nodules (12). A
previous study has indicated that TGF-β1 induced exfoliated
deciduous tooth stem cells to overexpress bone sialoprotein (BSP)
and osteocalcin (OCN) (13).
The aim of the present study was to assess the
effects of CGFe and TGF-β1 on hDPSC proliferation and osteogenic
differentiation in order to discover improved treatments for
periodontitis and chronic apical periodontitis. Specifically, the
effects of CGFe and TGF-β1 were examined on the enhancement of
alkaline phosphatase (ALP) activity and the expression levels of
BSP, Runt-related transcription factor 2 (RUNX2) and OCN in
hDPSCs.
Materials and methods
hDPSC isolation and culture
All experiments reported in the present study were
approved by the Ethics Committee of the Stomatological School of
Jilin University (Changchun, China). Dental pulp tissues were
obtained from healthy individuals (3 females and 3 males; age,
12-18 years; without systemic disease) with informed consent from
their parents, between March 2019 and December 2019. The dental
pulp tissue samples were collected from third molar extractions at
Jilin University Health Science Center (Changchun, China). Dental
pulp stem cells were isolated and cultured following procedures
described previously (14). The
dental pulp matrix was gently removed from the tooth, minced with
ophthalmic scissors and digested in a solution containing 3 mg/ml
collagenase type I (Sigma-Aldrich; Merck KGaA) and 4 mg/ml
Dispase® II (Roche Diagnostics GmbH) at 37˚C for 60 min.
The cells were purified using a mouse anti-STRO-1+
antibody (1:200; 4˚C; 3 h; cat. no. sc-4773; Santa Cruz
Biotechnology, Inc.) and goat anti-mouse IgG Dynabeads (1:500; 4˚C;
2 h; cat. no. 11201D; Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The purified hDPSCs
were cultured in α-minimum essential medium (α-MEM; Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) as well as 100 U/ml streptomycin and 100
mg/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.) at 37˚C in
5% CO2. The medium was changed every 3 days, and hDPSCs
were passaged by trypsinization (Gibco; Thermo Fisher Scientific,
Inc.) until they reached 80% confluence. hDPSCs between passages
3-6 were used for the experiments of the present study.
Immunocytochemistry staining
hDPSCs (1x105 cells) at passage 4 were
seeded into six-well plates covered in advance with coverslips, and
incubated for 72 h at 37˚C. The cells were then rinsed three times
with 0.01 M PBS and fixed with 4% paraformaldehyde for 15 min at
room temperature. Endogenous peroxidase activity was eliminated by
incubation with 3% H2O2 for 15 min at room
temperature. The cells were then incubated with anti-vimentin
(1:100; 2 h; cat. no. ab24525; Abcam) and anti-cytokeratin-15
primary antibodies (1:200; 2 h; cat. no. AM06387SU-N; OriGene
Technologies, Inc.) at 4˚C. hDPSCs were subsequently incubated with
secondary goat-anti-rabbit, goat-anti-mouse and goat anti-chicken
IgG were AlexaFluor 488 (cat. no. A-11008; Invitrogen; Thermo
Fisher Scientific, Inc.), 568 (cat. no. A-11004; Invitrogen; Thermo
Fisher Scientific, Inc.), and 647 (cat. no. A-21449; Invitrogen;
Thermo Fisher Scientific, Inc.) labeled, respectively; and used in
various combinations at a 1:1,000 dilution at 4˚C (15). The SP (no. SNM500; OriGene
Technologies, Inc.) immunohistochemistry assay kit (OriGene
Technologies, Inc.) was used for immunocytochemical staining
according to the manufacturer's protocol, and 3,3'-diaminobenzidine
(OriGene Technologies, Inc.) was used to stain positive cells,
which were imaged using a fluorescent microscope (IX73; Olympus
Corporation; magnification, x400;) (15). The blue color of the nuclei in
Fig. 1 was produced by hematoxylin
staining for 5 min at 25˚C.
Preparation of CGFe and TGF-β1
CGFe was obtained from four healthy male donors
(aged 22-30 years) visiting the outpatient clinic at the Health
Science Center of Jilin University (Changchun, China) between March
and September 2019, with their informed consent. According to an
existing protocol (15), a 5-ml
venous blood sample was collected from each donor, and the blood
samples were used to produce CGF and CGFe. The blood samples were
centrifuged at 750 x g for 12 min at 4˚C. A white CGF clot was
formed between acellular plasma and red blood cells (RBCs), which
was separated from the RBCs using scissors, placed on an endo box
and compressed by the endo box cover. The CGF clot was converted
into CGFe by applying pressure. CGFe was filtered using a 0.22-µm
sterile syringe filter unit (MilliporeSigma), and the pooled CGFe
samples were stored at -80˚C. The original concentration of CGFe
was defined as 100, 50 and 25% concentrations of CGFe were obtained
by dilution of the 100% CGFe with α-MEM. As observed in the present
study, the CGF membrane used in clinical treatment were completely
absorbed in 7 to 14 days, day 7 and 14 were selected as two
monitoring time points for reporting the results in the present
experiments.
TGF-β1 powder (PeproTech China) was dissolved in
distilled water according to the manufacturer's instructions, and
different dilutions of TGF-β1 were stored at -80˚C for subsequent
experiments.
MTT assay
MTT was used to quantify the effects of CGFe and
TGF-β1 on hDPSC viability. hDPSCs (3x103 cells/well)
were seeded into 96-well plates (Corning, Inc.) in 10% FBS complete
medium (α-MEM) and incubated for 24 h at 37˚C. hDPSCs were then
exposed to TGF-β1 (at concentrations of 0, 1, 5, 10 and 20 ng/ml)
for 7 days. After 7 days of culture, MTT reagent (10 µl) was added
to the culture medium of each well, followed by incubation at 37˚C
for 4 h. The medium was removed and the cells were washed twice
with 0.01 M PBS. DMSO (Sigma-Aldrich; Merck KGaA) was added into
each well to dissolve the formazan crystals. The optical density
(OD) values were measured using an automatic ELISA reader (ELx800;
BioTek Instruments, Inc.) at 490 nm. The assay was repeated three
times under the same conditions, and the data are presented as the
mean ± SD. hDPSCs in 10% FBS complete medium (α-MEM) were used as
the control group.
In another experiment, hDPSCs were divided into four
groups: i) TGF-β1 (1 ng/ml); ii) 25% CGFe + TGF-β1 (1 ng/ml); iii)
50% CGFe + TGF-β1 (1 ng/ml); and iv) 100% CGFe + TGF-β1 (1 ng/ml)
group, and subsequently incubated for 7 days. MTT assay was
performed as aforementioned.
ALP activity assay
hDPSCs (1x104 cells/well) were seeded
into 24-well plates (Corning, Inc.) and incubated for 24 h at 37˚C.
Subsequently, the cells were exposed to 100% CGFe, TGF-β1 (1 ng/ml)
or TGF-β1 (1 ng/ml) + 100% CGFe for 7 or 14 days. hDPSCs in 10% FBS
complete medium (α-MEM) were used as the control group. At the
given time points, the cells were lysed using 0.1% Triton X-100,
and the lysates were centrifuged at 8,000 x g for 10 min at 4˚C.
The supernatant was added to 96-well plates (50 µl/well), and ALP
activity was examined using the ALP assay kit (cat. no. A059-2;
Nanjing Jiancheng Bioengineering Institute). The OD values were
measured using an automatic microplate reader (Infinite 200 PRO;
Tecan Group, Ltd.) at 520 nm. The assay was repeated three times
under the same conditions, and the data were presented as the mean
± SD.
Reverse transcription-quantitative PCR
(RT-qPCR)
hDPSCs (1x105 cells/well) were seeded
into six-well plates with standard medium (α-MEM) until they
reached 60-70% confluence. The cells were treated with four
different media: The control group was treated with
osteogenesis-inducing medium [α-MEM supplemented with 50 µg/ml
ascorbic acid and 10 mM β-sodium glycerophosphate (Sigma-Aldrich;
Merck KGaA)]; the three experimental groups were treated with
osteogenesis-inducing medium supplemented with 100% CGFe, TGF-β1 (1
ng/ml) or 100% CGFe + TGF-β1 (1 ng/ml).
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse
transcribed into cDNA on the 7 and 14th day after treatment. cDNA
synthesis was performed with 1 µg total RNA using SuperScript™ II
Reverse Transcriptase and random hexamer primers (cat. no.
18064014; Invitrogen; Thermo Fisher Scientific, Inc.). The
following temperature protocol was used for reverse transcription:
Room temperature for 10 min, 37˚C for 60 min and 85˚C for 5
min.
An aliquot (2 µl) of each sample was used for qPCR
determination of the expression of the osteogenic genes BSP,
RUNX2 and OCN using the SYBR® Premix Ex
Taq™ II kit (Takara Bio, Inc.). qPCR was performed in a Rotor-Gene
Q thermocycler as follows: 95˚C for 3 min, followed by 40 cycles of
95˚C for 3 sec and 60˚C for 60 sec. Each experiment was repeated
three times, and the 2-∆∆Cq method was used to calculate
the fold differences in gene expression (16), using β-actin for normalization. The
primers used are listed in Table
I.
 | Table IPrimers used for reverse
transcription-quantitative PCR analysis. |
Table I
Primers used for reverse
transcription-quantitative PCR analysis.
| Gene | Forward primer
(5'-3') | Reverse primer
(5'-3') | PCR product
(bp) |
|---|
| β-actin |
AGAAAATCTGGCACCACACC |
GGGTGTTGAAGGTCTAAA | 139 |
| OCN |
GGCGCTACCTGTATCAATGG |
TCAGCCAACTCGTCACAGTC | 106 |
| RUNX2 |
CACCATGTCAGCAAAACTTCTT |
TCACGTCGCTCATTTTGC | 96 |
| BSP |
GCAGTAGTGACTCATCCGAAGAA |
GCCTCAGAGTCTTCATCTTCATTC | 121 |
Western blotting
hDPSC protein lysates were prepared using RIPA
buffer with pH 7.4 (10 mM Tris-Cl, 1 mM PMSF, 150 mM NaCl, 0.1%
SDS, 1% sodium deoxycholate and 1% Triton X-100). The cells were
treated with four different media [control, 100% CGFe, TGF-β1 (1
ng/ml) or 100% CGFe + TGF-β1 (1 ng/ml)] for 7 days. The cell
lysates were incubated on ice for 30 min, then clarified by
centrifugation at 6,000 x g at 4˚C for 10 min. The protein contents
were quantified with a BCA assay kit. Protein samples (20 µg) were
denatured and resolved by 10% SDS-PAGE and transferred onto PVDF
membranes (MilliporeSigma) at 300 mA for 2 h at 4˚C. The membranes
were blocked with 5% non-fat milk at room temperature for 1 h, and
subsequently incubated with the following primary antibodies:
Anti-RUNX2 (1:1,000; cat. no. 12556; Cell Signaling Technology,
Inc.), anti-BSP (1:1,000; cat. no. ab92920; Abcam), anti-OCN
(1:1,000; cat. no. ab93876; Abcam); anti-ERK1/2 (1:1,000; cat. no.
BS3628; Bioworld Technology, Inc.), anti-phosphorylated (p)-ERK1/2
(1:1,000; cat. no. BS4759; Bioworld Technology, Inc.), anti-JNK
(1:1,000; cat. no. BS3631; Bioworld Technology, Inc.), anti-p-JNK
(1:1,000; cat. no. BS4763; Bioworld Technology, Inc.), anti-p38
(1:1,000; cat. no. BS9851M Bioworld Technology, Inc.), anti-p-p38
(1:1,000; cat. no. BS4635; Bioworld Technology, Inc.) and
anti-β-actin (1:1,000; cat. no. 6609-1-Ig; Abgent Biotech Co.,
Ltd.) at 4˚C overnight. The membranes were washed in TBS containing
0.1% Tween-20 (TBST) three times and incubated with HRP-labeled
secondary antibody (cat. no. SA00001-2; 1:10,000; ProteinTech
Group, Inc.) for 1 h at 22˚C. Following three washes with TBST, the
protein bands were visualized using an ECL kit (GE Healthcare) and
exposed to an X-ray film. β-actin was used as internal reference,
and the experiment was performed in triplicates (ImageJ v1.8.0;
NIH). The protein bands (ERK1/2, p-ERK1/2, JNK, p-JNK, p38, and
p-p38) were recorded at the selected time points of 0, 30, 60 and
90 min.
Statistical analysis
Numerical results are presented as the mean ± SD of
three independent experiments. Statistical analyses were performed
using SPSS software (version 22.0; IBM Corp.). hDPSC viability,
RT-qPCR and ALP activity assays were analyzed via one-way ANOVA and
Tukey's multiple comparison test. Western blotting data was
analyzed using one-way ANOVA followed by Bonferroni's post-hoc test
for independent samples. P<0.05 was considered to indicate a
statistically significant difference.
Results
hDPSC characteristics
hDPSC morphology resembled that of fibroblast-like
cells, with an elongated cell body and the nucleus located in the
center. Under the light microscope, the clonal proliferating cells
were closely arranged, the cell morphology was uniform and the
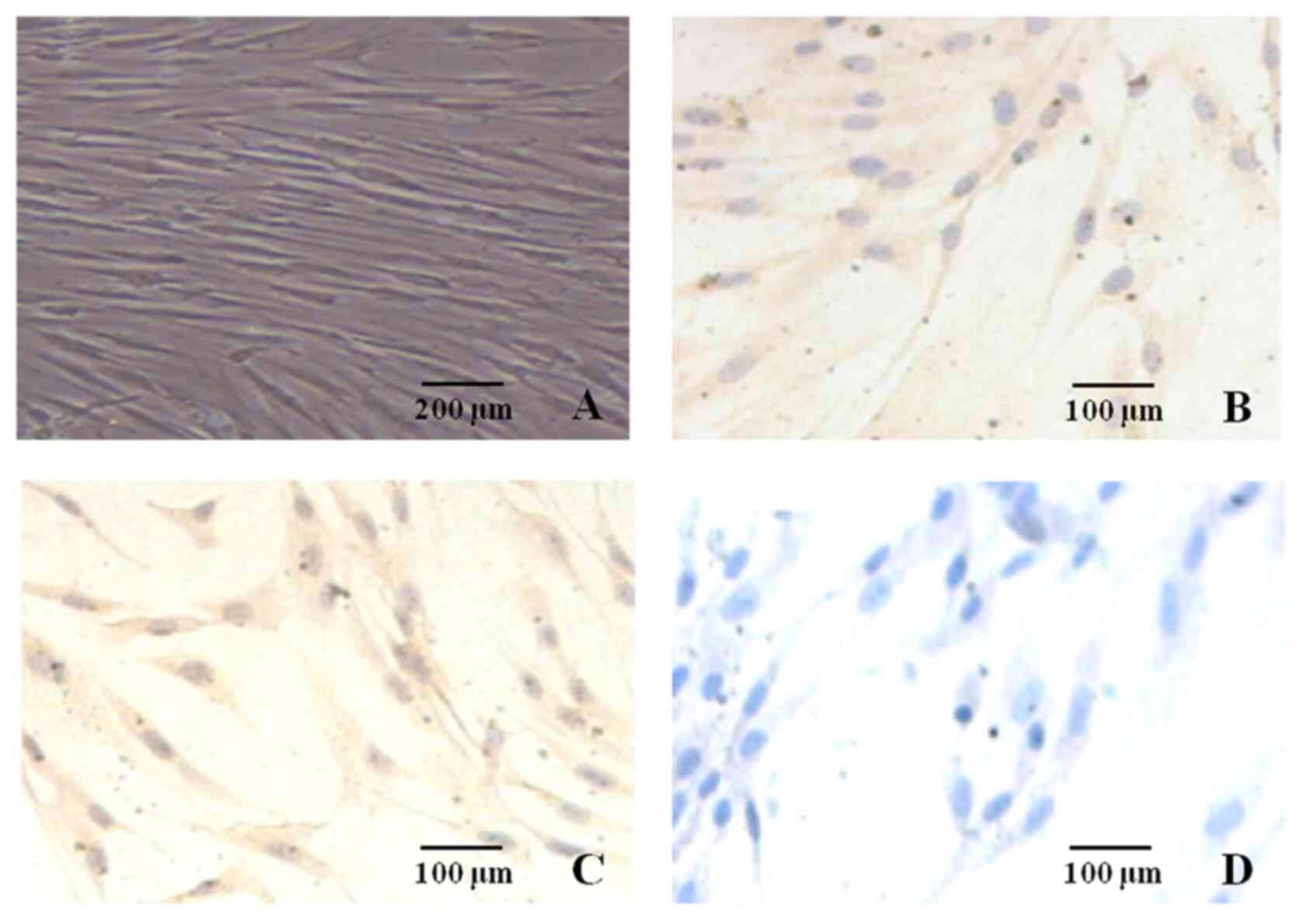
cytoplasm was abundant (Fig. 1A).
The nuclei were oval and contained distinct nucleoli.
Immunocytochemical staining of in vitro culture indicated
that the hDPSCs were positive for STRO-1 (Fig. 1B) and vimentin protein (Fig. 1C). The nuclei were colorless, and
anti-cytokeratin staining was negative (Fig. 1D). The cytosol was not stained,
which was consistent with the characteristics of mesenchymal tissue
derived cells.
Effects of CGFe and TGF-β1 on hDPSC
viability
The present study was designed to examine the impact
of TGF-β1 at a range of concentrations (0, 1, 5, 10 or 20 ng/ml) on
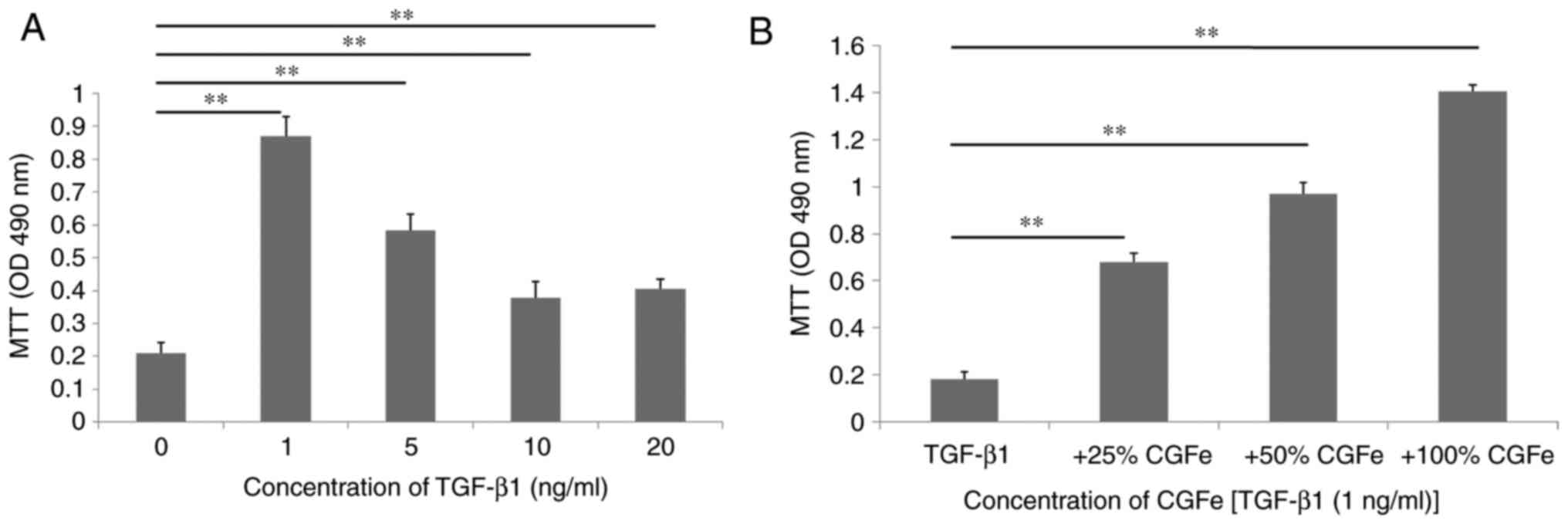
the viability of hDPSCs for 7 days in vitro (Fig. 2A). The viability of hDPSCs were
determined using an MTT assay. TGF-β1 at a concentration of 1 ng/ml
demonstrated the best promoting effects on the viability of hDPSCs.
The difference between the control and TGF-β1 (1 ng/ml) groups was
the highest (P<0.01), and the difference between the control and
TGF-β1 groups (5, 10 or 20 ng/ml) was also statistically
significant (all P<0.01). hDPSCs were exposed to TGF-β1 (1
ng/ml), 25% CGFe + TGF-β1 (1 ng/ml), 50% CGFe + TGF-β1 (1 ng/ml) or
100% CGFe + TGF-β1 (1 ng/ml) for 7 days (Fig. 2B). Compared with the TGF-β1 (1
ng/ml) group, the viability rate of the 100% CGFe + TGF-β1 (1
ng/ml) group increased significantly (P<0.01), and the 25% CGFe
+ TGF-β1 (1 ng/ml) and 50% CGFe + TGF-β1 (1 ng/ml) groups also
demonstrated notable rate increases (P<0.01).
ALP activity
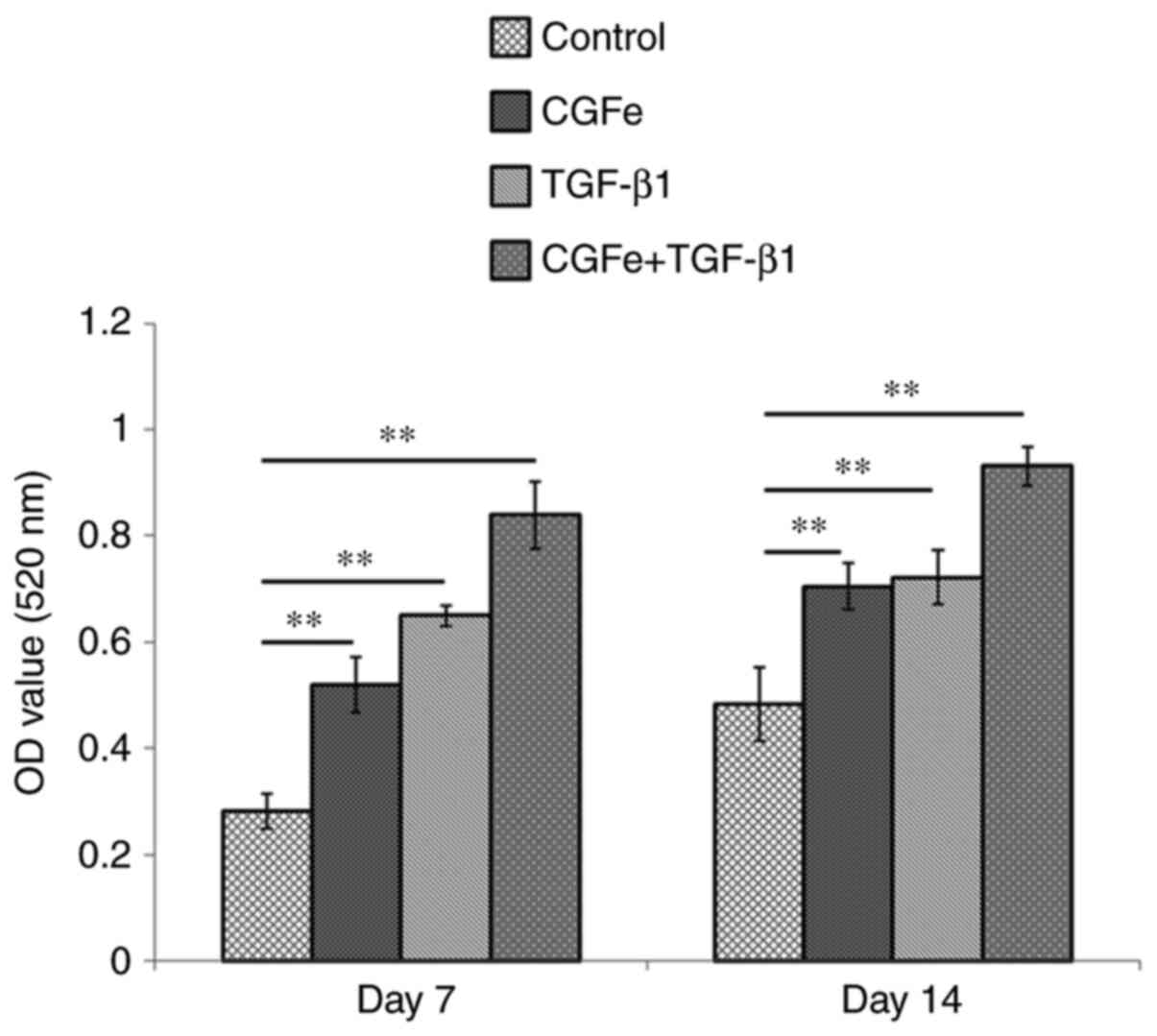
After 7 or 14 days of culture, hDPSCs cultured in
100% CGFe + TGF-β1 (1 ng/ml) demonstrated the highest levels of ALP
activity compared with the other experimental groups and the
control group (Fig. 3).
Furthermore, the ALP activity of hDPSCs in the 100% CGFe group and
the TGF-β1 (1 ng/ml) group on the 7 and 14th days was also
increased compared with the control group (all P<0.01).
Subsequent experiments were performed with the optimized
concentrations of CGFe (100%) and TGF-β1 (1 ng/ml).
Effects of CGFe and TGF-β1 on the
expression of osteogenesis-associated genes
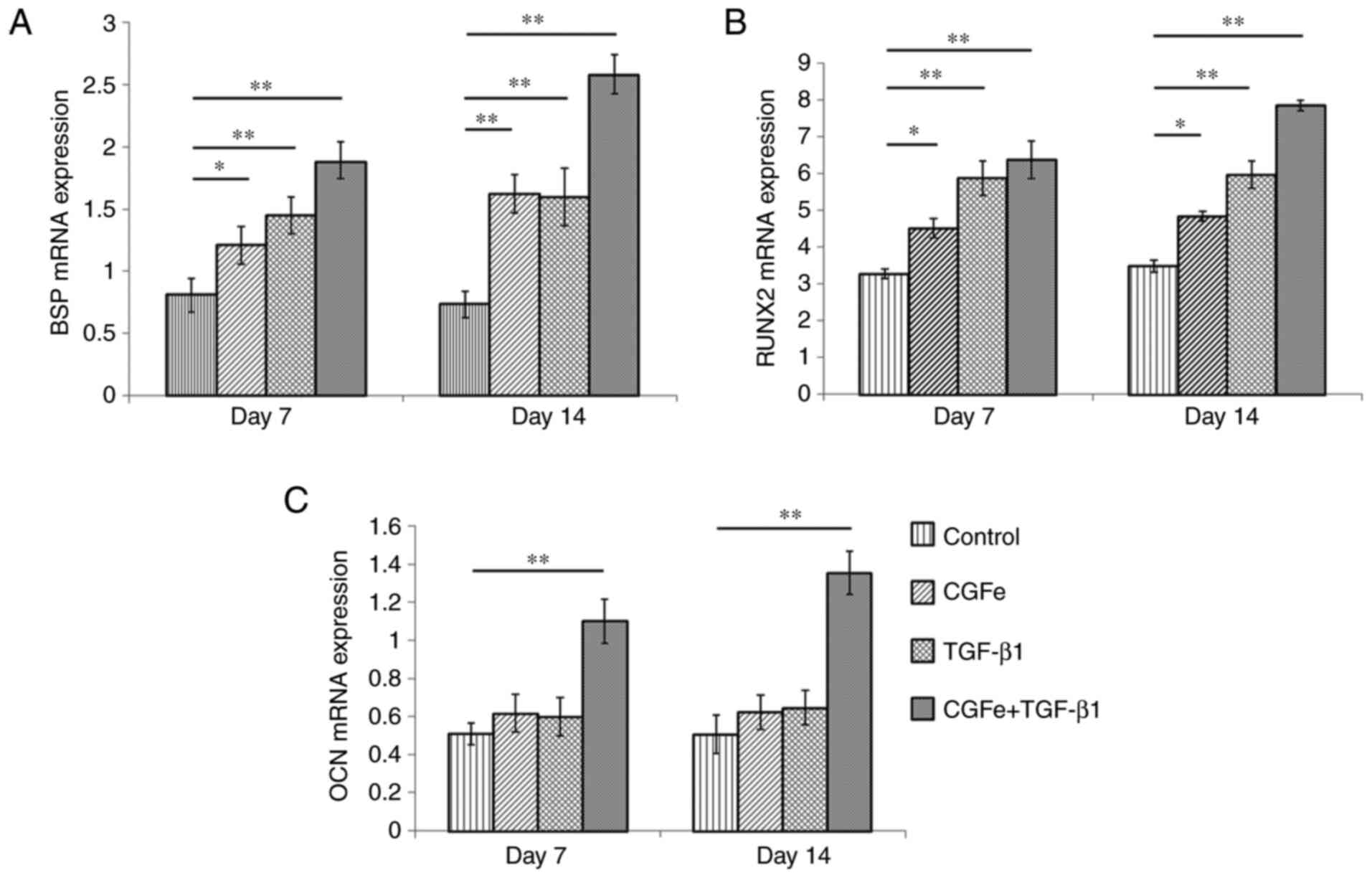
The expression levels of osteogenesis-associated
genes (RUNX2, BSP and OCN) were measured on
the 7 and 14th days after treatment. On the 7th day, the expression
levels of BSP and RUNX2 were both increased in the
CGFe, the TGF-β1 and the CGFe + TGF-β1 group, compared with the
control group, and the differences were significant (Fig. 4A and B; P<0.01). The expression levels of
BSP and RUNX2 were further increased on the 14th day,
compared with the control group (Fig.
4A and B; P<0.01). The gene
expression level of OCN in the CGFe and the TGF-β1 group was
not increased on day 7 and 14. However, the expression level of
OCN in the CGFe + TGF-β1 group was significantly increased
at both days 7 and 14 compared with the control group (Fig. 4C; P<0.01).
Effects of CGFe and TGF-β1 on
osteogenic proteins and MAPK signaling pathways
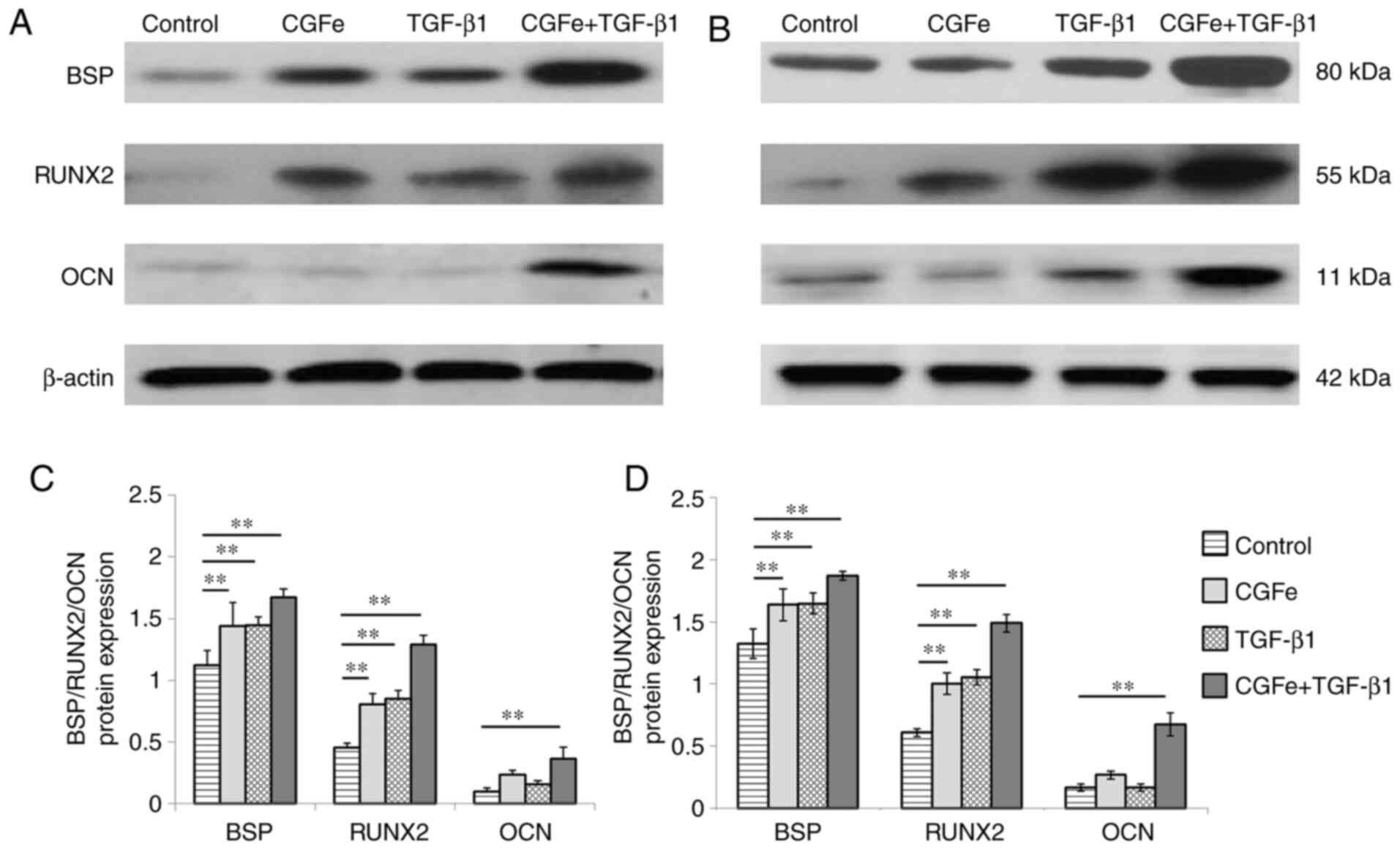
Western blotting was performed to examine the effect
of CGFe, TGF-β1 and CGFe + TGF-β1 on hDPSC differentiation, as well
as to confirm the RT-qPCR results at the protein level. hDPSCs were
cultured in the aforementioned four different media for 7 and 14
days.
As demonstrated in Fig.
5, compared with the control group, the protein levels of RUNX2
and BSP were increased in the CGFe, the TGF-β1 and the CGFe +
TGF-β1 group at different time points (all P<0.01). Compared
with the control group, the protein levels of OCN were increased in
the CGFe + TGF-β1 group (P<0.01), but were not significantly
increased (P>0.05) in the CGFe group and the TGF-β1 group.
As the present results illustrated that CGFe +
TGF-β1 treatment exhibited the most important effect on the
viability and differentiation of hDPSCs, this condition was
selected to examine the expression of MAPK pathway-related proteins
at different time points.
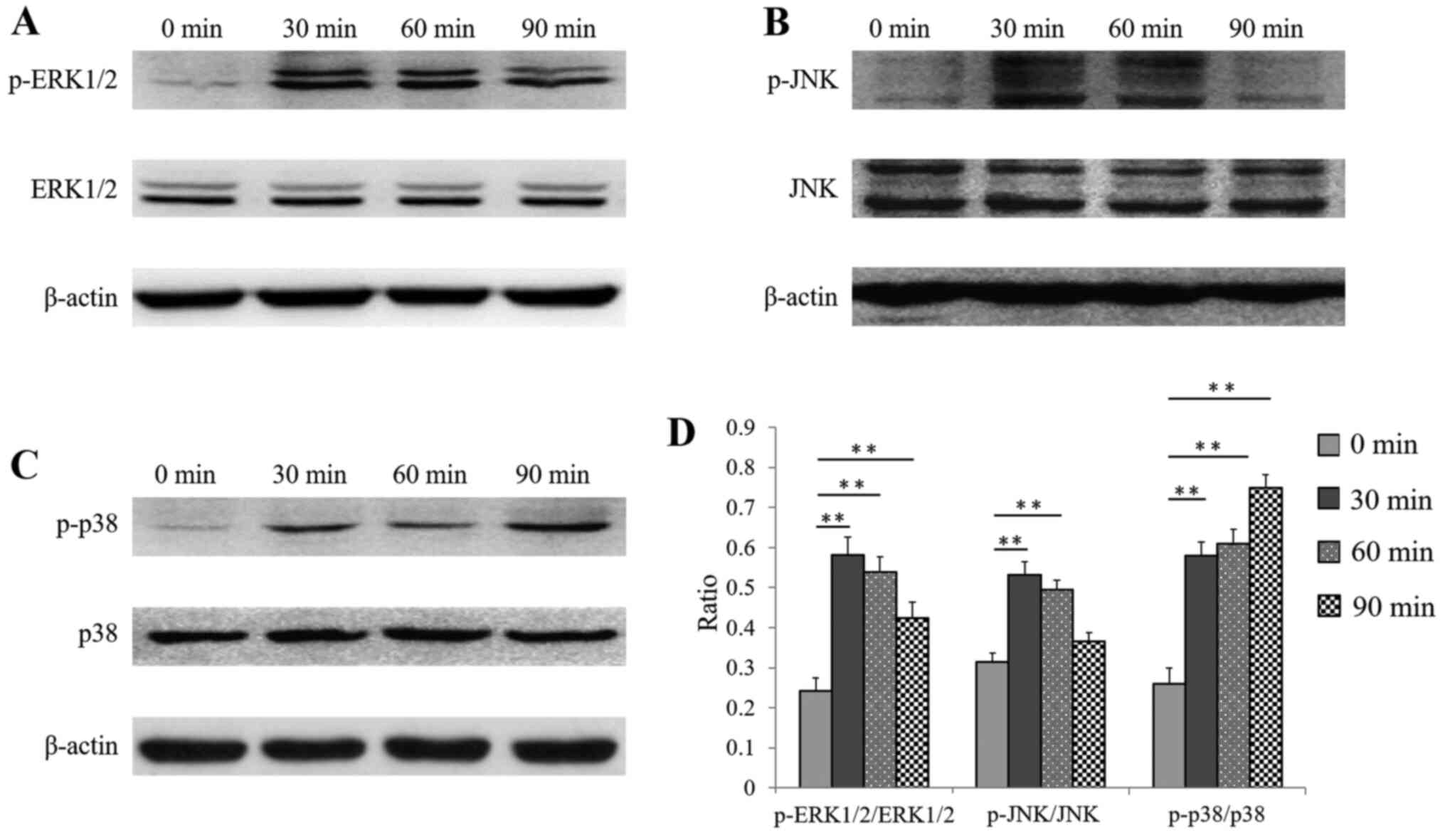
No significant difference in the total protein
expression of the three MAPK pathway proteins (ERK1/2, JNK and p38)
was noted at the different selected time points (Fig. 6A-C). The protein expression of
p-ERK1/2 in hDPSCs increased significantly after stimulation with
CGFe + TGF-β1 for 30 min, and decreased slightly after 60 and 90
min (Fig. 6A), compared with 0 min.
As presented in Fig. 6D, the ratios
of p-ERK1/2 to total ERK1/2 in hDPSCs after 30, 60 and 90 min were
all significantly higher than that at 0 min (all P<0.01).
The protein expression of p-JNK in hDPSCs was also
increased after 30 and 60 min, compared with 0 min, and then
decreased to its initial expression level after 90 min (Fig. 6B). As indicated in Fig. 6D, the ratio of p-JNK to JNK in
hDPSCs after 30 and 60 min was significantly higher than that at 0
min (both P<0.01), while no significant difference was observed
in hDPSCs after 90 min (P>0.05).
The protein expression of p-p38 in hDPSCs increased
progressively after stimulation with CGFe + TGF-β1 for 90 min, with
the largest increase at 90 min, compared with 0 min (Fig. 6C). As presented in Fig. 6D, the ratio of p-p38 to p38 in
hDPSCs at 30, 60 and 90 min was significantly higher than at 0 min
(all P<0.01).
In the 100% CGFe and the TGF-β1 groups, it was
observed that the ratios of p-ERK1/2 to ERK1/2, p-JNK to JNK and
p-p38 to p38 were almost identical at 0, 30, 60 and 90 min, and
there were no statistically significant changes observed (data not
shown).
Discussion
The main components of CGFe include epidermal growth
factor, PDGF, fibroblast growth factor, bone morphogenetic protein
and VEGF (17,18). These growth factors display
functions in accelerating the revascularization of injured tissues
and inducing the differentiation, proliferation and migration of
fibroblasts and osteoblasts (19,20).
In recent years, CGFe has been widely applied to the reconstruction
of bone tissue in dental practice (7,18,19).
Animal experiments indicated that TGF-β1 could
accelerate the healing of skull defects and tibia fractures, as
well as strengthen the new bone tissue (21,22).
Chitosan scaffolds carrying TGF-β1 were used in direct pulp capping
of canine teeth, and the results indicated that TGF-β1 promoted the
formation of regenerative dentin (23). The non-collagen components of the
extracellular matrix of bone tissue include core proteoglycan,
disaccharide chain proteoglycan, bone mucin, BSP, OCN and
osteopontin, among which BSP is the most important (24). TGF-β1 upregulates the expression of
BSP in dental pulp cells and promotes the formation of regenerative
dentin (24).
RUNX2 is major bone transcription factors necessary
for osteogenic differentiation (25). RUNX2 has been indicated to induce
osteogenic gene expression and biological mineral deposition in
primary dermal fibroblasts (26),
while directly regulating the expression of
craniosynostosis-associated genes and skeletal tissue-enriched
genes (26). Overexpression of
RUNX2 in adipose tissue-derived mesenchymal stem cells triggered
their osteoblastic differentiation (25). RUNX2 knockout mice demonstrated a
complete lack of bone formation due to the maturation arrest of
osteoblasts (25,26). OCN expression is used as a marker of
osteoblast metabolic activity and mineral deposition in osteoblast
cultures (26). In the present
study, the finding that TGF-β1 induced the upregulation of
RUNX2, BSP and OCN gene expression in hDPSCs
suggested that TGF-β1 acts as an important stimulatory factor
during osteogenesis and odontogenesis.
The present study reported that 100% CGFe + TGF-β1
induced the highest increase in hDPSC viability, compared with the
control group. Bone formation and odontogenic differentiation of
hDPSCs was also notably enhanced in the CGFe + TGF-β1 group, which
was evidenced by the increased ALP activity and the higher
expression of bone formation and odontogenic markers, compared with
the control group. It was observed that CGFe + TGF-β1 upregulated
the expression of p-ERK1/2, p-JNK and p-p38 in hDPSCs, indicating
that it activated MAPK pathways during the osteogenic and
odontogenic differentiation of hDPSCs. The present experiments
indicated that CGFe + TGF-β1 promoted the viability, as well as the
osteogenic and odontogenic differentiation of hDPSCs via the
activation of the MAPK pathway, indicating that the factors
included in CGFe and TGF-β1 played important roles during
osteogenic differentiation and could have clinical implications in
dental pulp regeneration and osteoporosis. The findings of the
present study suggested that CGF + TGF-β1-treated hDPSCs may
potentially be used for bone and tooth regeneration.
Subsequent studies may be carried out to explore
other pathway mechanisms involved in the differentiation of hDPSCs
stimulated by CGFe and TGF-β1. Further investigation of microRNA
expression during CGFe + TGF-β1-mediated differentiation may
facilitate the in vivo applications of CGFe and TGF-β1 in
bone and dental tissue engineering in the future.
In conclusion, the present study demonstrated that
CGFe together with TGF-β1 facilitated the viability and osteogenic
differentiation of hDPSCs through the activation of the MAPK
signaling pathway, suggesting that CGFe and TGF-β1 play important
roles in the osteogenic differentiation process. The present work
provided insights for the application of CGFe and TGF-β1 in
periodontal tissue regeneration and alveolar bone remodeling.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Shenzhen Fundamental
Research Program (grant no. JCYJ20180228164057158), the Longhua
District Health Bureau of Shenzhen Municipality (grant no. 2020017)
and the Guangzhou Health Science and technology project (grant no.
20211A011100).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL, HY, XD and ZY conceived and designed the study.
XL, ZY and YZ performed cell culture, immunostaining and viability
analysis. XL, XD and YZ performed the experimental procedures of
osteogenic differentiation induction, reverse
transcription-quantitative PCR and western blotting. HY, XD, BW and
JL provided reagents and interpreted the data. XL, HY, XD, ZY and
JL performed data analysis and wrote the manuscript. ZY and BW
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Stomatological School of Jilin University Health
Science Center (Changchun, China). Written informed consent was
obtained from all participants or their parents prior to
experimentation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen FM and Yan J: Periodontal tissue
engineering and regeneration: Current approaches and expanding
opportunities. Tissue Eng Part B Rev. 16:219–255. 2010.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bosshardt DD and Sculean A: Does
periodontal tissue regeneration really work? Periodontol.
5:208–219. 2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mey AH, Yu ZD and Huang TJ: Small
molecules affect human dental pulp stem cell properties via
multiple signaling pathways. Stem Cells Dev. 22:2402–2413.
2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yu J, Deng Z, Shi J, Zhai H, Nie X, Zhuang
H, Li Y and Jin Y: Differentiation of dental pulp stem cells into
regular-shaped dentin-pulp complex induced by tooth germ cell
conditioned medium. Tissue Eng. 12:3097–3105. 2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang Y, Li J, Song W and Yu J: Mineral
trioxide aggregate upregulates odonto/osteogenic capacity of bone
marrow stromal cells from craniofacial bones via JNK and ERK MAPK
signalling pathways. Cell Prolif. 47:241–248. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhang W, Shen X, Wan C, Zhao Q, Zhang L,
Zhou Q and Deng L: Effects of insulin and insulin-like growth
factor 1 on osteoblast proliferation and differentiation:
Differential signalling via Akt and ERK. Cell Biochem Funct.
30:297–302. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Kim TH, Kim SH, Sándor GK and Kim YD:
Comparison of platelet-rich plasma (PRP), platelet-rich fibrin
(PRF), and concentrated growth factor (CGF) in rabbit-skull defect
healing. Arch Oral Biol. 59:550–558. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Park HC, Kim SG, Oh JS, You JS, Kim JS,
Lim SC, Jeong MA, Kim JS, Jung C, Kwon YS and Ji H: Early bone
formation at a femur defect using CGF and PRF grafts in adult dogs:
A comparative study. Implant Dent. 25:387–393. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chin D, Boyle GM, Parsons PG and Coman WB:
What is transforming growth factor-beta (TGF-beta)? Br J Plast
Surg. 57:215–221. 2004.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kubiczkova L, Sedlarikova L, Hajek R and
Sevcikova S: TGF-β- an excellent servant but a bad master. J Transl
Med. 10(183)2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Tang Y, Wu X, Lei W, Pang L, Wan C, Shi Z,
Zhao L, Nagy TR, Peng X, Hu J, et al: TGF-beta1-induced migration
of bone mesenchymal stem cells couples bone resorption with
formation. Nat Med. 15:757–765. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Vahabi S, Torshabi M and Nejad AE: In
vitro comparison of the efficacy of TGF-β1 and PDGF-BB in
combination with freeze-dried bone allografts for induction of
osteogenic differention in MG-63 osteoblast-like cells. J Mater Sci
Mater Med. 27(182)2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Farea M, Husein A, Halim AS, Abdullah NA,
Mokhtar KI, Lim CK, Berahim Z and Mokhtar K: Synergistic effects of
chitosan scaffold and TGFβ1 on the proliferation and osteogenic
differentiation of dental pulp stem cells derived from human
exfoliated deciduous teeth. Arch Oral Biol. 59:1400–1411.
2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yu J, He H, Tang C, Zhang G, Li Y, Wang R,
Shi J and Jin Y: Differentiation potential of STRO-1+
dental pulp stem cells changes during cell passaging. BMC Cell
Biol. 11(32)2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li X, Yang H, Zhang Z, Yan Z, Lv H, Zhang
Y and Wu B: Concentrated growth factor exudate enhances the
proliferation of human periodontal ligament cells in the presence
of TNF-α. Mol Med Rep. 19:943–950. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Qiao J, Duan J, Zhang Y, Chu Y and Sun C:
The effect of concentrated growth factors in the treatment of
periodontal intrabony defects. Future Sci OA.
2(FS136)2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Rodella LF, Favero G, Boninsegna R,
Buffoli B, Labanca M, Scarì G, Sacco L, Batani T and Rezzani R:
Growth factors, CD34 positive cells, and fibrin network analysis in
concentrated growth factors fraction. Microsc Res Tech. 74:772–777.
2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Honda H, Tamai N, Naka N, Yoshikawa N and
Myoui A: Bone tissue engineering with bone marrow-derived stromal
cells integrated with concentrated growth factor in rattus
norvegicus calvaria defect model. J Artif Organs. 16:305–315.
2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Shinohara Y, Tsuchiya S, Hatae K and Honda
MJ: Effect of vitronectin bound to insulin-like growth factor-I and
insulin-like growth factor binding protein-3 on porcine enamel
organ-derived epithelial cells. Int J Dent.
2012(386282)2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sarahrudi K, Thomas A, Mousavi M, Kaiser
G, Köttstorfer J, Kecht M, Hajdu S and Aharinejad S: Elevated
transforming growth factor-beta 1 (TGF-β1) levels in human fracture
healing. Injury. 42:833–837. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Poniatowski LA, Wojdasiewicz P, Gasik R
and Szukiewicz D: . Transforming growth factor beta family: Insight
into the role of growth factors in regulation of fracture healing
biology and potential clinical application. Mediators Inflamm.
2015(137823)2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li F, Liu X, Zhao SL, Wu H and Xu HHK:
Porous chitosan bilayer membrane containing TGF-β1 loaded
microspheres for pulp capping and reparative dentin formation in a
dog model. Dent Mater. 30:172–181. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hwang YC, Hwang IN, Oh WM, Park JC, Lee DS
and Son HH: Influence of TGF-beta1 on the expression of BSP, DSP,
TGF-beta1 receptor I and smad proteins during reparative
dentinogenesis. J Mol Hist. 39:153–160. 2008.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chen Q, Liu WB, Sinha KM, Yasuda H and de
Crombrugghe B: Identification and characterization of microRNAs
controlled by the osteoblast-specific transcription factor osterix.
PLoS One. 8(e58104)2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jeong W, Song G, Bazer FW and Kim J:
Insulin-like growth factor I induces proliferation and migration of
porcine trophectoderm cells through multiple cell signaling
pathways, including protooncogenic protein kinase 1 and
mitogen-activated protein kinase. Mol Cell Endocrinol. 384:175–184.
2014.PubMed/NCBI View Article : Google Scholar
|