Introduction
Ischemic stroke, also known as cerebral ischemia, is
an acute cerebrovascular disease that is associated with high
disability and mortality rates (1,2).
According to statistics compiled by the American Heart Association
in collaboration with the National Institutes of Health and other
government agencies, stroke has become the third most common cause
of death worldwide after cancer and heart disease in recent years,
placing a huge psychological and financial burden on many families
(3).
Buyang Huanwu decoction (BYHWD) has been used to
promote blood circulation and was proposed by Wang Qingren in the
‘Correction on Errors in Medical Classic’. It is composed of Radix
Astragali, the dried roots of Astragalus membranaceus Bge;
Angelicae Sinensis Radix, the dried roots of Angelica
sinensis Diels; Paeoniae Radix Rubra, the dried roots of
Paeonia lactiflora Pall; Chuanxiong Rhizoma, the dried
rhizomes of Ligusticum chuanxiong Hort; Persicae Semen, the
dried seeds of Prunus persica Batsch; Carthami Flos, the
dried flowers of Carthamus tinctorius L; and Pheretima, the
dried bodies of Pheretima aspergillum (E.Perrier), in the
ratio of 120:6:4.5:3:3:3:3. This formula has been widely used in
Chinese clinical practice for the treatment and prevention of
ischemic cardio-cerebral vascular diseases (4). Previous studies have demonstrated that
BYHWD is efficacious in treating ischemic stroke and other vascular
diseases (5). However, the exact
mechanism of BYHWD in improving ischemic stroke and its active
components remain unclear. As such, there is a great interest in
identifying the active compounds and molecular targets in BYHWD
that act on ischemic stroke. The present study aimed to identify
the potential active constituents of BYHWD and explore its
mechanism of action in the amelioration of ischemic stroke.
BYHWD is a multi-component, multi-channel and
multi-target agent that is a product under the guidance of the
holistic view and dialectical theory (6). It has unique advantages for the
treatment of complex diseases, but due to the complex composition
of TCM compounds, the modernization process can be slow (7). Network pharmacology is a novel method
that combines system network analysis and pharmacology that can be
used to clarify the synergistic effects and underlying mechanisms
of compound-compound, compound-target and target-disease networks
at the molecular level. This helps to elucidate the interactions
among compounds, genes, proteins and diseases (8,9). Due
to its robust and systematic nature, which is consistent with the
overall view of TCM and the principle of dialectical theory, it has
been widely applied in TCM research (10). Therefore, the current study used
network pharmacology to study the mechanism of BYHWD in the
treatment of ischemic stroke, providing a reference and theoretical
basis for its experimental research and clinical application.
Materials and methods
Chemical database collection of
BYHWD-compound information
Information on the compounds included in BYHWD was
collected from two phytochemical databases, the TCM systems
pharmacology database (TCMSP, http://ibts.hkbu.edu.hk/LSP/tcmsp.php) and the TCM
Database@Taiwan (http://tcm.cmu.edu.tw/). The bidimensional chemical
structures were acquired from NCBI PubChem (http://pubchem.ncbi.nlm.nih.gov/).
Pharmacokinetic ADME evaluation
OB represents the ability of a compound to circulate
in the body after oral administration. OB can indicate whether the
active compounds in a formula can be delivered throughout the body
and produce a physicochemical effect (11). DL is an indicator for determining
the similarity or likeness of a compound and its physicochemical
properties with conventional drugs. DL can help determine if a
certain compound has a therapeutic effect (11). All compounds were selected using the
in silico integrative ADME model provided by the TCMSP
database. Chemicals without ADME information were removed from the
final list. Compounds were only retained if OB ≥30 and DL ≥0.18 to
satisfy the criteria suggested by the TCMSP database (12).
Target genes related to the identified
compounds
The TCMSP database is a unique pharmacological
database of traditional Chinese medicines describing the
relationships between drugs, targets and diseases. The name of the
identified compound was input into the TCMSP database version 2.3
(https://tcmspw.com/tcmsp.php) to obtain
its target name, after which the target name of each compound was
input into the UniProt (http://www.uniprot.org/) database to convert to the
target gene and confine the species to Homo sapiens. Subsequently,
the various IDs of the targets were transformed into UniProt IDs
(13).
Collection of the cerebral ischemia
target genes
Therapeutic Target Database (TTD) (https://db.idrblab.org/ttd/) explores therapeutic
protein and nucleic acid targets within a targeted disease
(14). DisGeNET (http://www.disgenet.org/web/DisGeNET/menu/home) is a
discovery platform containing one of the largest publicly available
collections of genes and variants associated with human diseases
(15). By searching the key word
‘cerebral ischemia’, the targets related to this disease were
collected from each database.
Construction of the networks and
pathway analyses
The constructed herbal-chemical-protein networks
were visualized using Cytoscape version 3.6.1 (http://www.cytoscape.org/). The relationship between
active ingredients and common targets of component diseases in
BYHED were summarized into a table and imported into Cytoscape
software to obtain a network of drug ingredient actions and disease
targets. The nodes in the networks represent herbs and chemicals,
and the edges indicated interactions between herbs and chemicals,
and between chemicals and target genes (16). The common targets of components and
diseases were imported into the ClueGo plug-in for Cytoscape
software. The main functional annotation clusters were ranked using
the Biocarta functional annotation cluster tool (http://amp.pharm.mssm.edu/Harmonizome/dataset/Biocarta+Pathways;
updated on July 3, 2016) (17). The
functional pathways of BYHWD associated with cerebral ischemia were
analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analysis (http://www.genome.jp/kegg/pathway.html). Gene Ontology
(GO) enrichment evaluation was determined using the DAVID database
version 6.8 (https://david.ncifcrf.gov/). Data with P<0.05 were
screened from the enrichment analysis results.
Protein-protein interaction (PPI)
analysis
Screened drug component-disease common targets were
imported into the STRING (https://string-db.org/) database to build the PPI
network model. The protein category was set as Homo sapiens. The
minimum interaction threshold was set as the medium highest
confidence (>0.9) and the PPI network was obtained using the
default setting of the other parameters. In the network, the size
of the nodes represented the degree size. The higher the degree,
the better the correlation between the protein and therapeutic
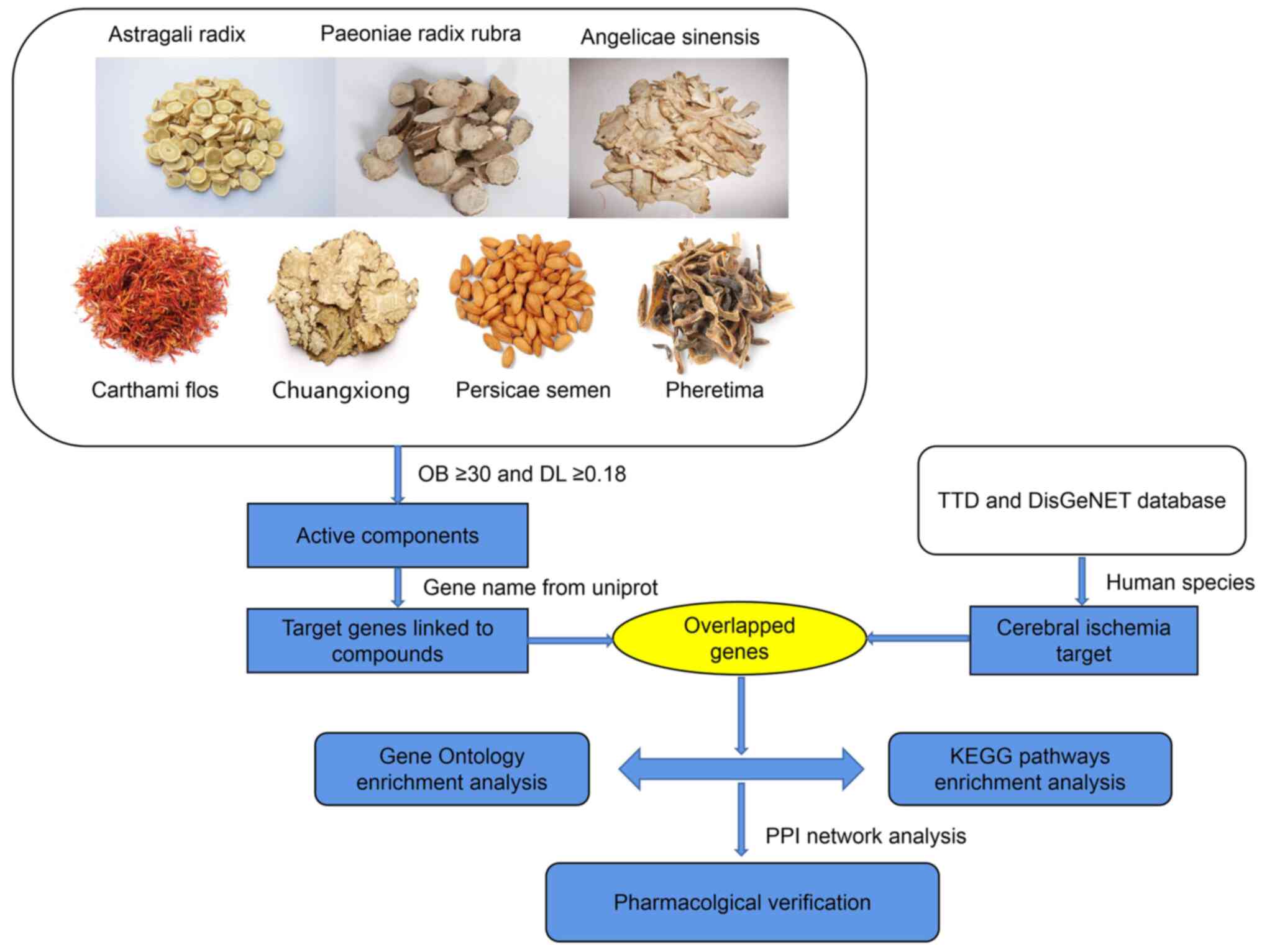
mechanism (18). Fig. 1 illustrated network pharmacology
analysis workflow.
Materials for pharmacological
experimental verification
BYHWD is a granule made of Astragali Radix,
Angelicae Sinensis Radix, Paeoniae Radix Rubra, Chuanxiong Rhizoma,
Persicae Semen, Carthami Flos and Pheretima according to the ratio
120:6:4.5:3:3:3:3. BYHWD was obtained from Guangdong Yifang
Pharmaceutical Co., Ltd. _batch no. 7070352). Rat brain
microvascular endothelial cells (BMECs) were purchased from the
Cell Biologics, Inc. (cat. no. C57-6023). Primary antibodies
against β-actin (cat. no. ab227387), VEGFA (cat. no. ab46154),
hypoxia-inducible-factor-1α (HIF-1α) (cat. no. ab179483) and IL6
(cat. no. ab9324) and secondary antibodies [HRP-conjugated goat
anti-rabbit (cat. no. ab7090) and HRP-conjugated goat anti-mouse
(cat. no. ab97040)] were purchased from Abcam.
Cell culture and treatments
BMECs were divided into three groups: Control,
oxygen-glucose deprivation (OGD) and OGD group treated with BYHWD
(80 µg/ml) groups. OGD was established as follows: Cells were
rinsed once with glucose-free DMEM (Gibco; Thermo Fisher
Scientific, Inc.) (19) and
transferred to an anaerobic chamber (Thermo Fisher Scientific,
Inc.) containing a gas mixture composed of 7% CO2 and
93% N2 for 6 h at 37˚C. The cells were then returned to
the normal culture conditions (37˚C). Control BMECs were cultured
in complete DMEM under normal conditions (37˚C) and did not receive
BYHWD treatment. BYHWD was dissolved in complete DMEM and treated
with a 0.22 µm membrane filter, after which BYHWD group cells were
treated (37˚C) with 80 µg/ml BYHWD for 12 h.
Western blotting
BMECs were lysed with cold radioimmunoprecipitation
assay buffer (Sigma-Aldrich; Merck KGaA) for 30 min and
protein concentrations were analyzed by the BCA method. Whole-cell
lysates (30 µg) were fractionated by 10% SDS-PAGE and transferred
onto polyvinylidene fluoride membranes (EMD Millipore). The
membranes were blocked with 5% bovine serum albumin (Beijing
Solarbio Science & Technology Co., Ltd.) at room temperature
for 30 min and incubated overnight at 4˚C with primary antibodies
against VEGFA (1:1,000; anti-rabbit), HIF-1α (1:1,000;
anti-rabbit), IL6 (1:1,000; anti-mouse), β-actin (1:1,000;
anti-rabbit). The membranes were subsequently incubated with a
secondary antibody (1:10,000; goat anti-rabbit or goat anti-mouse
antibody) at 37˚C for 1 h. The blots were visualized using an
ECL-Plus reagent (Santa Cruz Biotechnology, Inc.) and analyzed
using Quantity One System image analysis software version 4.6.2
(Bio-Rad Laboratories, Inc.).
Statistical analysis
All data in the present study were expressed as the
mean ± SD. One-way ANOVA followed by a Tukey's post hoc test was
used for analyzing differences between groups. P<0.05 was
indicated as the statistical significance.
Results
Compounds in the herbal medicines
BYHWD consists of seven herbal medicines, namely
Astragali Radix, Angelicae Sinensis Radix, Paeoniae Radix Rubra,
Chuanxiong Rhizoma, Persicae Semen, Carthami Flos and Pheretima.
All identified compounds were subjected to ADME screening. Of the
780 compounds assessed, 102 had an OB ≥30% and DL ≥0.18, which met
the suggested drug screening criteria. Of these, 21, 23, 4, 29, 9,
23 and 5 were Astragali Radix, Carthami Flos, Angelicae Sinensis
Radix, Paeoniae Radix Rubra, Chuanxiong Rhizoma, Persicae Semen and
Pheretima, respectively. There were 6 compounds that did not meet
the aforementioned requirements and were therefore considered to be
bioactive compounds: Hydroxysafflor yellow A, astragaloside IV,
ferulic acid, ligustrazine, Z-ligustilide and linoleic acid. These
compounds are the major components of BYHWD and their effects on
cerebral ischemia have been investigated previously (20-25).
Duplicate components and those with ambiguous targets were removed.
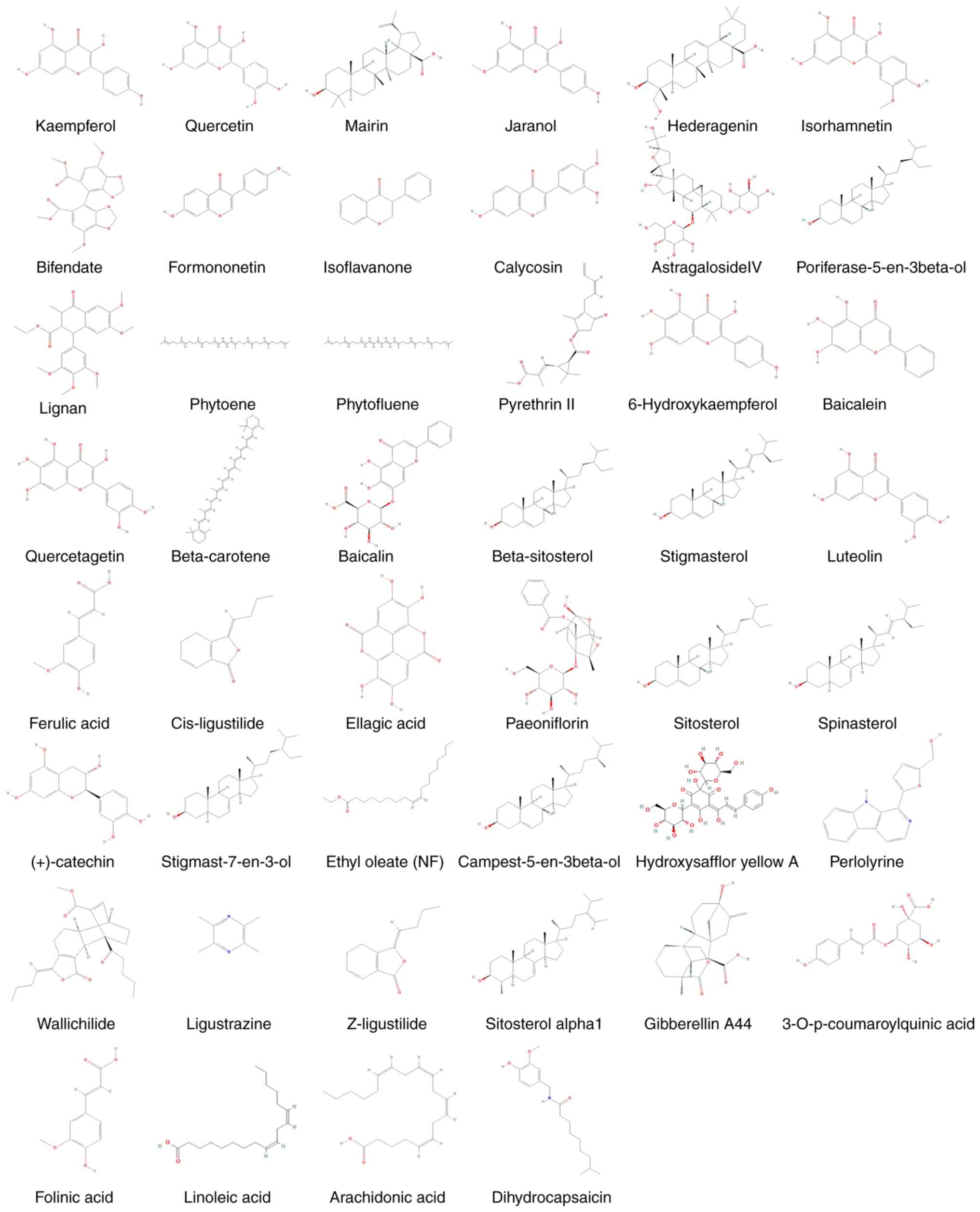
The compound information of BYHWD is presented in Table I, and the composition of each
structure is presented in Fig.
2.
 | Table IFinal selected compounds among the
seven herbal medicines of Buyang Huanwu Decoction. |
Table I
Final selected compounds among the
seven herbal medicines of Buyang Huanwu Decoction.
| No. | Compound | OB (%) | DL | Herbal
medicine |
|---|
| 1 | Kaempferol | 41.88 | 0.24 | Astragali Radix,
Carthami Flos |
| 2 | Quercetin | 46.43 | 0.28 | Astragali
Radix |
| 3 | Mairin | 55.38 | 0.78 | Astragali
Radix |
| 4 | Jaranol | 50.83 | 0.29 | Astragali
Radix |
| 5 | Hederagenin | 36.91 | 0.75 | Astragali Radix,
Persicae Semen |
| 6 | Isorhamnetin | 49.60 | 0.31 | Astragali
Radix |
| 7 | Bifendate | 31.10 | 0.67 | Astragali
Radix |
| 8 | Formononetin | 69.67 | 0.21 | Astragali
Radix |
| 9 | Isoflavanone | 109.99 | 0.30 | Astragali
Radix |
| 10 | Calycosin | 47.75 | 0.24 | Astragali
Radix |
| 11 |
AstragalosideIV | 17.74 | 0.15 | Astragali Radix,
Persicae Semen |
| 12 |
Poriferast-5-en-3beta-ol | 36.91 | 0.75 | Carthami Flos |
| 13 | Lignan | 43.32 | 0.65 | Carthami Flos |
| 14 | Phytoene | 39.56 | 0.50 | Carthami Flos |
| 15 | Phytofluene | 43.18 | 0.50 | Carthami Flos |
| 16 | Pyrethrin II | 48.36 | 0.35 | Carthami Flos |
| 17 |
6-Hydroxykaempferol | 62.13 | 0.27 | Carthami Flos |
| 18 | Baicalein | 33.52 | 0.21 | Carthami Flos,
Paeoniae Radix Rubra |
| 19 | Quercetagetin | 45.01 | 0.31 | Carthami Flos,
Paeoniae Radix Rubra |
| 20 | Beta-carotene | 37.18 | 0.58 | Carthami Flos |
| 21 | Baicalin | 40.12 | 0.75 | Carthami
Flos,Paeoniae Radix Rubra |
| 22 |
Beta-sitosterol | 36.91 | 0.75 | Carthami
Flos,Paeoniae Radix Rubra, Angelicae sinensis Radix |
| 23 | Stigmasterol | 43.83 | 0.76 | Carthami Flos,
Paeoniae Radix Rubra, Angelicae sinensis Radix |
| 24 | Luteolin | 36.16 | 0.25 | Carthami Flos |
| 25 | Ferulic acid | 39.56 | 0.06 | Angelicae sinensis
Radix |
| 26 |
Cis-ligustilide | 36.91 | 0.75 | Angelicae sinensis
Radix |
| 27 | Ellagic acid | 43.06 | 0.43 | Paeoniae Radix
Rubra |
| 28 | Paeoniflorin | 53.87 | 13.88 | Paeoniae Radix
Rubra |
| 29 | Sitosterol | 36.91 | 0.75 | Paeoniae Radix
Rubra, Chuangxiong Rhizoma |
| 30 | Spinasterol | 42.98 | 0.76 | Paeoniae Radix
Rubra |
| 31 | (+)-catechin | 54.83 | 0.24 | Paeoniae Radix
Rubra |
| 32 |
Stigmast-7-en-3-ol | 37.42 | 0.75 | Paeoniae Radix
Rubra |
| 33 | Ethyl oleate
(NF) | 32.40 | 0.19 | Paeoniae Radix
Rubra |
| 34 |
Campest-5-en-3beta-ol | 37.58 | 0.71 | Paeoniae Radix
Rubra |
| 35 | Hydroxysafflor
yellow A | 4.77 | 0.68 | Carthami Flos |
| 36 | Perlolyrine | 65.95 | 0.27 | Chuangxiong
Rhizoma |
| 37 | Wallichilide | 42.31 | 0.71 | Chuangxiong
Rhizoma |
| 38 | Ligustrazine | 20.01 | 0.03 | Chuangxiong
Rhizoma |
| 39 | Z-ligustilide | 53.72 | 0.07 | Chuangxiong
Rhizoma |
| 40 | Sitosterol
alpha1 | 36.91 | 0.75 | Persicae Semen |
| 41 | Gibberellin
A44 | 101.61 | 0.54 | Persicae Semen |
| 42 |
3-O-p-coumaroylquinic acid | 37.63 | 0.29 | Persicae Semen |
| 43 | Folinic acid | 68.96 | 0.71 | Persicae Semen |
| 44 | Linoleic acid | 41.90 | 0.14 | Pheretima |
| 45 | Arachidonic
acid | 45.57 | 0.20 | Pheretima |
| 46 |
Dihydrocapsaicin | 47.07 | 0.19 | Pheretima |
Combination of the compound-target and
cerebral ischemia target genes
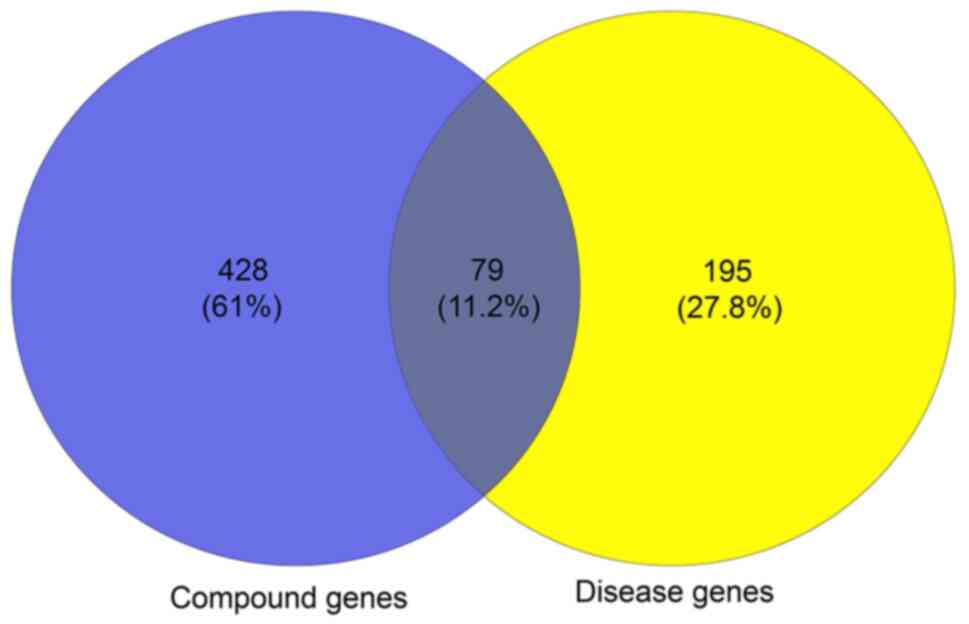
A total of 507 genes and compounds related to BYHWD
were obtained from the TCMSP database (Table SI). A total of 274 genes associated
with cerebral ischemia were retrieved from the TTD and DisGeNET
version 6.0 databases (Table SII).
A total of 79 overlapping genes were pinpointed by matching the 518
compound genes with the disease-associated genes (Fig. 3). Therefore, these target genes were
used as prediction targets of BYHWD in the treatment of stroke.
Potential target genes and network
analysis
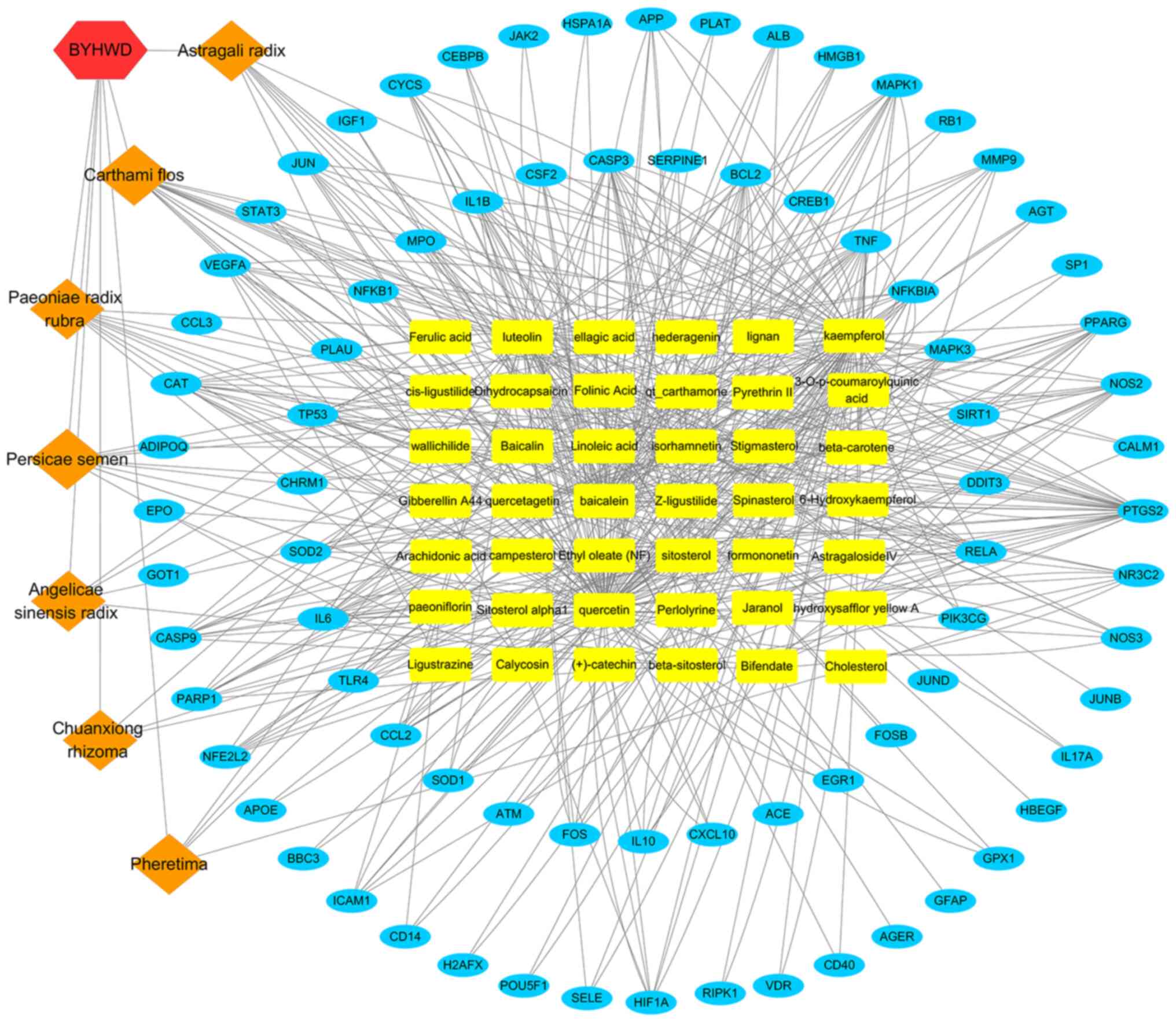
The constructed herbal-chemical-protein networks
were visualized using Cytoscape, which included 42 compounds, with
79 target genes, 129 nodes and 490 edges (Fig. 4). In particular, 16 of the compounds
(baicalein, beta-carotene, baicalin, kaempferol, luteolin,
quercetin, hydroxysafflor yellow A, isorhamnetin, bifendate,
formononetin, calycosin, astragaloside IV, stigmasterol,
sitosterol, Z-ligustilide and dihydrocapsaicin) were associated
with >5 genes. Additionally, 33 genes (PTGS2, MMP9, NOS2, MAPK1,
MAPK3, TNF, EGFR, APP, JUN, TP53, IL6, IL2, STAT3, NOS3, IL10,
CYCS, VEGFA, HIF-1α, BCL2, CASP9, CAT, PARP1, CASP3, CCL2, MPO,
NFE2L2, CSF2, RELA, ALB, NFKBIA, PPARG, NR3C2 and ICAM1) were
regulated by >4 compounds. The compound-target gene network
demonstrated intimate communication between several components and
multiple targets. This is helpful to better understand the
potential pharmacodynamic substances and targets of BYHWD in the
treatment of cerebral ischemia.
GO and KEGG enrichment analysis of
potential target genes
GO enrichment and KEGG pathway enrichment analyses
of the 79 potential targets were performed to determine the
underlying molecular mechanism of BYHWD in cerebral ischemia. GO
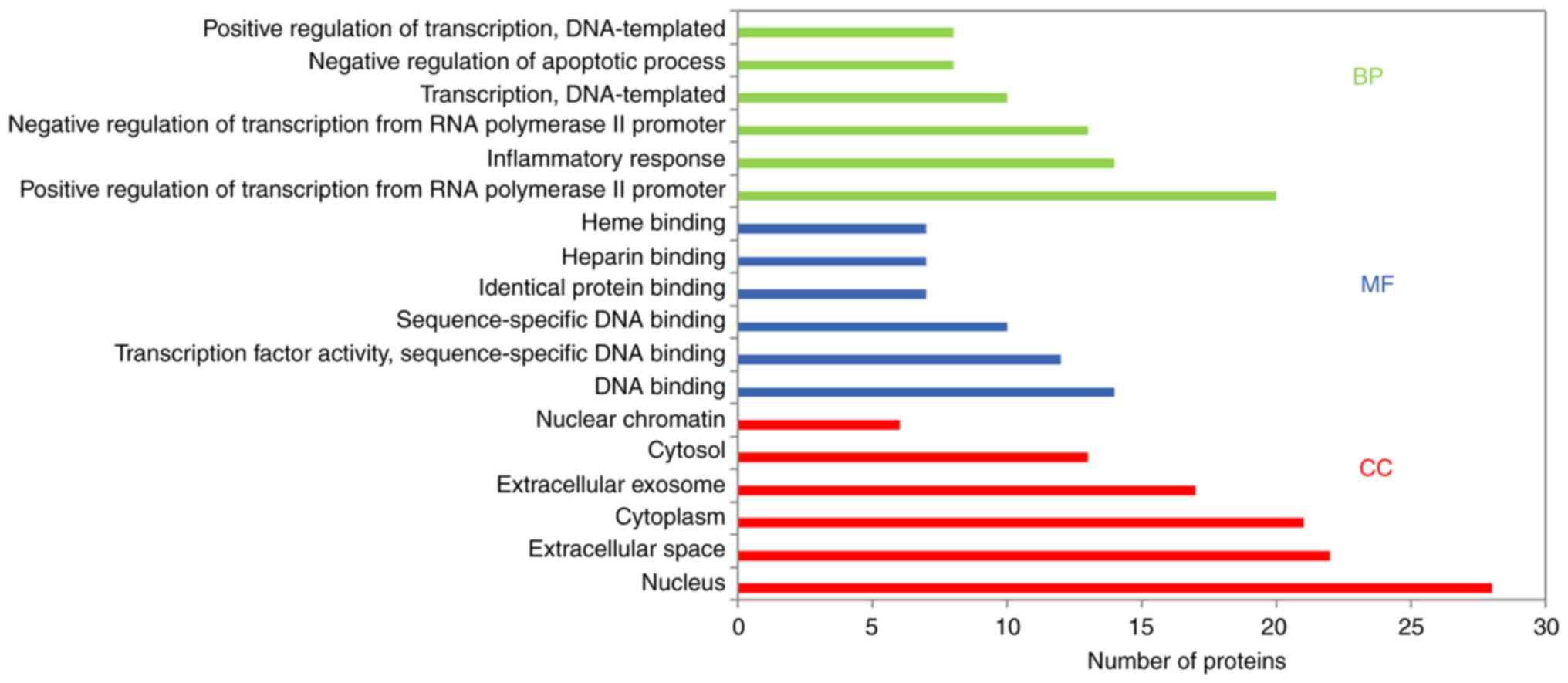
enrichment analysis provided biological process (BP), molecular
function (MF) and cellular component (CC) results. Using the DAVID
database (with P<0.05), the results revealed that the main BPs
were ‘positive regulation of transcription from RNA polymerase II
promoter’, ‘inflammatory response’, ‘negative regulation of
apoptosis’, ‘angiogenesis’, ‘positive regulation of NF-kB
transcription factor activity’, ‘apoptosis process’, ‘immune
response’ and ‘response to oxidative stress’. The MFs were mainly
associated with ‘DNA binding’, ‘transcription factor activity’,
‘heparin-binding’, ‘heme-binding’, sequence-specific DNA binding’
and ‘identical protein binding’. CC analysis revealed that there
was a higher proportion of protein in the nucleus and extracellular
space. Furthermore, GO analysis revealed the top six enriched
conditions in the BP, CC and MF categories (Fig. 5). Furthermore, the 20 BPs are listed
in Table II. To examine the
signaling pathways and functions of these target genes, KEGG
pathway functional enrichment analysis was performed. The signaling
pathways were obtained by screening their statistical significance
(P<0.05), and the resulting target genes were found to primarily
interact with the TNF, IL17, PI3K-Akt, toll-like receptor (TLR),
MAPK, NF-κB and HIF-1 signaling pathways. Therefore, these
signaling pathways appear to be closely associated with the
potential effects of BYHWD in cerebral ischemia. The identified
target genes are listed in Table
III. A KEGG bubble diagram was created using the top 20 signals
(Fig. 6A). Additionally, the main
functional annotation clusters were ranked by the Biocarta
functional annotation cluster tool and were presented in Fig. 6B.
 | Table IIBiological processes of potential
target genes based on GO enrichment analysis. |
Table II
Biological processes of potential
target genes based on GO enrichment analysis.
| GO ID | Biological
process | Genes | P-value |
|---|
| GO:0045944 | Positive regulation
of transcription from RNA polymerase II promoter | HMGB1, TNF, RELA,
PPARG, TP53, IGF1, NFKBIA, RB1, CD40, SIRT1, IL10, VDR, IL17A, APP,
HIF-1α, SP1, POU5F1, RIPK1, JUN and MAPK3 |
6.60x10-12 |
| GO:0006954 | Inflammatory
response | PIK3CG, HMGB1,
CCL2, PTGS2, RELA, TLR4, CD40, AGER, IL10, CXCL10, IL17A, JAK2,
NFE2L2 and CD14 |
7.35x10-11 |
| GO:0000122 | Negative regulation
of transcription from RNA polymerase II promoter | EGR1, VDR, TNF,
POU5F1, RELA, JUN, PPARG, TP53, NFKB1, RB1, SIRT1, DDIT3 and
EPO |
3.56x10-7 |
| GO:0006351 | Transcription,
DNA-templated | EGR1, VDR, HIF-1α,
CEBPB, JUN, MAPK3, PPARG, TP53 and STAT3 |
5.98x10-5 |
| GO:0043066 | Negative regulation
of apoptotic process | CASP3, ALB, BCL2,
TP53, IGF1, CAT, SIRT1 and STAT3 |
1.78x10-4 |
| GO:0045893 | Positive regulation
of transcription, DNA-templated | CEBPB, JUN, PPARG,
NFKB1, NFE2L2, STAT3 and EPO |
1.78x10-4 |
| GO:0031663 |
Lipopolysaccharide-mediated signaling
pathway | MAPK1, CCL2, TNF,
MAPK3, NFKBIA, NOS3 and TLR4 |
7.02x10-10 |
| GO:0001525 | Angiogenesis | PIK3CG, HIF-1α,
JUN, VEGFA, SERPINE, NOS3 and SIRT1 |
2.14x10-5 |
| GO:0042127 | Regulation of cell
proliferation | JUN, NFKBIA, JAK2,
NOS2, CD40, PLAU and CXCL10 |
8.12x10-5 |
| GO:0006357 | Regulation of
transcription from RNA polymerase II promoter | FOS, SP1, NFKB1,
RB1, NFE2L2, STAT3 and SOD2 |
1.22x10-3 |
| GO:0001666 | Response to
hypoxia | PLAT, VEGFA, NOS2,
AGER, PLAU and EPO |
1.96x10-5 |
| GO:0051092 | Positive regulation
of NF-kappaB transcription factor activity | TNF, RELA, RIPK1,
CAT, CD40 and AGER |
7.26x10-5 |
| GO:0006915 | Apoptotic
process | APP, RIPK1, CYCS,
TP53, RB1 and EPO |
2.98x10-4 |
| GO:0055114 | Oxidation-reduction
process | GPX1, NOS3, CAT,
NOS2, SOD1 and SOD2 |
1.43x10-3 |
| GO:0006955 | Immune
response | CSF2, IL6, JUN,
CD40, IL10 and CXCL10 |
6.95x10-3 |
| GO:0043491 | Protein kinase B
signaling | CSF2, CCL2, TNF,
IGF1 and CD40 |
1.21x10-5 |
| GO:0071356 | Cellular response
to lipopolysaccharide | JUN, SERPINE1,
NOS2, IL10 and CXCL10 |
1.82x10-4 |
| GO:0006979 | Response to
oxidative stress | GPX1, APP, PTGS2,
APOE and MPO |
2.91x10-4 |
| GO:0045766 | Positive regulation
of angiogenesis | HIF-1α, VEGFA,
SERPINE1, NOS3 and SIRT1 |
6.75x10-4 |
| GO:0043524 | Negative regulation
of neuron apoptotic process | CCL2, APOE, JUN,
BCL2 and SOD1 |
7.56x10-4 |
 | Table IIIPotential target gene function based
on Kyoto Encyclopedia of Genes and Genomes pathway analysis. |
Table III
Potential target gene function based
on Kyoto Encyclopedia of Genes and Genomes pathway analysis.
| KEGG ID | Pathway | Pathway genes | P-value |
|---|
| hsa05167 | Kaposi
sarcoma-associated herpesvirus infection |
PTGS2/CALM1/PIK3CG/RELA/RB1/MAPK1/MAPK3/
NFKBIA/CASP3/CSF2/CYCS/JUN/NFKB1/STAT3/TP53/
VEGFA/IL6/CASP9/ICAM1/FOS/HIF-1α /CREB1/JAK2 |
1.55x10-19 |
| hsa04668 | TNF signaling
pathway |
PTGS2/RELA/MMP9/MAPK1/MAPK3/NFKBIA/TNF/
CASP3/CEBPB/CSF2/IL1B/JUN/NFKB1/IL6/ICAM1/
CCL2/FOS/SELE/RIPK1/CXCL10/JUNB/CREB1 |
2.80x10-23 |
| hsa05163 | Human
cytomegalovirus infection |
PTGS2/CALM1/RELA/RB1/MAPK1/MAPK3/NFKBIA/
TNF/CASP3/CYCS/IL1B/NFKB1/STAT3/TP53/VEGFA/
IL6/CASP9/CCL2/RIPK1/SP1/CREB1/CCL3 |
1.83x10-16 |
| hsa04933 | AGE-RAGE signaling
pathway in diabetic complications |
RELA/MAPK1/MAPK3/TNF/BCL2/CASP3/IL1B/JUN/
NFKB1/STAT3/VEGFA/IL6/ICAM1/CCL2/SELE/
AGER/EGR1/NOS3/AGT/SERPINE1/JAK2 |
7.02x10-23 |
| hsa04657 | IL17 signaling
pathway |
PTGS2/RELA/MMP9/MAPK1/MAPK3/NFKBIA/TNF/
CASP3/CEBPB/CSF2/IL1B/JUN/NFKB1/IL6/CCL2/FOS/
CXCL10/FOSB/IL17A/JUND |
5.04x10-22 |
| hsa05161 | Hepatitis B |
RELA/MMP9/RB1/MAPK1/MAPK3/NFKBIA/TNF/BCL2/
CASP3/CYCS/JUN/NFKB1/STAT3/TP53/IL6/TLR4/CASP9/ FOS/CREB1/JAK2 |
6.08x10-17 |
| hsa05152 | Tuberculosis |
CALM1/NOS2/RELA/MAPK1/MAPK3/TNF/BCL2/CASP3/
CEBPB/CYCS/IL1B/NFKB1/IL6/TLR4/CASP9/CD14/VDR/ IL10/CREB1/JAK2 |
3.94x10-16 |
| hsa05145 | Toxoplasmosis |
NOS2/PIK3CG/RELA/MAPK1/MAPK3/NFKBIA/TNF/
BCL2/CASP3/CYCS/NFKB1/STAT3/TLR4/CASP9/IL10/ CD40/HSPA1A/JAK2 |
2.25x10-17 |
| hsa04210 | Apoptosis |
RELA/MAPK1/MAPK3/NFKBIA/TNF/BCL2/CASP3/
CYCS/JUN/NFKB1/TP53/CASP9/PARP1/BBC3/ATM/FOS/ RIPK1/DDIT3 |
6.85x10-16 |
| hsa05142 | Chagas disease
(American trypanosomiasis) |
NOS2/RELA/MAPK1/MAPK3/NFKBIA/TNF/IL1B/JUN/
NFKB1/IL6/TLR4/CCL2/FOS/IL10/ACE/ SERPINE1/CCL3 |
1.03x10-16 |
| hsa05418 | Fluid shear stress
and atherosclerosis |
CALM1/RELA/MMP9/TNF/BCL2/IL1B/JUN/NFKB1/TP53/
VEGFA/NFE2L2/ICAM1/CCL2/FOS/SELE/ NOS3/PLAT |
1.87x10-14 |
| hsa05202 | Transcriptional
misregulation in cancer |
PPARG/RELA/MMP9/CEBPB/CSF2/IGF1/MPO/NFKB1/
PLAU/TP53/IL6/ATM/CD14/CD40/SP1/DDIT3/PLAT |
2.41x10-12 |
| hsa05169 | Epstein-Barr virus
infection |
RELA/RB1/NFKBIA/TNF/BCL2/CASP3/CYCS/JUN/
NFKB1/STAT3/TP53/IL6/CASP9/ICAM1/RIPK1/CD40/ CXCL10 |
8.51x10-12 |
| hsa05166 | Human T-cell
leukemia virus 1 infection |
RELA/RB1/MAPK1/MAPK3/NFKBIA/TNF/CSF2/JUN/
NFKB1/TP53/IL6/ICAM1/ATM/FOS/CD40/EGR1/CREB1 |
3.38x10-11 |
| hsa04151 | PI3K-Akt signaling
pathway |
PIK3CG/RELA/MAPK1/MAPK3/BCL2/IGF1/NFKB1/TP53/
VEGFA/IL6/TLR4/CASP9/NOS3/CREB1/JAK2/EPO/ CHRM1 |
5.39x10-8 |
| hsa04620 | Toll-like receptor
signaling pathway |
RELA/MAPK1/MAPK3/NFKBIA/TNF/IL1B/JUN/NFKB1/
IL6/TLR4/CD14/FOS/RIPK1/CD40/CXCL10/CCL3 |
2.89x10-15 |
| hsa05162 | Measles |
RELA/NFKBIA/BCL2/CASP3/CYCS/IL1B/JUN/NFKB1/
STAT3/TP53/IL6/TLR4/CASP9/BBC3/FOS/HSPA1A |
2.81x10-13 |
| hsa05164 | Influenza A |
RELA/MAPK1/MAPK3/NFKBIA/TNF/CASP3/CYCS/IL1B/
NFKB1/IL6/TLR4/CASP9/ICAM1/CCL2/CXCL10/JAK2 |
5.60x10-12 |
| hsa05170 | Human
immunodeficiency virus 1 infection |
CALM1/RELA/MAPK1/MAPK3/NFKBIA/TNF/BCL2/
CASP3/CYCS/JUN/NFKB1/TLR4/CASP9/ATM/FOS/RIPK1 |
2.12x10-10 |
| hsa04010 | MAPK signaling
pathway |
RELA/MAPK1/MAPK3/TNF/CASP3/IGF1/IL1B/JUN/
NFKB1/TP53/VEGFA/CD14/FOS/JUND/DDIT3/HSPA1A |
2.60x10-8 |
| hsa05133 | Pertussis |
CALM1/NOS2/RELA/MAPK1/MAPK3/TNF/CASP3/IL1B/
JUN/NFKB1/IL6/TLR4/CD14/FOS/IL10 |
1.55x10-19 |
| hsa04064 | NF-kappa B
signaling pathway |
PTGS2/RELA/NFKBIA/TNF/BCL2/IL1B/NFKB1/PLAU/
TLR4/PARP1/ICAM1/ATM/CD14/RIPK1/CD40 |
2.80x10-23 |
| hsa04066 | HIF-1 signaling
pathway |
NOS2/RELA/MAPK1/MAPK3/BCL2/IGF1/NFKB1/STAT3/
VEGFA/IL6/TLR4/HIF-1α/NOS3/SERPINE1/EPO |
1.83x10-16 |
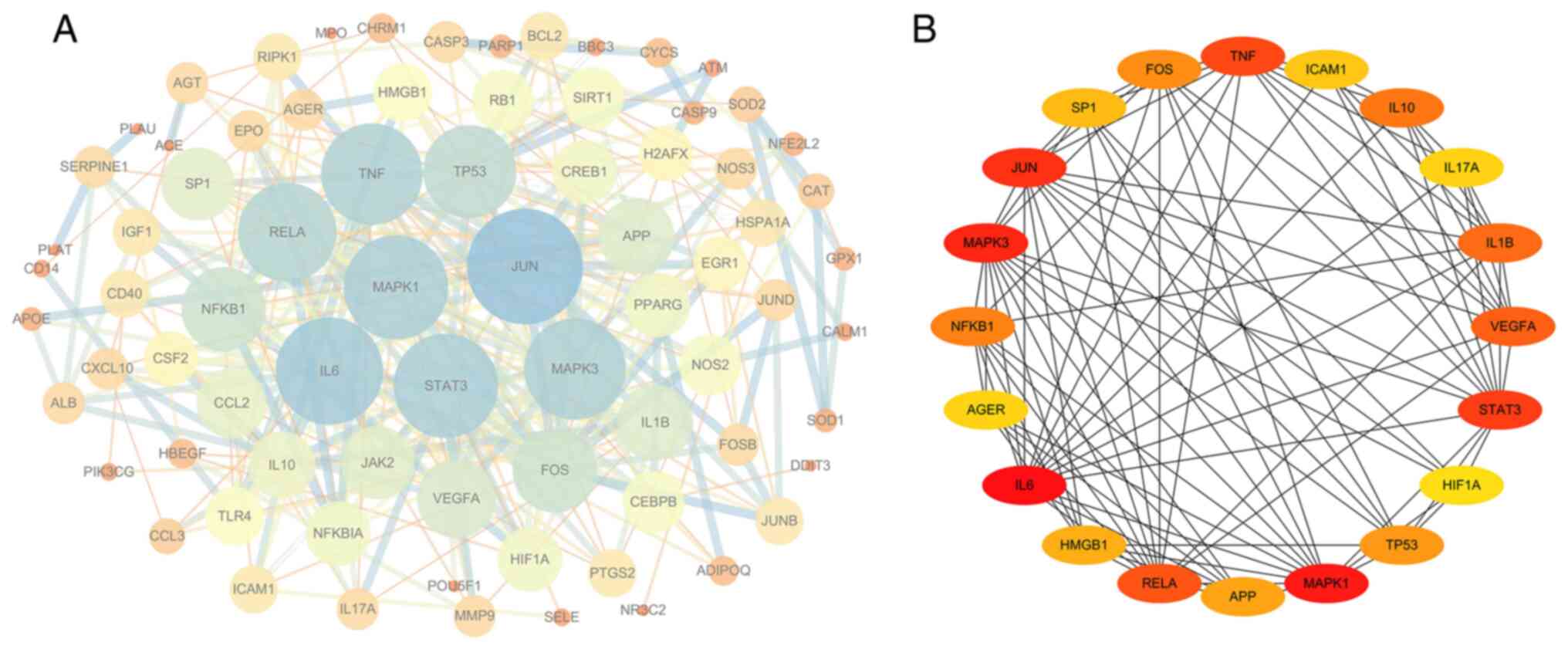
PPI network analysis of the target
genes
The drug component-disease common target was
imported into the STRING database to construct the PPI network.
When using an interaction score of >0.9 as a cut-off, the PPI
network contained 79 nodes and 327 edges, with an average node
degree of 8.28 and PPI enrichment P-value of
<1.0x10-16 (Fig. 7A).
Cytoscape version 3.6.1 was then used to analyze the degree, PPI
network topological eigenvalues, degree of node color, size
reaction center, edge thickness and color depth to determine a
combined score. The top 20 target genes with a high degree of
connectivity were selected as the hub genes of BYHWD for cerebral
ischemia (Fig. 7B). The core genes
in this network were IL6, TNF, VEGFA, HIF-1α, MAPK1, MAPK3, JUN,
STAT3, IL1B and IL10.
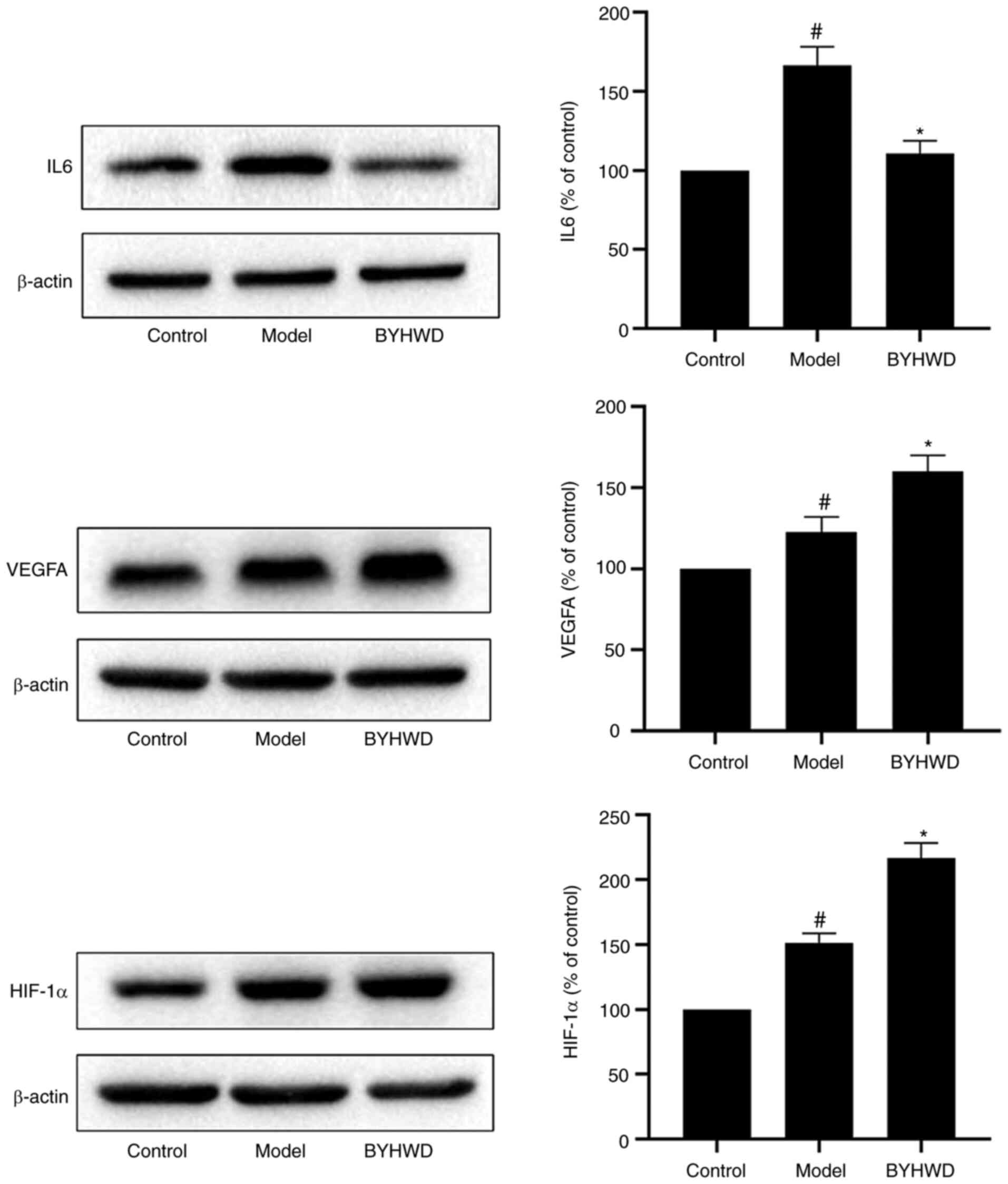
Experimental validation
To confirm the results of the network analysis and
verify the key targets of BYHWD, three key targets (IL6, VEGFA and
HIF-1α) were selected for pharmacological validation (Fig. 8). Western blotting results revealed
that, when compared with the control group, BYHWD significantly
inhibited the expression of IL6 and increased the expression of
HIF-1α and VEGFA (P<0.05).
Discussion
Ischemic stroke is one of the leading causes of
death and disability worldwide. Previous studies have indicated
that the injury mechanism of ischemic stroke includes apoptosis,
necrosis, inflammation, immune regulation and oxidative stress
(26). However, no effective
treatment has been found to prevent damage to the brain, except for
tissue plasminogen activator; however, this single compound or
single target drug has limited efficacy. Therefore, it is thought
that a promising treatment approach for ischemic stroke should
utilize multiple-component agents with multiple targets (27).
BYHWD activates blood circulation. It is a classic
prescription that has been used by doctors for many generations.
TCM cures disease using a multi-approach, multi-target and integer
concept. It has unique advantages for the treatment of complex
diseases, but due to the complex composition of TCM, its
application in modern medicine has been slow. Furthermore, network
pharmacology has provided a novel method of determining the
pharmacological mechanisms of TCMs. Scholars have attempted to
apply network pharmacology to evaluate the ingredients, targets and
mechanisms of herbal formulas (8).
In the present study, network pharmacology was
performed to investigate the pharmacological mechanisms of BYHWD in
relation to cerebral ischemia. The network pharmacological analysis
of BYHWD identified seven herbs, 42 compounds and 79 target
gene-regulated pathways associated with cerebral ischemia.
Additionally, 16 compounds (baicalein, beta-carotene, baicalin,
kaempferol, luteolin, quercetin, hydroxysafflor yellow A,
isorhamnetin, bifendate, formononetin, calycosin, astragaloside IV,
stigmasterol, sitosterol, Z-ligustilide and dihydrocapsaicin) were
associated with >5 genes. Previous studies have reported that
baicalein has potent neuroprotective properties under in
vitro and in vivo systems (28). Beta-carotene serves as an
antioxidant, inhibiting free radical production. It may also
regulate cell growth and death (29). Baicalin inhibits microglial cell
activation and reduces inflammation, oxidative damage and brain
edema (30). In addition,
kaempferol has strong anti-inflammatory and antioxidant effects.
Numerous scientific reports have revealed that it serves a
beneficial role in different inflammatory-related diseases, such as
cardiovascular and neurodegenerative diseases. Luteolin suppresses
inflammation in the brain tissue and regulates different cell
signaling pathways (31). Quercetin
has antioxidant stress and neuroprotective effects (32). Moreover, hydroxysafflor yellow A
protects BMECs against OGD/reoxygenation-induced injury by
inhibiting autophagy via the Class IPI3K/Akt/mTOR signaling pathway
(33). Treatment of experimental
stroke mice with isorhamnetin attenuated cerebral edema, improved
blood-brain barrier function and upregulated the gene expression of
certain tight junction proteins, including occludin, zonula
occluden-1 and claudin-5(34).
Furthermore, calycosin protected the rat brain against ischemic
injury by inhibiting calpain activation (35). Dihydrocapsaicin-treated cerebral
ischemia-reperfusion rats demonstrated attenuated cerebral and
blood-brain barrier damage by inhibiting oxidative stress and
inflammatory pathways (36). These
findings suggest that the main components of BYHWD are effective
for treating cerebral ischemia. However, as the compounds in the
database may be incomplete, the predicted active ingredients of
BYHWD may also be incomplete, which is a limitation of network
pharmacology.
Genes with high degrees of differential articulation
were acquired as a result of the PPI system analysis of the current
study. Of these, IL6, TNF, VEGFA, HIF-1α, MAPK1, MAPK3, JUN, STAT3,
IL1B and IL10 were recognized as the center genes. IL6 is a
multifunctional cytokine with a wide range of biological
activities, including regulation of the immune system and
generation of acute phase reactions (37). VEGF is a pleiotropic growth factor
that is crucially involved in neurovascular remodeling in the
ischemic brain. VEGF promotes angiogenesis, protects ischemic
neurons from injury, has potent anti-inflammatory actions and
promotes brain plasticity (38).
HIF-1α regulates the expression of gene encoding molecules that
participate in erythropoiesis, cell proliferation and energy
metabolism, and is closely associated with the regulation of
neuronal survival in ischemia (39,40).
HIF-1α can upregulate the expression of proteins associated with
the vascular system and can promote the angiogenesis of VEGF and
its receptors to increase blood flow and reduce ischemic injury
(41). TNF is a typical cytokine
involved in the acute phase of systemic inflammation and is closely
associated with the severity of cerebral ischemia (42). IL10 is a potent anti-inflammatory
mediator and, if overexpressed, can suppress neuronal degeneration
(43).
GO and KEGG pathway analyses were used to further
understand the interaction and action pathways of target genes. GO
analysis revealed that target genes were strongly associated with
the following BPs: ‘positive regulation of transcription including
ribonucleic acid (RNA) polymerase II promoter’, ‘inflammatory
response’, ‘transcription’, ‘DNA-templated’, ‘negative regulation
of the apoptotic process’, ‘positive regulation of transcription’,
‘angiogenesis’ and ‘response to hypoxia’. The enriched MF
ontologies were ‘DNA binding transcription factor activity’,
‘sequence-specific DNA binding’, ‘identical protein binding’,
‘heparin-binding’ and ‘heme-binding’. The KEGG pathway analysis
primarily pertained to TNF, IL17, apoptosis, PI3K-Akt, TLR, MAPK,
NF-κB and the HIF-1 signaling pathway. The results indicated that
these pathways may interact to exert their combined effects against
cerebral ischemia, which could explain the apparent effects of
BYHWD.
Cerebral ischemia results in decreased cerebral
blood flow and decreased oxygen supply, which leads to HIF-1
signaling pathway activation and upregulated HIF-1α expression.
This helps to recover blood circulation in the penumbra after
cerebral ischemia, transport glucose and mediate hypoxia adaptation
after hypoxia, serving a protective role in promoting cell survival
and inhibiting brain tissue apoptosis (44). The MAPK and PI3K/Akt signaling
pathways are the main pathways related to apoptosis after cerebral
ischemia. Currently, it is hypothesized that MAPK signaling serves
a dual role in the process of cell apoptosis, while PI3K/Akt
signaling is important to the cell survival signaling pathway
(45). Multiple neurotrophic
factors inhibit apoptosis by activating the PI3K/Akt signaling
pathway, thus playing a protective role in the brain (45). The PI3K/Akt signaling pathway is
involved in the regulation of various intracellular signaling
pathways and serves a key role in promoting cell survival and
proliferation, anti-apoptosis, regulating glucose metabolism and
protein synthesis (46). TLRs, as
inflammatory signal receptors, serve an important role in the
inflammatory cascade reaction triggered by cerebral ischemia and
are closely related to the expression of various inflammatory
mediators (47). Therefore, it is
of great significance to intervene in the TLR signaling pathway
during the initial stage of the inflammatory response to
effectively reduce inflammatory injury in the acute stage of
ischemic stroke (47).
The current study verified three key targets (IL6,
VEGFA and HIF-1α) using western blotting, which were predicted in
the network. Therefore, the pharmacological mechanism of BYHWD in
the treatment of ischemic stroke can be more clearly verified. The
results revealed that when compared with the control group, BYHWD
significantly inhibited the expression of IL6 and increased the
expression of HIF-1α and VEGFA.
In conclusion, the network pharmacology analysis of
BYHWD identified seven herbs, 42 compounds and 79 target genes that
were associated with ischemic stroke. The current study did not
validate the identified active components, which was a limitation
of the study. As the results of network pharmacology analysis
revealed that baicalein, beta-carotene, baicalin, kaempferol,
luteolin, quercetin, hydroxysafflor yellow A, isorhamnetin,
bifendate, formononetin, calycosin, astragaloside IV, stigmasterol,
sitosterol, Z-ligustilide and dihydrocapsaicin were the main
effective components of BYHWD in the treatment of ischemic stroke,
subsequent studies should first verify these results. Based on the
pathway enrichment results of the present study, it was determined
that the effects of BYHWD against ischemic stroke may be due to
some of the ingredients that can simultaneously target multiple
pathways, such as the TNF, IL17, apoptosis, PI3K-Akt, TLR, MAPK,
NF-κB and HIF-1 signaling pathways. Furthermore, genes with high
degrees of differential articulation were identified from PPI
system analysis. IL6, TNF, VEGFA, HIF-1α, MAPK1, MAPK3, JUN, STAT3,
IL1B and IL10 were recognized as center genes. The results
indicated that compound-target gene networks can reveal close
interactions between multi-components and multi-targets, enhancing
understanding of the potential effects of BYHWD in ischemic
stroke.
Supplementary Material
Target genes and compounds associated
with Buyang Huanwu Decoction.
Target genes associated with cerebral
ischemia.
Acknowledgements
Not applicable.
Funding
Funding: This research was funded by the National Natural
Science Foundation of China (grant nos. 81503280 and 81573549).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KW, LL, JC and YQ designed the experiments. KW and
LL conducted the experiments and researched the literature. KW, JC,
YQ and RL collected and analyzed the data. KW wrote the manuscript.
KW, JC and ZY revised the manuscript. JD, ZF, YD and YM interpreted
the data. ZY and EZ obtained funding and designed the study. KW and
LL confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Del Sette M, Chiti A and Dinia L:
Intraarterial treatment for acute ischemic stroke. N Engl J Med.
372(1177)2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang P, Shao BZ, Deng Z, Chen S, Yue Z and
Miao CY: Autophagy in ischemic stroke. Prog Neurobiol.
163-164:98–117. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Writing Group Members. Mozaffarian D,
Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de
Ferranti S, Després JP, et al: Heart disease and stroke
statistics-2016 update: A report From the American Heart
Association. Circulation. 133:e38–e360. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hung IL, Hung YC, Wang LY, Hsu SF, Chen
HJ, Tseng YJ, Kuo CE, Hu WL and Li TC: Chinese herbal products for
ischemic stroke. Am J Chin Med. 43:1365–1379. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Li JH, Liu AJ, Li HQ, Wang Y, Shang HC and
Zheng GQ: Buyang huanwu decoction for healthcare: Evidence-based
theoretical interpretations of treating different diseases with the
same method and target of vascularity. Evid Based Complement
Alternat Med. 2014(506783)2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Huang J, Guo W, Cheung F, Tan HY, Wang N
and Feng Y: Integrating network pharmacology and experimental
models to investigate the efficacy of Coptidis and
Scutellaria containing Huanglian Jiedu decoction on
hepatocellular carcinoma. Am J Chin Med. 48:161–182.
2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhang J, Liu X, Wu J, Zhou W, Tian J, Guo
S, Jia SS, Meng Z and Ni M: A bioinformatics investigation into the
pharmacological mechanisms of the effect of the Yinchenhao
decoction on hepatitis C based on network pharmacology. BMC
Complement Med Ther. 20(50)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lee AY, Park W, Kang TW, Cha MH and Chun
JM: Network pharmacology-based prediction of active compounds and
molecular targets in Yijin-tang acting on hyperlipidaemia and
atherosclerosis. J Ethnopharmacol. 221:151–159. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li XK, Yang HJ, Xiao JC, Zhang J, Zhang J,
Liu M, Zheng Y and Ma L: Network pharmacology based investigation
into the bioactive compounds and molecular mechanisms of
Schisandrae chinensis Fructus against drug-induced liver
injury. Bioorg Chem. 96(103553)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Xu T, Li S, Sun Y, Pi Z, Liu S, Song F and
Liu Z: Systematically characterize the absorbed effective
substances of Wutou Decoction and their metabolic pathways in rat
plasma using UHPLC-Q-TOF-MS combined with a target network
pharmacological analysis. J Pharm Biomed Anal. 141:95–107.
2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kim SK and Lee S, Lee MK and Lee S: A
systems pharmacology approach to investigate the mechanism of
Oryeong-san formula for the treatment of hypertension. J
Ethnopharmacol. 244(112129)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang W, Liu T, Yang L, Ma Y, Dou F, Shi L,
Wen A and Ding Y: Study on the multi-targets mechanism of
Triphala on cardio-cerebral vascular diseases based on
network pharmacology. Biomed Pharmacother.
116(108994)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang J, Liang R, Wang L and Yang B:
Effects and mechanisms of Danshen-Shanzha herb-pair for
atherosclerosis treatment using network pharmacology and
experimental pharmacology. J Ethnopharmacol. 229:104–114.
2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xu T, Ma C, Fan S, Deng N, Lian Y, Tan L,
Du W, Zhang S, Liu S, Ren B, et al: Systematic understanding of the
mechanism of baicalin against Ischemic stroke through a network
pharmacology approach. Evid Based Complement Alternat Med.
2018(2582843)2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Piñero J, Bravo À, Queralt-Rosinach N,
Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz
F and Furlong LI: DisGeNET: A comprehensive platform integrating
information on human disease-associated genes and variants. Nucleic
Acids Res. 45:D833–D839. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhang J, Yang B, Wang L and Liang R:
Effects and mechanisms of Danshen-Shanzha herb-pair for
atherosclerosis treatment using network pharmacology and
experimental pharmacology. J Ethnopharmacol. 229:104–114.
2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Rouillard A, Gundersen G, Fernandez N,
Wang Z, Monteiro CD, McDermott MG and Ma'ayan A: The harmonizome: A
collection of processed datasets gathered to serve and mine
knowledge about genes and proteins. Database (Oxford).
2016(baw100)2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res. 41
(Database issue):D808–D815. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Qiao P, Yan H and Wang J: EGb761 protects
brain microvascular Endothelial cells against oxygen-glucose
deprivation-induced injury through lncRNA Rmst/miR-150 axis.
Neurochem Res. 45:2398–2408. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sun Y, Xu DP, Qin Z, Wang PY, Hu BH, Yu
JG, Zhao Y, Cai B, Chen YL, Lu M, et al: Protective cerebrovascular
effects of hydroxysafflor yellow A (HSYA) on ischemic stroke. Eur J
Pharmacol. 818:604–609. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yin F, Zhou H, Fang Y, Li C, He Y, Yu L,
Wan H and Yang J: Astragaloside IV alleviates ischemia
reperfusion-induced apoptosis by inhibiting the activation of key
factors in death receptor pathway and mitochondrial pathway. J
Ethnopharmacol. 248(112319)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fritsche KL: Linoleic acid, vegetable oils
& inflammation. Mo Med. 111:41–43. 2014.PubMed/NCBI
|
|
23
|
Zdunska K, Dana A, Kolodziejczak A and
Rotsztejn H: Antioxidant properties of ferulic acid and its
possible application. Skin Pharmacol Physiol. 31:332–336.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li J, Yu J, Ma H, Yang N, Li L, Zheng DD,
Wu MX, Zhao ZL and Qi HY: Intranasal pretreatment with
Z-Ligustilide, the main volatile component of Rhizoma
Chuanxiong, confers prophylaxis against cerebral ischemia via
Nrf2 and HSP70 signaling pathways. J Agric Food Chem. 65:1533–1542.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wei Y, Liu J, Zhang H, Du X, Luo Q, Sun J,
Liu F, Li M, Xu F, Wei K and Dong J: Ligustrazine attenuates
inflammation and the associated chemokines and receptors in
ovalbumine-induced mouse asthma model. Environ Toxicol Pharmacol.
46:55–61. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Pandya RS, Mao L, Zhou H, Zhou S, Zeng J,
Popp AJ and Wang X: Central nervous system agents for ischemic
stroke: Neuroprotection mechanisms. Cent Nerv Syst Agents Med Chem.
11:81–97. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang WW, Xu F, Wang D, Ye J and Cai SQ:
Buyang Huanwu Decoction ameliorates ischemic stroke by modulating
multiple targets with multiple components: In vitro evidences. Chin
J Nat Med. 16:194–202. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sowndhararajan K, Deepa P, Kim M, Park SJ
and Kim S: Baicalein as a potent neuroprotective agent: A review.
Biomed Pharmacother. 95:1021–1032. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Palozza P: Can beta-carotene regulate cell
growth by a redox mechanism? An answer from cultured cells. Biochim
Biophys Acta. 1740:215–221. 2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Shi X, Fu Y, Zhang S, Ding H and Chen J:
Baicalin attenuates subarachnoid hemorrhagic brain injury by
modulating blood-brain barrier disruption, Inflammation, and
oxidative damage in mice. Oxid Med Cell Longev.
2017(1401790)2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Nabavi SF, Braidy N, Gortzi O,
Sobarzo-Sanchez E, Daglia M, Skalicka-Woźniak K and Nabavi SM:
Luteolin as an anti-inflammatory and neuroprotective agent: A brief
review. Brain Res Bull. 119:1–11. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Barreca D, Bellocco E, D'Onofrio G, Nabavi
SF, Daglia M, Rastrelli L and Nabavi SM: Neuroprotective effects of
quercetin: From chemistry to medicine. CNS Neurol Disord Drug
Targets. 15:964–975. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yang G, Wang N, Seto SW, Chang D and Liang
H: Hydroxysafflor yellow a protects brain microvascular endothelial
cells against oxygen glucose deprivation/reoxygenation injury:
Involvement of inhibiting autophagy via class I PI3K/Akt/mTOR
signaling pathway. Brain Res Bull. 140:243–257. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhao JJ, Song JQ, Pan SY and Wang K:
Treatment with isorhamnetin protects the brain against ischemic
injury in mice. Neurochem Res. 41:1939–1948. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Guo C, Ma Y, Ma S, Mu F, Deng J, Duan J,
Xiong L, Yin Y, Wang Y, Xi M and Wen A: The role of TRPC6 in the
neuroprotection of calycosin against cerebral ischemic injury. Sci
Rep. 7(3039)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Janyou A, Wicha P, Jittiwat J, Suksamrarn
A, Tocharus C and Tocharus J: Dihydrocapsaicin attenuates blood
brain barrier and cerebral damage in focal cerebral
ischemia/reperfusion via oxidative stress and inflammatory. Sci
Rep. 7(10556)2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Robson-Ansley P, Cockburn E, Walshe I,
Stevenson E and Nimmo M: The effect of exercise on plasma soluble
IL-6 receptor concentration: A dichotomous response. Exerc Immunol
Rev. 16:56–76. 2010.PubMed/NCBI
|
|
38
|
Ma Y, Zechariah A, Qu Y and Hermann DM:
Effects of vascular endothelial growth factor in ischemic stroke. J
Neurosci Res. 90:1873–1882. 2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ciurea AV, Palade C, Voinescu D and Nica
DA: Subarachnoid hemorrhage and cerebral vasospasm - literature
review. J Med Life. 6:120–125. 2013.PubMed/NCBI
|
|
40
|
Guo Y: Role of HIF-1a in regulating
autophagic cell survival during cerebral ischemia reperfusion in
rats. Oncotarget. 8:98482–98494. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chen ZZ, Gong X, Guo Q, Zhao H and Wang L:
Bu Yang Huan Wu Decoction prevents reperfusion injury following
ischemic stroke in rats via inhibition of HIF-1 α, VEGF and
promotion β-ENaC expression. J Ethnopharmacol. 228:70–81.
2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Oliveira DMG, Aguiar LT, de Oliveira
Limones MV, Gomes AG, da Silva LC, de Morais Faria CDC and Scalzo
PL: Aerobic training efficacy in inflammation, neurotrophins, and
function in chronic stroke persons: A randomized controlled trial
protocol. J Stroke Cerebrovasc Dis. 28:418–424. 2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Nakajima M, Nito C, Sowa K, Suda S,
Nishiyama Y, Nakamura-Takahashi A, Nitahara-Kasahara Y, Imagawa K,
Hirato T, Ueda M, et al: Mesenchymal stem cells overexpressing
interleukin-10 promote neuroprotection in experimental acute
ischemic stroke. Mol Ther Methods Clin Dev. 6:102–111.
2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Singh N, Sharma G and Mishra V: Hypoxia
inducible factor-1: Its potential role in cerebral ischemia. Cell
Mol Neurobiol. 32:491–507. 2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Franke TF: PI3K/Akt: Getting it right
matters. Oncogene. 27:6473–6488. 2008.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Gao X, Zhang H, Takahashi T, Hsieh J, Liao
J, Steinberg GK and Zhao H: The Akt signaling pathway contributes
to postconditioning's protection against stroke; the protection is
associated with the MAPK and PKC pathways. J Neurochem.
105:943–955. 2008.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Eltzschig HK and Eckle T: Ischemia and
reperfusion-from mechanism to translation. Nat Med. 17:1391–1401.
2011.PubMed/NCBI View Article : Google Scholar
|