Introduction
Liver cancer is a great threat to people's lives.
Over 30,000 people die of liver and intrahepatic bile duct cancer
each year, with more than 40,000 new patients annually (1). As a great killer, the major therapies,
including surgical resection, chemotherapy and radiotherapy are not
effective for all patients, particularly those with recurrence or
distant metastasis (2).
Approximately 18% of patients are diagnosed with distant metastasis
and 27% with regional metastasis (1). Only 44% of patients are diagnosed at
the localized stage. As a result, the total five-year survival rate
for patients with liver cancer is just 18% (1). In fact, the prognosis for patients
with liver cancer is disappointing and requires further
investigation.
Lack of knowledge regarding the mechanism underlying
liver cancer is one of the major reasons causing its poor clinical
outcome. In addition, heterogeneity further increases the
complexity and difficulty involved in curing patients with liver
cancer (3). By analyzing the
reports from the PubMed database, a list of genes contributing
toward the development and progression of liver cancer may be
constructed. Among them, some are dominant genes, while others
serve auxiliary function. Due to heterogeneity, the dominant gene
is different in a particular cohort of patients, leading toward
different responses to specialized drugs (3,4). For
example, sorafenib is a small molecular inhibitor targeting Raf-1
and has been applied in the clinic for the treatment of recurrent
or metastatic liver cancer (5).
However, resistance to sorafenib has been reported extensively and
poses a novel challenge to patients with liver cancer (6). In fact, application of sorafenib in
clinics is limited greatly in certain patients. Therefore, it is
very important to investigate the molecular mechanism underlying
liver cancer.
NEIL3, also called nei-like DNA glycosylase
3, is mapped at chromosome 4q34.3 and encodes a protein belonging
to the DNA glycosylase family (7).
NEIL3 was reported to initiate the DNA base excision repair
process and cleave the bases damaged by radiation or oxidative
response (7-9).
NEIL3 has served important roles in brain development and
neurogenesis or protection (10-12).
Additionally, Skarpengland et al (13) reported that NEIL3 regulated
lipid metabolism and prevented atherosclerosis in mice.
NEIL3 is also involved in the progression of cerebral
ischemia, autoimmunity, Huntington's disease and HIV replication
(14-17).
NEIL3 also serves important roles in cancer (18). For example, Kim et al
(19) reported that polymorphisms
in NEIL3 increased the risk of prostate cancer. In glioblastoma,
loss of NEIL3 increased replication-associated double strand
breaks in DNA strands (20).
Aberration in NEIL3 was associated with short survival times
in patients with colorectal cancer and astrocytoma (21,22).
In breast, ovarian and prostate cancer, NEIL3 affected
cancer progression (23-25).
NEIL3 may repair telomere damage during the mitosis process
(26). Telomeres are critical
components in the maintenance of cell life and maybe candidate
targets in cancer therapy (27,28).
Using bioinformatics analysis, Zhang et al (29) identified SNPs in NEIL3 in
liver cancer. However, no additional data regarding the role of
NEIL3 in liver cancer was found.
The present study aimed to investigate the role of
NEIL3 in liver cancer through decreasing the expression of
NEIL3 in HepG2 cells. Next, cell growth, proliferation,
migration, invasion, cycle transition and apoptosis were analyzed.
In addition, the clinical value of NEIL3 in liver cancer and
the preliminary molecular mechanism was investigated.
Materials and methods
Cell lines and cell culture
Human liver cancer HepG2 and Huh-7cells were
purchased from American Type Cell Culture collection and cultured
in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 1% antibiotics (penicillin and streptomycin)
in an atmosphere with 5% CO2 at 37˚C. All cells had been
authenticated using the STR method (30).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cancer cell lines using
RNAeasyTM kit (Beyotime Institute of Biotechnology),
according to the manufacturer's protocols, and the concentration of
RNA was determined on an ultraviolet spectrophotometer (NanoDrop
2000; Thermo Fisher Scientific, Inc.). Next, 1 µg RNA was reverse
transcribed (42˚C for 50 min) into first-strand cDNA using the
BeyoRTTM III cDNA kit (Beyotime Institute of
Biotechnology). Next, qPCR was performed on an ABI7000 (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using SYBR-Green qPCR
mix (Shanghai Yeasen Biotechnology Co., Ltd.). β-actin was the
internal control. The primers for qPCR are listed in Table I. The thermocycling conditions were
as follows: 95˚C for 2 min; (95˚C for 10 sec; 60˚C for 15 sec) for
40 cycles. The relative mRNA levels of target genes were calculated
using the 2-∆∆Cq method (31).
 | Table IPrimers for quantitative polymerase
chain reaction amplification of NEIL3 and β-actin. |
Table I
Primers for quantitative polymerase
chain reaction amplification of NEIL3 and β-actin.
| Gene | Primer sequence
(5'-3') |
|---|
| NEIL3 | Forward |
TGGAAGTGCAGCTCACCAAA |
| | Reverse |
AGCACATCACCTAGCATCCG |
| β-actin | Forward |
GTGCTATCCCTGTACGCCTC |
| | Reverse |
AGGTAGTCAGTCAGGTCCCG |
Western blotting
Total proteins were extracted from cancer cells
using a protein extraction kit (Beyotime Institute of
Biotechnology), according to the manufacturer's protocols. Next,
equal amounts of 10 µg of protein were analyzed by 12% SDS-PAGE and
transferred onto PVDF membranes (Beyotime Institute of
Biotechnology). Following blocking with 5% skimmed milk for 1 h at
room temperature, primary antibodies against phosphorylated PI3K
(cat. no. ab138364; dilution, 1:1,000; Abcam), phosphorylated mTOR
(cat. no. ab109268; dilution, 1:2,500; Abcam), phosphorylated Akt
(cat. no. ab38449; dilution, 1:500; Abcam) and GAPDH(cat. no.
ab181602; dilution, 1:10,000; Abcam) were added to the vessel and
co-incubated overnight at 4˚C. Subsequently, the PVDF membranes
were washed with PBS (Beyotime Institute of Biotechnology) and
incubated with HRP-conjugated goat anti-rabbit IgG antibody (cat.
no. ab7090; dilution, 1:5,000; Abcam) for 2 h at room temperature.
Finally, target proteins were detected using chemiluminescence
assay kits (Beyotime Institute of Biotechnology) and images were
captured.
Construction of the expression
plasmid, pcDNA3.1-NEIL3, siRNA synthesis and transfection
The human NEIL3 gene was searched in the
NCBI-gene database (https://www.ncbi.nlm.nih.gov/nuccore/NM_018248.3),
synthesized and cloned into the expression plasmid, pcDNA3.1. Next,
the recombinant expression plasmid, pcDNA3.1-oeNEIL3 (oeNEIL3), was
confirmed by DNA sequencing. siRNA oligonucleotide targeting the
human NEIL3 gene (siNEIL3) was designed and synthesized. A
random sequence was used as negative control. Next, Lipofectamine
3000 reagent (Invitrogen; Thermo fisher Scientific, Inc.) was used
to transfectsi NEIL3 (#1forward, GAGCAGAAAGUGAAGUUAATT and reverse,
UUAACUUCACUUUCUGCUCTT; #2 forward, GCUCAAGAGUGAAGAAAAUTT and
reverse, AUUUUCUUCACUCUUGAGCTT) or oeNEIL3 (sequence listed on
https://www.ncbi.nlm.nih.gov/nuccore/NM_018248.3)
into HepG2 cancer cells. In brief, 50 pmol siNEIL3 or 0.2 µg
oeNEIL3 with 1 µl Lipofectamine 3000 was mixed for 20 min and
co-cultured with HepG2 cancer cells for 6 h at 37˚C. Next, the
supernatant was replaced with fresh DMEM and cultured at 37˚C with
5% CO2. The time interval between transfection and
subsequent experiment was 48 h.
Cell Counting kit-8 (CCK-8)
HepG2 cells treated with siNEIL3 or scramble were
seeded into a 96-well plate at 3x103 cells/well and
cultured for 4 days. At designated time points of 24, 48, 72 and 96
h, 10 µl CCK-8 (Beyotime Institute of Biotechnology) was added, and
cells were cultured for another 2 h. The absorbance value was
determined at 450 nm wavelength using a microplate reader (Bio-Rad
Laboratories, Inc.).
Plate-colony formation assay
HepG2 cells treated with siNEIL3 or scramble were
seeded onto 24-well plates at 1x103 cells/well and
cultured for ten days at 37˚C in a humid environment with 5%
CO2. Next, cell colonies were fixed in 4%
paraformaldehyde (Beyotime Institute of Biotechnology) at room
temperature for 30 min, washed with cold PBS, and stained with 0.5%
crystal violet (Beyotime Institute of Biotechnology) at room
temperature for 15 min. Next, positively-stained cancer cells were
counted under a light inverted microscope (NikonTS100; Nikon
Corporation) at x100 magnification. The relative colony formation
ability was evaluated by counting the positively-stained clones in
each well and the number of clones was presented as a bar
graph.
Wound-healing assay
A total of 2x105 siNEIL3- or
scramble-treated cells per well were seeded onto 24-well plates and
cultured for 24 h. When cell confluence arrived at 90%, a scratch
was produced by a 10 µl sterile tip. The debris was washed gently,
and the width of each scratch was recorded and set as the 0 h time
point (W0h). Next, fresh DMEM with no serum was added
and cells were cultured for an additional 24 h. Subsequently, the
width of each scratch was recorded and set as the 24 h time point
(W24h). The relative migration rate was detected under a
light inverted microscope (NikonTS100; Nikon Corporation) at x100
magnification and calculated as follows:
R=(W24h-W0h)/W0h.
Cell invasion assay (chamber room
method)
Chamber rooms with 8-µm-pore size membranes (Corning
Incorporated) were used to detect the invasion of liver cancer
cells. Chamber rooms were pretreated with Matrigel (BD Biosciences)
for 6 h at 37˚C and 2x104 cells/well in DMEM with no
serum were seeded into the upper chamber (the insert). In the
bottom chamber (below the insert), DMEM with 10% FBS was added.
After 24 h at 37˚C, cells on the top surface of the membrane of the
insert were removed and cells in the bottom surface of the membrane
of the insert were fixed in 4% paraformaldehyde at room temperature
for 30 min, washed and stained with 0.5% crystal violet at room
temperature for 15 min. Next, positively-stained cells were counted
under a light inverted microscope ((NikonTS100; Nikon Corporation)
at x100 magnification.
Distribution of the cell cycle (PI
dying method)
A total of 3x105 siNEIL3- or
scramble-treated cells per well were seeded onto 6-well plates and
cultured for 48 h. Next, the cells were collected, washed and fixed
in 75% cold alcohol for 24 h at 4˚C. Following centrifugation at
300 x g for 5 min at room temperature and washing with cold PBS,
500 µl staining buffer with 10 µl PI and 10 µl RNaseA solution
(Shanghai Yeasen Biotechnology Co., Ltd.) was used to re-suspend
the cells and cells were cultured at 37˚C for 30 min. Next, cells
were analyzed using a flow cytometer (FACSCelesta; BD Biosciences).
The edition number of software was BD FACSDiva Software
v8.0.1.1.
Detection of cell apoptosis (Annexin
V/FITC-PI dye)
A total of 3x105 siNEIL3- or
scramble-treated cells per well were seeded onto 6-well plates and
cultured for 48 h. Next, cells were collected, washed and stained
using an Apoptosis Detection kit (Shanghai Yeasen Biotechnology
Co., Ltd.), according to the manufacturer's protocols. In brief,
staining buffer with 5 µl FITC and 10 µl PI solution was used to
re-suspend cancer cells and samples were placed on ice for 15 min.
Next, cell apoptosis was detected using a flow cytometer
(FACSCelesta, BD Biosciences). The edition number of software was
BD FACSDiva Software v8.0.1.1.
Statistical analysis
All data were statistically analyzed using SPSS 16.0
software (SPSS, Inc.). The unpaired Student's t-test method was
used to evaluate the significance between two groups while the
one-way analysis of variance method followed by Tukey's test was
used to estimate the difference among multiple groups. All data are
presented as the mean ± standard deviation. *P<0.05
was considered to indicate a statistically significant
difference.
Results
NEIL3 is associated with short
survival time in liver cancer
TCGA is a public database that consists of the
clinical information of patients affected by cancer. Based on the
TCGA data (URL:http://ualcan.path.uab.edu/cgi-bin/ualcan-res.pl),
the expression of the NEIL3 gene was analyzed in liver
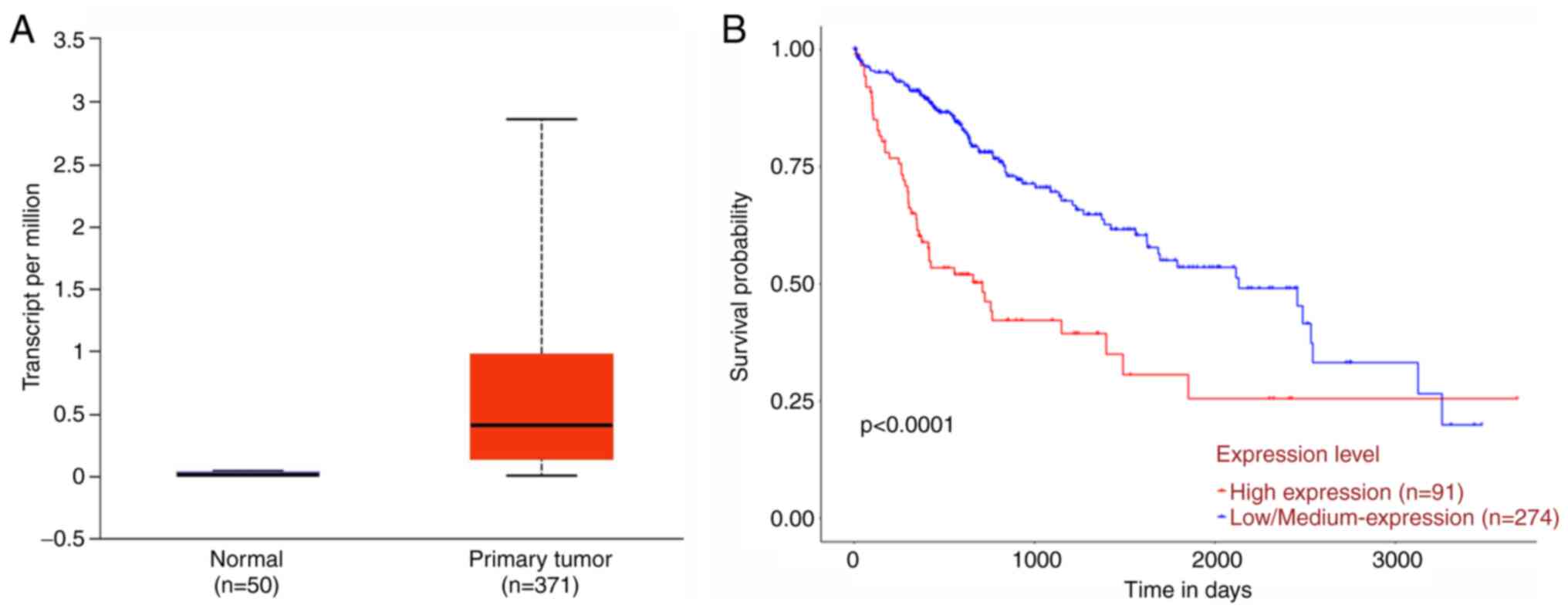
cancer tissues (n=371) and normal control tissues (n=50). As shown
in Fig. 1A, the mean expression
value of NEIL3 in liver cancer tissues was ~28 times as much
as that in the normal control group. Clinicopathological analysis
indicated that NEIL3 expression was correlated with tumor
grade (Table II). However, no
association was found with age, gender, race and weight. Next, by
Kaplan-Meier method (32), patients
with high NEIL3 expression (n=91) displayed poorer10-year
survival probability than patients with low NEIL3 expression
(n=274; P<0.001; Fig. 1B).
Therefore, NEIL3 was clinically associated with survival
time and may serve important roles in liver cancer.
 | Table IIClinical pathological analysis of
NEIL3 expression with tumor grade. |
Table II
Clinical pathological analysis of
NEIL3 expression with tumor grade.
| Comparison | Statistical
significance |
|---|
| Normal (n=50) vs.
grade 1 (n=54) |
7.57x10-4 |
| Normal (n=50) vs.
grade 2 (n=173) |
1.25x10-12 |
| Normal (n=50) vs.
grade 3 (n=118) |
2.67x10-12 |
| Normal (n=50) vs.
grade 4 (n=12) |
3.04x10-3 |
NEIL3 deficiency inhibits cell
proliferation and growth in liver cancer
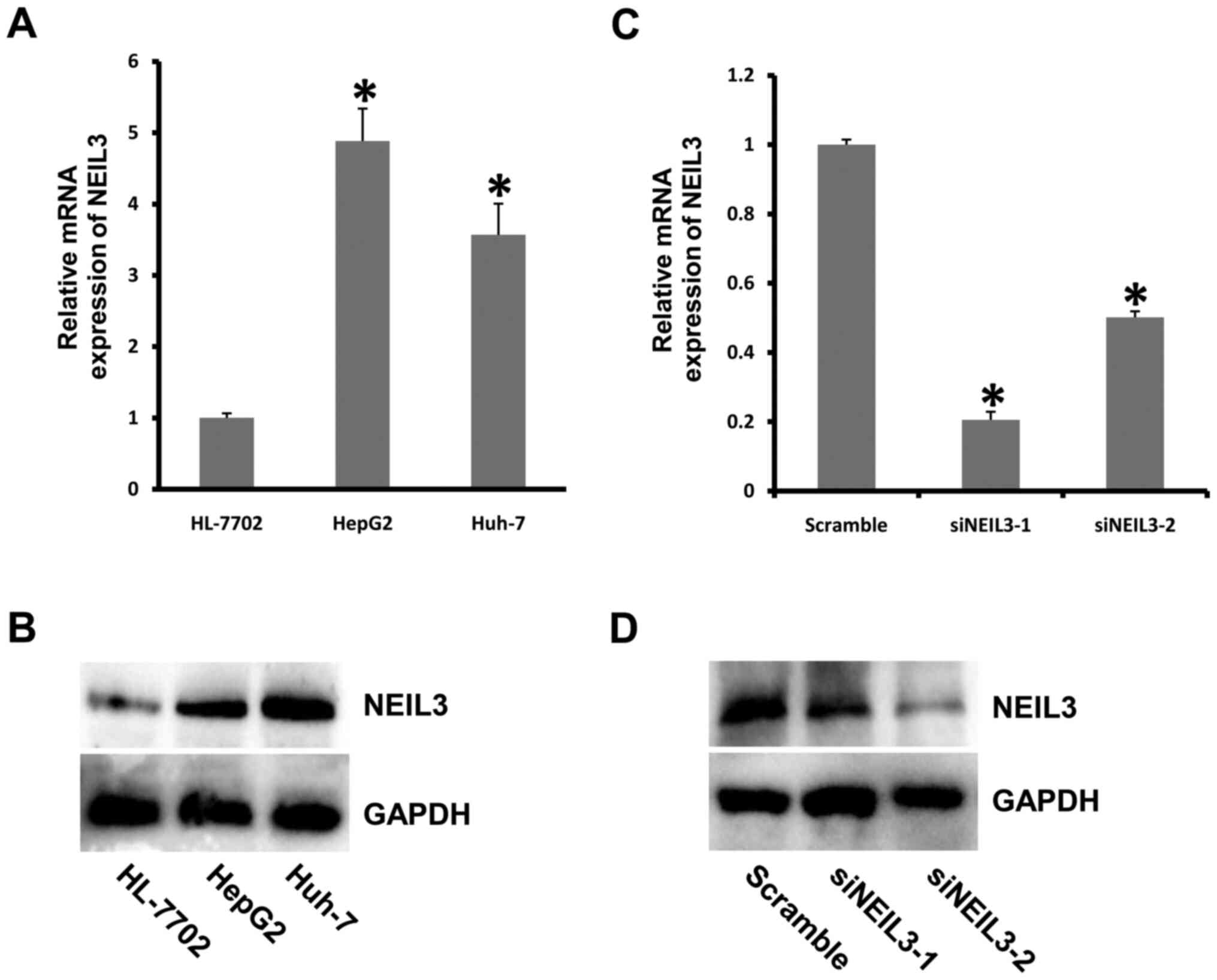
To investigate the role of NEIL3 in liver
cancer, the expression of NEIL3 was detected in liver cancer
cell lines. NEIL3 displayed much higher expression in HepG2
and Huh-7 cells than in the control group at both the mRNA and
protein levels (Fig. 2A and
B). Next, the mRNA expression of
NEIL3 in HepG2 cells was decreased by RNAi technology. The
knockdown efficiency was ~80% in cells treated with siNEIL3-1
oligonucleotide (Fig. 2C and
D). To detect the effect of
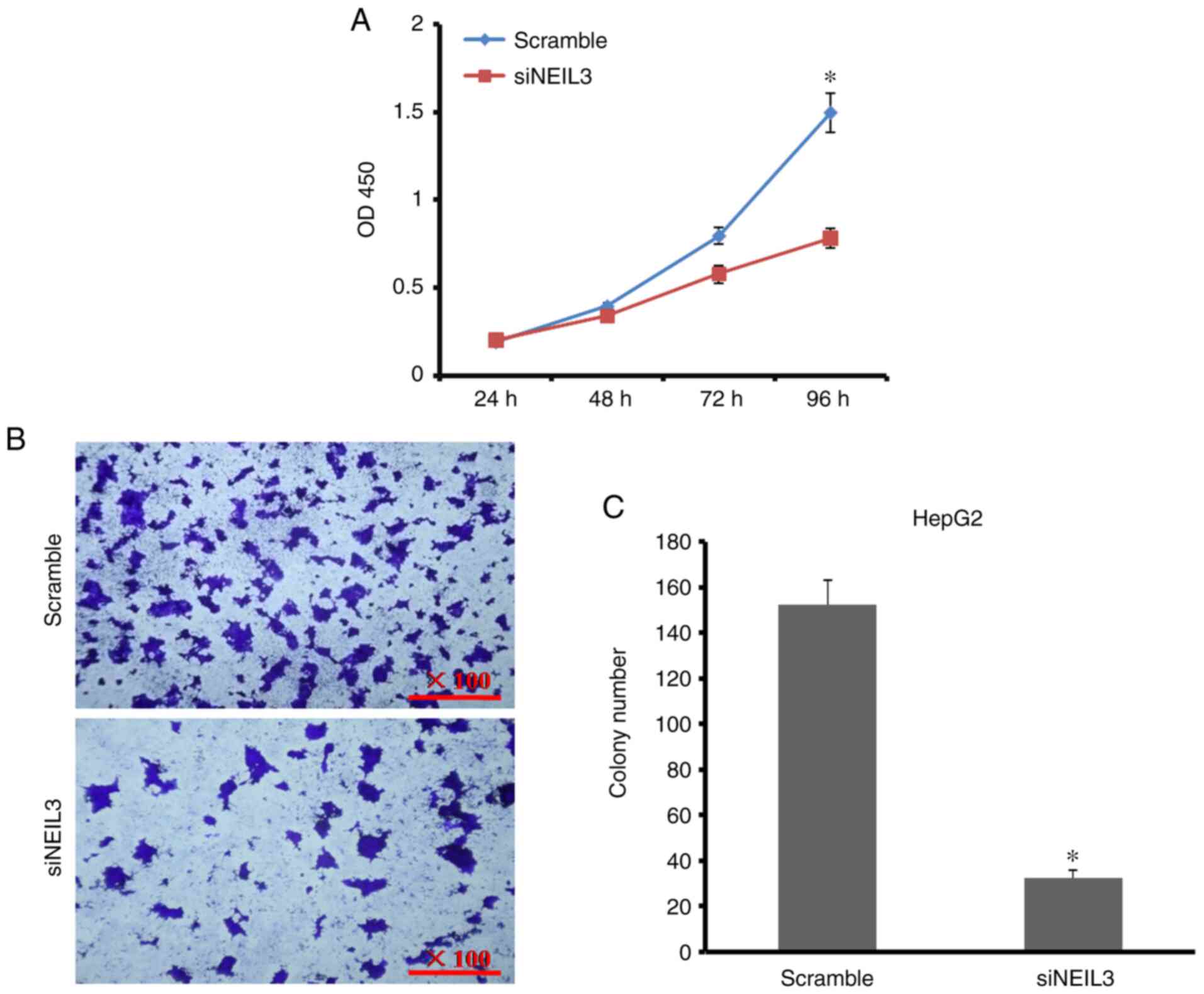
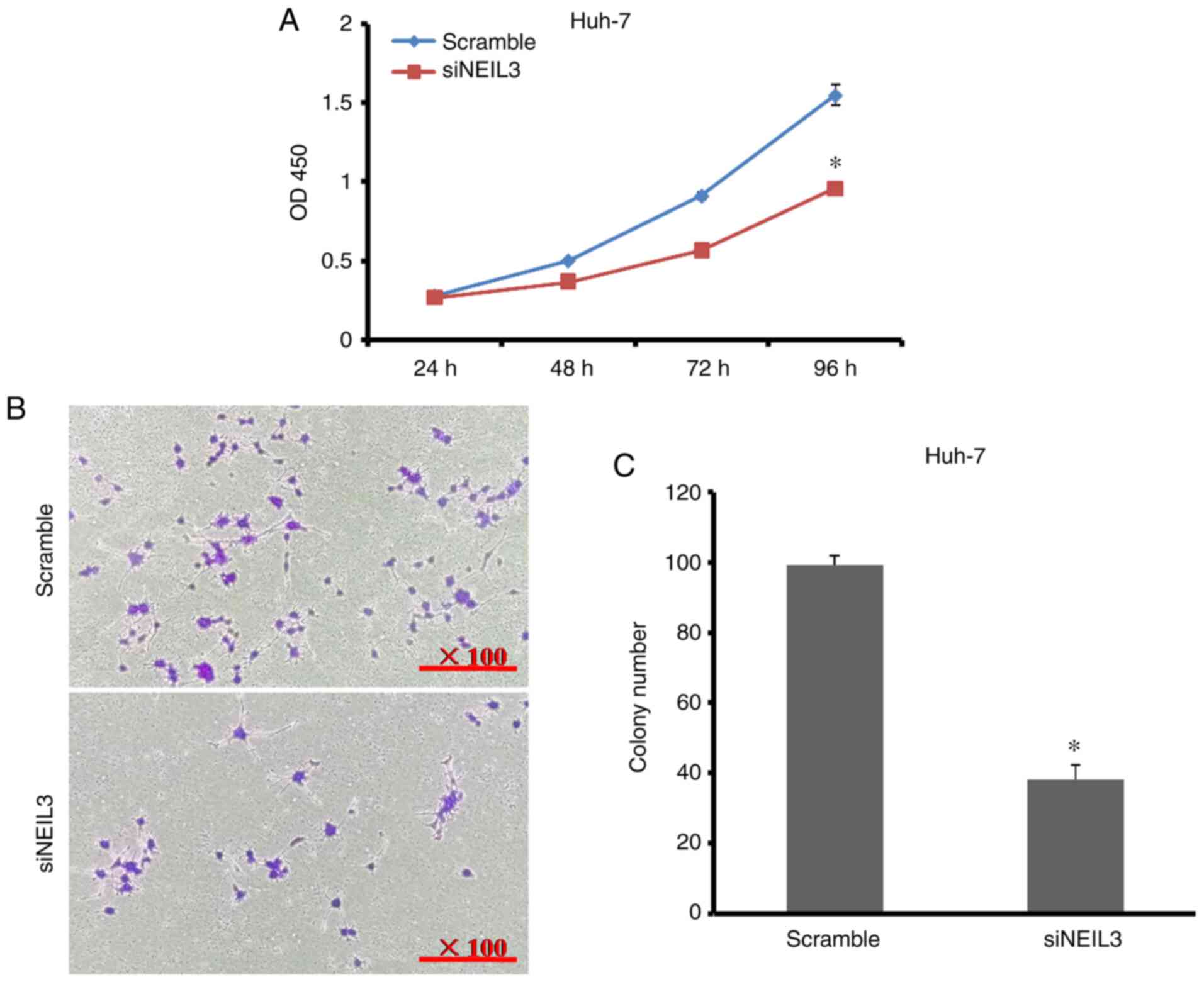
NEIL3-knockdown on cell growth and proliferation, CCK-8 and
plate-colony formation assays were performed. As shown in Fig. 3A, the cell proliferation rate
decreased by 61.5% at 96 h after NEIL3 gene expression was
decreased in HepG2 cells. The number of cell colonies decreased by
52.1% compared with the control (Fig.
3B and C). In Huh-7 cells, the
cell proliferation rate decreased by 38% when the number of cell
colonies decreased by 61.6% compared with the control (Fig. 4A-C). It is clear that NEIL3
contributes toward cell growth and proliferation in liver
cancer.
NEIL3 deficiency suppresses cell
migration and invasion in liver cancer
To detect the effects of NEIL3-knockdown on
cell migration and invasion in liver cancer cells, wound-healing
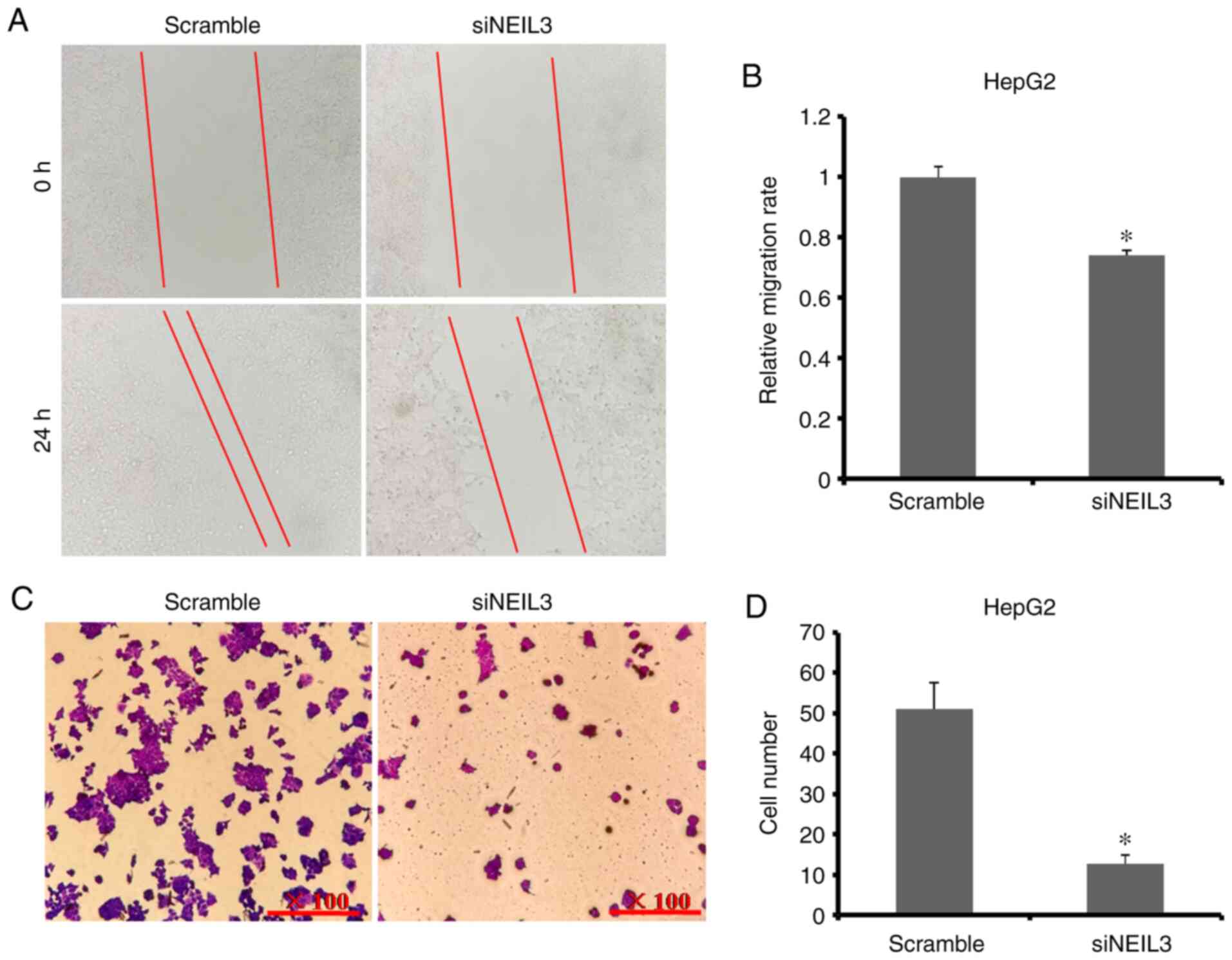
and Transwell assays were performed. As shown in Fig. 5A and B, the relative cell migration rate of
HepG2 cells was decreased by ~27% in NEIL3-knockdown cells,
which suggested that siNEIL3 suppressed cell migration. The average
cell number transferred through the chamber membrane in
siNEIL3-treated HepG2 cells was significantly lower than that in
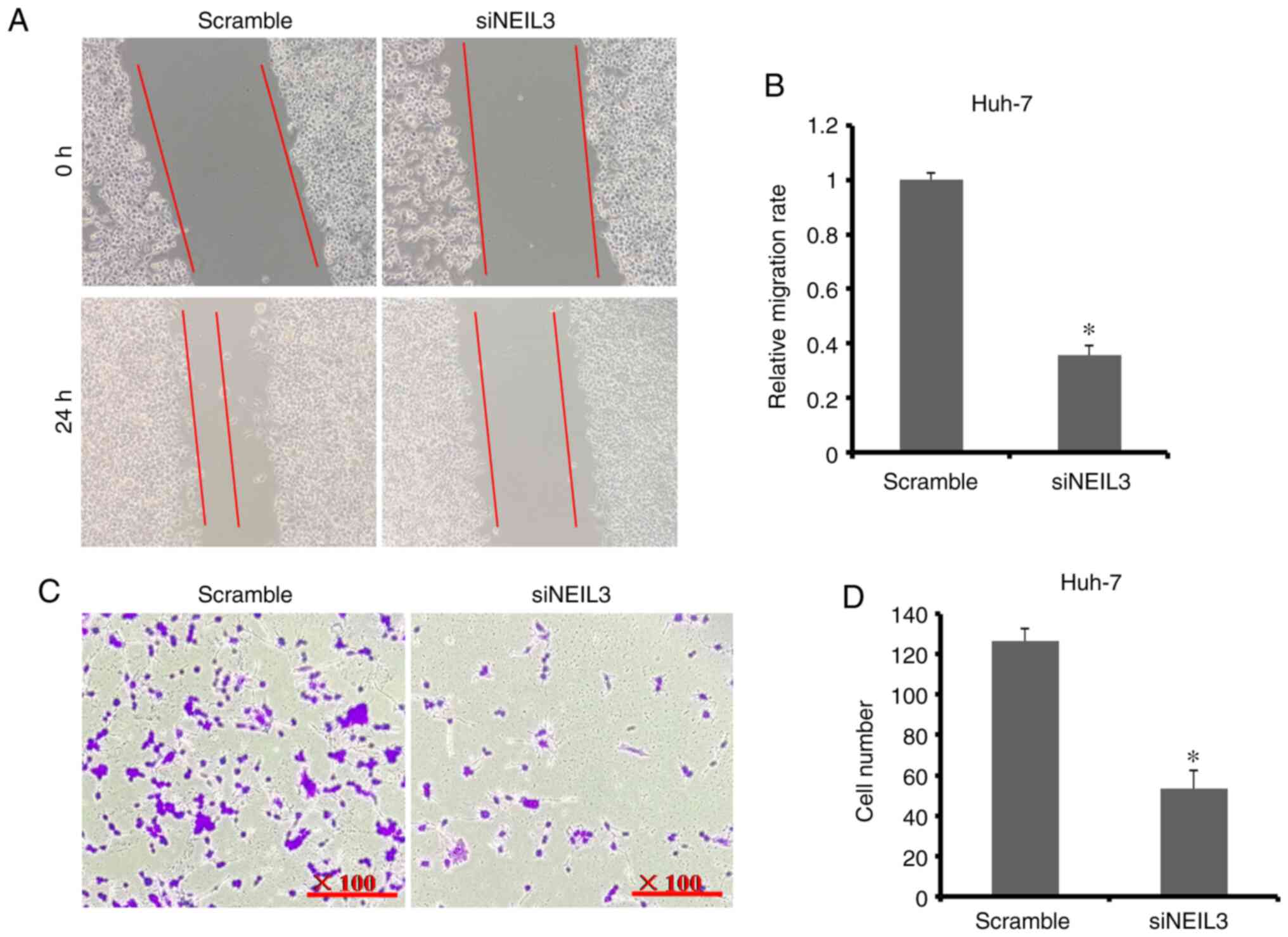
the scramble group (11 versus 52; P<0.05; Fig. 5C and D). In Huh-7 cells, the relative cell
migration rate in the NEIL3-knockdown group was 35.9%
compared with the scramble group (Fig.
6A and B). The average number
of Huh-7 cells transferred through the chamber membrane in the
NEIL3-knockdown group was 53, compared with126 in the
scramble control group (Fig. 6C and
D). These data suggested that
NEIL3 deficiency suppresses cell migration and invasion in
liver cancer.
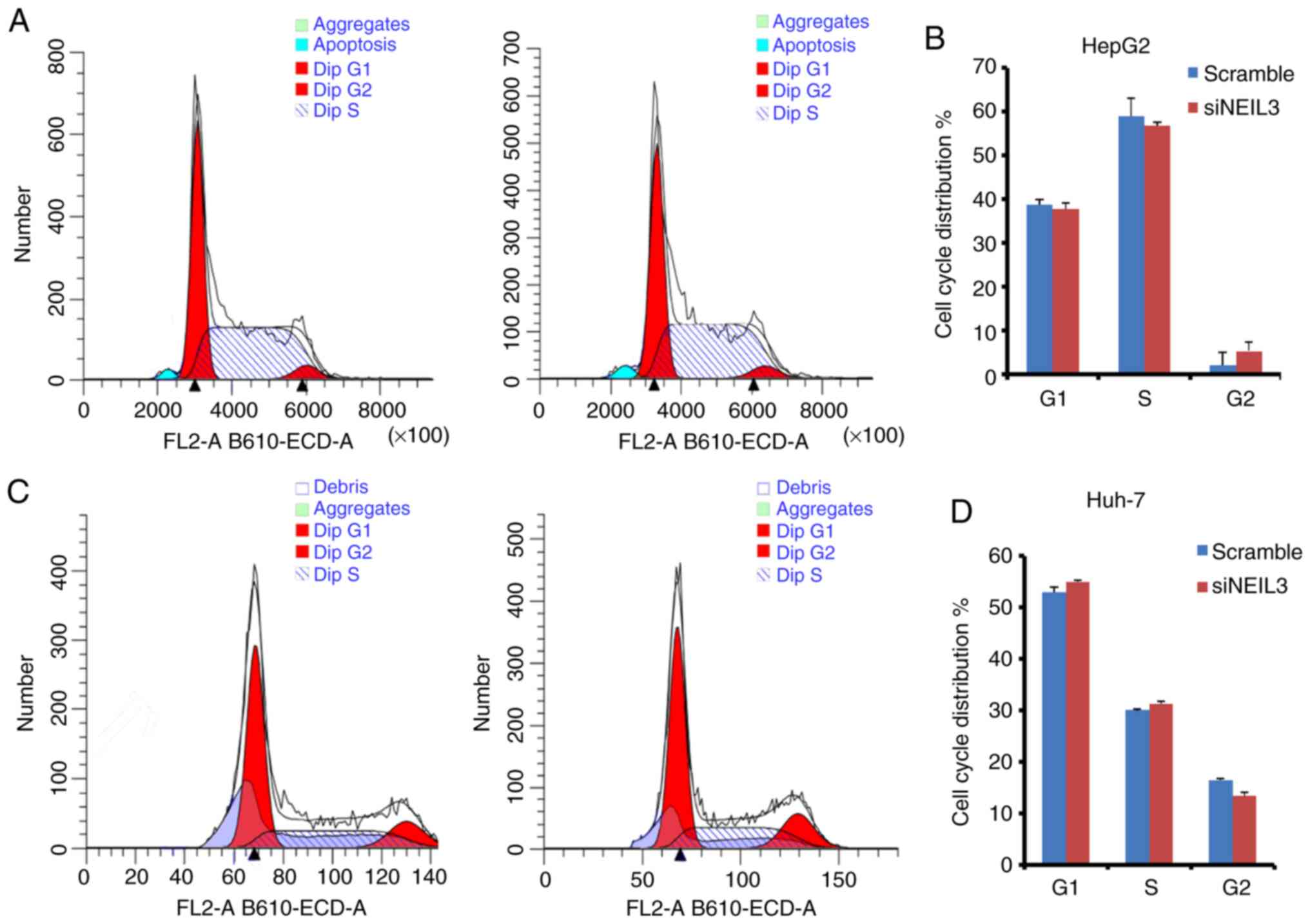
The cell cycle is affected by NEIL3
deficiency in liver cancer
The cell cycle is accelerated in cancer. FACS
analysis with PI staining was performed to detect the effects of
NEIL3 on cell cycle distribution in HepG2 and Huh-7 cells.
The ratio of cells in the G2 phase was increased from 2.2 to 5.3%
in the siNEIL3 group compared with the scramble control. The ratio
of cells in the G1 and S phases was slightly decreased (Fig. 7A and B). In Huh-7 cells, cells in the G1 and S
phases increased slightly, while cells in the G2 phase decreased
slightly (Fig. 7C and D). However, none of the differences were
significant (P>0.05), which suggests that the cell cycle is not
the target of NEIL3 in liver cancer.
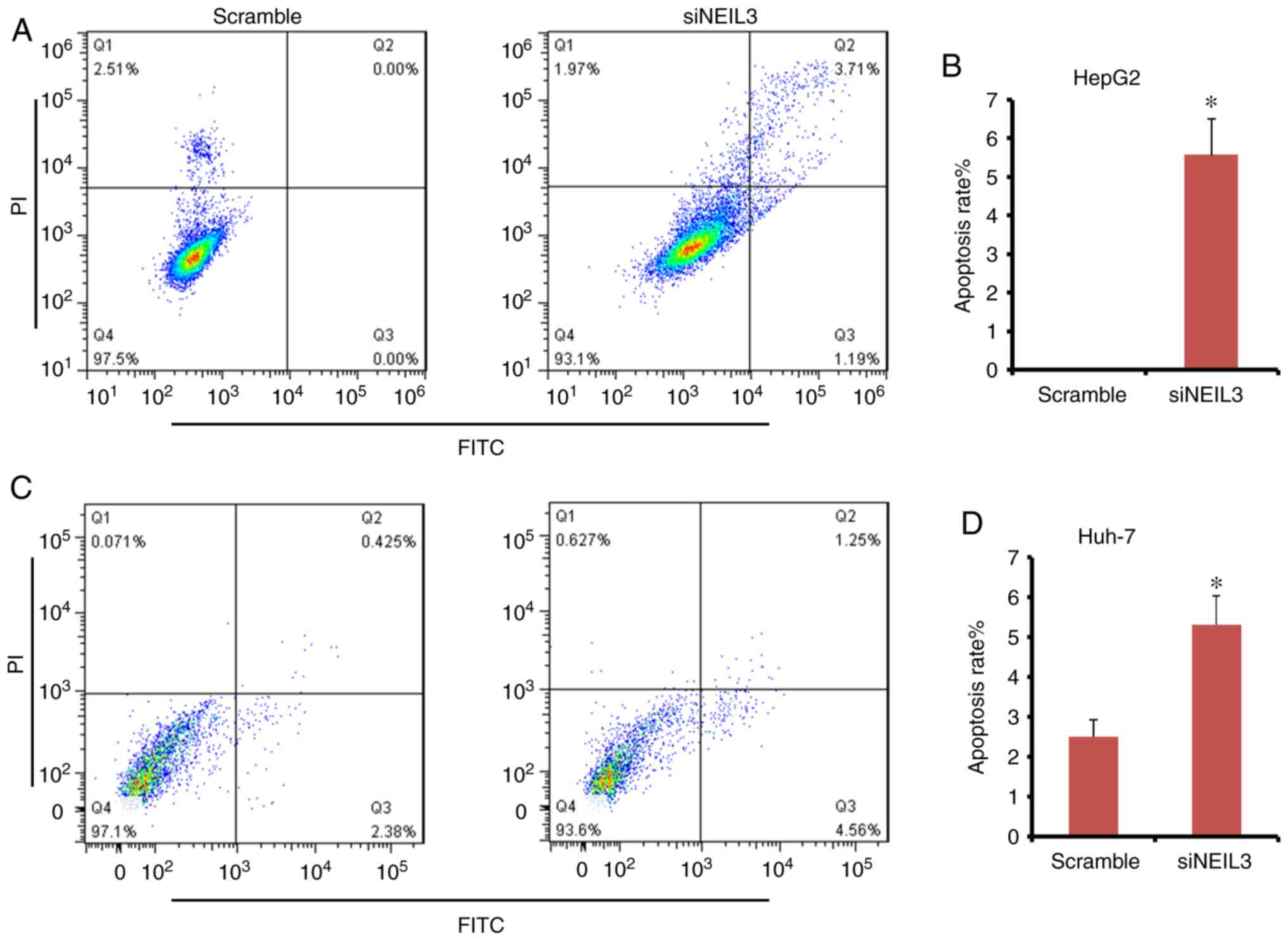
NEIL3 deficiency induces cell
apoptosis in liver cancer
Suppression of programmed cell death (PCD) or cell
apoptosis is adopted by nearly all tumors. By contrast, inducing
cell apoptosis is the major mechanism of chemotherapy drugs in the
clinic. Double-dye staining with Annexin V-FITC/PI solution was
performed to detect the effect of siNEIL3 on cell apoptosis. In
HepG2 cells, the mean apoptosis rate in the siNEIL3 group was 5.5%
following NEIL3-knockdown, but it was 0% in the scramble
group (Fig. 8A and B). The difference between the two groups
was significant (P<0.05). In Huh-7 cells, NEIL3-knockdown
induced cell apoptosis ranging between 2.8 and 5.8% compared with
the scramble control (P<0.05; Fig.
8C and D). Therefore,
NEIL3 deficiency induces apoptosis in liver cancer.
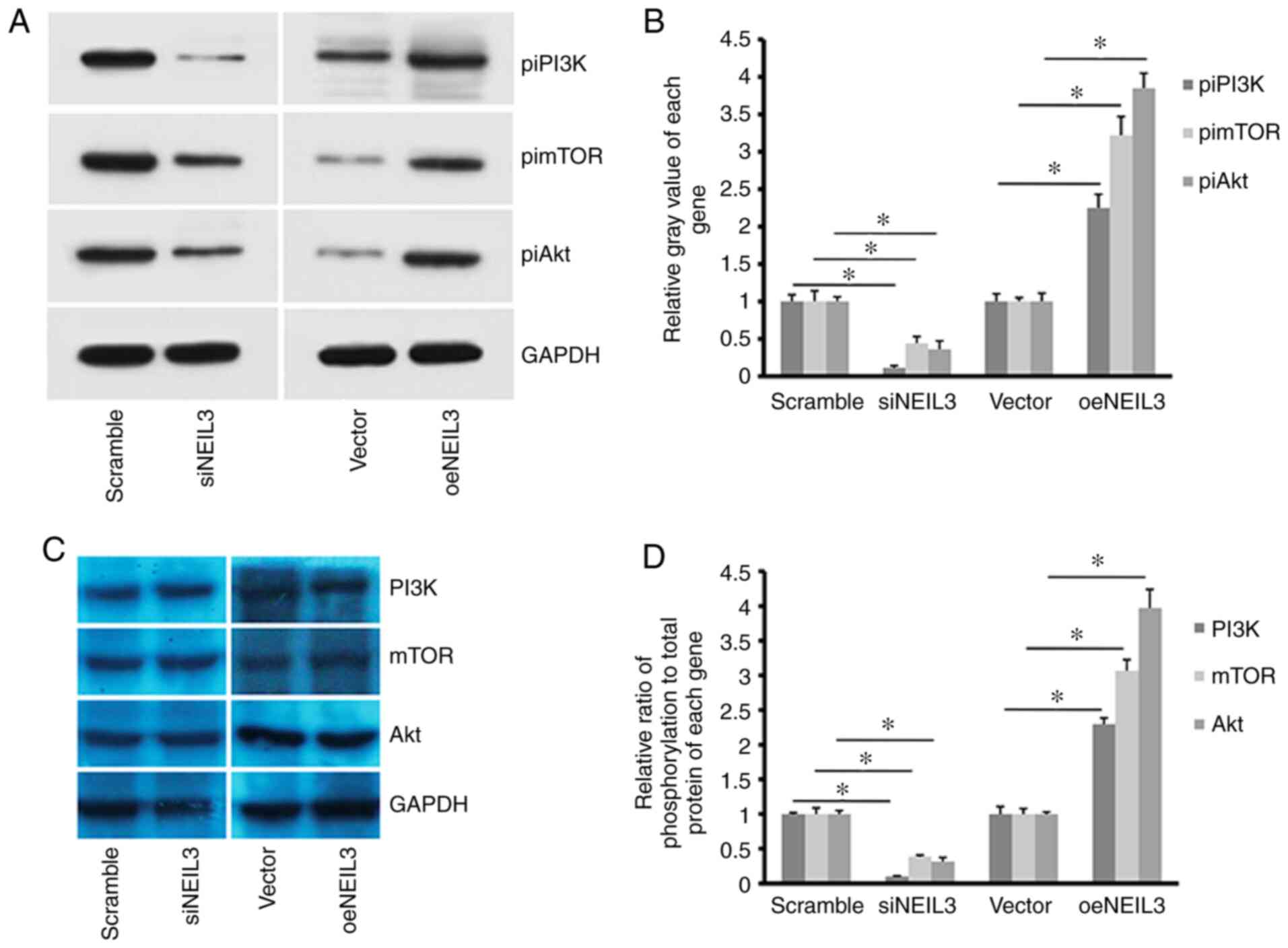
NEIL3 regulates PI3K/Akt/mTOR
signaling in liver cancer
NEIL3 was reported to regulate the expression
of the BRCA1/2oncogene in cancer (33). The present study reported that the
phosphorylation level of PI3K (piPI3K) was decreased when
NEIL3 was decreased in HepG2 cells (Fig. 9A and B). In addition, the phosphorylation levels
of downstream molecules of the PI3K signaling pathway, including
Akt (piAkt) and mTOR (pimTOR), were also decreased. By contrast,
overexpression of NEIL3 enhanced the phosphorylation level
of PI3K, Akt and mTOR in HepG2 cells. However, the background
expression levels of PI3K, Akt and mTOR did not exhibit clear
alterations regardless of NEIL3-knockdown in HepG2 cells (Fig. 9C). Therefore, NEIL3 at least
partially regulates the activation of the PI3K/Akt/mTOR signaling
pathway in liver cancer.
Discussion
Liver cancer is one of the ten most malignant cancer
types in humans. Thousands of individuals die of liver cancer each
year and the number of deaths keeps growing steadily (1). One major reason is the absence of
knowledge of the cause and the molecular mechanism underlying liver
cancer. In the clinic, liver cancer is heterogeneous, and patients
display different responses to particular therapies or drugs
(3). DNA sequencing technology
suggests that genetic variation serves critical roles in the
formation and progression of liver cancer (34). Genetic variations, including gene
mutations, alterations, truncations and fusions are major causes of
heterogeneity in cancer and have been considered in precision
medicine (35). For example, PD-L1
is a tumor antigen that is expressed on the surface of cancer cells
(36). Opadivo, a specific antibody
drug against PD-1, was developed and displayed great success in the
clinic for the treatment of liver cancer (37). In fact, dozens of antibody drugs
have been approved to treat cancer over recent years. However,
genetic variations have prolonged the process to fight cancer.
TCGA is a public database containing a large
quantity of information from patients with cancer and it has
greatly advanced research in cancer (38). In the present study, by extracting
the genetic data regarding liver cancer from TCGA, it was found
that NEIL3 was overexpressed in patients with liver cancer
and was associated with tumor stage. Furthermore, higher expression
of NEIL3 predicted poorer survival, suggesting that
NEIL3 may serve very important roles in liver cancer in the
clinic. This was consistent with the role of NEIL3 in
colorectal cancer and astrocytoma (21,22).
Indeed, a list of genes from TCGA database was proven to be
involved in the formation and/or progression of liver cancer. An
in vitro assay further supported the significance of
NEIL3 in liver cancer. As described earlier, knockdown of
NIEL3 inhibited cell growth, proliferation, migration,
invasion and cell cycle transition, and induced cell apoptosis in
HepG2 and Huh-7 cells. Cancer cells are characterized by potent
abilities of proliferation and invasion, accelerated cell division
and suppressed cell apoptosis (39). In physiological conditions, normal
cells show contact-inhibitory activity and communicate with each
other to survive in a limited environment (40). By contrast, cancer cells lost this
limitation and evolve to have potent proliferation abilities, which
results in unlimited tumor expansion (40). Accelerated cell life also
contributes toward cell expansion. The entire life of a cell can be
divided into G1, S, G2 and M phase. Additionally, cell cycle
transition is controlled strictly by checkpoint mechanisms
(41). However, cancer cells evolve
a novel mechanism to circumvent the checkpoints and accelerate cell
cycle transition, which promotes cell division (42). In the present study, NEIL3
affected cell cycle transition, but not significantly, suggesting
that NEIL3mayregulate other behaviors in liver cancer.
Apoptosis is an important mechanism for homeostasis but is
suppressed in cancer (43,44). NEIL3-knockdown in HepG2 and
Huh-7 cells induced apoptosis, which suggested that NEIL3
was a negative factor for apoptosis in liver cancer. This finding
further suggests that NEIL3 promotes progression in liver
cancer. NEIL3 was also reported to promote progression in
breast, ovarian and prostate cancer (23-25).
As a result, it is hypothesized that NEIL3 contributes
toward the development and/or progression of liver cancer.
The PI3K/Akt and mTOR signaling pathways serve
important roles in normal development and regulate a variety of
physiological processes, including cell proliferation, migration,
survival and differentiation (45).
The PI3K family consists of heterodimeric lipid kinases and maybe
classified into three classes based on substrate specificity and
sequence homology (46). Of them,
class 1 PI3Ks are responsible for catalyzing PIP2 into the
secondary messenger, PIP3, which recruits Akt to the inner membrane
and activates Akt by phosphorylating its serine/threonine kinase
sites (46). Tuberous sclerosis
complex (TSC) negatively regulates the activation of mTORC1
signaling, while activated Akt inhibits TSC complex activity and
initiates the mTOR signaling pathway followed by phosphorylated
activation of eIF4E and 4EBP1 (47,48).
Next, cell growth and proliferation are switched on. However,
abnormal activation of PI3K/Akt/mTOR signaling often leads to
severe diseases, including cancer. PI3K/Akt/mTOR signaling shows
markedly increased activity in cancer. For example, Chen and Costa
(49) reported that activation of
PI3K/Akt/mTOR signaling caused a number of cancer types. In
glioblastoma, inhibitors targeting PI3K/Akt/mTOR signaling provided
a promising way to fight the disease (50). In liver cancer, the PI3K/Akt/mTOR
signaling pathway was a promising target for screening effective
drugs (51). The present study
proved, by western blotting, that NEIL3 mayregulate the
phosphorylation levels of PI3K, Akt and mTOR, which suggests that
NEIL3 regulates the PI3K/Akt/mTOR signaling pathway in liver
cancer. However, further investigation is necessary to confirm
this. Furthermore, the clinical significance of NEIL3 in
liver cancer needs to be evaluated in a large cohort of
patients.
In summary, NEIL3 contributes toward cell
growth, proliferation, migration and invasion, but inhibits
apoptosis in liver cancer. NEIL3 regulates PI3K/Akt/mTOR
signaling in liver cancer and is associated with prognosis in
patients with liver cancer.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WW performed the main experiments and wrote the
manuscript. QY analyzed and interpreted the data from TCGA database
and in the experiment. SG performed part of the experiment and
analyzed the experiment data. JW designed the whole study and
reviewed the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Liu CY, Chen KF and Chen PJ: Treatment of
liver cancer. Cold Spring Harb Perspect Med.
5(a021535)2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li L and Wang H: Heterogeneity of liver
cancer and personalized therapy. Cancer Lett. 379:191–197.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Marin JJG, Cives-Losada C, Asensio M,
Lozano E, Briz O and Macias RIR: Mechanisms of anticancer drug
resistance in hepatoblastoma. Cancer (Basel).
11(407)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Keating GM: Sorafenib: A review in
hepatocellular carcinoma. Target Oncol. 12:243–253. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhu YJ, Zheng B, Wang HY and Chen L: New
knowledge of the mechanisms of sorafenib resistance in liver
cancer. Acta Pharmacol Sin. 38:614–622. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu MM, Bandaru V, Bond JP, Jaruga P, Zhao
X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M and Wallace SS:
The mouse ortholog of NEIL3 is a functional DNA glycosylase in
vitro and in vivo. Proc Natl Acad Sci USA. 107:4925–4930.
2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bandaru V, Sunkara S, Wallace SS and Bond
JP: A novel human DNA glycosylase that removes oxidative DNA damage
and is homologous to Escherichia coli endonuclease VIII. DNA Repair
(Amst). 1:517–529. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liu M, Doublie S and Wallace SS: Neil3,
the final frontier for the DNA glycosylases that recognize
oxidative damage. Mutat Res. 743-7444–11. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jalland CM, Scheffler K, Benestad SL,
Moldal T, Ersdal C, Gunnes G, Suganthan R, Bjørås M and Tranulis
MA: Neil3 induced neurogenesis protects against prion disease
during the clinical phase. Sci Rep. 6(37844)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sejersted Y, Hildrestrand GA, Kunke D,
Rolseth V, Krokeide SZ, Neurauter CG, Suganthan R, Atneosen-Åsegg
M, Fleming AM, Saugstad OD, et al: Endonuclease VIII-like 3 (Neil3)
DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia.
Proc Natl Acad Sci USA. 108:18802–18807. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Regnell CE, Hildrestrand GA, Sejersted Y,
Medin T, Moldestad O, Rolseth V, Krokeide SZ, Suganthan R, Luna L,
Bjørås M and Bergersen LH: Hippocampal adult neurogenesis is
maintained by Neil3-dependent repair of oxidative DNA lesions in
neural progenitor cells. Cell Rep. 2:503–510. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Skarpengland T, Holm S, Scheffler K,
Gregersen I, Dahl TB, Suganthan R, Segers FM, Østlie I, Otten JJ,
Luna L, et al: Neil3-dependent base excision repair regulates lipid
metabolism and prevents atherosclerosis in Apoe-deficient mice. Sci
Rep. 6(28337)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yang LX, Zhang X and Zhao G: Ginsenoside
Rd attenuates DNA damage by increasing expression of DNA
glycosylase endonuclease VIII-like proteins after focal cerebral
ischemia. Chin Med J (Engl). 129:1955–1962. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Massaad MJ, Zhou J, Tsuchimoto D, Chou J,
Jabara H, Janssen E, Glauzy S, Olson BG, Morbach H, Ohsumi TK, et
al: Deficiency of base excision repair enzyme NEIL3 drives
increased predisposition to autoimmunity. J Clin Invest.
126:4219–4236. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Smatlikova P, Askeland G, Vaskovicova M,
Klima J, Motlik J, Eide L and Ellederová Z: Age-related oxidative
changes in primary porcine fibroblasts expressing mutated
huntingtin. Neurodegener Dis. 19:22–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhou H, Xu M, Huang Q, Gates AT, Zhang XD,
Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ and Espeseth
AS: Genome-scale RNAi screen for host factors required for HIV
replication. Cell Host Microbe. 4:495–504. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shinmura K, Kato H, Kawanishi Y, Igarashi
H, Goto M, Tao H, Inoue Y, Nakamura S, Misawa K, Mineta H and
Sugimura H: Abnormal expressions of DNA glycosylase genes NEIL1,
NEIL2, and NEIL3 are associated with somatic mutation loads in
human cancer. Oxid Med Cell Longev. 2016(1546392)2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kim YS, Kim Y, Choi JW, Oh HE and Lee JH:
Genetic variants and risk of prostate cancer using pathway analysis
of a genome-wide association study. Neoplasma. 63:629–634.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Klattenhoff AW, Thakur M, Chu CS, Ray D,
Habib SL and Kidane D: Loss of NEIL3 DNA glycosylase markedly
increases replication associated double strand breaks and enhances
sensitivity to ATR inhibitor in glioblastoma cells. Oncotarget.
8:112942–112958. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jiraskova K, Hughes DJ, Brezina S,
Gumpenberger T, Veskrnova V, Buchler T, Schneiderova M, Levy M,
Liska V, Vodenkova S, et al: Functional polymorphisms in DNA repair
genes are associated with sporadic colorectal cancer susceptibility
and clinical outcome. Int J Mol Sci. 20(97)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
de Sousa JF, Torrieri R, Searfim RB, Di
Cristofaro LF, Escanfella DF, Ribeiro R, Zanette DL, Paçó-Larson
ML, da Silva WA Jr, Tirapelli DP, et al: Expression signatures of
DNA repair genes correlate with survival prognosis of astrocytoma
patients. Tumour Biol. 39(1010428317694552)2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Matta J, Morales L, Dutil J, Bayona M,
Alvarez C and Suarez E: Differential expression of DNA repair genes
in Hispanic women with breast cancer. Mol Cancer Biol.
1(54)2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Nwani NG, Condello S, Wang Y, Swetzig WM,
Barber E, Hurley T and Matei D: A novel ALDH1A1 inhibitor targets
cells with stem cell characteristics in ovarian cancer. Cancers
(Basel). 11(502)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Barry KH, Koutros S, Berndt SI, Andreotti
G, Hoppin JA, Sandler DP, Burdette LA, Yeager M, Freeman LE, Lubin
JH, et al: Genetic variation in base excision repair pathway genes,
pesticide exposure, and prostate cancer risk. Environ Health
Perspect. 119:1726–1732. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou J, Chan J, Lambele M, Yusufzai T,
Stumpff J, Opresko PL, Thali M and Wallace SS: NEIL3 repairs
telomere damage during S phase to secure chromosome segregation at
mitosis. Cell Rep. 20:2044–2056. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
McNally EJ, Luncsford PJ and Armanios M:
Long telomeres and cancer risk: The price of cellular immortality.
J Clin Invest. 130:3474–3481. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen X, Tang WJ, Shi JB, Liu MM and Liu
XH: Therapeutic strategies for targeting telomerase in cancer. Med
Res Rev. 40:532–585. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhang H, Ma H, Wang Q, Chen M, Weng D,
Wang H, Zhou J, Li Y, Sun J, Chen Y, et al: Analysis of loss of
heterozygosity on chromosome 4q in hepatocellular carcinoma using
high-throughput SNP array. Oncol Rep. 23:445–455. 2010.PubMed/NCBI
|
|
30
|
Masters JR, Thomson JA, Daly-Burns B, Reid
YA, Dirks WG, Packer P, Toji LH, Ohno T, Tanabe H, Arlett CF, et
al: Short tanderm repeat profiling provides an international
reference standard for human cell lines. Proc Natl Acad Sci USA.
98:8012–8017. 2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bland JM and Altman DG: Survival
probabilities (the Kaplan-Meier method). BMJ.
317(1572)1998.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tran OT, Tadesse S, Chu C and Kidane D:
Overexpression of NEIL3 associated with altered genome and poor
survival in selected types of human cancer. Tumour Biol.
42(1010428320918404)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fujimoto A, Furuta M, Totoki Y, Tsunoda T,
Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, et
al: Whole-genome mutational landscape and characterization of
noncoding and structural mutations in liver cancer. Nat Genet.
48:500–509. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ma G, Yang X, Liang Y, Wang L, Li D, Chen
Y, Liang Z, Wang Y and Niu H: Precision medicine and bladder cancer
heterogeneity. Bull Cancer. 105:925–931. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Constantinidou A, Alifieris C and Trafalis
DT: Targeting programmed cell death-1 (PD-1) and ligand (PD-L1): A
new era in cancer active immunotherapy. Pharmacol Ther. 194:84–106.
2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tella SH, Mahipal A, Kommalapati A and Jin
Z: Evaluating the safety and efficacy of Nivolumab in patients with
advanced hepatocellular carcinoma: Evidence to date. Onco Targets
Ther. 12:10335–10342. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Blum A, Wang P and Zenklusen JC: SnapShot:
TCGA-analyzed tumors. Cell. 173(530)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Fouad YA and Aanei C: Revisiting the
hallmarks of cancer. Am J Cancer Res. 7:1016–1036. 2017.PubMed/NCBI
|
|
40
|
Ribatti D: A revisited concept: Contact
inhibition of growth. From cell biology to malignancy. Exp Cell
Res. 359:17–19. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Barnum KJ and Q'Connell MJ: Cell cycle
regulation by checkpoints. Methods Mol Biol. 1170:29–40.
2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wenzel ES and Singh ATK: Cell-cycle
checkpoints and aneuploidy on the path to cancer. In Vivo. 32:1–5.
2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Tuzlak S, Kaufmann T and Villunger A:
Interrogating the relevance of mitochondrial apoptosis for
vertebrate development and postnatal tissue homeostasis. Genes Dev.
30:2133–2151. 2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Goldar S, Khaniani MS, Derakhshan SM and
Baradaran B: Molecular mechanisms of apoptosis and roles in cancer
development and treatment. Asian Pac J Cancer Prev. 16:2129–2144.
2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Czech MP: PIP2 and PIP3: Complex roles at
the cell surface. Cell. 100:603–606. 2000.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Dowling RJ, Topisirovic I, Fonseca DB and
Sonenberg N: Dissecting the role of mTOR: Lessons from mTOR
inhibitors. Biochim Biophys Acta. 1804:433–439. 2010.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Hay N and Sonenberg N: Upstream and
downstream of mTOR. Genes Dev. 18:1926–1945. 2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Chen QY and Costa M: PI3K/Akt/mTOR
signaling pathway and the biphasic effect of arsenic in
carcinogenesis. Mol Pharmacol. 94:784–792. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li X, Wu C, Chen N, Gu H, Yen A, Cao L,
Wang E and Wang L: PI3K/Akt/mTOR signaling pathway and targeted
therapy for glioblastoma. Oncotarget. 7:33440–33450.
2016.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Kattan SW, Nafie MS, Elmgeed GA, Alelwani
W, Badar M and Tantawy MA: Molecular docking, anti-proliferative
activity and induction of apoptosis in human liver cancer cells
treated with androstane derivatives: Implication of PI3K/AKT/mTOR
pathway. J Steroid Biochem Mol Biol. 198(105604)2020.PubMed/NCBI View Article : Google Scholar
|