Introduction
Leukemia is a common hematological malignant tumor
with an incidence rate of 5-8/100,000 per year, endangering human
health and life. Chronic myelogenous leukemia (CML) represents one
type of leukemia caused by the dysfunction of pluripotent
hematopoietic cells in patients resulting in indefinite
proliferation of these cells. The development of CML can be divided
into chronic, accelerated and acute phases, with a slow progression
in the early stage (1). Although
treatments such as chemotherapy, interferon therapy and targeted
therapy have progressed in recent years, multiple drug resistance
(MDR) and relapse still constitute major challenges for curing CML
(2). For example, MDR CML is highly
resistant to chemotherapeutic drugs (3,4). Some
patients can gradually become resistant to imatinib which is a
tyrosine kinase inhibitor targeting the Bcr-Abl gene (5). New mechanisms associated with MDR have
been found and further investigated in recent years (6), such as autophagy (7) and the Warburg effect (8), providing novel therapeutic targets for
CML.
The Warburg effect, which is also called aerobic
glycolysis, is an aberrant metabolic process during which
glycolysis occurs in cancer cells even in the presence of oxygen.
Cancer cells with the Warburg effect exhibit higher glucose uptake
and lactate generation (9,10). It has been increasingly recognized
that the Warburg effect plays an important role in drug resistance
and can be utilized as a potential target for drug development
(11,12). The Warburg effect has been reported
in different types of cancer, including hepatic carcinoma (13), breast cancer (14), esophageal cancer (15) and leukemia. It has been demonstrated
that glucose transporter-1 protein (GLUT) is upregulated and the
level of lactate is elevated in pediatric acute lymphoblastic
leukemia cells and that these cells are sensitive to glycolytic
inhibitors (16). GLUT expression
on the surface of Bcr-Abl-positive CML cells can be repressed by
imatinib, resulting in decreased glucose uptake (17). Targeting the Warburg effect provides
potential therapeutic opportunities for leukemia (18). A clinical trial has been carried out
to change the metabolic process in order to enhance drug
sensitivity (19).
Hypoxia-inducible factor 1α (HIF-1α) plays a key role in the
Warburg effect. Glycolytic gene expression can be enhanced by
HIF-1α through interaction with hypoxia-responsive elements of
glycolytic gene promoters (20). In
addition, overexpression of HIF-1α is closely associated with
cancer cell proliferation, apoptosis inhibition, cancer cell
infiltration and MDR. Activation of HIF-1α usually indicates poor
prognosis of patients with tumors (21). The current study hypothesized that
inhibiting HIF-1α in leukemia cells, such as the
Adriamycin-resistant CML cell line K562/ADM, may reverse the
Warburg effect and improve drug sensitivity.
MicroRNAs (miRNAs/miRs) are short non-coding RNAs
with a length of 20-25 nt that participate in regulating various
biological effects, such as proliferation, apoptosis and invasion
of cancer cells. It has been increasingly demonstrated that miRNAs
play regulatory roles in different types of cancer (22,23).
miRNAs constitute potential candidates for leukemia diagnosis and
therapy (24). miR-18a-5p belongs
to the miR-17-92 family (25).
Aberrant expression of miR-18a-5p has been reported in esophageal
squamous cell carcinoma, pancreatic cancer and thyroid cancer, and
miR-18a-5p may function as an oncogene or a tumor suppressor
(26,27). Previous studies mainly focused on
the impact of miR-18a-5p on proliferation, apoptosis, invasion and
migration of cancer cell (28,29).
The present study aimed to explore the regulatory role of
miR-18a-5p in the Warburg effect in K562/ADM cells, which will
provide new insight into the mechanism of miR-18a-5p regulation in
leukemia cells and potential targets for CML treatment.
Materials and methods
Materials
RPMI-1640 medium was from Hyclone; Cytiva. FBS,
trypsin and penicillin-streptomycin solution were from Thermo
Fisher Scientific, Inc. Chronic myelogenous leukaemia cell lines
K562 and K562/ADM were from American Type Culture Collection.
Primary human normal lymphocyte culture was kindly supplied by Dr
Shaohua Wang from the Department of Pharmacology at the Medical
School of Southeast University and maintained at the Department of
Clinical Laboratory in The First Affiliated Hospital of Kunming
Medical University. Cell Counting Kit-8 (CCK-8) reagent was from
Dojindo Molecular Technologies, Inc. Adriamycin was from
Sigma-Aldrich; Merck KGaA. K562/ADM cells were treated with 5 µg/ml
Adriamycin and incubated at 37˚C with 5% CO2 for 48 h.
The coding sequence of HIF-1α (National Center for Biotechnology
Information reference sequence, NM_001530.3), HIF-1α small
interfering RNA (siHIF-1α: 5'-CTACTCAGGACACAGATTTAGACTTGGAG-3') and
the scramble siRNA (5'-CATCCTGAACAGACTTAAGACGTGGTAGT-3') were
synthesized and cloned into pcDNA-3.1 vectors for HIF-1α
overexpression and knockdown by Sangon Biotech Co., Ltd. miRNAs
that could specifically target HIF-1α were screened using
Targetscan 7.2 (http://www.targetscan.org/vert_72/) and Miranda 2.0
(http://www.miranda.org/) software. miR-18a-5p
mimics (5'-GAUAGACGUGAUCUACGUGGAU-3') and miRNA negative control
(miR-NC, 5'-UUCUCCGAACGUGUCACGUACG-3') were synthesized by Sangon
Biotech Co., Ltd. Lipofectamine® 2000 was from
Invitrogen; Thermo Fisher Scientific, Inc. RNAiso Plus™ was from
Takara Biotechnology Co., Ltd. RevertAid™ Reverse Transcriptase was
from Thermo Fisher Scientific, Inc. All reverse
transcription-quantitative PCR (RT-qPCR) primers were synthesized
by Sangon Biotech Co., Ltd. miRNA qPCR Master Mix for miRNA
quantification and 2X SGExcel FastSYBR Mixture for mRNA
quantification were from Sangon Biotech Co., Ltd. HIF-1α, glucose
transporter type 1 (GLUT1), hexokinase 2 (HK2), pyruvate kinase M2
(PKM2), lactate dehydrogenase A (LDHA) and β-actin antibodies were
from Abcam.
Cell culture and transfection
K562 cells or K562/ADM cells were incubated in
RPMI-1640 medium containing 10% fetal calf serum and 100 U/ml
penicillin and 100 mg/ml streptomycin (Thermo Fisher Scientific,
Inc.) in a 37˚C, 5% CO2 incubator. 5x105
cells/well were seeded into a 6-well plate. When the K562/ADM cell
confluency reached >80%, miRNAs, siRNA or overexpression vectors
were transfected using Lipofectamin®2000 according to
the manufacturer's instructions. miRNAs, siRNA or overexpression
plasmid vectors (Sangon Biotech Co., Ltd.) were diluted to a final
concentration of 100 nM in 250 µl of FBS-free RPMI-1640 medium and
mixed gently. Lipofectamin®2000 (5 µl) was gently mixed,
diluted in 250 µl FBS-free RPMI-1640 medium and incubated for 5 min
at room temperature. Following gentle mixing, samples were further
incubated for 20 min at room temperature. The
DNA/lipofectamine®2000 complexes were then added to
cells and incubated at 37˚C in a CO2 incubator for 48
h.
Cell proliferation analysis
CCK-8 assay was used to analyze cell proliferation.
A total of 24 h after miRNA transfection, cells were seeded in a
96-well plate at a density of 5x104 cells/well. A total
of 1, 2, 3 and 4 days after plating, the CCK-8 reagent was added to
the cells at a final concentration of 10% and the cells were
incubated in a 5%CO2, 37˚C incubator for 2 h. The
absorbance at 450 nm was measured and recorded at each time point.
Finally, the proliferative curve was analyzed to compare the
proliferation rate among groups.
Apoptosis analysis
Apoptosis was evaluated using an Annexin V-FITC/PI
cell apoptosis detection kit (Nanjing KeyGen Biotech Co., Ltd.).
K562/ADM cells were collected and stained with PI and Annexin V for
15 min at 25˚C away from light and tested by a BD flow cytometer
(BD Biosciences) and analyzed using FlowJo™ software version 7.6.5
(FlowJo LLC).
Reverse transcription-qPCR
(RT-qPCR)
Total RNA was purified by RNAiso Plus reagent. For
miRNA quantification, the RNA was reverse transcribed using a
miR-18a-5p-specific primer
(5'-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAUGGAAU-3') and a
U6-specific primer (5'-AACGCTTCACGAATTTGCGT-3'). A total of 500 ng
RNA, 20 pmol miR-18a-5p-specific primer, 20 pmol U6-specific
primer, 4 µl 5X reaction buffer, 0.5 µl Ribolock RNase inhibitor, 2
µl dNTP mix and 1 µl RevertAid Reverse Transcriptase (Thermo Fisher
Scientific, Inc.) were added to a 20 µl mixture. The temperature
protocol of reverse transcription protocol was as follows: 42˚C for
60 min, followed by 72˚C for 10 min. miRNA quantification was
performed using the miRNA qPCR master mix. U6 was used as a
reference gene. The qPCR primer pair for miR-18a-5p was as follows:
Forward, GGGGAUAGACGUGAUCUA and reverse, GTGCAGGTCCGAGGT. The
primer pair for U6 was as follows: Forward, CTCGCTTCGGCAGCACA and
reverse, AACGCTTCACGAATTTGCGT. A total of 10 µl 2X miRNA qPCR
master mix, 0.5 µl forward primer, 0.5 µl reverse primer and 2 µl
template were mixed to a total volume of 20 µl. The thermocycling
conditions for PCR were as follows: 95˚C for 30 sec; 95˚C for 5 sec
and 60˚C for 30 sec for 40 cycles. HIF-1α mRNA quantification was
performed using 2X SGExcel FastSYBR Mixture (Sangon Biotech Co.,
Ltd.). The RNA was reverse transcribed using the oligo (dT) 15
primer. A total of 500 ng RNA, 20 pmol oligo (dT) 15 primer, 4 µl
5X reaction buffer, 0.5 µl Ribolock RNase inhibitor, 2 µl dNTP mix
and 1 µl RevertAid Reverse Transcriptase were added to a total
mixture of 20 µl. The reverse transcription protocol was as
follows: 42˚C for 60 min, followed by 72˚C for 10 min. β-actin was
used as a reference gene. The primer pair for HIF-1α was as
follows: Forward, GGACAAGTCACCACAGGACA and reverse,
GGGAGAAAATCAAGTCGTGC. The primer pair for β-actin was as follows:
Forward, CACCTTCTACAATGAGCTGCGTGTG and reverse,
ATAGCACAGCCTGGATAGCAACGTAC. 25 µl 2 x SGExel FastSYBR Mixture (with
ROX), 1 µl forward primer, 1 µl reverse primer and 2 µl template
were mixed in 50 µl mixture. The PCR protocol for mRNA
quantification is as follows: 95˚C for 20 sec;
95˚C for 3 sec and 60˚C for 30 sec with 35
cycles. The relative expression of miR-18a-5p and HIF-1α mRNA was
calculated according to the 2-ΔΔCq method (30).
Western blotting
K562/ADM cells were lysed and total proteins were
extracted using RIPA buffer with protease and phosphatase
inhibitors (Thermo Fisher Scientific, Inc.). Total protein
concentration was determined using a BCA protein assay (Thermo
Fisher Scientific, Inc.). A total of 20 µg protein was loaded per
lane, separated by 5% SDS-PAGE and transferred to a PVDF membrane.
Primary antibodies for targeted molecules were added and incubated
at 4˚C for 12 h. The following primary antibodies were used (all
from Abcam): Caspase-3 and C-Caspase-3 (cat. no. ab13847; 1:200),
HIF-1α (cat. no. ab179483; 1:500), GLUT1 (cat. no. ab115730;
1:200), HK2 (cat. no. ab209847; 1:600), PKM2 (cat. no. ab137852;
1:500), LDHA (cat. no. ab101562; 1:300) and β-actin (cat. no.
ab8226; 1:500). A rabbit IgG-HRP antibody (Abcam; cat. no.
ab270144; 1:800) was used as the secondary antibody and was
incubated at 25˚C for 1 h. β-actin antibody (cat. no. ab6276;
1:1,000; Abcam) was used as an internal reference. Membranes were
stripped with RestoreTM Plus Western Blot Stripping
Buffer (Thermo Fisher Scientific, Inc.) for 30 min at room
temperature. The blots were re-blocked in 5% w/v milk, 1X TBS +
0.1% v/v Tween-20 at 25˚C for 1 h and re-probed. Target protein
expression levels were normalized to the β-actin level. The density
of respective bands was semi-quantified using a densitometer with
Alpha View Software 3.0 (ProteinSimple).
Luciferase reporter assay
The wild type 3'-UTR and mutated 3'-UTR of HIF-1α
were cloned into Dual-Glo Luciferase assay plasmids (Promega
Corporation). K562/ADM cells cultured in a 6-well plate were
transfected with 1 µg plasmids and 50 pmol miR-18a-5p mimics or
miR-NC using Lipofectamine®2000. The pRL-CMV Renilla
luciferase vector (Promega Corporation) was used to normalize cell
numbers and transfection efficiency. After 24 h, the luciferase
assay was performed using a luciferase assay kit (Promega
Corporation).
ATP analysis
ATP was quantified using an ATP Bioluminescence
Assay kit Cls II (Roche Diagnostics) according the manufacturer's
instruction. Briefly, 2x106 K562/ADM cells were
collected and mixed with 400 µl ATP buffer [100 mM Tris, 4 mM EDTA
(pH 7.75)] and the mixture incubated at 100˚C for 2 min. After
centrifugation, the supernatant was collected and the protein was
quantified by the BCA assay. Luciferase reagent at a volume of 100
µl was added into 100-µl sample. Fluorescence was detected by an
ELx808 Absorbance Microplate Reader (BioTek Instruments, Inc.).
After subtracting the blank values from the raw data, the ATP
concentration was calculated from the standard curve data.
Glucose, lactic acid and pyruvate
measurement
For glucose uptake analysis, 2x105
K562/ADM cells were seeded into a 96-well plate 48 h after
transfection and further incubated at 37˚C in a 5% CO2
incubator for 48 h. 2-NBDG (Invitrogen; Thermo Fisher Scientific,
Inc.) was added into the cells at a final concentration of 5 mM.
The cells were incubated for another 15 min at 37˚C and the
absorbance at 488 nm was recorded and normalized to the cell
number. The relative glucose uptake for all groups was expressed as
the absorbance value normalized to that of the untreated group.
For lactic acid analysis, 1x105 K562/ADM
cells were seeded into a 12-well plate and incubated at 37˚C for 10
h. The media were replaced with FBS-free media. After incubation at
37˚C for 1 h, the supernatant was collected for lactic acid
measurement using a lactate assay kit (BioVision, Inc.) according
to the manufacturer's protocol. Absorbance at 450 nm was recorded
and normalized to the cell number. The relative lactic acid level
was expressed as the absorbance value normalized to that of the
untreated group.
For pyruvate analysis, 5x105 K562/ADM
cells were treated with Pyruvate Kinase Assay Buffer (BioVision,
Inc.). The extract was collected by centrifugation at 3,000 x g at
4˚C for 30 min and measured by Pyruvate Kinase Activity Assay kit
(BioVision, Inc.) according to the manufacturer's protocol.
Absorbance at 570 nm was recorded and normalized to the cell
number. The relative pyruvate level was expressed as the absorbance
value normalized to that of the untreated group.
Extracellular acidification rate
(ECAR) and oxygen consumption rate (OCR) assays
ECAR and OCR were analyzed by Seahorse XF Glycolysis
Stress Test kit and Seahorse XF Cell Mito Stress Test kit (Agilent
Technologies), respectively, using Seahorse XFe 96 Extracellular
Flux Analyzer (Seahorse Bioscience; Agilent Technologies).
Transfected cells (1x105) were inoculated into a
microplate and incubated at 37˚C for 10 h followed by ECAR and OCR
tests. For ECAR analysis, glucose, oligomycin (oxidative
phosphorylation inhibitor) and 2-DG (glycolytic inhibitor) were
added into the cells according to the manufacturer's protocol. For
OCR analysis, oligomycin, carbonyl
cyanide-4-(trifluoromethoxy)phenylhydrazone (reversible inhibitor
of oxidative phosphorylation) and the mitochondrial complex I
inhibitor rotenone combined with the mitochondrial complex III
inhibitor antimycin A were sequentially added according to the
manufacturer's protocol. Data were analyzed by Seahorse XF96 Wave
software (version 2.6; Seahorse Bioscience; Agilent
Technologies).
Statistical analysis
Statistical analyses were performed using SPSS
software version 16.0 (SPSS, Inc.). Data are presented as the mean
± standard deviation. Every experiment was carried out
independently three times. Statistical significance was evaluated
by one-way ANOVA with Turkey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-18a-5p targets and downregulates
HIF-1α in K562/ADM cells
Using bioinformatics software Targetscan and
Miranda, miRNAs that could specifically target HIF-1α were firstly
screened. It was shown that miR-18a-5p has a binding site in the
3'-UTR in HIF-1α (Fig. 1A).
miR-18a-5p expression was further quantified and compared among
normal lymphocytes, and K562 and K562/ADM cells by RT-qPCR.
miR-18a-5p level was significantly downregulated in K562/ADM cells
compared with the K562 cell group and normal lymphocyte group.
miR-18a-5p level in K562 cells was significantly lower than that in
the normal lymphocyte group, but higher than that in the K562/ADM
group (Fig. 1B). Subsequently,
miR-18a-5p mimics were transfected into K562/ADM cells and the
level of miR-18a-5p increased up to 8-fold compared with that in
the miR-NC group (Fig. 1C). In
addition, HIF-1α vector transfection could efficiently upregulate
the HIF-1α mRNA expression level and siHIF-1α vector transfection
could efficiently downregulate the HIF-1α mRNA expression level in
K562/ADM cells (Fig. 1D).
Overexpression of miR-18a-5p significantly downregulated HIF-1α
expression on both mRNA (Fig. 1E)
and protein levels (Fig. 1F).
Luciferase assay showed that miR-18a-5p significantly attenuated
the relative luciferase activity of the wild type 3'-UTR construct
but not the mutated 3'-UTR construct (Fig. 1G), indicating that miR-18a-5p
suppressed HIF-1α expression by directly targeting the 3'-UTR in
HIF-1α.
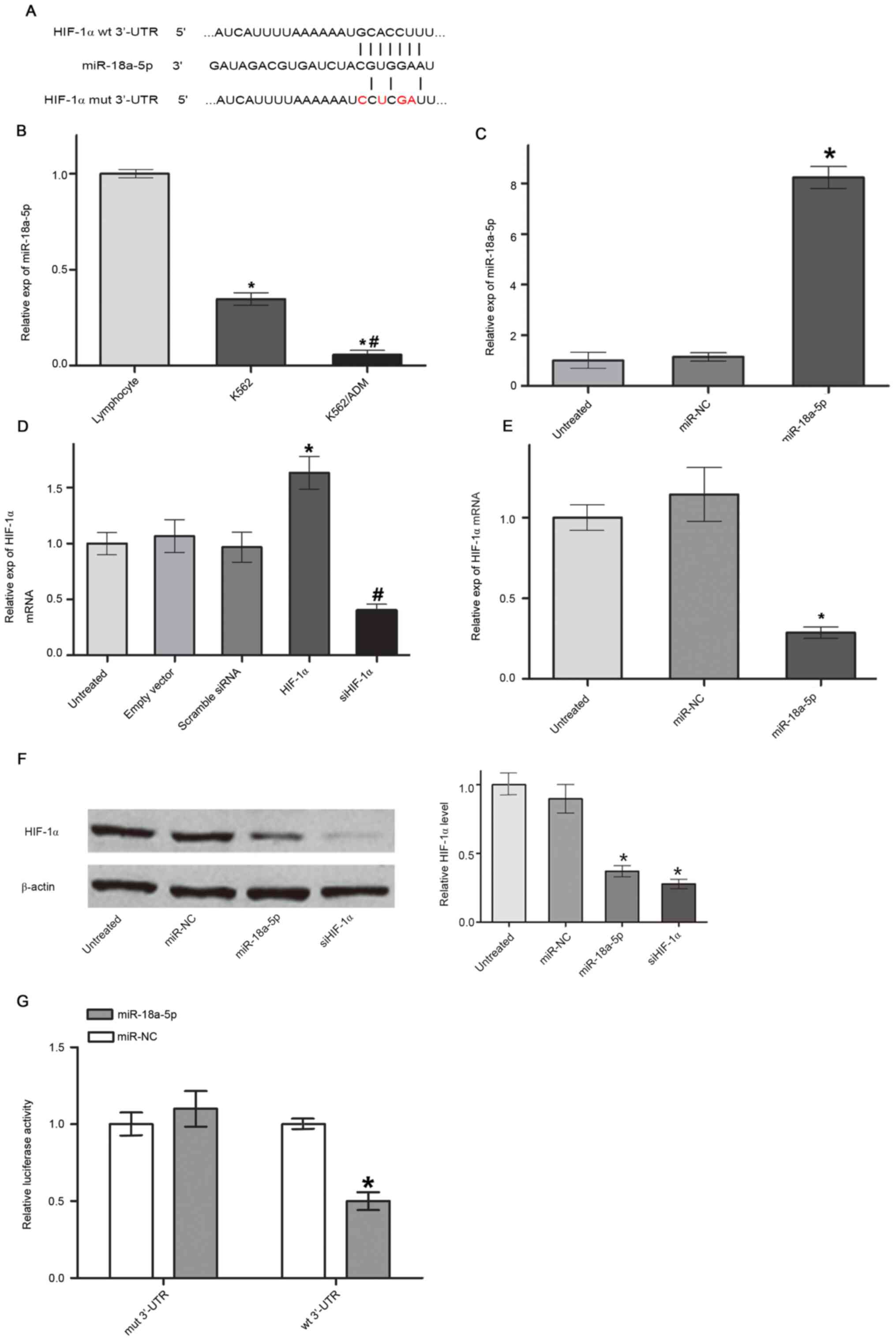 | Figure 1miR-18a-5p downregulates HIF-1α by
binding to the 3'-UTR in HIF-1α. (A) Binding site of miR-18a-5p to
the wt 3'-UTR and mut 3'-UTR in HIF-1α. The red color denotes the
mutated nucleotides. (B) miR-18a-5p level in K562 cells, K562/ADM
cells and normal lymphocytes. *P<0.05 vs. lymphocyte;
#P<0.01 vs. K562. (C) miR-18a-5p expression levels in
K562/ADM cells transfected with miR-NC and miR-18a-5p mimics,
quantified by RT-qPCR. *P<0.05 vs. miR-NC. (D) HIF-1α
mRNA expression levels in K562/ADM cells transfected with an empty
vector, HIF-1α, scrambled siRNA and siHIF-1α, quantified by
RT-qPCR. *P<0.05 vs. empty vector;
#P<0.05 vs. scrambled siRNA. (E) HIF-1α mRNA level in
K562/ADM cells transfected with miR-NC and miR-18a-5p mimics,
evaluated by RT-qPCR. *P<0.05 vs. miR-NC. (F) HIF-1α
protein expression evaluated by western blot analysis and
normalized to β-actin. *P<0.05 vs. miR-NC. (G)
Luciferase reporter assays of K562/ADM cells co-transfected with
mut 3'-UTR plasmids or wt 3'-UTR plasmids and miR-18a-5p mimics or
miR-NC. *P<0.05 vs. miR-NC (n=3). miR, microRNA; UTR,
untranslated region; mut, mutated; wt, wild-type; HIF-1α,
hypoxia-inducible factor 1α; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; si, small interfering RNA. |
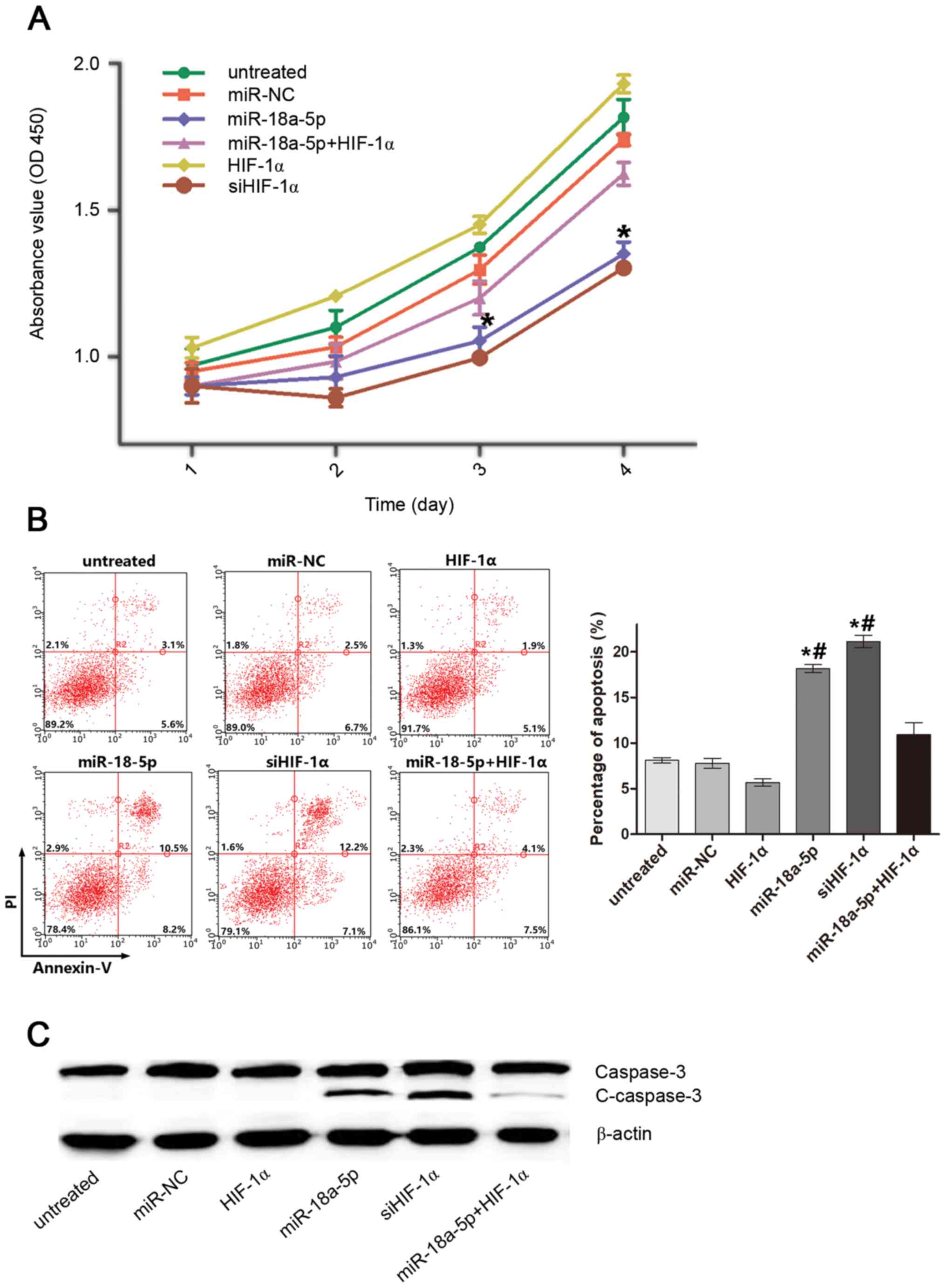
miR-18a-5p inhibits proliferation and
induces apoptosis of K562/ADM cells by targeting HIF-1α
The biological effects, including cell proliferation
and apoptosis, that miR-18a-5p exerted on K562/ADM cells by
targeting HIF-1α were investigated. As shown in Fig. 2A, transfection with miR-18a-5p
mimics or siHIF-1α markedly inhibited cell proliferation compared
with the untreated group, miR-NC group and HIF-1α overexpressing
group. Furthermore, overexpression of HIF-1α ameliorated the
inhibitory effect of miR-18a-5p on cell proliferation. Similarly,
both miR-18a-5p mimics and siHIF-1α efficiently induced cell
apoptosis, and restoration of HIF-1α attenuated the apoptosis
induced by miR-18a-5p (Fig. 2B).
Accordingly, the expression level of apoptosis-associated protein
C-caspase-3 was markedly upregulated by miR-18a-5p and restored by
HIF-1α overexpression (Fig. 2C).
The results indicated that HIF-1α was a target of miR-18a-5p to
inhibit proliferation and induce apoptosis of K562/ADM cells.
miR-18a-5p suppresses the warburg
effect by targeting HIF-1α
HIF-1α has been previously shown to play a key role
in the Warburg effect (31).
Therefore, it was hypothesized that miR-18a-5p may act as a
potential Warburg effect regulator. However, it remained to be
elucidated whether miR-18a-5p may participate in aerobic glycolysis
in leukemia cells. To analyze the influence of miR-18a-5p on the
Warburg effect by targeting HIF-1α, miR-18a-5p mimics were
transfected or co-transfected with HIF-1α-overexpressing plasmids
into K562/ADM cells. miR-18a-5p level was confirmed by RT-qPCR
(Fig. 3A) and HIF-1α protein
expression was confirmed by western blotting (Fig. 3B). Glucose uptake, lactate
production, pyruvate production and ATP synthesis, which are the
characteristics of the Warburg effect, were analyzed after
miR-18a-5p mimics, siHIF-1α or miR-18a-5p mimics/HIF-1α
overexpression vector transfection. The results showed that
miR-18a-5p or siHIF-1α transfection significantly inhibited glucose
uptake (Fig. 3C), lactate
production (Fig. 3D), pyruvate
level (Fig. 3E) and ATP synthesis
(Fig. 3F) in K562/ADM cells.
However, the effects were reversed by HIF-1α overexpression. In
addition, ECAR was significantly repressed (Fig. 3G) and OCR was significantly
decreased (Fig. 3H) by
overexpression of miR-18a-5p, indicative of a decreased overall
glycolytic flux and an enhanced mitochondrial respiration,
respectively. These effects were reversed by HIF-1α overexpression.
Key molecules involved in the Warburg effect, including GLUT1, HK2,
PKM2 and LDHA, were also analyzed by western blotting. It was shown
that miR-18a-5p mimic transfection inhibited the expression of
these molecules (Fig. 3I). The
inhibitory effect of miR-18a-5p was abrogated by overexpression of
HIF-1α. It was confirmed by these results that miR-18a-5p could
reverse the Warburg effect by targeting HIF-1α.
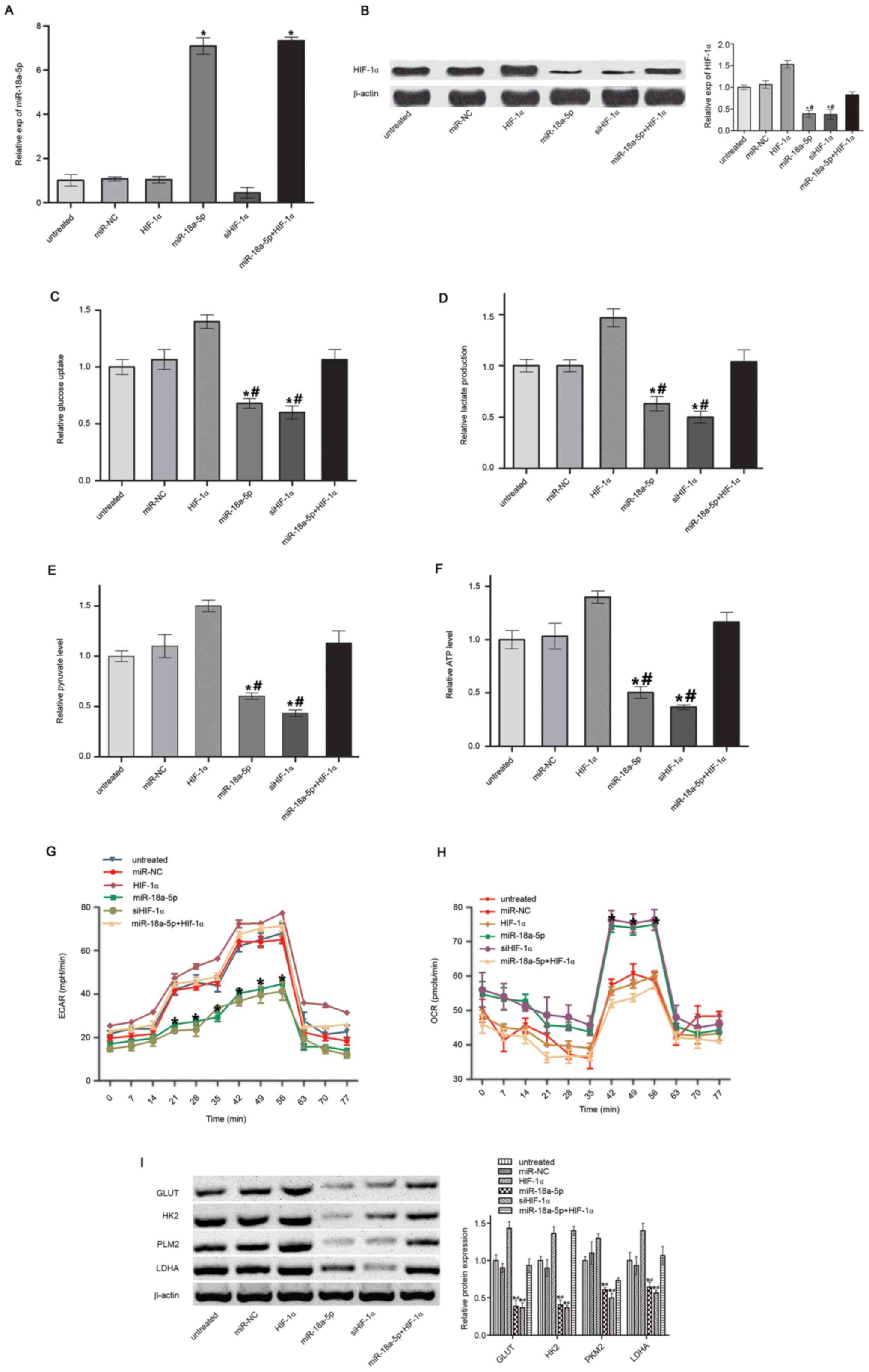 | Figure 3Warburg effect analysis. (A)
miR-18a-5p level. (B) HIF-1α protein level. (C) Glucose uptake. (D)
Lactate production. (E) Pyruvate level. (F) ATP synthesis. (G)
ECAR. (H) OCR. (I) Expression of Warburg effect-associated proteins
in K562/ADM cells. *P<0.05 vs. miR-NC;
#P<0.05 vs miR-18a-5p + HIF-1α. n=3. miR, microRNA;
HIF-1α, hypoxia-inducible factor 1α; NC, negative control; ECAR,
extracellular acidification rate; OCR, oxygen consumption rate;
GLUT, glucose transporter-1 protein; HK2, hexokinase 2; PKM2,
pyruvate kinase M2; LDHA, lactate dehydrogenase A; si, small
interfering RNA. |
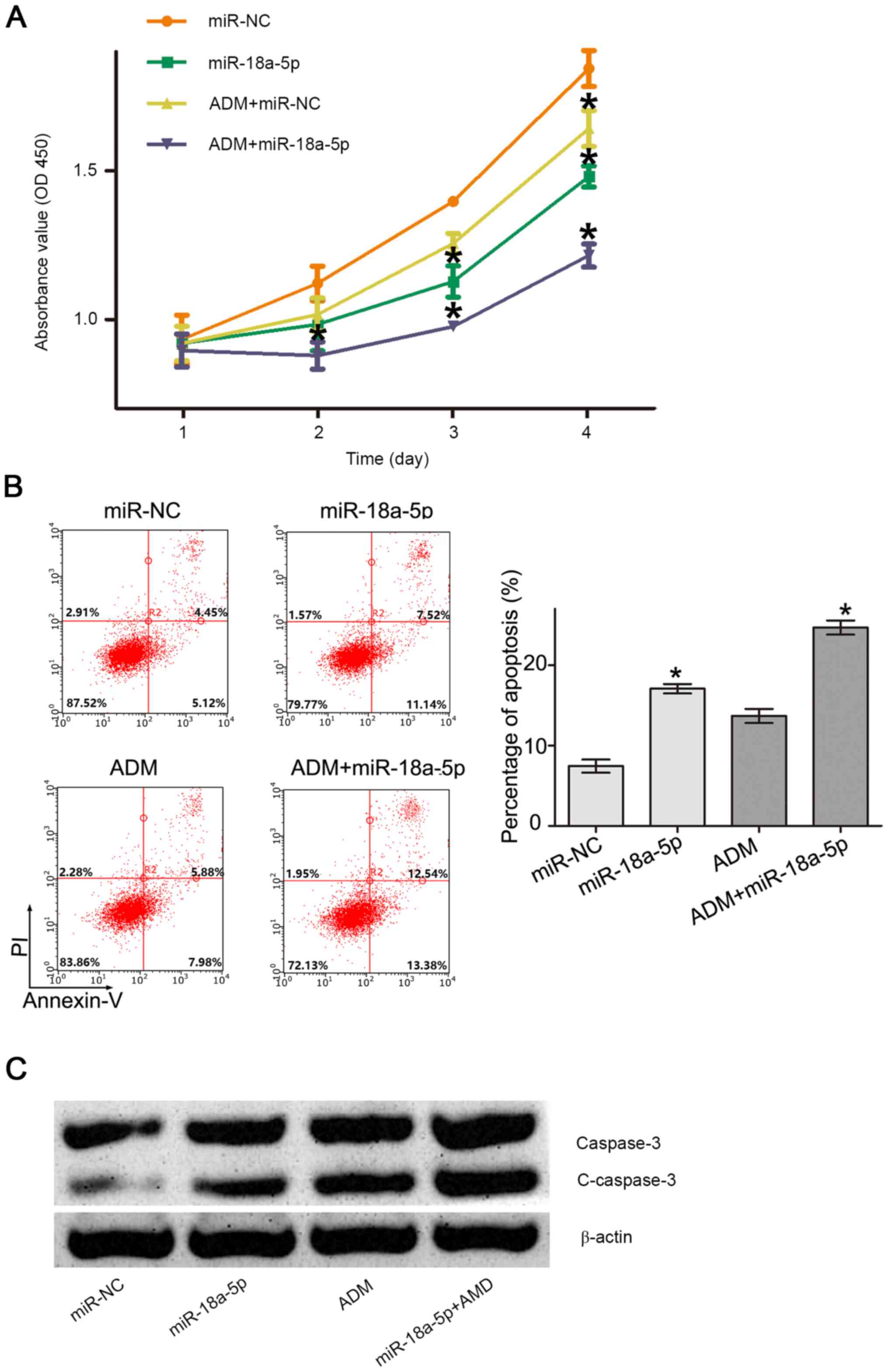
miR-18a-5p enhances sensitivity of
K562/ADM cells to Adriamycin by reversing the Warburg effect
The Warburg effect was previously shown to be
associated with drug resistance (32). The present study aimed to determine
whether inhibiting the Warburg effect by miR-18a-5p could improve
the sensitivity of K562/ADM cells to Adriamycin. Firstly, the CCK-8
assay was performed to determine cell proliferation. As shown in
Fig. 4A, cell proliferation was
inhibited significantly by Adriamycin at day 4 and miR-18a-5p at
days 3 and 4 compared with the miR-NC group, whereas the combined
administration of Adriamycin and miR-18a-5p reduced cell
proliferation more significantly at days 2, 3 and 4 compared with
miR-18a-5p or Adriamycin alone. In addition, Adriamycin combined
with miR-18a-5p resulted in a significantly increased apoptosis
rate in K562/ADM cells compared with miR-18a-5p or Adriamycin alone
(Fig. 4B). Accordingly, the
expression of apoptosis-associated protein C-caspase-3 was markedly
upregulated by miR-18a-5p, ADM and miR-18a-5p + ADM, and the level
of C-caspase-3 in miR-18a-5p + ADM group was markedly higher than
miR-18a-5p and ADM groups (Fig.
4C). These results indicated that miR-18a-5p enhanced the
sensitivity of K562/ADM cells to Adriamycin by reversing the
Warburg effect.
Discussion
The Warburg effect is a hallmark of cancer cells
exhibiting an increased glucose uptake, lactate fermentation,
increased glycolysis rate and decreased respiration, even in the
presence of oxygen. Using this aberrant metabolic process, cancer
cells can acquire enough energy for proliferation, migration and
survival (33). It has been widely
recognized that the transcription factor HIF-1α plays an essential
role in the Warburg effect concerning the glucose uptake, lactate
production, pyruvate metabolism and respiration, providing a
valuable target for cancer treatment and insight into the mechanism
conducive to novel drug discovery (34). The present study investigated the
Warburg effect on a cellular level and a strategy of improving the
sensitivity of leukemia cells to Adriamycin by targeting
HIF-1α.
miRNAs take part in a series of biological process
of cancer cells, including proliferation, differentiation and
apoptosis (35). According to the
complementary base pairing principle, miRNAs bind and degrade
target genes at the post-transcriptional level (36). An array of miRNAs have been
identified to be involved in regulating aerobic glycolysis. For
example, miRNAs can inhibit the Warburg effect through targeting
the 3'-UTR of LDHA or HK2(37). Zhu
et al (38) reported that
expression of miRNA-31-5p was positively associated with the
Warburg effect in two lung cancer cell lines by specifically
targeting HIF-1α inhibitor mRNA, which indicated the important
regulatory role of HIF-1α in the Warburg effect. In the present
study, miR-18a-5p was identified as a direct inhibitor of HIF-1α.
Hence, it was hypothesized that miR-18a-5p may play a functional
role in the Warburg effect. Aberrant expression of miR-18a-5p has
been observed in cancer cell lines and tumor tissues, and its
regulatory mechanism may be varied in different types of cancer. In
HCT116 colon cancer cells, miR-18a-5p can upregulate autophagy and
ataxia telangiectasia mutated gene, with the potential for colon
cancer inhibition (39). However,
miR-18a-5p is upregulated in non-small cell lung cancer (NSCLC)
tissues and NSCLC cell lines and functions as an oncogene by
directly targeting interferon regulatory factor 2(28). Downregulating miR-18a-5p using long
non-coding RNA FENDRR also inhibits prostate cancer progression
(29).
The present study demonstrated that the level of
miR-18a-5p in the K562/ADM cell line was significantly lower than
that in the normal lymphocytes, indicating that miR-18a-5p may act
as a suppressor in CML. Bioinformatics analysis revealed that
HIF-1α may be a potential target of miR-18a-5p. The miR-18a-5p
expression level in K562/ADM cells was significantly downregulated
compared with that in the K562 cells and normal lymphocytes.
However, the miR-18a-5p expression level in K562 cells was
relatively high compared with that in K562/ADM cells. The function
of miR-18a-5p in K562 cells, which is a limitation of the present
study, will be explored in our future work. The present study
indicated that the mRNA and protein levels of HIF-1α can be
decreased by miR-18a-5p overexpression. Transfection of miR-18a-5p
mimics inhibited cell proliferation and induced apoptosis in
K562/ADM cells, and restoration of HIF-1α expression reversed the
biological effects of miR-18a-5p. The above results indicated that
miR-18a-5p can inhibit K562/ADM cells by targeting HIF-1α. The
Warburg effect associated with miR-18a-5p was further explored in
the present study. miR-18a-5p could inhibit the ATP level, glucose
uptake, lactate production and pyruvate level in K562/ADM cells.
ECAR and OCR assays were performed to analyze the overall
glycolytic flux and mitochondrial oxidative respiration changes
(40,41). The results revealed that ECAR was
decreased and OCR was enhanced by miR-18a-5p mimics. Furthermore,
expression of downstream molecules of HIF-1α, including GLUT1, HK2,
PKM2 and LDHA, which are closely associated with the Warburg
effect, was affected by miR-18a-5p transfection. However,
overexpression of HIF-1α could counteract the effects on the
Warburg effect exerted by miR-18a-5p mimics. The results indicated
that miR-18a-5p was able to inhibit the Warburg effect by targeting
HIF-1α. The Warburg effect has been shown to be involved in drug
resistance by increasing drug efflux, DNA damage repair, drug
inactivation, and epigenetic alterations or mutations in drug
targets (42). Therapeutic
strategies, such as RNA interference targeting the Warburg effect
combined with chemotherapeutic drugs, are expected to combat
chemoresistance of tumors. The present study aimed to sensitize
K562/ADM cells, which are commonly utilized as the MDR CML cell
model, to Adriamycin using miR-18a-5p mimics. The results suggested
that miR-18a-5p combined with Adriamycin significantly inhibited
proliferation and induced apoptosis of K562/ADM cells.
In conclusion, the present study demonstrated that
miR-18a-5p can suppress the Warburg effect by downregulating HIF-1α
in K562/ADM cells, revealing a novel regulatory mechanism of
miR-18a-5p and providing potential targets for CML treatment.
Further studies will analyze the relationship between miR-18a-5p
and clinical leukemia samples and explore the therapeutic
application of miR-18a-5p in an animal model.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Joint Special
Fund of Yunnan Province (grant no. 2017FE468), and Health Science
and Technology Program of Yunnan Province (grant nos. 2016NS047 and
2018NS0129).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
KW and YZ conceived and designed the study. SJC, CG
and YZ analyzed and interpreted the data, and drafted the
manuscript. YXL, BN, JRY, QZ, SJC and YHL performed the
experiments. All authors participated in the writing and revision
of the manuscript. YZ and BN supervised the project. All authors
read and approved the final manuscript. KW and CG confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang B, Li L, Ho Y, Li M, Marcucci G,
Tong W and Bhatia R: Heterogeneity of leukemia-initiating capacity
of chronic myelogenous leukemia stem cells. J Clin Invest.
126:975–991. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Valera ET, Scrideli CA, Queiroz RG, Mori
BM and Tone LG: Multiple drug resistance protein (MDR-1), multidrug
resistance-related protein (MRP) and lung resistance protein (LRP)
gene expression in childhood acute lymphoblastic leukemia. Sao
Paulo Med J. 122:166–171. 2004.PubMed/NCBI View Article : Google Scholar
|
|
3
|
O'Hare T, Corbin AS and Druker BJ:
Targeted cml therapy: Controlling drug resistance, seeking cure.
Curr Opin Genet Dev. 16:92–99. 2006.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Gora-Tybor J: Emerging therapies in
chronic myeloid leukemia. Curr Cancer Drug Targets. 12:458–470.
2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Baccarani M, Deininger MW, Rosti G,
Hochhaus A, Soverini S, Apperley JF, Cervantes F, Clark RE, Cortes
JE, Guilhot F, et al: European LeukemiaNet recommendations for the
management of chronic myeloid leukemia: 2013. Blood. 122:872–884.
2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Luqmani YA: Mechanisms of drug resistance
in cancer chemotherapy. Med Prin Pract. 14 (Suppl 1):S35–S48.
2005.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang X, Chen B, Zhao L, Zhi D, Hai Y, Song
P, Li Y, Xie Q, Inam U, Wu Z, et al: Autophagy enhanced antitumor
effect in k562 and k562/ADM cells using realgar transforming
solution. Biomed Pharmacother. 98:252–264. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Dyshlovoy SA, Pelageev DN, Hauschild J,
Borisova KL, Kaune M, Krisp C, Venz S, Sabutskii YE, Khmelevskaya
EA, Busenbender T, et al: Successful targeting of the warburg
effect in prostate cancer by glucose-conjugated
1,4-Naphthoquinones. Cancers (Basel). 11(1690)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Koppenol WH, Bounds PL and Dang CV: Otto
Warburg's contributions to current concepts of cancer metabolism.
Nat Rev Cancer. 11:325–337. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Liberti MV and Locasale JW: The warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bhattacharya B, Mohd Omar MF and Soong R:
The warburg effect and drug resistance. Br J Pharmacol.
173:970–979. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li L, Liang Y, Kang L, Liu Y, Gao S, Chen
S, Li Y, You W, Dong Q, Hong T, et al: Transcriptional regulation
of the warburg effect in cancer by six1. Cancer Cell.
33:368–385.e7. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ni Z, He J, Wu Y, Hu C, Dai X, Yan X, Li
B, Li X, Xiong H, Li Y, et al: Akt-mediated phosphorylation of
ATG4B impairs mitochondrial activity and enhances the warburg
effect in hepatocellular carcinoma cells. Autophagy. 14:685–701.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fu D, Li J, Wei J, Zhang Z, Luo Y, Tan H
and Ren C: HMGB2 is associated with malignancy and regulates
Warburg effect by targeting LDHB and FBP1 in breast cancer. Cell
Commun Signal. 16(8)2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
de Andrade Barreto E, de Souza Santos PT,
Bergmann A, de Oliveira IM, Wernersbach Pinto L, Blanco T, Rossini
A, Pinto Kruel CD, Mattos Albano R and Ribeiro Pinto LF:
Alterations in glucose metabolism proteins responsible for the
warburg effect in esophageal squamous cell carcinoma. Exp Mol
Pathol. 101:66–73. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 44:646–674. 2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Barnes K, Mcintosh E, Whetton AD, Daley
GQ, Bentley J and Baldwin SA: Chronic myeloid leukaemia: An
investigation into the role of Bcr-Abl-induced abnormalities in
glucose transport regulation. Oncogene. 24:3257–3267.
2005.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang YH and Scadden DT: Targeting the
warburg effect for leukemia therapy: Magnitude matters. Mol Cell
Oncol. 2(e981988)2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Brittain EL: Clinical trials targeting
metabolism in pulmonary arterial hypertension. Adv Pulm Hypertens.
17:110–114. 2018.
|
|
20
|
Yeung SJ, Pan J and Lee MH: Roles of p53,
MYC and HIF-1 in regulating glycolysis-the seventh hallmark of
cancer. Cell Mol Life Sci. 65:3981–3999. 2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Szakács G, Paterson JK, Ludwig JA,
Booth-Genthe C and Gottesman MM: Targeting multidrug resistance in
cancer. Nat Rev Drug Discov. 5:219–234. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Zhu W, Huang Y, Pan Q, Xiang P, Xie N and
Yu H: MicroRNA-98 suppress warburg effect by targeting HK2 in colon
cancer cells. Dig Dis Sci. 62:660–668. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang S, Pei M, Li Z, Li H, Liu Y and Li
J: Double-negative feedback interaction between DNA
methyltransferase 3A and microRNA-145 in the warburg effect of
ovarian cancer cells. Cancer Sci. 109:2734–2745. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Fathullahzadeh S, Mirzaei H, Honardoost
MA, Sahebkar A and Salehi M: Circulating microRNA-192 as a
diagnostic biomarker in human chronic lymphocytic leukemia. Cancer
Gene Ther. 23:327–332. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ichihara A, Wang Z, Jinnin M, Izuno Y,
Shimozono N, Yamane K, Fujisawa A, Moriya C, Fukushima S, Inoue Y
and Ihn H: Upregulation of miR-18a-5p contributes to epidermal
necrolysis in severe drug eruptions. J Allergy Clin Immunol.
133:1065–1074. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xu XL, Jiang YH, Feng JG, Su D, Chen PC
and Mao WM: MicroRNA-17, microRNA-18a, and microRNA-19a are
prognostic indicators in esophageal squamous cell carcinoma. Ann
Thorac Surg. 97:1037–1045. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Takakura S, Mitsutake N, Nakashima M,
Namba H, Saenko VA, Rogounovitch TI, Nakazawa Y, Hayashi T, Ohtsuru
A and Yamashita S: Oncogenic role of miR-17-92 cluster in
anaplastic thyroid cancer cells. Cancer Sci. 99:1147–1154.
2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liang C, Zhang X, Wang HM, Liu XM, Zhang
XJ, Zheng B, Qian GR and Ma ZL: MicroRNA-18a-5p functions as an
oncogene by directly targeting IRF2 in lung cancer. Cell Death Dis.
8(e2764)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhang G, Han G, Zhang X, Yu Q, Li Z, Li Z
and Li J: Long non-coding RNA FENDRR reduces prostate cancer
malignancy by competitively binding miR-18a-5p with RUNX1.
Biomarkers. 23:435–445. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li B, He L, Zuo D, He W, Wang Y, Zhang Y,
Liu W and Yuan Y: Mutual Regulation of MiR-199a-5p and HIF-1α
modulates the warburg effect in hepatocellular carcinoma. J Cancer.
8:940–949. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Icard P, Shulman S, Farhat D, Steyaert JM,
Alifano M and Lincet H: How the Warburg effect supports
aggressiveness and drug resistance of cancer cells? Drug Resist
Updates. 38:1–11. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lévy P and Bartosch B: Metabolic
reprogramming: A hallmark of viral oncogenesis. Oncogene.
35:4155–4164. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Semenza GL: HIF-1 mediates the warburg
effect in clear cell renal carcinoma. J Bioenerg Biomembr.
39:231–234. 2007.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Rothschild SI: microRNA therapies in
cancer. Mol Cell Ther. 2(7)2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4(e05005)2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li L, Kang L, Zhao W, Feng Y, Liu W, Wang
T, Mai H, Huang J, Chen S, Liang Y, et al: miR-30a-5p suppresses
breast tumor growth and metastasis through inhibition of
LDHA-mediated Warburg effect. Cancer Lett. 400:89–98.
2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhu B, Cao X, Zhang W, Pan G, Yi Q, Zhong
W and Yan D: MicroRNA-31-5p enhances the Warburg effect via
targeting FIH. FASEB J. 33:545–556. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Qased AB, Yi H, Liang N, Ma S, Qiao S and
Liu X: MicroRNA-18a upregulates autophagy and ataxia telangiectasia
mutated gene expression in HCT116 colon cancer cells. Mol Med Rep.
7:559–564. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Faubert B, Boily G, Izreig S, Griss T,
Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, et
al: AMPK is a negative regulator of the warburg effect and
suppresses tumor growth in vivo. Cell Metab. 17:113–124.
2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Xian S, Shang D, Kong G and Tian Y: FOXJ1
promotes bladder cancer cell growth and regulates Warburg effect.
Biochem Biophys Res Commun. 495:988–994. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Raz S, Sheban D, Gonen N, Stark M, Berman
B and Assaraf YG: Severe hypoxia induces complete antifolate
resistance in carcinoma cells due to cell cycle arrest. Cell Death
Dis. 5(e1067)2014.PubMed/NCBI View Article : Google Scholar
|