Introduction
Plasmacytoma is a malignant disease characterized by
plasma cell proliferation in bone, soft tissue, or bone marrow,
with an overproduction of monoclonal protein, called M-protein,
which causes severe bone pain and bone fractures (1). Accumulation of malignant plasma cells
in bone marrow leads to impaired hematopoiesis, recurrent
infections, kidney damage, and is associated with a poor prognosis
(2). The advanced stage of
plasmacytoma is often called multiple myeloma. Homing of
plasmacytoma to bone marrow is essential to cell survival and drug
resistance (3,4). According to a model described by
Butcher and Picker, homing of plasmacytoma cells into the bone
marrow is likely to be mediated by a multistep process, including
rolling, firm adhesion, and transmigration into the tissue.
Within our present research, DHMEQ was found to
inhibit the expression of KISS1 receptor (KISS1R). The KISS1 gene
encodes KISS1, a protein that is rapidly processed in serum into
smaller but biologically active peptides, called kisspeptins
(5). KISS1 and the kisspeptins send
the signal via the G-protein coupled receptor KISS1R (also called
GPR54) (6). KISS1 was first found
to suppress metastasis of malignant melanoma (7). KISS1 was also found to be a metastasis
suppressor in many other cancers, such as bladder cancer (8), colorectal cancer (9), ovary cancer (10), and prostate cancer (11). However, besides working as a
suppressor of tumorigenesis and metastasis, growing evidence has
shown that the KISS1/KISS1R axis plays a cancer-promoting role
depending on the cancer type. In triple-negative breast cancer,
KISS1R was reported to be overexpressed and to promote drug
resistance (12). In addition to
playing a promoting role in breast cancer metastasis, KISS1 and
KISS1R appear to promote hepatic cell carcinoma (13).
Patients with plasmacytoma often progress to having
multiple myeloma with a median survival of 3-5 years (14). Conventional therapies with
alkylating agents, such as melphalan, cyclophosphamide,
anthracyclines, and corticosteroids, including high-dose therapy
followed by autologous transplantation, have shown improved
outcomes. However, such therapies cause numerous side effects
(15,16). Moreover, the remission rate remains
low and patients often develop resistance to chemotherapy (17).
NF-κB is reported to be constitutively activated in
plasmacytoma or multiple myeloma cells (18,19).
Recent studies have shown that elevated NF-κB activity promotes
growth and drug resistance in plasmacytoma cells, and these effects
can be inhibited by blocking NF-κB (20,21).
Dehydroxymethylepoxyquinomicin (DHMEQ, Fig. 1A) is a specific inhibitor of NF-κB.
It has inhibited various solid tumors and leukemias in animal
models without any toxicity (22).
DHMEQ was reported to induce apoptosis in multiple myeloma cells
and reduce tumor growth in vivo (23). Moreover, DHMEQ enhanced the
antitumor effect of fludarabine in chronic lymphocytic leukemia
cells (24). In the present study,
we researched the effect of DHMEQ on the cellular invasion of mouse
plasmacytoma SP2/0 cells. In addition, we studied the effect of
DHMEQ on sensitivity to melphalan.
In our research we have found that DHMEQ inhibits
constitutively activated NF-κB, and inhibits the cellular invasion
by suppressing KISS1R expression. It also enhances apoptotic
sensitivity to melphalan, possibly by down-regulated expression of
anti-apoptotic proteins.
Materials and methods
Chemicals
DHMEQ was synthesized as previously described
(25). Melphalan was purchased from
Sigma-Aldrich; Merck KGaA. DHMEQ and melphalan were dissolved in
dimethyl sulfoxide (DMSO), which was diluted by the medium.
Cell culture
The murine plasmacytoma SP2/0 (RIKEN BioResource
Center) and human myeloma KMS-11 (Japanese Collection of Research
Bioresources Cell Bank) and RPMI-8226 (Japanese Collection of
Research Bioresources Cell Bank) cells were cultured in RPMI-1640
medium (Sigma Chemical) supplemented with 10% fetal bovine serum
(FBS; Invitrogen; Thermo Fisher Scientific), and maintained at 37˚C
in humidified 95% air, with a 5% CO2 atmosphere.
NF-κB-DNA binding assay in vitro
SP2/0 cells in complete medium (3x106
cells) were grown in 60-mm dishes. The next day, the nuclear
extracts were prepared with Nuclear Extract Kit (Active Motif)
according to the instructions of the manufacturer. Briefly, the
cells were collected in ice-cold PBS in the presence of phosphatase
inhibitors. Then, the cells were suspended in a hypotonic buffer
and incubated on ice for 15 min. After incubation, detergent was
added and the samples were centrifuged. The supernatants were
discarded. The pellets were suspended in TransAM lysis buffer and
shaken for 30 min at 4˚C. Then, suspensions were centrifuged for 10
min at 14,000 g at 4˚C. Supernatants that contained nuclear
extracts were collected. Protein contents were determined via the
BCA assay. Nuclear extracts were treated with or without DHMEQ for
15 min. The DNA binding activity of NF-κB in nuclear extracts was
measured with the TransAM NF-κB p65 Transcription Factor Assay kit
(Active Motif). Briefly, nuclear extracts and Jurkat nuclear extract
for positive control were incubated in 96-well plates coated with
an immobilized oligonucleotide containing the NF-κB consensus
(5'-GGGACTTTCC-3') for 1 h. Activated NF-κB binding to the target
oligonucleotide was detected by 1 h incubation with a primary
antibody specific for the activated form of p65, followed by 1 h
incubation with a horseradish peroxidase-conjugated secondary
antibody. Then, developing solution was added. Reactions were
measured by a colorimetric method at 450 nm, and data were shown as
the percent of the positive control signal.
Cell viability assay
Cell viability was evaluated by the MTT assay. Cell
suspensions at a density of 3x105 cells per ml were
seeded in a 96-well plate. The next day, cells were treated with
the desired concentrations of DHMEQ for 24 h, and at each end-point
2 µl MTT was added to each well and incubated for additional 1 h.
For the combined activity study, cells were treated with DHMEQ for
30 min, then melphalan was added at the indicated concentrations
and incubated at the indicated times. After incubation, MTT reagent
was added to each well and incubated for 1 h. Absorbance was
measured at 570 nm with a microplate reader. Cell viability was
expressed as a percentage of the control samples.
Inhibition of NF-κB in cultured
cells
SP2/0 cells in complete medium (3x106
cells) were grown in 60-mm dishes. The next day, cells were treated
with the desired concentrations of DHMEQ for 2 h. The nuclear
extracts were prepared with a Nuclear Extract kit (Active Motif),
as described previously. Protein contents were determined via the
BCA assay. Then, the DNA binding activity of NF-κB in nuclear
extracts was measured with the TransAM NF-κB p65 Transcription
Factor Assay kit (Active Motif), as described previously.
Matrigel invasion assay
The cells were harvested and washed with PBS. After
washing, 3x105 cells were resuspended in 500 µl
serum-free medium and treated with different concentrations of
DHMEQ. Cells were then transferred to the top of Matrigel-coated
invasion chambers (24-well insert; Corning Inc.), and 750 µl of 20%
FBS-RPMI-1640 was added to the bottom chamber. After 24 h of
incubation, the non-invading cells were removed, and the invading
cells that were attached to the bottom were fixed with methanol for
10 min and stained with Diff-Quick solution (Sysmex). Invading
cells were photographed under the microscope at x10 magnification
and counted.
PCR analysis
SP2/0 cells were treated with DHMEQ for 6 h. Total
RNA was extracted from cells using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific) following the manufacturer's
instructions. Reverse transcription was carried out at 37˚C for 120
min with a High-Capacity cDNA Reverse Transcription kit (Applied
Biosystems). The cDNA was used for PCR amplification with rTaq DNA
polymerase. The sequence-specific primers were as follows: GAPDH
sense, 5'-ACCCAGAAGACTGTGGATGG-3', antisense,
5'-GGATGCAGGGATGATGTTCT-3'; Bcl-XL sense
5'-AGTAAACTGGGGTCGCATCG-3', antisense, 5'-GGGTGTACCTCCACTCACAC-3';
Bfl-1 sense 5'-GCTCATGCATATCCACTCCCT-3', antisense,
5'-GTAGCACTCTGCATGCTTGG-3'; c-FLIP sense 5'-AGAACCTGGCTGCACCTAAC-3,
antisense, 5'-GAGAAGGTCAAACCGCCTCA-3'; KISS1R sense,
5'-ATGTCCTACAGCAACTCGGC-3', antisense, 5'-AGAGTGAGGCAGTGCGTTC-3'.
PCR products were electrophoresed in 2% agarose gels, stained with
ethidium bromide, and visualized with a UV illuminator.
Knockdown by siRNA transfection
siKISS1R (sc-60748), and control siRNA (sc-37007)
were purchased from Santa Cruz Biotechnology Inc. siKISS1R
knockdown was performed using Lipofectamine® RNAiMax
transfection reagent (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. The efficiency of transfection was
determined by mRNA expression.
Western blotting
Detection of cleaved caspase-3 induced by DHMEQ and
melphalan was performed by western blot analysis. Antibodies used
in these experiments were as follows: Rabbit polyclonal antibody
for cleaved caspase-3 (Asp175) (Cell Signaling Technology, Inc.),
rabbit polyclonal antibody for caspase-3 (Cell Signaling
Technology, Inc.), and tubulin (Santa Cruz Biotechnology, Inc.).
Cells were lysed in lysis buffer (RIPA). Cell lysates were
subjected to electrophoresis on an SDS polyacrylamide gel and
transferred onto nitrocellulose membranes. The membranes were
blocked with TBS containing 0.05% Tween and 5% nonfat milk at 4˚C
overnight. Subsequently, they were incubated for 1 h with
anti-caspase-3 or anti-tubulin antibodies. The membranes were then
incubated for 1 h with peroxidase-labeled anti-rabbit or anti-mouse
secondary antibody and developed using an enhanced
chemiluminescence system. ImageJ software-based analysis was used
to quantify the intensity of the bands obtained by the Western blot
assay.
PCR array
Total RNA was extracted from SP2/0 cells using
RNeasy Mini kit (Qiagen GmbH). RT2 First Strand kit
(Qiagen, Inc.) was used for reverse transcription. The cDNA was
added to the qPCR Master Mix and the aliquot mixture across the
Human Tumor Metastasis PCR Array (Qiagen, Inc.). The comparative CT
method was used for data analysis.
Annexin V assay
The cells were seeded in 24-well culture plates
(5x105 cells/well). Next, the cells were incubated with
the melphalan (0.3 µg/ml) and/or DHMEQ (10 µg/ml) for 24 h,
followed by incubation with annexin V (Ax)-FITC (MBL) and PI (10
µg/ml) at 25˚C room temperature for 15 min. Finally, fluorescence
intensities were determined by fluorescence-activated cell sorting
(FACS) using a FACSCantoII (BD Biosciences).
Statistical analysis
All results were expressed as mean ± standard
deviation (SD). One-way analysis of variance (ANOVA) followed by
Dunnett's post hoc test was used for statistical analysis by EXCEL
software version 2016. P<0.05 was considered to indicate a
statistically significant difference.
Results
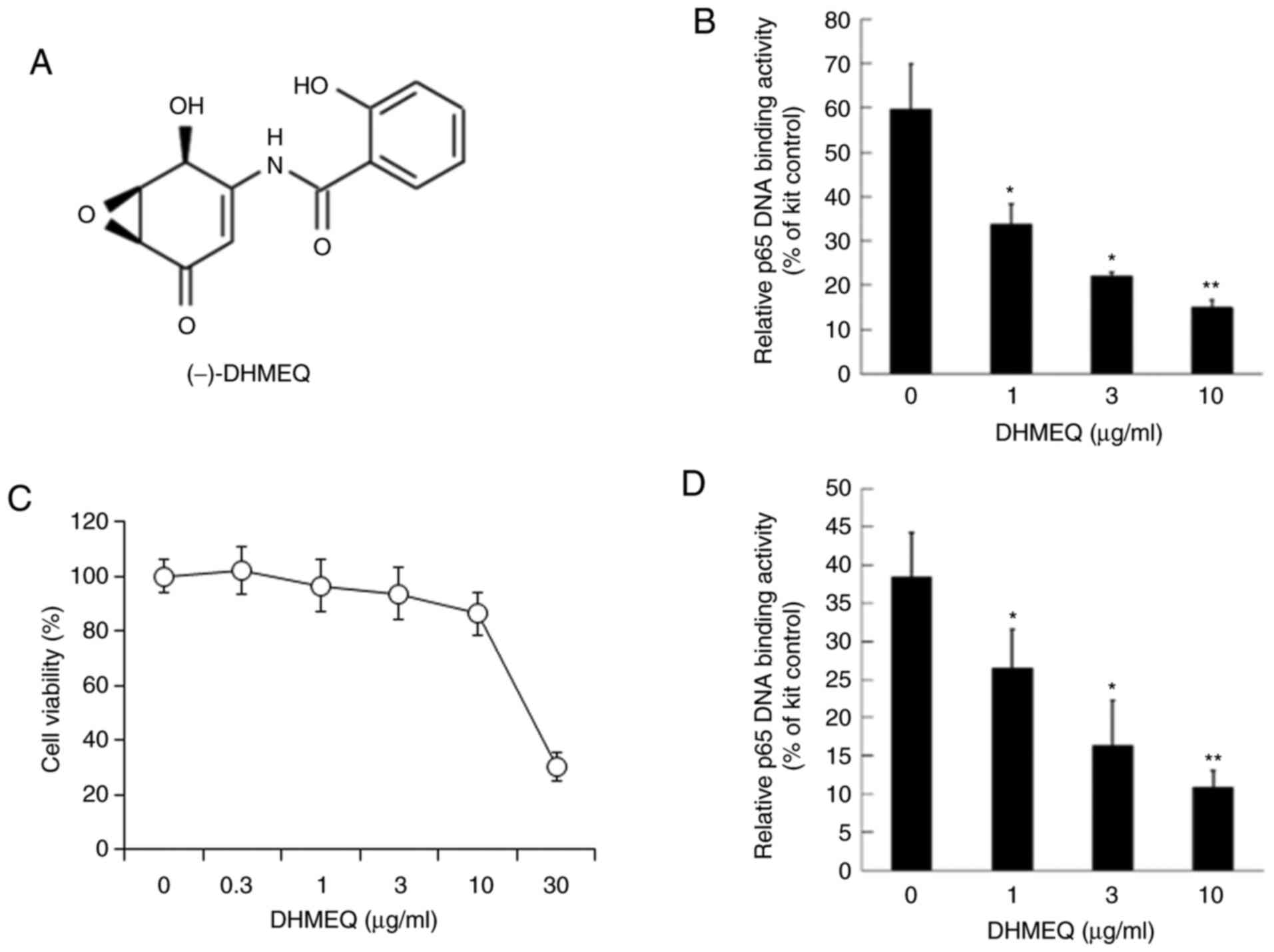
Inhibition of NF-κB by DHMEQ
The nuclear fraction of SP2/0 cells was used for
NF-κB, including p65. The in vitro binding of p65 and κB
sequence DNA was inhibited by DHMEQ (Fig. 1A) at 1-10 µg/ml, as shown in
Fig. 1B. Next, the effect on
viability was studied. DHMEQ did not show prominent cytotoxicity
below 10 µg/ml in 24 h, as shown in Fig. 1C. Then, DHMEQ was added to the
cultured SP2/0 cells for 2 h. DHMEQ at 1-10 µg/ml inhibited the
NF-κB activity in cultured SP2/0 cells (Fig. 1D).
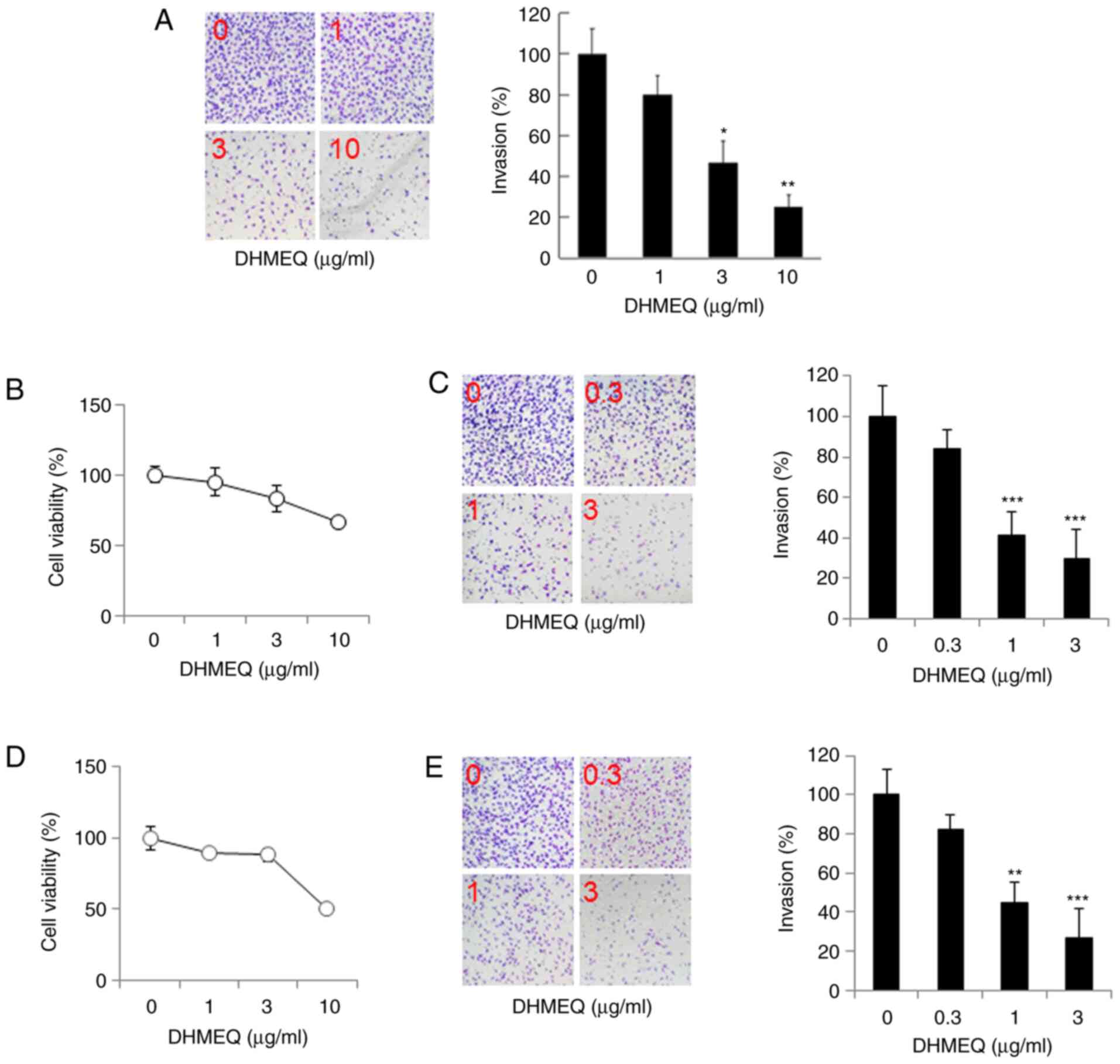
Inhibition of cellular invasion by
DHMEQ
Next, we studied the effect of DHMEQ on the cellular
invasion of SP2/0 cells using a Matrigel chamber. DHMEQ inhibited
the invasion at 1-10 µg/ml dose-dependently, as shown in Fig. 2A. We then studied the effect of
DHMEQ on human myeloma cell lines KMS-11 and RPMI-8226. DHMEQ was
not toxic at 3 µg/ml, and inhibited cellular invasion below 3 µg/ml
in KMS-11 cells (Fig. 2B and
C) and in RPMI-8226 cells (Fig. 2D and E). Thus, DHMEQ inhibited cellular invasion
in SP2/0 cells and human myeloma KMS-11 and RPMI-8226 cells at
nontoxic concentrations.
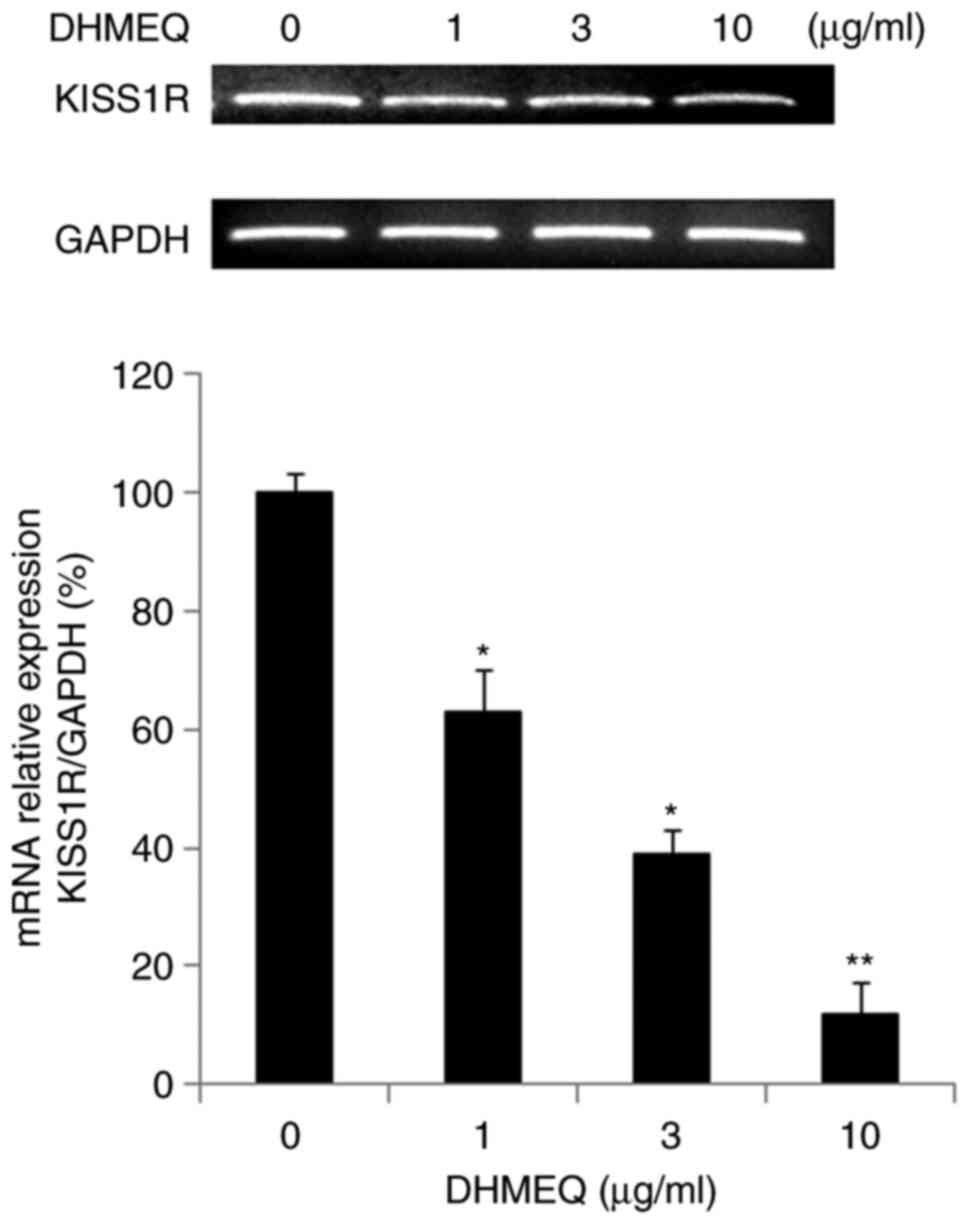
Inhibition of KISS1R expression by
DHMEQ in mouse metastasis PCR array
We employed a mouse metastasis PCR array to study
the mechanism of inhibition by DHMEQ. The results of the PCR array
are shown in Table SI. Several
gene expressions were activated or inhibited by DHMEQ. Among them,
we selected KISS1R for further study because its function in cancer
progression is not fully understood. We confirmed the effect of
DHMEQ on KISS1R expression by independent PCR. DHMEQ inhibited the
KISS1R expression as shown in Fig.
3.
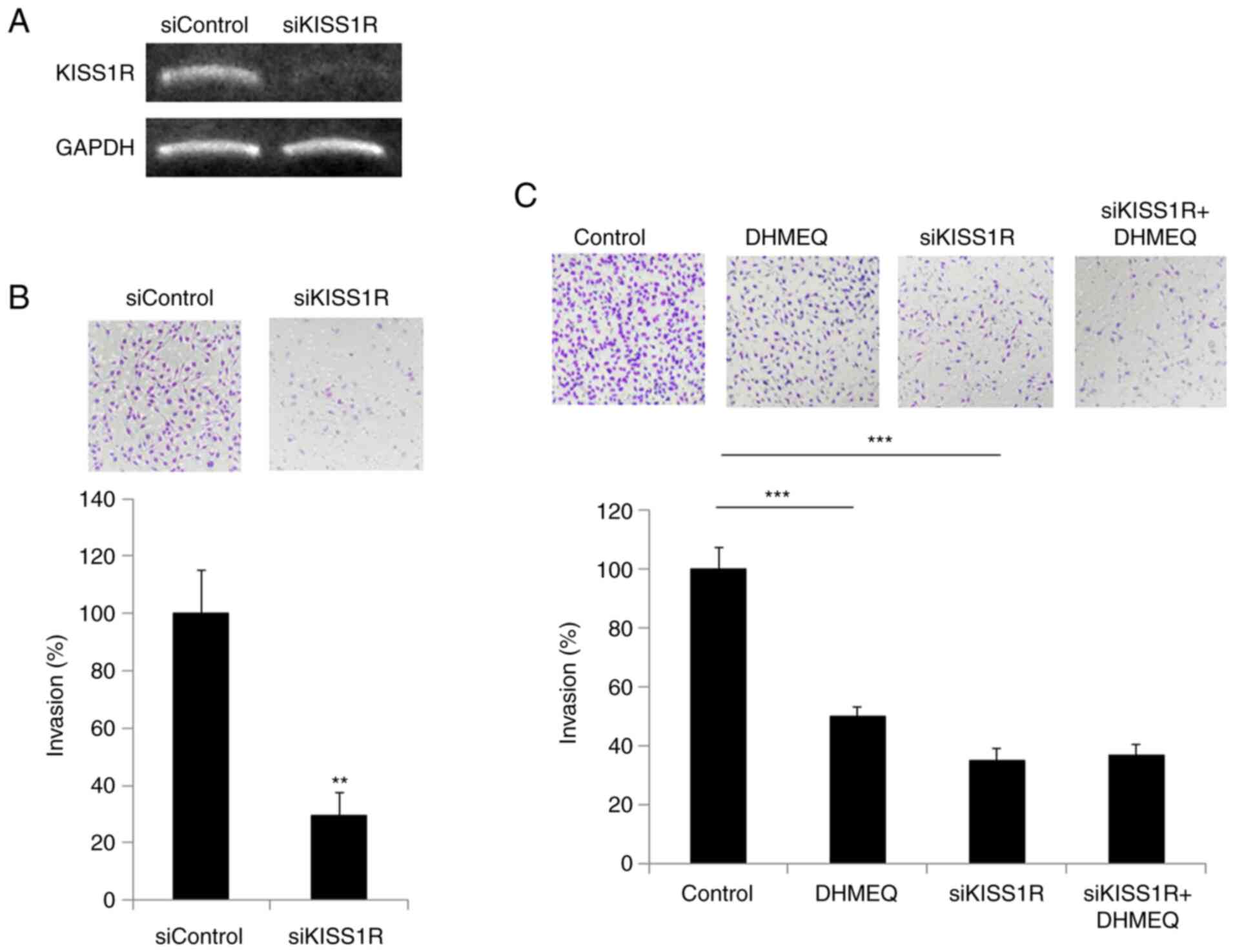
Inhibition of cellular invasion by
KISS1R knockdown
Next, we prepared KISS1R-knockdown SP2/0 cells using
siRNA (Fig. 4A). The knockdown of
KISS1R inhibited the invasion, as shown in Fig. 4B. DHMEQ showed no prominent
additional effect on invasion (Fig.
4C), which indicates KISS1R is mainly involved in the effect of
DHMEQ on inhibition of invasion.
Increase of anticancer drug
sensitivity by DHMEQ
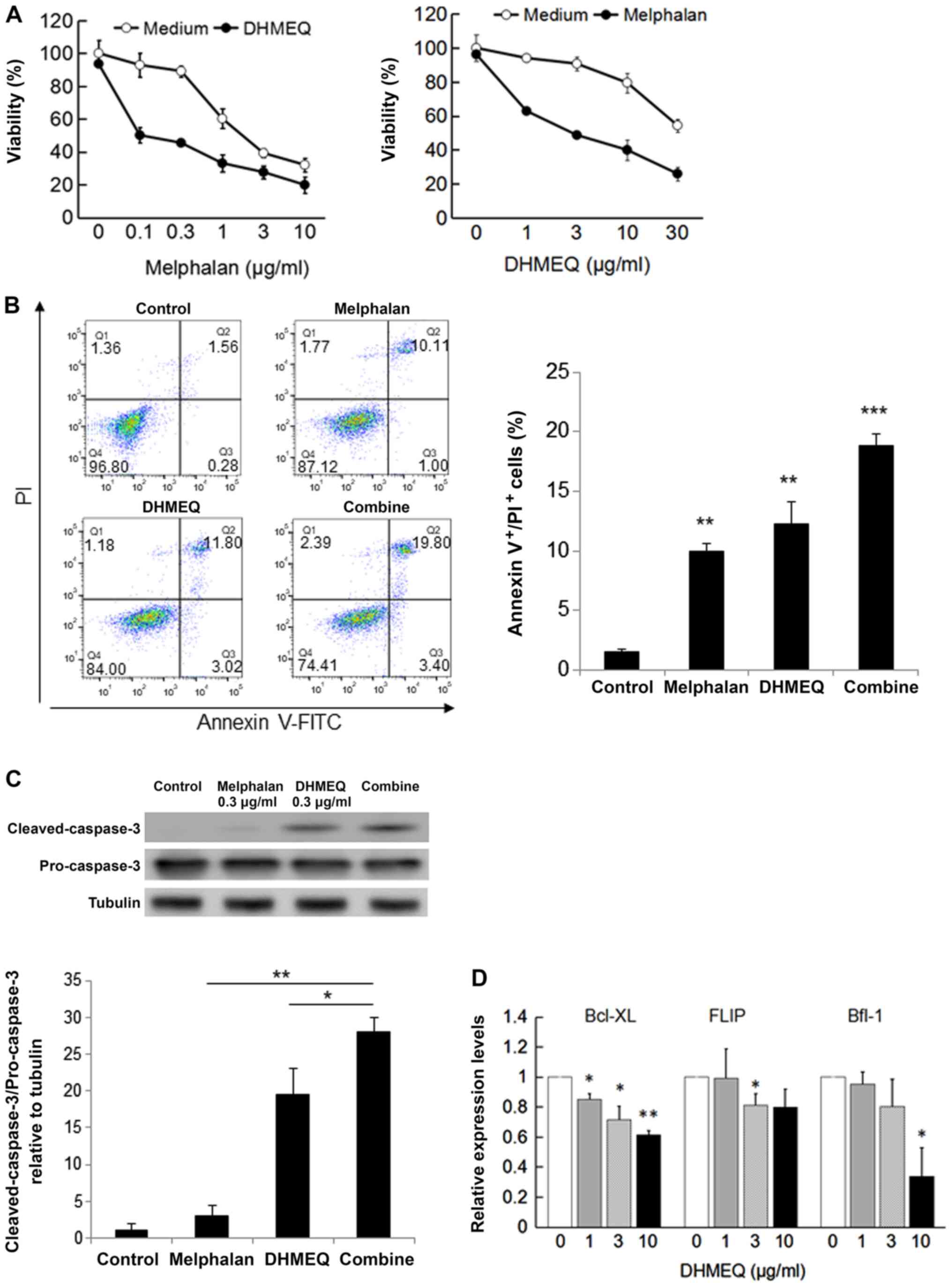
Melphalan is an alkylating agent often used for the
treatment of plasmacytoma and multiple myeloma. We found that DHMEQ
enhanced the cytotoxicity of melphalan in SP2/0 cells (Fig. 5A). Apoptosis was measured by annexin
V-FITC/PI and cleaved caspase-3 expression. DHMEQ increased the
melphalan-induced apoptosis synergistically (Fig. 5B and C). To understand the mechanism of
synergistic apoptosis induction, we studied the effect on
NF-κB-dependent apoptosis inhibitory proteins. As shown in Fig. 5D, DHMEQ inhibited the expression of
Bcl-XL, Flip, and Bfl-1 in SP2/0 cells. Thus, DHMEQ was shown to
increase melphalan sensitivity, possibly by decreasing apoptosis
inhibitory proteins.
Discussion
NF-κB is constitutively activated and plays an
important role in the suppression of apoptosis in plasmacytoma or
multiple melanoma cells (26,27).
Over-activation of NF-κB is known to contribute to tumor
progression and metastasis (28).
Also, drug resistance is a serious problem for plasmacytoma
chemotherapy. In the present research, we studied the cellular
anti-metastatic activity and the effect on drug sensitivity of an
NF-κB inhibitor, DHMEQ.
DHMEQ inhibited cellular invasion by lowering the
expression of KISS1R. The KISS1/KISS1R axis has been reported as
both a metastasis suppressor and metastasis promoter. Our results
indicate that the KISS1/KISS1R axis would act as a metastasis
promoter in plasmacytoma cells, and DHMEQ could inhibit the
invasion and metastasis via suppression of KISS1R expression. We
could not locate a report describing the existence of a κB-site in
the promoter of KISS1R. Therefore, it is considered that DHMEQ
inhibits KISS1R expression indirectly.
Melphalan is a DNA alkylating agent often used in
combination with other anticancer agents or steroids for
plasmacytoma patients. For newly diagnosed patients not eligible
for a transplant the standard drug combination is melphalan,
prednisone, and thalidomide, while an alternate combination is
melphalan, prednisone, and bortezomib. However, the patients
receive either therapy often experience drug-mediated side effects,
such as neutropenia, thrombocytopenia, and infections (16). Bortezomib, a proteasome inhibitor,
inhibits NF-κB by blocking proteasomal degradation of the
inhibitory protein, I-κBα. Even though bortezomib shows remarkable
anti-tumor activity, it has been associated with possible
off-target toxicities and the development of drug resistance
(29,30). In the present study, we demonstrate
that DHMEQ enhances the cytotoxicity of melphalan on SP2/0 cells.
DHMEQ synergistically increases the melphalan-induced apoptosis,
possibly by inhibition of antiapoptotic protein expression.
Compared to other NF-κB inhibitors, DHMEQ is distinctive by
covalently binding to the specific cysteine residue of the Rel
family proteins, which is essential for DNA binding (31). It should be noted that DHMEQ alone
did not significantly decrease the viability of SP2/0 cells. This
finding supports the notion that DHMEQ has no toxicity. Hence, it
should be combined with melphalan. On the other hand, we believe
that the effect of DHMEQ on bortezomib would not be prominent. This
would be because both compounds inhibit the NF-κB activating
pathway (32). DHMEQ would also not
increase the sensitivity to dexamethasone since this compound is an
anti-inflammatory steroid and would affect the NF-κB activity
(33).
In conclusion, targeting NF-κB activity is a logical
strategy to treat plasmacytoma and prevent progression to multiple
myeloma. We also wish to investigate further how DHMEQ inhibits
KISS1R expression by searching for the κB sequence in the promoter
region.
Supplementary Material
Changes in gene expression in
DHMEQ-treated SP2/0 cells.
Acknowledgements
The authors would like to thank Dr Sivasundaram
Karnan (Department of Biochemistry, Aichi Medical University,
Nagakute, Japan) for providing us with the human myeloma KMS-11 and
RPMI-8226 cell lines.
Funding
Funding: The present study was supported in part by JSPS Kakenhi
(grant no. 17K01967) and AMED (grant no. JP18fk0310118).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author's contributions
YL and KS did the majority of experiments. YL and KS
confirmed the authenticity of all the raw data. KU, NK and TK
designed the experiments. JM prepared DHMEQ. KU, KS and YL prepared
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
Department of Molecular Target Medicine is a
fund-donated laboratory (the authors YL, KS and KU are affiliated
with this laboratory). It is supported financially by Shenzhen
Wanhe Pharmaceutical Company (the author JM is affiliated with this
company), Shenzhen, China, Meiji Seika Pharma, Tokyo, Japan, Fukuyu
Medical Corporation, Nisshin, Japan, and Brunaise Co., Ltd.,
Nagoya, Japan.
References
|
1
|
Dimopoulos MA, Moulopoulos LA, Maniatis A
and Alexanian R: Solitary plasmacytoma of bone and asymptomatic
multiple myeloma. Blood. 96:2037–2044. 2000.PubMed/NCBI
|
|
2
|
Laubach J, Richardson P and Anderson K:
Multiple Myeloma. Annu Rev Med. 62:249–264. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Radbruch A, Muehlinghaus G, Luger EO,
Inamine A, Smith KGC, Dörner T and Hiepe F: Competence and
competition: The challenge of becoming a long-lived plasma cell.
Nat Rev Immunol. 6:741–750. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Azab AK, Runnels JM, Pitsillides C, Moreau
AS, Azab F, Leleu X, Jia X, Wright R, Ospina B, Carlson AL, et al:
CXCR4 inhibitor AMD3100 disrupts the interaction of multiple
myeloma cells with the bone marrow microenvironment and enhances
their sensitivity to therapy. Blood. 113:4341–4351. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mead EJ, Maguire JJ, Kuc RE and Davenport
AP: Kisspeptins: A multifunctional peptide system with a role in
reproduction, cancer and the cardiovascular system. Br J Pharmacol.
151:1143–1153. 2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kotani M, Detheux M, Vandenbogaerde A,
Communi D, Vanderwinden JM, Poul EL, Brézillon S, Tyldesley R,
Suarez-Huerta N, Vandeput F, et al: The metastasis suppressor Gene
KiSS-1 encodes Kisspeptins, the natural ligands of the orphan g
protein-coupled receptor GPR54. J Biol Chem. 276:34631–34636.
2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lee JH, Miele ME, Hicks DJ, Phillips KK,
Trent JM, Weissman BE and Welch DR: KiSS-1, a novel human malignant
melanoma metastasis-suppressor gene. J Natl Cancer Inst.
88:1731–1737. 1996.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Takeda T, Kikuchi E, Mikami S, Suzuki E,
Matsumoto K, Miyajima A, Okada Y and Oya M: Prognostic role of
KiSS-1 and possibility of therapeutic modality of metastin, the
final peptide of the KiSS-1 gene, in urothelial carcinoma. Mol
Cancer Ther. 11:853–863. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhu C, Takasu C, Morine Y, Bando Y,
Ikemoto T, Saito Y, Yamada S, Imura S, Arakawa Y and Shimada M:
KISS1 associates with better outcome via inhibiting matrix
metalloproteinase-9 in colorectal liver metastasis. Ann Surg Oncol.
22:1516–1523. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang Y, Berk M, Singh LS, Tan H, Yin L,
Powell CT and Xu Y: KiSS1 suppresses metastasis in human ovarian
cancer via inhibition of protein kinase C alpha. Clin Exp
Metastasis. 22:369–376. 2005.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang H, Jones J, Turner T, He QP, Hardy S,
Grizzle WE, Welch DR and Yates C: Clinical and biological
significance of KISS1 expression in prostate cancer. Am J Pathol.
180:1170–1178. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Blake A, Dragan M, Tirona RG, Hardy DB,
Brackstone M, Tuck AB, Babwah AV and Bhattacharya M: G
protein-coupled KISS1 receptor is overexpressed in triple negative
breast cancer and promotes drug resistance. Sci Rep.
7(46525)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Schmid K, Wang X, Haitel A, Sieghart W,
Peck-Radosavljevic M, Bodingbauer M, Rasoul-Rockenschaub S and Wrba
F: KiSS-1 overexpression as an independent prognostic marker in
hepatocellular carcinoma: An immunohistochemical study. Virchows
Archiv. 450:143–149. 2007.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Rajkumar SV: Multiple myeloma: 2013 update
on diagnosis, risk-stratification, and management. Am J Hematol.
88:226–235. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hideshima T, Richardson P and Anderson KC:
Novel therapeutic approaches for multiple myeloma. Immunolol Rev.
194:164–176. 2003.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Gay F and Palumbo A: Management of
disease- and treatment-related complications in patients with
multiple myeloma. Med Oncol. 27:43–52. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Mahindra A, Laubach J, Raje N, Munshi N,
Richardson PG and Anderson K: Latest advances and current
challenges in the treatment of multiple myeloma. Nat Rev Clin
Oncol. 9:135–143. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Keats JJ, Fonseca R, Chesi M, Schop R,
Baker A, Chng WJ, Wier SV, Tiedemann R, Shi CX, Sebag M, et al:
Promiscuous mutations activate the noncanonical NF-kappaB pathway
in multiple myeloma. Cancer Cell. 12:131–144. 2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Annunziata CM, Davis RE, Demchenko Y,
Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W,
et al: Frequent engagement of the classical and alternative
NF-kappaB pathways by diverse genetic abnormalities in multiple
myeloma. Cancer Cell. 12:115–130. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Berenson JR, Ma HM and Vescio R: The role
of nuclear factor-kappaB in the biology and treatment of multiple
myeloma. Semin Oncol. 28:626–633. 2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Landowski TH, Olashaw NE, Agrawal D and
Dalton WS: Cell adhesion-mediated drug resistance (CAM-DR) is
associated with activation of NF-kappa B (RelB/p50) in myeloma
cells. Oncogene. 22:2417–2421. 2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Umezawa K: Possible role of peritoneal
NF-κB in peripheral inflammation and cancer: Lessons from the
inhibitor NF-κB (Review). Biomed Pharmacother. 65:252–259.
2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tatetsu H, Okuno Y, Nakamura M, Matsuno F,
Sonoki T, Taniguchi I, Uneda S, Umezawa K, Mitsuya H and Hata H:
Dehydroxymethylepoxyquinomicin, a novel nuclear factor-kappa B
inhibitor, induces apoptosis in multiple myeloma cells in an
IkappaBalpha-independent manner. Mol Cancer Ther. 4:1114–1120.
2005.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Horie R, Watanabe M, Okamura T, Taira M,
Shoda M, Motoji T, Utsunomiya A, Watanabe T, Higashihara M and
Umezawa K: DHMEQ, a new NF-kappaB inhibitor, induces apoptosis and
enhances fludarabine effects on chronic lymphocytic leukemia cells.
Leukemia. 20:800–806. 2006.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Suzuki Y, Sugiyama C, Ohno O and Umezawa
K: Preparation and biological activities of optically active
dehydroxymethylepoxyquinomicin, a novel NF-κB inhibitor.
Tetrahedron. 60:7061–7066. 2004.
|
|
26
|
Ni H, Ergin M, Huang Q, Qin JZ, Amin HM,
Martinez RL, Saeed S, Barton K and Alkan S: Analysis of expression
of nuclear factor B (NF-kappa B) in multiple myeloma:
Down-regulation of NF-kappa B induces apoptosis. Br J Haematol.
115:279–286. 2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Barkette M and Gilmore TD: Control of
apoptosis by Rel/NF-kappaB transcription factors. Oncogene.
18:6910–6924. 1999.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Pahl HL: Activators and target genes of
Rel/NF-kappaB transcription factors. Oncogene. 18:6853–6866.
1999.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Richardson PG, Sonneveld P, Schuster MW,
Irwin D, Stadtmauer EA, Facon T, Harousseau JL, Ben-Yehuda D,
Lonial S, Miguel JF, et al: Safety and efficacy of bortezomib in
high risk and elderly patients with relapsed multiple myeloma. Br J
Haematol. 137:429–435. 2007.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lonial S, Waller EK, Richardson PG,
Jagannath S, Orlowski RZ, Giver CR, Jaye DL, Francis D, Giusti S,
Torre C, et al: Risk factors and kinetics of thrombocytopenia
associated with bortezomib for relapsed, refractory multiple
myeloma. Blood. 106:3777–3784. 2005.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yamamoto M, Horie R, Takeiri M, Kozawa I
and Umezawa K: Inactivation of NF-kappaB components by covalent
binding of (-)-dehydroxymethylepoxyquinomicin to specific cysteine
residues. J Med Chem. 51:5780–5788. 2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Vega MI, Martinez-Paniagua M, Jazirehi AR,
Huerta-Yepez S, Umezawa K, Martinez-Maza O and Bonavida B: The
NF-kappaB inhibitors (bortezomib and DHMEQ) sensitise
rituximab-resistant AIDS-B-non-Hodgkin lymphoma to apoptosis by
various chemotherapeutic drugs. Leuk Lymphoma. 49:1982–1994.
2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ando Y, Sato Y, Kudo A, Watanabe T,
Hirakata A, Okada AA, Umezawa K and Keino H: Anti-inflammatory
effects of the NF-κB inhibitor dehydroxymethylepoxyquinomicin on
ARPE-19 cells. Mol Med Rep. 22:582–590. 2020.PubMed/NCBI View Article : Google Scholar
|