Introduction
Kashin-Beck disease (KBD) is a chronic and endemic
osteoarthropathy characterized by chondrocyte necrosis in growth
plates and articular cartilage (1).
The geographical distribution of KBD includes southeastern Siberia
in Russia, the northern region of North Korea and a long narrow
zone from northeast to southwest China (2). Patients with KBD often exhibit
clinical features such as arthralgia, restricted mobility and an
enlarged metaphysis (1). In severe
cases, short stature and dwarfism may occur (3). The pathogenesis of KBD has yet to be
fully elucidated, although it is generally considered that its
cause is multifactorial, including such contributory factors as
selenium deficiency, iodine deficiency, food contaminated with
mycotoxins and drinking water contaminated with humic acid
(4).
Mycotoxins are a group of toxic secondary
metabolites produced by fungal species. According to present
statistics, ~25% of the world's food crops are contaminated with
mycotoxins (5). T-2 toxin is a
mycotoxin widely found in grains in the geographical regions where
KBD is prevalent, and has been shown to induce apoptosis of human
chondrocytes, oxidative stress and mitochondrial damage (6). When compared with normal subjects, the
expression levels of apoptosis-associated molecules, including
Bcl-2, Bax, Fas and inducible nitric oxide synthase, in the
articular cartilage of patients with KBD have been shown to be
elevated (7). In addition, T-2
toxin contamination and selenium deficiency have been shown to be
widespread in drinking water and cereals in KBD-endemic areas
(8). Selenium is an essential
biological trace element that has previously been used in the
treatment of KBD (9).
The 2019 Nobel Prize in Physiology or Medicine was
awarded to scientists for their discovery of how cells sense and
adapt to oxygen supply, and also for their contribution towards
understanding of the molecular machinery that regulates the
activity of genes in response to varying levels of oxygen (10). Previous studies have demonstrated
that hypoxia and hypoxia-associated signaling pathways exert an
important role in the progression of KBD disease (11,12).
Under low-O2 (hypoxic) conditions, cells activate
various adaptive responses to match the oxygen demands of
metabolic, bioenergetic and redox processes (13). Hypoxia-inducible factor (HIF) is
considered to be a key regulator of the transcriptional response to
hypoxic stress. HIF is a heterodimeric transcription factor that
consists of either HIF-1α or HIF-2α and HIF-1β/ARNT subunits
(14). It is a key transcription
factor that is activated in a hypoxic environment, subsequently
regulating the expression of a series of genes that are responsible
for cell metabolism, migration, proliferation, angiogenesis and
inflammation (15). Previous
studies have shown that T-2 toxin is able to induce the production
of reactive oxygen species in chondrocytes, and activate the
expression of both NF-κB and HIF-2α (16-18).
Known HIF-1 signaling pathways include the
PI3K-Akt/HIF-1α, MAPK/HIF-1α and HIF-1α/vascular endothelial growth
factor A (VEGFA) signaling pathways (19-22).
VEGFA is a member of the VEGF platelet-derived growth factor family
of structurally related mitogens (23). Previous studies have shown that VEGF
in the articular cartilage of patients with KBD may be abnormally
expressed (24,25). Moreover, HIF has been shown to
regulate the expression levels of several hundred genes, and VEGF
is one of the primary target genes (26). In the past decade, extensive
research has clarified the key role of HIF and VEGF in controlling
the survival of hypoxic cartilage [for a review on this topic, see
(27)]. It was therefore possible
to hypothesize that the HIF-1α/VEGFA signaling pathway may be
associated with the pathogenesis and progression of KBD.
In the present study, the Comparative Toxicogenomics
Database (CTD) was used to identify genes associated with KBD, T-2
toxin and selenium. After identifying which of the genes were
intersecting, those genes were further selected for subsequent
enrichment analysis and protein-protein interaction (PPI) network
construction, and western blotting was then performed to verify the
expression levels of HIF-1α and VEGFA in chondrocytes treated with
T-2 toxin.
Materials and methods
Target identification using the
CTD
The CTD (http://ctdbase.org) is a useful public resource
featuring extensive information regarding exposure to numerous
types of chemicals and human health (28). The database includes in excess of 38
million toxicogenomic relationships that may be explored further in
terms of analytical investigations and the development of
scientific hypotheses. In the present study, all key genes
associated with KBD, T-2 toxin and selenium were predicted using
CTD database that was updated to June 2020 (inference score,
>3.07).
Gene Ontology (GO) and pathway
enrichment analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; https://david.ncifcrf.gov) is an online bioinformatics
tool designed to identify the functions of a large number of genes
or proteins (11). In the present
study, KBD-associated key genes were uploaded, and GO enrichment
results were collected (http://geneontology.org/), including biological
processes (BP), cellular component (CC), molecular function (MF)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) database was
used for pathway enrichment analysis (https://www.kegg.jp/). P<0.05 was considered to
indicate a statistically significant result in this analysis.
PPI network analysis
Information regarding PPIs may be evaluated using an
online tool, the Search Tool for the Retrieval of Interacting Genes
(STRING; https://string-db.org) (29). To estimate the interactions of
KBD-associated key genes, these genes were first analyzed by
STRING, and subsequently Cytoscape software (version 3.6.1;
https://cytoscape.org) was used to construct a
PPI network. The Molecular Complex Detection (MCODE; http://apps.cytoscape.org/apps/mcode)
plug-in for Cytoscape was used to investigate modules of the PPI
network (degree cutoff=2; maximum depth=100; k-core=2; and node
score cutoff=0.2). Similarly, the STRING database and Cytoscape
software were used to construct a PPI network of genes associated
with the HIF-1 signaling pathway.
Chondrocyte culture and experimental
protocol
Human C28/I2 normal chondrocytes were purchased from
the BeNa Culture Collection and cultured in DMEM Nutrient Mixture
F-12 (DMEM/F12; 1:1) (Gibco; Thermo Fisher Scientific) supplemented
with 10% FBS (Hyclone; Cytiva) in a humidified incubator containing
5% CO2 at 37˚C. All cells were used for subsequent
experiments between the fifth and tenth passages. Cells were
cultured in 6-well plates and used for protein extraction once the
cell density had reached 6x104 cells/well. The medium
was replaced every other day. T-2 toxin was provided by
MedChemExpress and dissolved in DMSO to make up a working solution
with a concentration of 100 µg/ml T-2 toxin. The cells were plated
and incubated for 24 h to allow them to adhere prior to treatment
with T-2 toxin. Subsequently, cells were exposed to fresh medium
containing various doses of T-2 toxin (0, 0.001, 0.005, 0.01, 0.02
and 0.05 µg/ml). Proteins were then extracted by ultrasonic
disruption after the C28/I2 chondrocytes had been incubated at 37˚C
with T-2 toxin for 3 days, as detailed in previous studies
(30,31). Incubation with each concentration of
T-2 toxin was repeated five times, and DMSO-treated cells were used
as a control.
Western blot analysis
The protein concentration was determined by using
the BCA method (Biyuntian Biotechnology). SDS-PAGE loading buffer
was added and the protein sample boiled in 100˚C water. Protein
samples of chondrocytes treated with different concentrations of
T-2 toxin were separated using SDS-PAGE (10% gels) and transferred
to PVDF membranes (EMD Millipore). After blocking with 5% skimmed
milk diluted in TBS containing 0.1% Tween-20 (TBST) overnight at
room temperature, the membranes were incubated with primary
antibodies, as detailed below, and then incubated with the
secondary antibodies conjugated with horseradish peroxidase. Blots
were visualized by using a hypersensitivity ECL chemiluminescence
detection kit (Biyuntian Biotechnology). The anti-HIF-1α (1:1,000;
cat. no. ab82832) and anti-VEGFA (1:1,000; cat. no. ab46154)
antibodies were purchased from Abcam, whereas the rabbit
anti-β-actin (1:3,000; cat. no. bs-0061R) and goat anti-rabbit IgG
(1:3,000; cat. no. bs-0295G-HRP) antibodies were purchased from
BIOSS. The primary antibodies were incubated for 30 min at 37˚C and
then overnight at 4˚C. The membrane was subsequently incubated with
a secondary antibody for 1 h at room temperature after washing
three times in TBST. The relative intensities of the blots
featuring the target proteins of interest were calculated by
normalizing against β-actin. Densitometric analysis of western
blots was performed using ImageJ software (v1.52; National
Institutes of Health).
Transcription factor prediction
IRegulon (http://apps.cytoscape.org/apps/iregulon) was developed
using a genome-wide ranking-and-recovery approach as a Cytoscape
plug-in for the purpose of detecting enriched transcription factor
motifs and their optimal sets of direct targets (32). This technology was used to perform
the enrichment of transcription factor motifs in target sequences
with a position matrix method to identify transcription factors
that were associated with HIF-1α, VEGFA and the HIF-1 signaling
pathway. A minimum identity between orthologous genes was defined
as 0.05, and the maximum false discovery rate (FDR) value of motif
similarity was set to 0.001. Associations with the normalized
enrichment score (NES) were used for further analysis.
Statistical analysis
The results are expressed as the mean ± SD for
experiments performed in triplicate. Statistical analysis was
performed using one-way ANOVA and the means were compared by
Dunnett's post hoc test using GraphPad Prism statistical software
(version 8.01; GraphPad Software, Inc.). P<0.05 was considered
to indicate a statistically significant difference.
Results
Identification of genes associated
with KBD, T-2 toxin and selenium
Using the CTD database, 3,676 genes were identified
to be associated with KBD, 2,299 to be associated with T-2 toxin
and 1,669 to be associated with selenium. In order to obtain the
KBD-associated key genes, the above genes were selected for
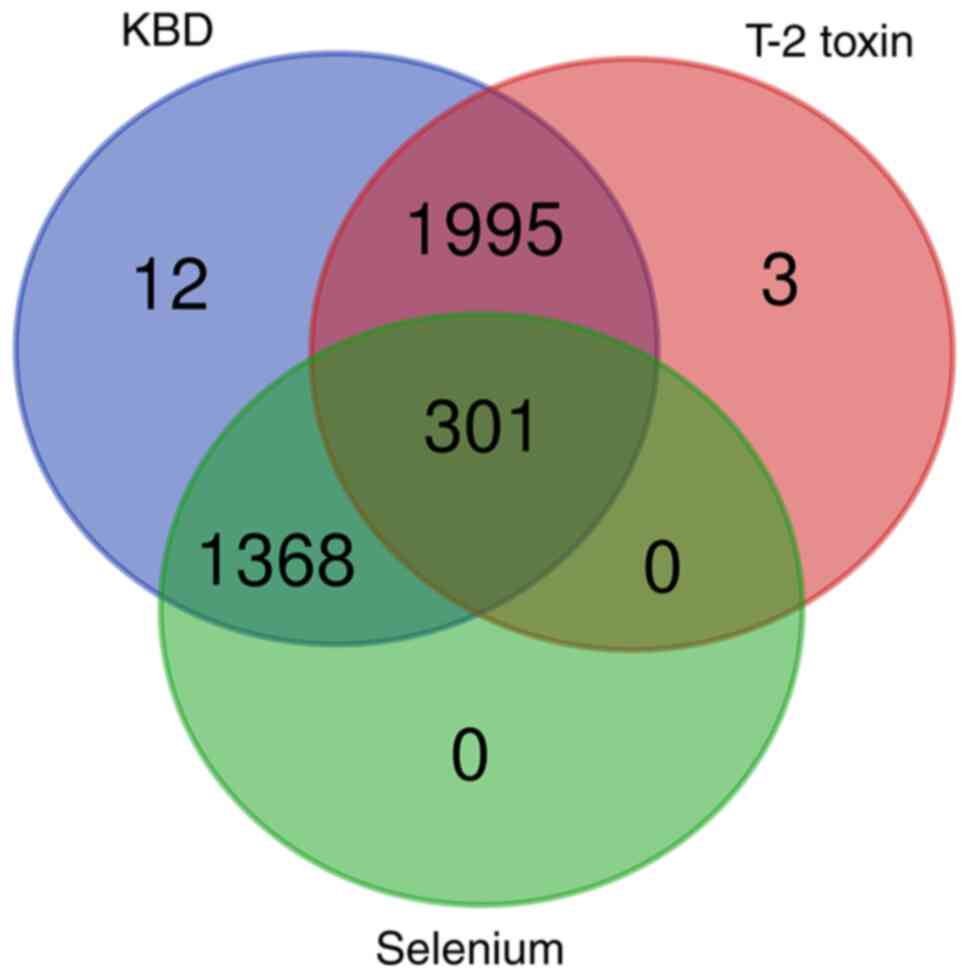
intersection analysis. A set of 301 key genes were revealed to be
held in common among the genes of the KBD, T-2 toxin and selenium
groups, as shown by the Venn diagram in Fig. 1.
Functional enrichment analysis of
KBD-associated key genes
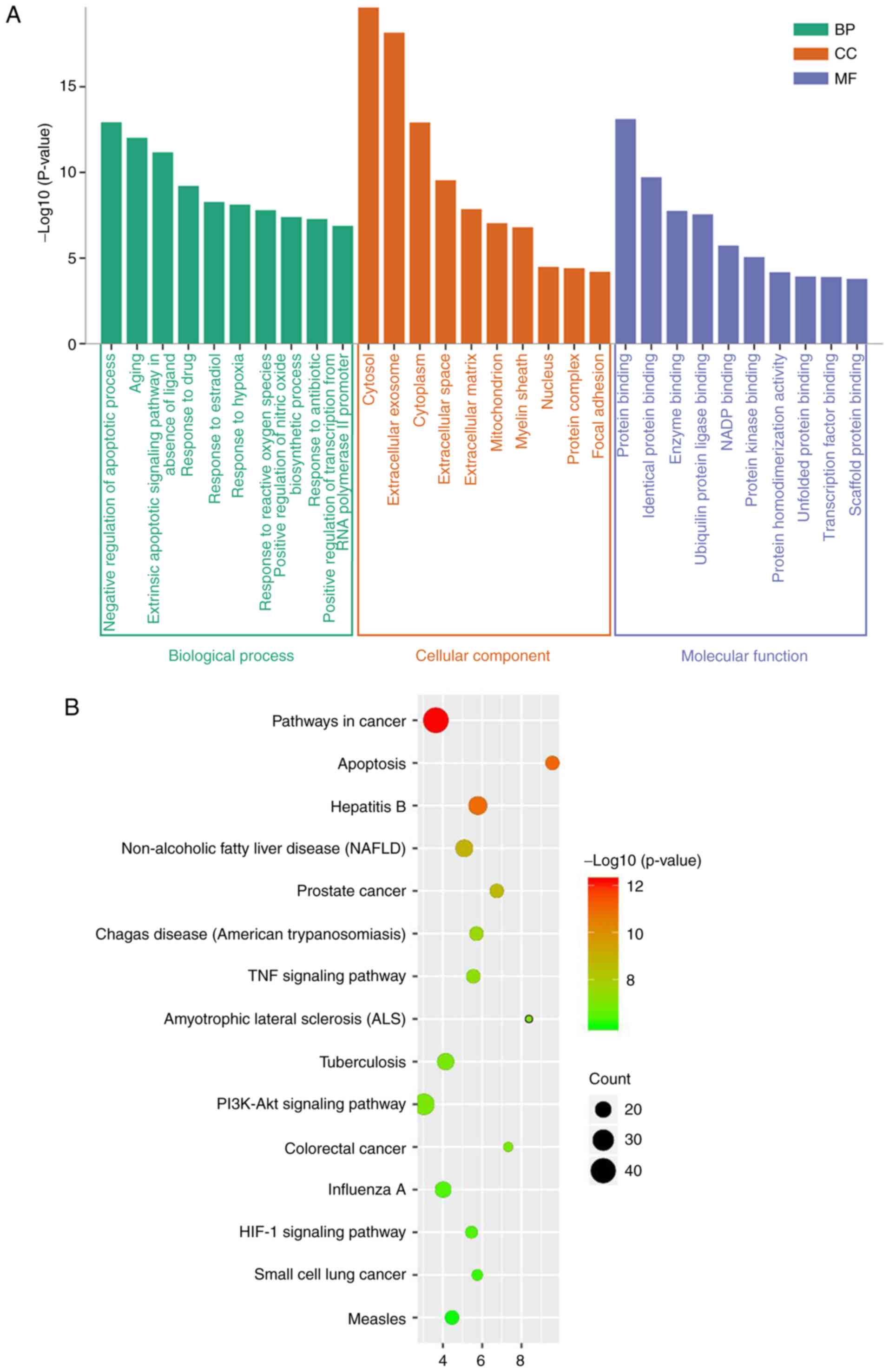
The results revealed that the majority of key genes
included in the GO enrichment analysis were associated with the BP
component term ‘negative regulation of apoptotic process’ (GO:
0043066; Fig. 2A). The BP terms
‘response to hypoxia’ and ‘response to reactive oxygen species’
were also in the top 10 results of the GO enrichment analysis. The
majority of key genes were associated with the CC parameter term
‘cytosol’ (GO: 0005829) and MF parameter term ‘protein binding’
(GO: 0005515). Furthermore, KEGG enrichment analysis of the key
genes found the term ‘pathways in cancer’ (hsa05200) to be most
significantly enriched (Fig. 2B).
In addition, signaling pathways known to be associated with KBD
were identified among the top 15 enrichment terms, including
‘apoptosis’ (hsa04210), and the ‘TNF signaling pathway’ (hsa04668),
‘PI3K-AKT signaling pathway’ (hsa04151) and ‘HIF-1 signaling
pathway’ (hsa04066).
Construction of the PPI network for
KBD-associated key genes
A PPI network of key genes was constructed in the
online database STRING, and contained 299 nodes and 2,830 edges.
Subsequently, the interaction pairs were entered into Cytoscape
software to construct multiple PPI networks. The core network
module was then selected using the Cytoscape MCODE plug-in. The
first-ranked module was extracted under the default parameters and
contained 42 nodes and 716 edges (Fig.
3). The top 10 genes with MCODE scores in this module were
revealed to be BCL2L1, MMP9, CASP8,
HSP90AA1, IL10, HSPA4, CXCL8,
CASP9, MTOR and TNF.
Downregulation of HIF-1 α and VEGFA in
T-2 toxin-treated chondrocytes
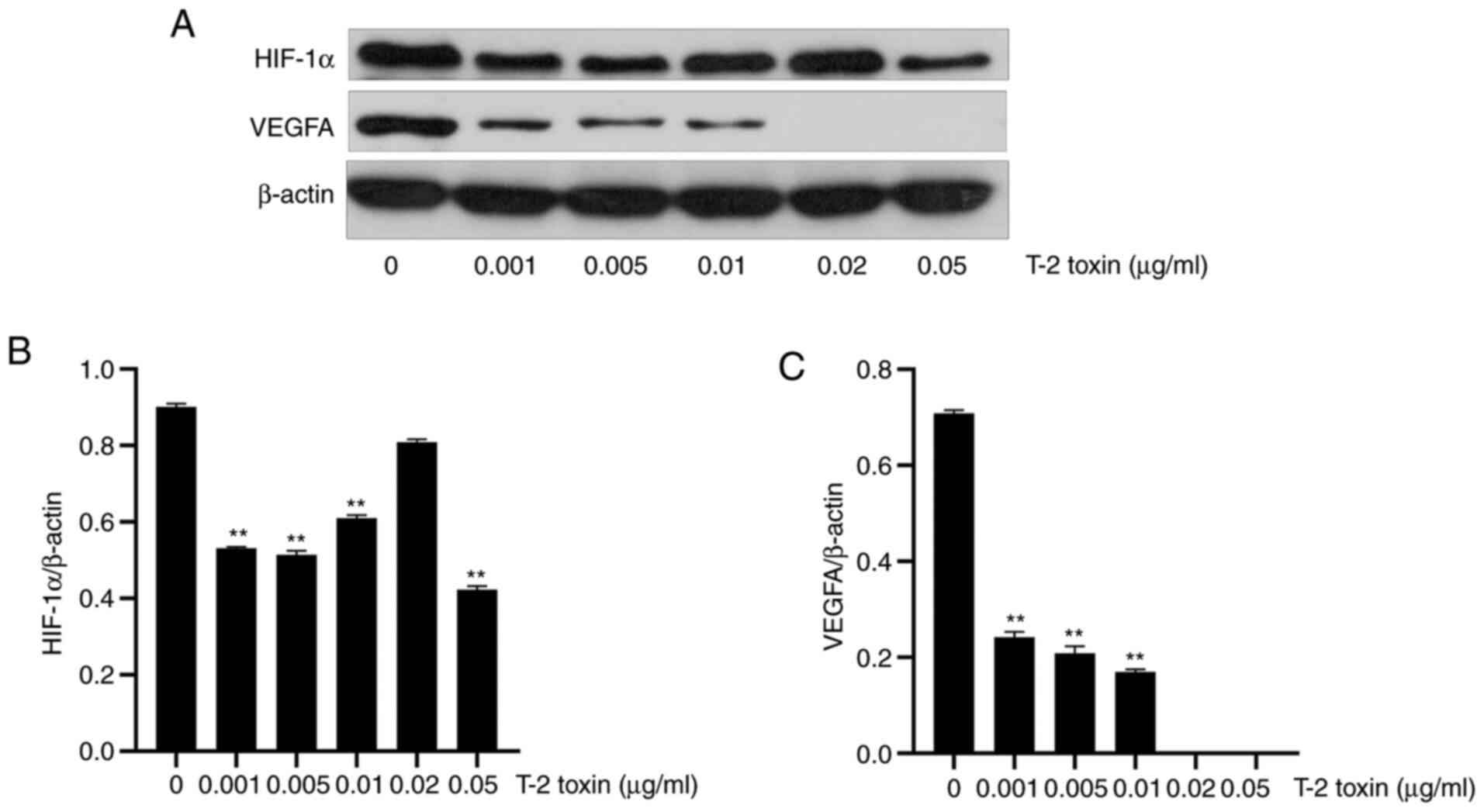
In order to study the expression of HIF-1α and VEGFA
in KBD, different concentrations of T-2 toxin were used to treat
chondrocytes for 3 days. Western blotting results revealed that the
expression levels of HIF-1α and VEGFA in the T-2 toxin-treated
group were significantly reduced in comparison with the control
group. As the concentration of T-2 toxin was increased, the
expression levels of HIF-1α and VEGFA were gradually reduced.
However, when the concentration of the T-2 toxin added was 0.01 and
0.02 µg/ml, the expression level of HIF-1α showed a tendency to
increase (Fig. 4).
Construction of the PPI network
associated with the HIF-1 signaling pathway and prediction of
transcription factors
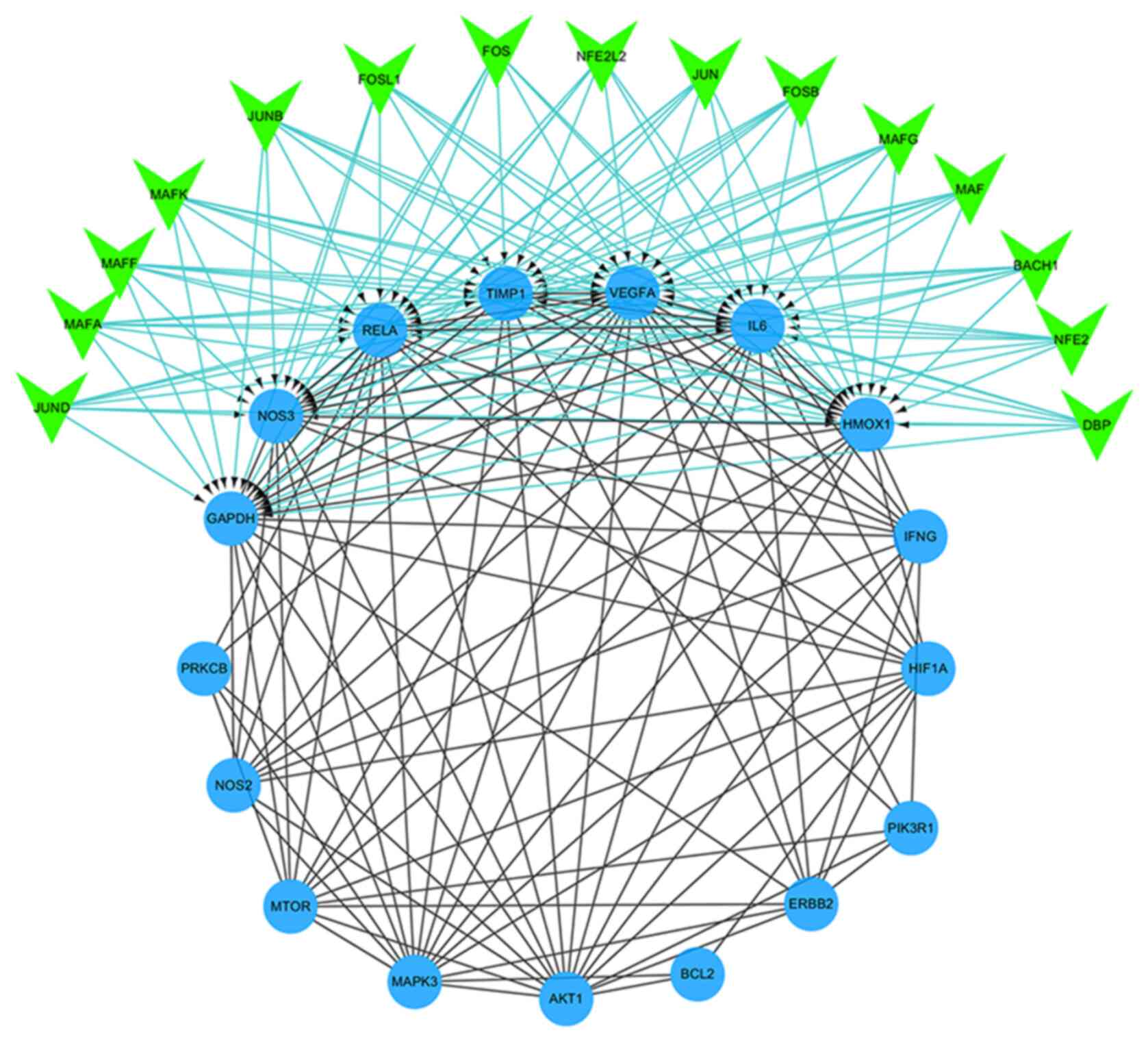
Including HIF-1α and VEGFA, all molecules of the
HIF-1 signaling pathway identified by KEGG enrichment analysis were
used to construct a PPI network (Fig.
5). Subsequently, the constructed network was imported into
Cytoscape, and the iRegulon plug-in was used to predict the
transcription factors that may regulate these target genes
(NES=7.152). The results obtained showed that 7 targets
(NOS3, VEGFA, IL6, RELA, TIMP1,
GAPDH and HMOX1) in the PPI network composed of 17
genes were regulated by the predicted 15 transcription factors
(MAFK, MAFG, NFE2, NFE2L2, MAFF,
BACH1, MAFA, JUNB, FOS, JUND,
FOSL1, JUN, FOSB, MAF and
DBP).
Discussion
KBD is a chronic and severe progressive bone and
joint degenerative disease of unknown etiology. An elevated
prevalence of KBD has been shown in populations living in
geographic areas with low selenium abundance and high exposure to
mycotoxins (8). An accumulating
body of evidence, together with recent scientific discoveries, have
indicated that mycotoxins, including T-2 toxin, have the potential
to trigger cell hypoxia (9,10).
In the present study, the target genes associated
with KBD, T-2 toxin and selenium were first obtained from the CTD
database. These targets were associated with the GO BP terms of
‘apoptosis’ and ‘hypoxia’, as well as the ‘TNF signaling pathway’,
‘PI3K-AKT signaling pathway’ and ‘HIF-1 signaling pathway’. These
results suggested that TNF-α was associated with inflammation and
apoptosis in KBD; this is in agreement with previous studies that
indicated that the levels of TNF-α in the serum and cartilage of
patients with KBD were markedly higher compared with those of
healthy controls (33,34). PI3K-AKT is the main signaling
pathway for chondrocyte survival and apoptosis, and the core hub
for transmitting external signals (35). A previous study indicated that
oxidative stress-induced chondrocyte apoptosis may be mediated via
upregulation of the PI3K-AKT signaling pathway (36). An additional study indicated that
the regulation of HIF-1α by components of the PI3K-AKT signaling
pathway may directly regulate the stability of HIF-1α protein via
its downstream effects (37).
In addition to the PI3K-AKT/HIF-1α pathway, HIF-1
signaling also includes the MAPK/HIF-1α signaling pathway and the
HIF-1α/VEGFA signaling pathway (38). A previous study revealed that
excessive apoptosis of chondrocytes and oxidative stress served a
crucial role in the pathophysiology of KBD (39). Earlier research also indicated that
under hypoxic or normoxic conditions, the level of apoptosis of
HIF-1α-deficient chondrocytes was significantly increased in
osteoarthritis (40,41). Therefore, it was hypothesized that
T-2 toxin-induced chondrocyte apoptosis was associated with HIF-1α,
and experiments were devised to assess the expression of HIF-1α in
KBD. The results indicated that HIF-1α expression was reduced in
chondrocytes treated with different concentrations of T-2 toxin in
a dose-dependent manner. By contrast, VEGF-A expression was shown
to be reduced in a dose-dependent manner following treatment with
different concentrations of T-2 toxin. However, the precise
mechanism via which HIF-1α regulates VEGFA in KBD requires further
exploration.
Through the PPI network constructed by the key genes
associated with KBD and the MCODE plug-in of Cytoscape, 10
molecules were screened out that may be associated with KBD. The
genes CASP8 and MTOR have been previously associated
with KBD (42-44).
Using Cytoscape software and its plugin to analyze the HIF-1
signaling pathway led to the prediction of 15 transcription factors
and 7 target genes. Of these, the genes IL6 (45), RELA (46), TIMP1 (47) and HMOX1 (48), as well as the transcription factors
NFE2L2 (48), JUNB
(49), FOS (50), JUND (49) and JUN (47), have been reported to be associated
with KBD.
There are several limitations of the present study
that should be acknowledged. Follow-up experiments on selenium
deficiency were not performed. Whether selenium deficiency or
selenium deficiency combined with T-2 toxin is able to affect the
expression of HIF-1α and VEGFA following treatment of the
chondrocytes requires further experimental verification. In
addition, further experiments, such as inhibitor studies or
transfection experiments, are required to determine the association
between HIF-1α and VEGFA.
In conclusion, a total of 10 core genes and 15
transcription factors associated with KBD were identified in the
present study. The results also indicated that the expression
levels of HIF-1α and VEGFA in T-2 toxin-treated chondrocytes were
downregulated. Therefore, the results of the present study
suggested that the HIF-1α/VEGFA signaling pathway is involved in
KBD, and this knowledge may help to both further elucidate the
pathogenesis of KBD and provide possible avenues for treatment of
KBD in the future.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by grants from the Program on
Health Research funded under the Shaanxi Health and Family Planning
Commission (grant no. 2018A018) and the Subject Innovation Team of
the Second Affiliated Hospital of Shaanxi University of Chinese
Medicine (grant no. 2020XKTD-C07).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WL and GW conceived and designed the experiments,
performed the experiments, analyzed the data, contributed materials
and analytical tools, prepared the figures and authored and
reviewed drafts of the paper. BX and HH analyzed the data and
authored and reviewed drafts of the paper. All authors have read
and approved the final manuscript. BX and WL confirmed the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xiong G: Diagnostic, clinical and
radiological characteristics of Kashin-Beck disease in Shaanxi
Province, PR China. Int Orthop. 25:147–150. 2001.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang K, Yu J, Liu H, Liu Y, Liu N, Cao Y,
Zhang X and Sun D: Endemic Kashin-Beck disease: A food-sourced
osteoarthropathy. Semin Arthritis Rheum. 50:366–372.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yang L, Zhang J, Li X, Xu C, Wang X and
Guo X: Expression profiles of selenium-related genes in human
chondrocytes exposed to T-2 toxin and deoxynivalenol. Biol Trace
Elem Res. 190:295–302. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sudre P and Mathieu F: Kashin-Beck
disease: From etiology to prevention or from prevention to
etiology? Int Orthop. 25:175–179. 2001.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Gutleb AC, Morrison E and Murk AJ:
Cytotoxicity assays for mycotoxins produced by Fusarium strains: A
review. Environ Toxicol Pharmacol. 11:309–320. 2002.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lei Y, Guanghui Z, Xi W, Yingting W, Xialu
L, Fangfang Y, Goldring MB, Xiong G and Lammi MJ: Cellular
responses to T-2 toxin and/or deoxynivalenol that induce cartilage
damage are not specific to chondrocytes. Sci Rep.
7(2231)2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang SJ, Guo X, Zuo H, Zhang YG, Xu P,
Ping ZG, Zhang Z and Geng D: Chondrocyte apoptosis and expression
of Bcl-2, Bax, Fas, and iNOS in articular cartilage in patients
with Kashin-Beck disease. J Rheumatol. 33:615–619. 2006.PubMed/NCBI
|
|
8
|
Lei R, Jiang N, Zhang Q, Hu S, Dennis BS,
He S and Guo X: Prevalence of selenium, T-2 toxin, and
deoxynivalenol in kashin-beck disease areas in qinghai province,
Northwest China. Biol Trace Elem Res. 171:34–40. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Jirong Y, Huiyun P, Zhongzhe Y, Birong D,
Weimin L, Ming Y and Yi S: Sodium selenite for treatment of
Kashin-Beck disease in children: A systematic review of randomised
controlled trials. Osteoarthritis Cartilage. 20:605–613.
2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wu Q, Wu W and Kuca K: From hypoxia and
hypoxia-inducible factors (HIF) to oxidative stress: A new
understanding of the toxic mechanism of mycotoxins. Food Chem
Toxicol. 135(110968)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang F, Guo X, Wang W, Yan H and Li C:
Genome-wide gene expression analysis suggests an important role of
hypoxia in the pathogenesis of endemic osteochondropathy
Kashin-Beck disease. PLoS One. 6(e22983)2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wu W, He A, Wen Y, Xiao X, Hao J, Zhang F
and Guo X: Comparison of microRNA expression profiles of
Kashin-Beck disease, osteoarthritis and rheumatoid arthritis. Sci
Rep. 7(540)2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Majmundar AJ, Wong WJ and Simon MC:
Hypoxia-inducible factors and the response to hypoxic stress. Mol
Cell. 40:294–309. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Schönenberger MJ, Krek W and Kovacs WJ:
EPAS1/HIF-2α is a driver of mammalian pexophagy. Autophagy.
11:967–969. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Gao RY, Wang M, Liu Q, Feng D, Wen Y, Xia
Y, Colgan SP, Eltzschig HK and Ju C: Hypoxia-inducible factor-2α
reprograms liver macrophages to protect against acute liver injury
through the production of interleukin-6. Hepatology. 71:2105–2117.
2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tian J, Yan J, Wang W, Zhong N, Tian L,
Sun J, Min Z, Ma J and Lu S: T-2 toxin enhances catabolic activity
of hypertrophic chondrocytes through ROS-NF-κB-HIF-2α pathway.
Toxicol In Vitro. 26:1106–1113. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xu J, Jiang C, Zhu W, Wang B, Yan J, Min
Z, Geng M, Han Y, Ning Q, Zhang F, et al: NOD2 pathway via RIPK2
and TBK1 is involved in the aberrant catabolism induced by T-2
toxin in chondrocytes. Osteoarthritis Cartilage. 23:1575–1585.
2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shi M, He Y, Zhang Y, Guo X, Lin J, Wang W
and Chen J: LncRNA MIAT regulated by selenium and T-2 toxin
increases NF-κB-p65 activation, promoting the progress of
Kashin-Beck disease. Hum Exp Toxicol. 40:869–881. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang T, Zhu X, Wu H, Jiang K and Zhao G:
Targeting the ROS/PI3K/AKT/HIF-1α/HK2 axis of breast cancer cells:
Combined administration of polydatin and 2-Deoxy-d-glucose. J Cell
Mol Med. 23:3711–3723. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Guan R, Wang J, Li Z, Ding M, Li D, Xu G,
Wang T, Chen Y, Yang Q, Long Z, et al: Sodium tanshinone IIA
sulfonate decreases cigarette smoke-induced inflammation and
oxidative stress via blocking the activation of MAPK/HIF-1α
signaling pathway. Front Pharmacol. 9(263)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang Z, Deng M, Huang J, Wu J, Li Z, Xing
M, Wang J, Guo Q and Zou W: Microglial annexin A3 downregulation
alleviates bone cancer-induced pain through inhibiting the
Hif-1α/vascular endothelial growth factor signaling pathway. Pain.
161:2750–2762. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Carmeliet P: VEGF as a key mediator of
angiogenesis in cancer. Oncology. 69 (Suppl 3):S4–S10.
2005.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Guo X, Zuo H, Cao CX, Zhang Y, Geng D,
Zhang ZT, Zhang YG, von der Mark K and von der Mark H: Abnormal
expression of Col X, PTHrP, TGF-beta, bFGF, and VEGF in cartilage
with Kashin-Beck disease. J Bone Miner Metab. 24:319–328.
2006.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang F, Guo X, Duan C, Wu S, Yu H and
Lammi M: Identification of differentially expressed genes and
pathways between primary osteoarthritis and endemic osteoarthritis
(Kashin-Beck disease). Scand J Rheumatol. 42:71–79. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Balamurugan K: HIF-1 at the crossroads of
hypoxia, inflammation, and cancer. Int J Cancer. 138:1058–1066.
2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Maes C, Carmeliet G and Schipani E:
Hypoxia-driven pathways in bone development, regeneration and
disease. Nat Rev Rheumatol. 8:358–366. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Davis AP, Grondin CJ, Johnson RJ, Sciaky
D, McMorran R, Wiegers J, Wiegers TC and Mattingly CJ: The
comparative toxicogenomics database: Update 2019. Nucleic Acids
Res. 47:D948–D954. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang X, Ning Y, Zhang P, Yang L, Wang Y
and Guo X: Chondrocytes damage induced by T-2 toxin via
Wnt/β-catenin signaling pathway is involved in the pathogenesis of
an endemic osteochondropathy, Kashin-Beck disease. Exp Cell Res.
361:141–148. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu YN, Jiang ZC, Li SY, Li ZZ, Wang H,
Liu Y, Liao YC, Han J and Chen JH: Integrin α2β1 is involved in T-2
toxin-induced decrease of type II collagen in C28/I2 chondrocytes.
Toxicon. 186:12–18. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Janky R, Verfaillie A, Imrichová H, Van de
Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval
Sanchez M, Potier D, et al: iRegulon: From a gene list to a gene
regulatory network using large motif and track collections. PLoS
Comput Biol. 10(e1003731)2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wang WZ, Guo X, Duan C, Ma WJ, Zhang YG,
Xu P, Gao ZQ, Wang ZF, Yan H, Zhang YF, et al: Comparative analysis
of gene expression profiles between the normal human cartilage and
the one with endemic osteoarthritis. Osteoarthritis Cartilage.
17:83–90. 2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Duan C, Guo X, Zhang XD, Yu HJ, Yan H, Gao
Y, Ma WJ, Gao ZQ, Xu P and Lammi M: Comparative analysis of gene
expression profiles between primary knee osteoarthritis and an
osteoarthritis endemic to Northwestern China, Kashin-Beck disease.
Arthritis Rheum. 62:771–780. 2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wu Q, Qin Z, Kuca K, You L, Zhao Y, Liu A,
Musilek K, Chrienova Z, Nepovimova E, Oleksak P, et al: An update
on T-2 toxin and its modified forms: Metabolism, immunotoxicity
mechanism, and human exposure assessment. Arch Toxicol.
94:3645–3669. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Du XA, Wang HM, Dai XX, Kou Y, Wu RP, Chen
Q, Cao JL, Mo XY and Xiong YM: Role of selenoprotein S
(SEPS1)-105G>A polymorphisms and PI3K/Akt signaling pathway in
Kashin-Beck disease. Osteoarthritis Cartilage. 23:210–216.
2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Movafagh S, Crook S and Vo K: Regulation
of hypoxia-inducible factor-1a by reactive oxygen species: New
developments in an old debate. J Cell Biochem. 116:696–703.
2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Feng FB and Qiu HY: Effects of artesunate
on chondrocyte proliferation, apoptosis and autophagy through the
PI3K/AKT/mTOR signaling pathway in rat models with rheumatoid
arthritis. Biomed Pharmacother. 102:1209–1220. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wang W, Wei S, Luo M, Yu B, Cao J, Yang Z,
Wang Z, Goldring MB and Chen J: Oxidative stress and status of
antioxidant enzymes in children with Kashin-Beck disease.
Osteoarthritis Cartilage. 21:1781–1789. 2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhang FJ, Luo W and Lei GH: Role of HIF-1α
and HIF-2α in osteoarthritis. Joint Bone Spine. 82:144–147.
2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Yudoh K, Nakamura H, Masuko-Hongo K, Kato
T and Nishioka K: Catabolic stress induces expression of
hypoxia-inducible factor (HIF)-1 alpha in articular chondrocytes:
Involvement of HIF-1 alpha in the pathogenesis of osteoarthritis.
Arthritis Res Ther. 7:R904–R914. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
42
|
Wang Y, Guo X, Zhang ZT, Wang M and Wang
SJ: Expression of Caspase-8 and Bcl-2 in the cartilage loose bodies
in patients with Kashin-Beck disease. Nan Fang Yi Ke Da Xue Xue
Bao. 31:1314–1317. 2011.PubMed/NCBI(In Chinese).
|
|
43
|
Wen Y, Li P, Hao J, Duan C, Han J, He A,
Du Y, Liu L, Liang X, Zhang F and Guo X: Integrating genome-wide
DNA methylation and mRNA expression profiles identified different
molecular features between Kashin-Beck disease and primary
osteoarthritis. Arthritis Res Ther. 20(41)2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wu C, Liu H, Zhang F, Shao W, Yang L, Ning
Y, Wang S, Zhao G, Lee BJ, Lammi M and Guo X: Long noncoding RNA
expression profile reveals lncRNAs signature associated with
extracellular matrix degradation in kashin-beck disease. Sci Rep.
7(17553)2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ning Y, Wang X, Lammi MJ and Guo X:
Changes in the NF-κB signaling pathway in juvenile and adult
patients with Kashin-Beck disease. Exp Cell Res. 379:140–149.
2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Xiong YM, Mo XY, Zou XZ, Song RX, Sun WY,
Lu W, Chen Q, Yu YX and Zang WJ: Association study between
polymorphisms in selenoprotein genes and susceptibility to
Kashin-Beck disease. Osteoarthritis Cartilage. 18:817–824.
2010.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Chang Y, Wang X, Sun Z, Jin Z, Chen M,
Wang X, Lammi MJ and Guo X: Inflammatory cytokine of IL-1β is
involved in T-2 toxin-triggered chondrocyte injury and metabolism
imbalance by the activation of Wnt/β-catenin signaling. Mol
Immunol. 91:195–201. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Li Y, Mo X and Xiong Y: The study on
polymorphism of TrxR and Nrf2/HO-1 signaling pathway in
Kaschin-Beck disease. Biol Trace Elem Res. 190:303–308.
2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wu R, Zhang R, Xiong Y, Sun W, Li Y, Yang
X, Liu J, Jiang Y, Guo H, Mo X and Cao J: The study on
polymorphisms of Sep15 and TrxR2 and the expression of AP-1
signaling pathway in Kashin-Beck disease. Bone. 120:239–245.
2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Han L, Yang X, Sun W, Li Z, Ren H, Li B,
Zhang R, Zhang D, Shi Z, Liu J, et al: The study of GPX3
methylation in patients with Kashin-Beck disease and its mechanism
in chondrocyte apoptosis. Bone. 117:15–22. 2018.PubMed/NCBI View Article : Google Scholar
|