Introduction
Hepatitis B virus (HBV) infection is a global
epidemic. According to the World Health Organization, ~2 billion
people worldwide have been infected with HBV, and chronic HBV
infections account for 240 million of these cases (1). The incidence of HBV infection is high
in China. The HBV carrying rate in the general Chinese population
is 9.9%, which closely correlates with the occurrence of chronic
hepatitis, cirrhosis and liver cancer (2,3).
Mother-to-child transmission is the main transmission route of
chronic HBV infection (4,5). Mechanisms include intrauterine
infection, intrapartum infection and puerperal infection (6). The latter two mechanisms can be
prevented by treating the infant with the hepatitis B vaccine and
hepatitis B immunoglobulin immediately after birth (7). Despite these measures, 5-10% of
infants fail to acquire immunity (8-10).
This is mainly attributed to intrauterine infection. However, the
mechanisms underlying HBV intrauterine infection remain
unclear.
Intrauterine infection via the placenta is a growing
concern among clinical workers and scientific researchers. It is
believed that mother-to-foetus HBV infection may be caused by
introduction of HBV into circulating foetal blood from peripheral
blood mononuclear cells through the placenta (11). HBV can integrate into placental
trophoblastic cells, where it can replicate and produce
intrauterine infection (12).
Studies have demonstrated that hepatitis B surface antigen,
hepatitis B core antigen and HBV DNA are distributed in all
placental cell layers. The rate of HBV infection was demonstrated
to gradually reduce in the placenta from the mother's side to the
fetus' side (13,14). Studies also indicate that in
placental HBV infection, there is a gradual decreasing trend of
trophoblast cells from the uterine interface to the villus vascular
endothelial cells; however, the odds ratio (OR) value of foetal
intrauterine infection gradually increases following the same
pattern. This suggests that HBV can infect a foetus from the
placenta via cell-to-cell transmission (15,16).
Hepatitis B virus X (HBx) is a multi-functional
regulatory protein with a wide range of trans-activating functions
and can bind to a variety of intracellular factors involved in
transcription and gene regulation (17,18).
Studies have demonstrated that HBx can promote viral transcription
and replication in hepatoma cells (19,20).
HBx can stimulate different signalling pathways in cells,
triggering a variety of biochemical and behavioural changes. It has
been found in hepatoma cells that the primary function of HBx is to
degrade structural maintenance of chromosomes 5/6, which restricts
HBV replication by inhibiting HBV gene expression (21-23).
In liver cells, HBx was revealed to activate viral and cell
promoters and regulate signal transduction pathways (18). Previous research revealed that HBV
can reach the placenta through maternal blood during pregnancy,
where it infects placental trophoblast cells, producing HBx protein
(24). HBx protein is associated
with inhibition of trophoblast apoptosis by intracellular PI3K and
the AKT signalling pathway (24,25).
HBx influences the activation of EGFR and other EGFR family members
(26,27).
The aim of the present study was to elucidate the
mechanism underlying EGFR activation by hepatitis B virus X antigen
(HBxAg) in placental trophoblast cells. The effect of EGFR
activation on HBV replication was investigated by collecting serum
samples from pregnant women with different HBV DNA titres. The
samples were used to infect trophoblasts and HBxAg expression was
detected. Wild-type plasmid containing the full length HBV genome
and HBx deletion mutant (ΔHBx) plasmid were transfected into
trophoblasts to mechanistically study the effects of HBxAg on HBV
replication. Dual-luciferase activity assay and shRNA techniques
were employed to study the effects of HBx protein on the EGFR
promoter, EGFR/PI3K/phosphorylated (p)-AKT signalling and
inhibition of trophoblast apoptosis, thereby establishing the role
of HBxAg in HBV intrauterine infection.
Materials and methods
Blood sample collection
HBV serum was collected from four pregnant women
diagnosed with chronic hepatitis B, who had been admitted to the
First Affiliated Hospital of Xi'an Jiaotong University (Shaanxi,
China), between February 2018 and March 2018. Informed written
consent was obtained from all study participants following a
detailed explanation of the study at the time of blood and serum
collection. The study protocol was approved by the Institutional
Review Board of the First Affiliated Hospital, Xi'an Jiaotong
University.
Serum HBV DNA titres were obtained from each donor
respectively: i) 4.05x102 IU/ml; ii) 7.65x103
IU/ml; iii) 3.28x105 IU/ml; and iv) 8.99x107
IU/ml. The donors were 28±4 weeks pregnant and aged between
27.5±2.5 years old. Control serum was collected in the same date
range as the HBV serum samples from healthy pregnant women admitted
to the First Affiliated Hospital of Xi'an Jiaotong University. The
control donors were 28±4 weeks and aged between 27.5±2.5 years.
These patients had no history of hepatitis B or other diseases.
Construction and validation of a
full-length HBV vector expression plasmid with HBx gene
deletion
To create an HBx deletion mutation in the pTriEx-1.1
HBV vector (gifted from Professor Xu Dongping, 5th Medical Center
of the Chinese PLA General Hospital) encoding the entire HBV
sequence, the CAA codon encoding the eighth glutamine in the HBx
coding sequence was mutated to the termination codon TAA. The
forward and reverse primers (Beijing Tianyi Huiyuan Biotechnology
Co., Ltd.) encoding the mutation were designed as follows: ptxup,
5'-TAACTGGATCCTGCGCGGGACGTCCT-3'; ptxlow,
5'-GCGCAGGATCCAGTTAGCAGCACATC-3'. Site-directed mutagenesis was
accomplished using a Fast Mutagenesis System kit (TransGen Biotech
Co., Ltd.) according to manufacturer's instructions. Reaction
conditions were as follows: Initial denaturation at 94˚C for 5 min;
25 cycles of 94˚C for 20 sec, 60˚C for 20 sec and 72˚C for 3 min;
and a final extension at 72˚C for 10 min. Next, 1 µl DMT enzyme
(modified DpnI restriction endonuclease; Beijing TransGen
Biotech Co., Ltd.) was added to the PCR products and digested at
37˚C for 1 h. The product was directly translated into the DMT
chemically competent cells (cat. no. CD511-01; TransGen Biotech
Co., Ltd.), coated on the Luria-Bertani (LB) tablet containing
ampicillin (50 µg/ml; Gibco; Thermo Fisher Scientific, Inc.) and
incubated at 37˚C overnight. Five white colonies were randomly
selected for culture and the bacterial solution was used for PCR.
Specific steps were as follows: A total of 11 µl sterile water were
added to the PCR tube, a single white colony was picked and lightly
rinsed several times in the PCR tube, the toothpick was removed and
lightly streaked on an LB tablet, and a replication plate was
prepared. The PCR tube was placed on ice immediately after
incubation for 10 min at 94˚C. After the bacterial cell was lysed,
DNA was released as a template for amplification and
electrophoresis identification. The reaction conditions were as
follows: Initial denaturation at 94˚C for 3 min; 20 cycles of 94˚C
for 40 sec, 60˚C for 100 sec and 72˚C for 4 min; and a final
extension at 72˚C for 10 min. Mutated plasmid samples were sent for
sequencing (Beijing Tianyi Huiyuan Biotechnology Co., Ltd.).
Cell culture and transfection
Human choriocarcinoma cell line JEG-3
(ATCC® HTB-36™; Shanghai Life Sciences
Research Institute) was cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 0.1 mg/ml streptomycin
(Gibco; Thermo Fisher Scientific, Inc.). Human trophoblast cell
line, HTR-8/SVneo (ATCC CRL-3271™; Laboratory of
Obstetrics and Gynecology, Tangdu Hospital) was cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and streptomycin (Gibco; Thermo Fisher Scientific,
Inc.). The HTR-8/SVneo cell line was developed by Graham et
al (28). It was generated
using freshly isolated extravillous cytotrophoblasts from first
trimester placenta and transfected with a plasmid containing the
simian virus 40 large T antigen (SV40). The results of a previous
study demonstrated that this cell line contains two populations,
one of epithelial and one of mesenchymal origin (29). All cell lines were incubated at 37˚C
in 5% CO2. Cells were inoculated in 12-well plates until
the cell density reached 50-60% confluence. Cell culture medium was
discarded, and cells were incubated with 30% HBV DNA serum (HBV DNA
titres: 1x102, 1x103, 1x105 and
1x107) and 30% control human serum at 37˚C for 72 h. In
subsequent experiments, 2 µg of plasmids (full length wild-type 1.1
HBV plasmid, HBx deletion mutant plasmid; pGFP-HBx plasmid, pGFP
empty vector) were mixed with 4 µl X-tremeGENE HP (Roche
Diagnostics) at room temperature for 20 min when cell densities
reached 50-60% confluence. The mixtures were added to culture media
and incubated for 72 h. The experiments were repeated at least
three times independently. The optimal time point was determined by
detecting 24, 48, 72 and 96 h (data not shown).
HBV DNA quantitation in cells and
supernatants
Following 72 h transfection, cells were collected,
cleaved on 0.5% NP-40 at 4˚C for 30 min and supernatant was
extracted by centrifugation (13,000 x g, 7 min, 4˚C). DNase I (New
England Biolabs, Inc.) was added to digest DNA at 37˚C for 5 h and
proteinase K (Merck KGaA) was then added to digest in a 42˚C water
bath overnight. Equal volumes of phenol/chloroform/isoamyl alcohol
mixture were added (total volume, 660 µl). Following centrifugation
(10,000 x g, 15 min, 25˚C), the supernatant was transferred to a
new tube, 1/10 volume 3 M NaAc and isopropyl alcohol were added and
mixed at room temperature for 5 min. The mixture was then
centrifuged at 13,000 x g for 30 min at 4˚C and washed once with
500 µl 75% ethanol. The mixture was centrifuged again at 13,000 x g
for 15 min at 4˚C and dried. Finally, HBV DNA was dissolved with 30
µl double-distilled H2O. The supernatant and viral DNA
samples were sent for HBV DNA quantification (Beijing NaGene
Diagnosis Reagent Co., Ltd.).
Western blotting
Cells were lysed with 150 µl RIPA (CoWin
Biosciences) buffer and 1.5 µl PMSF (CoWin Biosciences) was added.
The protein concentration was determined using a BCA protein assay
kit and 20 µg of protein from each sample was separated using 12%
SDS-PAGE before transferring to PVDF membranes for immunoblotting.
The membranes were rinsed for 3 min with 1X TBST (0.05% Tween-20;
Thermo Fisher Scientific, Inc.) on a shaker. The membranes were
subsequently blocked with 5% skimmed milk (Thermo Fisher
Scientific, Inc.) on a shaker at 25˚C for 2 h. The following
primary antibodies and dilutions were used (incubated at 4˚C for 16
h): Rabbit anti-HBx antibody (1:300; cat. no. ab2741; Abcam),
rabbit anti-structural maintenance of chromosomes (Smc)5 antibody
(1:500; cat. no. ab154103; Abcam), rabbit anti-Smc6 antibody
(1:500; cat. no. ab155495; Abcam), rabbit anti-EGFR antibody
(1:1,000; cat. no. ab52894; Abcam), rabbit anti-PI3K antibody
(1:500; cat. no. 4249; Cell Signaling Technology, Inc.), rabbit
anti-AKT antibody (1:800; cat. no. 4691T; Cell Signaling
Technology, Inc.), rabbit anti-p-AKT antibody (1:500; cat. no.
4060; Cell Signaling Technology, Inc.) and mouse anti-human β-actin
antibody (1:1,000; cat. no. CW0096; CoWin Biosciences). After
incubation with HRP-labelled goat anti-mouse/rabbit IgG secondary
antibody at 25˚C for 1 h (1:2,000; cat. no. CW0102/0103; CoWin
Biosciences), proteins were detected using a ChemiDoc™
XRS imaging system (Bio-Rad Laboratories, Inc.). The experiments
were repeated at least three times independently.
ELISA
The cells infected with HBV serum in vitro
were collected, and HBxAg was detected using Diagnostic kit for
hepatitis B virus X antigen (ELISA) (cat. no. DM00914; Shanghai
Duma Biological Technology Co., Ltd.). The cells transfected with
full length wild-type 1.1 HBV plasmid and HBx deletion mutant
plasmid were collected, and HBeAg was detected using Diagnostic kit
for hepatitis B virus e antigen (ELISA) (cat. no. HBeAg-96T;
Beijing Wantai Biopharmaceutical Co., Ltd.). A total of 100 µl/well
samples and enzyme conjugate were added in 96-well ELISA plates for
1 h incubation at 37˚C. The board was washed with washing solution
from the kit for 5 times and patted dry. Developer A and B liquid
was added to each hole and incubated at 37˚C, avoiding light for 15
min to aid color development. The termination liquid was added and
shaken gently to mix. The absorbance was read at 450 nm within 30
min.
pgRNA quantification
Total RNA was extracted from cells using an EasyPure
Viral DNA/RNA kit (TransGen Biotech Co., Ltd.) and RNase inhibitors
were regularly sprayed throughout the experiment (DNase I and
RNase-free; Thermo Fisher Scientific, Inc.). DNA was digested,
DNase was inactivated, reverse transcriptase primers were added and
cDNA was synthesised using reverse transcriptase kits (Transcriptor
First Strand cDNA Synthesis Kit; Roche Diagnostics). cDNA, primers,
probes and MIX [2xRealStar Power Probe Mixture UNG; Beijing Kangrun
Chengye Biotechnology Co., Ltd. (GenStar)] were proportionally
mixed to determine pgRNA concentrations using RT-qPCR. The results
were quantified as described previously (30,31).
The linear regression equation was obtained by taking the logarithm
value of the concentration of the standard and that of the Ct
value. The logarithm value of the concentration according to the Ct
value of the sample was calculated, and finally the concentration
of pgRNA was calculated. Primers and probes (Beijing Tianyi Huiyuan
Biotechnology Co., Ltd.) were as follows: HR-F2:
5'-AGACCACCAAATGCCCCT-3'; HR-R2: 5'-TCACACCGTAACACACGACAC-3';
HR-RT2: 5'-TCTCACACCGTAACACACGACACAGGCGAGGGAGTTCTTCTTCTA-3'; Probe
HR: 5'-CAACACTTCCGGARACTACTGTTGTTAGACG-3'. Reaction conditions were
as follows: 50˚C for 5 min; 94˚C for 10 min; 45 cycles of 94˚C for
15 sec and 58˚C for 40 sec.
Immunofluorescence assays
Cells were seeded onto µ-Slide 8-well chamber slides
(4.5x104 cells/well; Ibidi GmbH) and transfected with
plasmids (full length wild-type 1.1 HBV plasmid, HBx deletion
mutant plasmid; pGFP-HBx plasmid, pGFP empty vector). After
incubation at 37˚C for 48 h, the cells were fixed in 4%
paraformaldehyde at 25˚C for 20 min, permeabilized in 0.1% Triton
X-100 for 10 min and blocked with 3% BSA (Gibco; Thermo Fisher
Scientific, Inc.) for 2 h at room temperature. Cells were incubated
with primary antibodies before staining with Cy3-conjugated goat
anti-rabbit IgG (1:500; cat. no. CW0159; CoWin Biosciences) in the
dark for 40 min at 37˚C. The following primary antibodies and
dilutions were used (incubated at 37˚C for 2 h): Anti-HBx (1:150;
cat. no. ab2741; Abcam), anti-Smc5 (1:200; cat. no. ab154103;
Abcam), anti-Smc6 (1:200; cat. no. ab155495; Abcam), anti-EGFR
(1:500; cat. no. ab52894; Abcam), anti-PI3K (1:200; cat. no. 4249;
Cell Signaling Technology, Inc.), anti-AKT (1:400; cat. no. 4691T;
Cell Signaling Technology, Inc.) and anti-p-AKT (1:200; cat. no.
4060; Cell Signaling Technology, Inc.). Cell nuclei were stained
with DAPI at 25˚C for 10 min. The cells were washed three times
with 1 x PBS between each step. Images were captured using an
Olympus FV1000 confocal microscope (Olympus Corporation). All
confocal images were captured using the same imaging parameters and
≥10 images for each sample were examined. Fluorescence intensity
was analyzed by Image-Pro Plus 6.0 software (Media Cybernetics,
Inc.).
Dual-luciferase reporter assay
Cells were seeded into 24-well plates at
3x105 cells/well and incubated for 16-18 h. pGL3-EGFR
promoter luciferase expression vector (BK328 pGL3-basic-EGFR;
Umibio (Shanghai) Co. Ltd.) and the internal reference vector
pRL-TK (Promega Corporation) were co-transfected with X-tremeGENE
HP (Roche Diagnostics) at room temperature for 20 min when cell
densities reached 50-60% confluence, at a ratio of 30:1 (expression
vector 0.3 µg/well and internal reference 0.01 µg/well). pGL3-EGFR
promoter luciferase expression vector and pGFP-HBx (Addgene, Inc.)
were co-transfected as the experimental group, pGL3-EGFR promoter
luciferase expression vector and pGFP empty vector (Addgene, Inc.)
were co-transfected as the control group, pGL3-Basic and pGFP-HBx
were co-transfected as a negative control (NC) and pGL3-Control was
used as a positive control (Promega Corporation). Cells were lysed
48 h later and assayed for luciferase activity. Firefly luciferase
activity was normalized to internal reference Renilla
luciferase activity. A single-tube multifunctional detection
reagent was used to detect luciferase activity according to kit
instructions (Dual-Luciferase Reporter Assay; Promega
Corporation).
Construction of cell lines stably
expressing EGFR
Cells were seeded into 24-well plates at
9.5x104 cells/well and incubated at 37˚C for 16-18 h.
Spent culture medium was removed before adding fresh complete
culture medium. A titred solution of EGFR overexpressing lentivirus
particles [GeneCopoeia, Inc.; lentivirus volume (unit: ml)=the
number of cells exposed to lentivirus x MOI/lentivirus titre (unit:
TU/ml)] was added to the cells and gently mixed. Culture medium
containing lentivirus particles was harvested after 12-16 h of
infection at 37˚C and fresh complete culture medium was added to
the culture plate to continue culturing the cells. The negative
control (NC) group was infected with irrelevant control lentiviral
vector. GFP fluorescence was observed at 72-96 h post infection.
After 72 h of infection, 2 µg/ml of puromycin was added to select
for infected cells. In subsequent experiments, when cell densities
reached 50-60% confluence, 2 µg of EGFR short hairpin (sh)RNA
(Vigene Bioscience Inc.) and shRNA control vector (Vigene
Bioscience Inc.) were mixed with 4 µl X-tremeGENE HP (Roche
Diagnostics) at room temperature for 20 min, respectively. The
mixtures were added to culture media and incubated at 37˚C for 72
h. As shown in Fig. S1, EGFR
expression in the overexpressing cells was knocked down using EGFR
short hairpin (sh)RNA.
Detection of cell apoptosis using flow
cytometry
Cell apoptosis was detected using a cell apoptosis
kit from Nanjing KeyGen Biotech Co., Ltd. Following transfection at
37˚C for 48 h (EGFR overexpressing cells), the medium was removed
and cells (2.0x105 cells/well) were washed twice with
pre-cooled PBS. The supernatant was removed, and cells were
suspended in 300 µl of 1X binding buffer (at a density of
5x105-5x106 cells/ml). A total of 5 µl
Annexin V FITC and 5 µl PI were added into the suspension and the
mixture was incubated at 25˚C for 10 min in the dark. Each sample
was analyzed using flow cytometry (Gallios; Beckman Coulter, Inc.)
within 1 h. The results of apoptosis were analysed by FlowJo v10
software (Becton, Dickinson and Company). The experiments were
repeated at least three times independently. The proportion of
apoptosis was calculated from the sum of Q3 and Q2.
Statistical analysis
Band intensities in scanned western blots were
quantitated using ImageJ v1.8.0.112 software (National Institutes
of Health). Fluorescence intensities were analysed using Image-Pro
Plus software (Media cybernetics Inc.). Data were expressed as the
mean ± standard deviation. Differences among variables were
examined using unpaired Student's t-test or one-way ANOVA with
Bonferroni's multiple comparison correction. Statistical analyses
were performed using SPSS version 18.0 (SPSS, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
HBxAg expression in placental
trophoblast cells infected with HBV serum in vitro
The serum of patients with HBV demonstrated an
ability to infect JEG-3 and HTR-8 cells in vitro (Fig. 1A). When the concentration of
HBV-containing serum was 30% in the culture medium, HBxAg
expression gradually increased with increasing amounts of HBV DNA
in patient serum. No HBxAg was detected in the no serum control
group or the HBV DNA negative serum group.
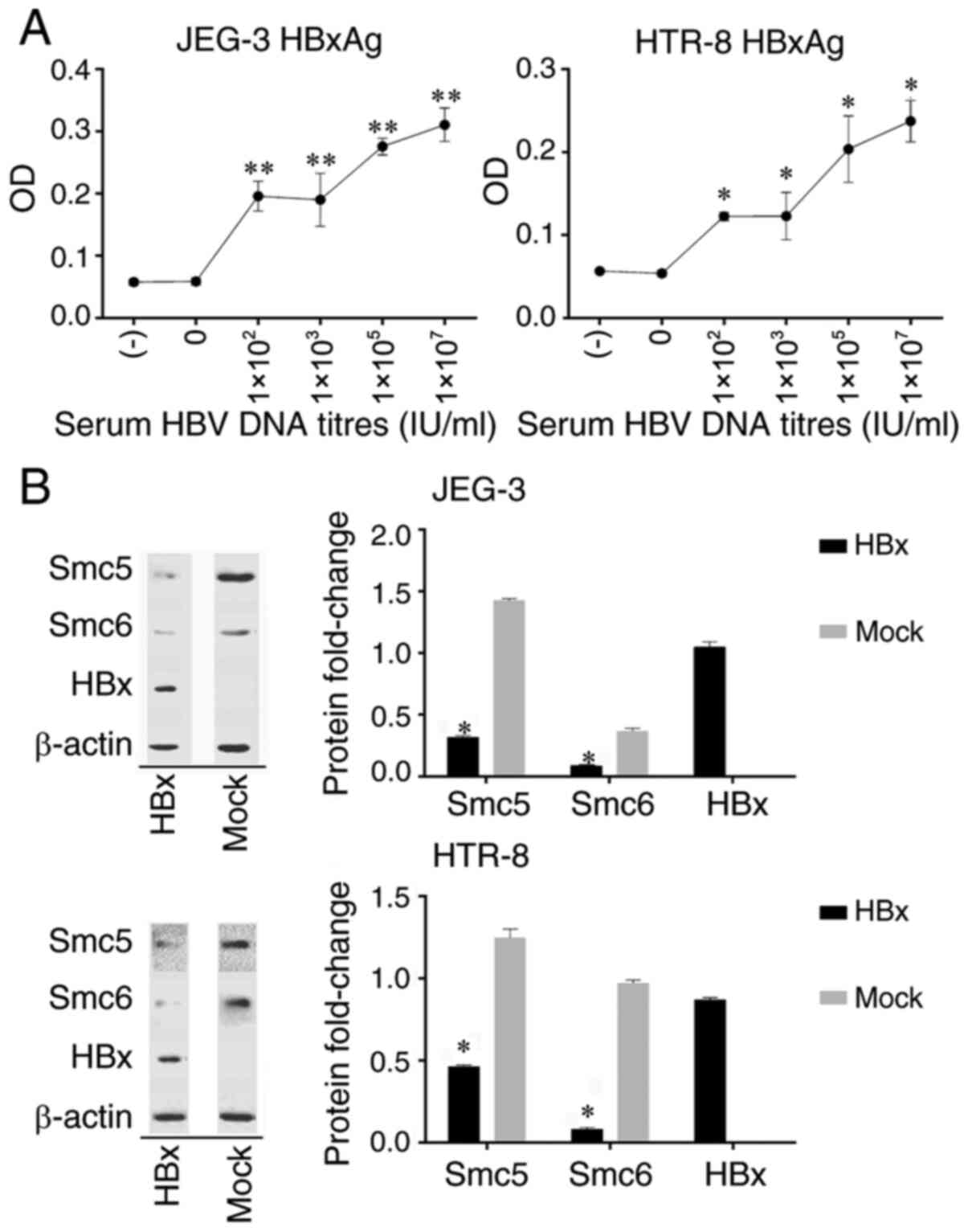 | Figure 1(A) HBxAg expression in trophoblasts
infected with HBV serum in vitro. (-), no serum control
group; 0, HBV DNA negative serum group; 1x102,
1x103, 1x105 and 1x107 are the
serum HBV DNA titres. *P<0.05, **P<0.01
vs. (-). (B) Western blot analysis of HBx and Smc5/6 protein
expression. Protein fold-change is displayed as the ratio of HBx
and Smc5/6 to β-actin. The blots/different groups were run on the
same membrane. Mock, pGFP empty vector; HBx, pGFP-HBx plasmid.
*P<0.05 vs. Mock. HBxAg, hepatitis B virus X antigen;
HBV, hepatitis B virus; Smc5/6, structural maintenance of
chromosomes 5/6; HBx, hepatitis B virus X. |
HBxAg expression in each group was compared in JEG-3
cells. With the exception of the HBV DNA negative serum group,
HBxAg expression was significantly higher than in the no serum
control group. There was no difference in HBxAg expression between
group 1x102 and group 1x103. HBxAg expression
of group 1x105 was significantly higher than that of
group 1x103 (31.05%). Furthermore, HBxAg expression of
group 1x107 increased by 11.26% compared with that of
group 1x105.
HBxAg expression in each group of HTR-8 cells was
compared. With the exception of the HBV DNA negative serum group,
HBxAg expression in all other groups was significantly higher than
in the no serum control group. There was no difference in HBxAg
expression between group 1x102 and group
1x103. HBxAg expression of group 1x105
significantly increased (39.68%) compared with that of group
1x103. Further, HBxAg expression of group
1x107 increased by 14.21% compared with that of group
1x105, but the difference was not significant.
Western blot analysis of HBx and
Smc5/6 protein expression
As demonstrated in Fig.
1B, HBx expression was observed in JEG-3 and HTR-8 cells
transfected with HBx plasmid. Smc5/6 expression was significantly
higher in green fluorescent protein (GFP) empty vector
(pGFP)-transfected cells than in cells in which HBx protein was
expressed.
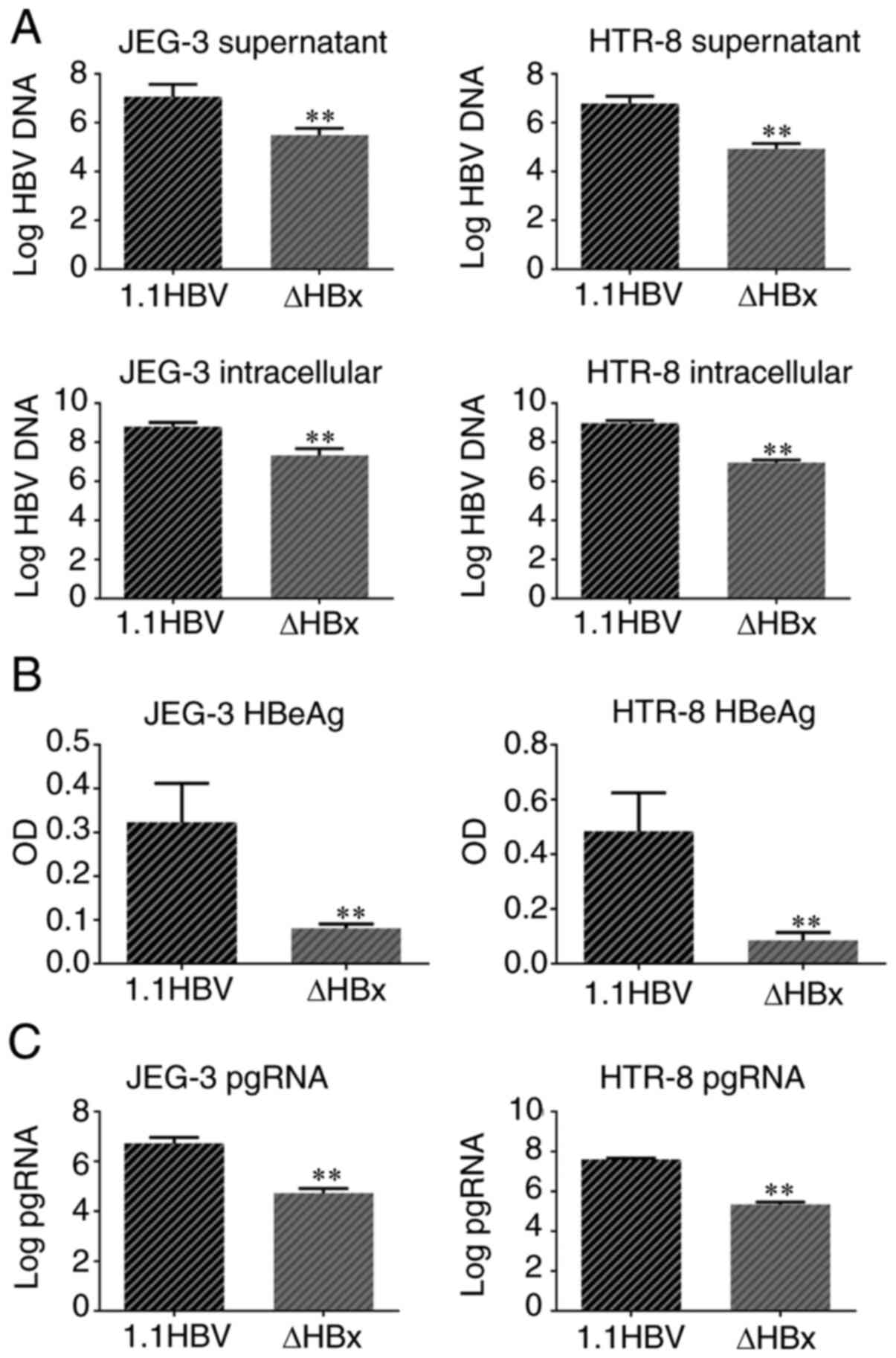
Comparison of HBV DNA replication
levels in placental trophoblast cells
HBV DNA replication in JEG-3 cell supernatant and
JEG-3 cells in the ΔHBx group was significantly reduced by 98.1 and
96.1% respectively, compared with that in the wild-type HBV group
(Fig. 2A). HBV DNA replication in
HTR-8 cell supernatant and HTR-8 cells in the ΔHBx group was
significantly decreased by 98.8 and 99.0% respectively, compared
with that in the wild-type HBV group.
Comparison of HBeAg expression in
placental trophoblast cells
The OD values for HBeAg in the ΔHBx group were
significantly lower in JEG-3 and HTR-8 cells than in the 1.1 HBV
group (Fig. 2B).
Comparison of pgRNA in placental
trophoblast cells
The pgRNA levels in JEG-3 cells transfected with
ΔHBx plasmid were 99.0% lower than in wild-type HBV-transfected
cells. pgRNA levels in ΔHBx-transfected HTR-8 cells were 99.4%
lower than in wild-type HBV-transfected cells (Fig. 2C).
Confocal laser microscopy analysis of
HBx and Smc5 protein expression and localization
As revealed in Fig.
3A, HBx protein expression was detected when JEG-3 and HTR-8
cells were transfected with HBx and 1.1 HBV plasmids. HBx protein
expression was not detected in cells transfected with ΔHBx or pGFP.
HBx protein localized to the cytoplasm in both cell types. Smc5
protein was mainly expressed in the nucleus and Smc5 expression was
higher in ΔHBx-transfected or pGFP-transfected cells than in cells
in which HBx protein was expressed. Protein levels were consistent
with western blotting results.
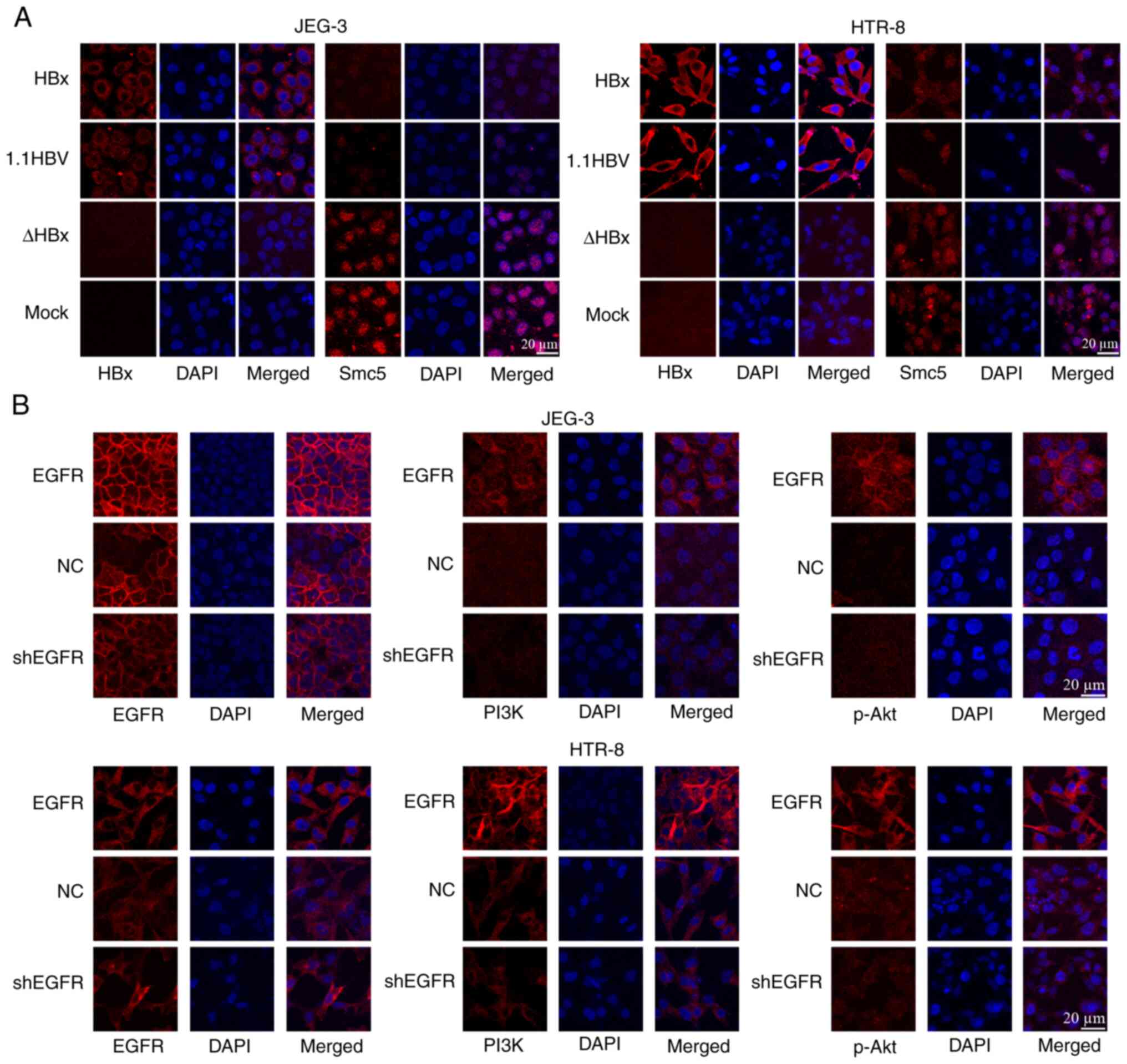 | Figure 3(A) Confocal laser microscopy
analysis of HBx and Smc5 protein expression and localization. The
nuclei stained with DAPI showed blue fluorescence, the HBx and Smc5
proteins showed red fluorescence and the merged showed two
overlapping fluorescence images. (B) Confocal laser microscopy
analysis of EGFR/PI3K/p-AKT protein expression and localization.
The nuclei stained with DAPI showed blue fluorescence, the
EGFR/PI3K/p-AKT proteins showed red fluorescence and the merged
showed two overlapping fluorescence images. Magnification, x400.
Scale bar, 20 µm. Smc5, structural maintenance of chromosomes 5;
HBx, hepatitis B virus X; EGFR, EGFR overexpressing cells; NC,
negative control; sh, short hairpin; HBx, pGFP-HBx plasmid; 1.1
HBV, full length wild type 1.1 HBV plasmid; ΔHBx, HBx deletion
mutant plasmid; mock, pGFP empty vector. |
Dual-luciferase reporter gene assay of
HBx activity on the EGFR promoter
As demonstrated in Fig.
4A, co-transfection of HBx and EGFR promoter plasmids in JEG-3
and HTR-8 cells significantly elevated EGFR promoter driven
luciferase expression relative to the control group, which was
co-transfected with EGFR promoter and pGFP empty vector.
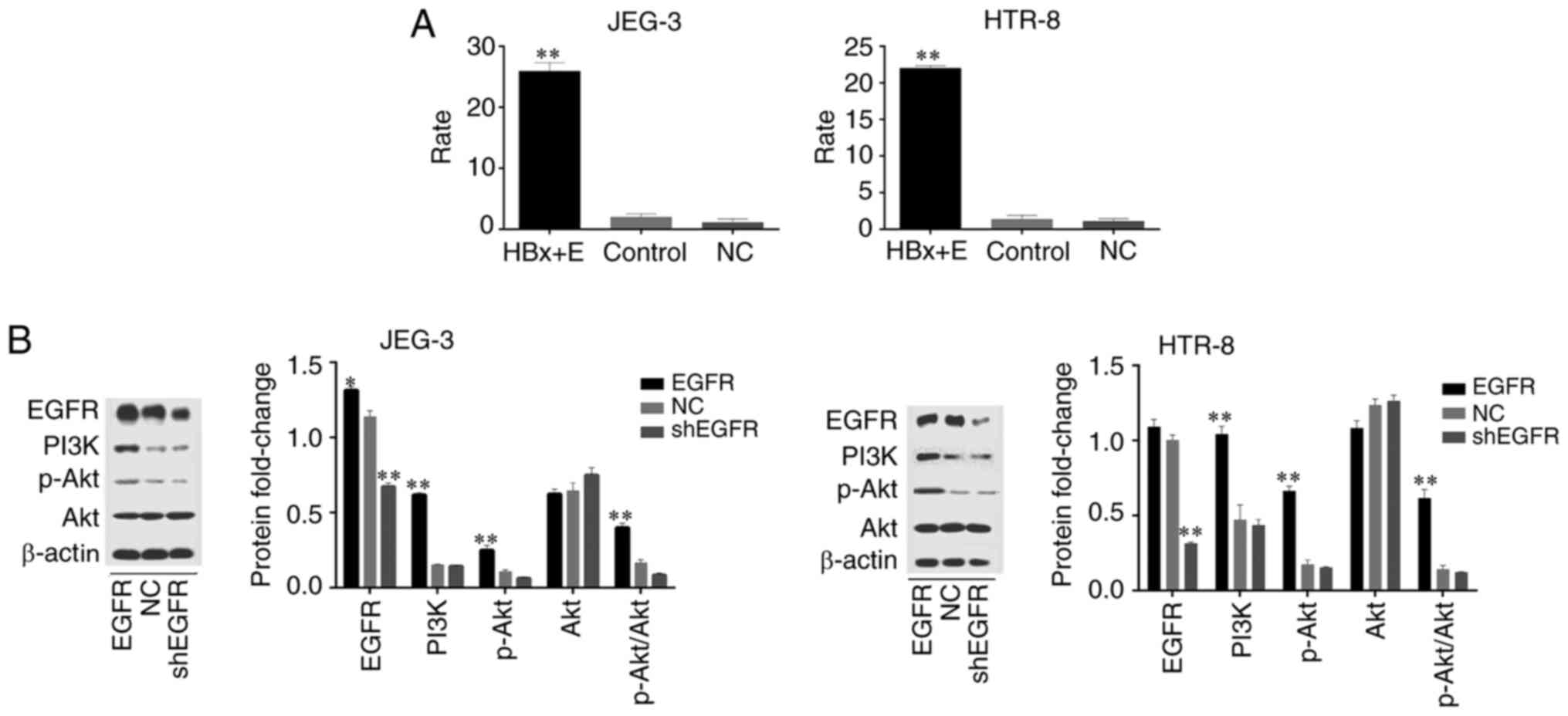 | Figure 4(A) Dual-luciferase reporter gene
assay of HBx activity on the EGFR promoter. HBx + E,
co-transfection of pGFP-HBx and pGL3-EGFR promoter plasmids;
control, co-transfection of pGFP empty vector and pGL3-EGFR
promoter plasmids; NC, co-transfection of pGFP-HBx and pGL3-basic
empty vector. Rate, firefly luciferase and internal reference
Renilla luciferase ratio. **P<0.01 vs.
control. (B) Western blot analysis of EGFR/PI3K/p-AKT expression.
Protein fold change is displayed as the ratio of the
EGFR/PI3K/p-Akt/Akt protein band to the β-actin protein band;
p-AKT/AKT, the ratio of p-AKT protein band to the AKT protein band.
*P<0.05, **P<0.01 vs. NC. EGFR, EGFR
overexpressing cells; NC, negative control; sh, short hairpin; HBx,
hepatitis B virus X; p-, phosphorylated. |
Western blot analysis of
EGFR/PI3K/p-AKT expression
EGFR protein expression was higher in the EGFR
overexpressing cells than that in the shEGFR-transfected cells or
the NC group (JEG-3; Fig. 4B). When
the EGFR overexpressing cells were transfected with shEGFR,
intracellular EGFR expression was significantly decreased in both
cell types as compared with that in EGFR overexpressing cells. When
EGFR was overexpressed, PI3K and p-AKT levels were significantly
higher in both cell types than in shEGFR-transfected cells and NC
group. There was no difference in AKT expression between
groups.
Confocal laser microscopy analysis of
EGFR/PI3K/p-AKT protein expression and localization
EGFR, PI3K and p-AKT were found in the cytoplasm. As
highlighted in Fig. 3B, EGFR
protein expression in the stable EGFR-overexpressing cells was
higher than in the shEGFR-expressing group and NC group. When
stable EGFR-overexpressing cells were transfected with shEGFR,
intracellular EGFR expression in both cell types was lower than in
the parent EGFR overexpressing cells. EGFR overexpression in both
cell types increased PI3K and p-AKT levels as compared with those
in shEGFR-transfected cells and NC group. Protein levels were
consistent with western blotting results.
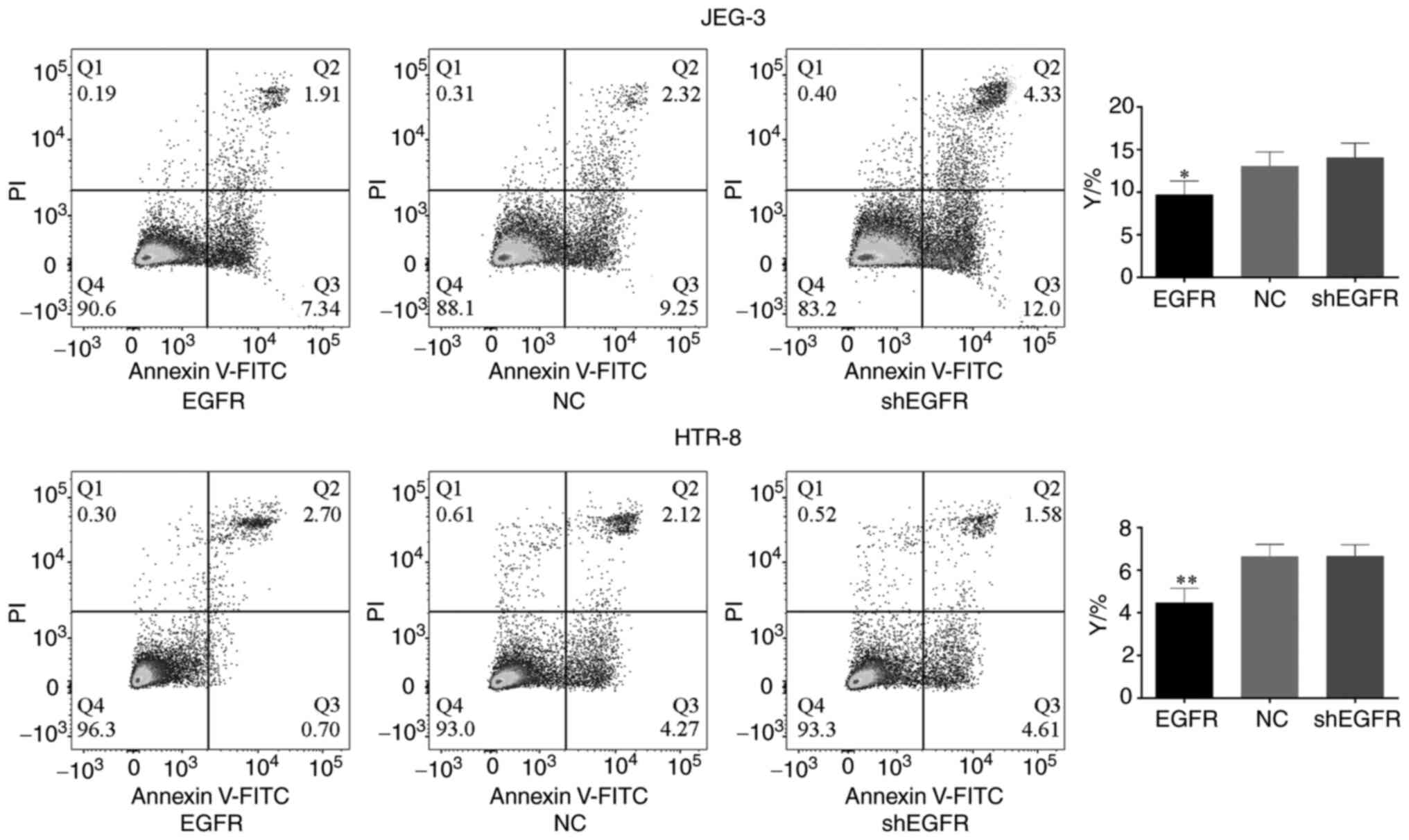
Association between EGFR expression
and apoptosis
As indicated in Fig.
5, the apoptosis percentage in HTR-8 and JEG-3 cells was
significantly lower than in NC group when EGFR was overexpressed.
When each type of stable EGFR-overexpressing cell was transfected
with shEGFR, the percentage of apoptotic cells increased
significantly. The percentage of apoptotic cells was negatively
associated with EGFR/PI3K/p-AKT expression.
Discussion
According to the cell transmission theory, the HBV
infection rate gradually decreases from the maternal side of the
placenta to the foetal side, whereby the virus infects the placenta
in the following order: i) Decidua cell; ii) trophoblast cell; iii)
villi interstitial cell; and iv) villi capillary endothelial cell.
On the other hand, the OR value of intrauterine infection gradually
increases, suggesting that HBV can infect a foetus from the
placenta via cell-to-cell transmission (13,15).
However, the underlying mechanisms responsible for this remain
unclear. Since trophoblast cells form the outermost placental
structure, direct contact with maternal blood during pregnancy is
the first barrier to HBV passage through the placenta (32,33).
Therefore, studying the role of HBV-infected placental trophoblasts
is key to clarifying intrauterine HBV infection mechanisms.
High HBV DNA load and HBeAg (+) are risk factors for
intrauterine infection (34-36).
A previous study found that HBxAg levels were significantly higher
in placental tissues of pregnant women with HBV DNA titres
>1x103 copies/ml than in tissues with HBV DNA titres
<1x103 copies/ml (24). In the present study, HBV DNA
expression was significantly higher in cells stably transfected
with full length wild-type HBV than in cells transfected with ΔHBx.
Expression levels of HBeAg and pgRNA in cells stably expressing
full length wild-type HBV were significantly higher than those in
ΔHBx-expressing cells. These results indicate that, HBV DNA
replication, HBeAg and pgRNA levels are significantly decreased
when HBx is deleted. The present results confirm that HBx protein
promotes HBV replication in placental trophoblast cells, HBx
protein expression in trophoblast cells increases with increasing
HBV DNA titre in maternal blood and HBx protein expression is
regulated by a positive feedback mechanism associated with HBV
replication in the host.
The covalently closed circular (ccc)DNA is the
template of transcription for 5 viral RNAs necessary for the
production of the viral antigens and for viral replication, the
latter of which takes place in the cytoplasm after reverse
transcription of an overlength pgRNA within newly formed
nucleocapsids (37). The
transcriptional activity of the cccDNA can be determined by
measuring pgRNA contents (38). In
the current study, the expression of pgRNA decreased significantly
when HBx was absent in the placenta trophoblasts transfected with
an HBV plasmid, which indicated that the transcription activity of
cccDNA in trophoblasts decreased and the replication of HBV DNA was
limited. The current study also demonstrated that HBx protein can
upregulate HBeAg expression, which further increases the risk of
HBV intrauterine infection.
Smc5/6 is a complex that directly binds DNA and is
required for chromosome remodelling and stability (39,40).
Smc5/6 has been extensively studied in yeast, but less so in
mammals. It has been revealed to serve a role in homologous
recombination and in resolving replication-induced DNA supercoiling
(41,42). Besides chromosome maintenance,
certain data suggest that Smc5/6 binds episomes (including cccDNA)
and blocks episome transcription (43). In placental trophoblast cells
transfected with HBx plasmid, Smc5/6 expression was significantly
lower compared with in mock-transfected cells. In addition,
confocal laser microscopy showed that HBx protein was mainly
located in the cytoplasm, whereas Smc5 protein was mainly located
in the nucleus. These results are consistent with other published
results of studies conducted in hepatoma cells (21,23).
It was hypothesized that the HBx protein promotes Smc5/6
degradation in placental trophoblast cells, thereby increasing the
transcription of cccDNA, promoting pgRNA synthesis and increasing
HBV DNA replication (Fig. S2).
Previous studies found that the expression of
PI3K/p-AKT was upregulated and trophoblast apoptosis was
significantly inhibited in vitro and in vivo under
the condition of high replication (HBV DNA >1x103
copies/ml) (24,25). HBx protein may inhibit trophoblast
apoptosis by activating PI3K/p-AKT (44). However, the exact mechanism by which
HBx protein activates the PI3K/p-AKT signalling pathway remains
unclear, thus indicating the requirement for further study. The HBx
gene integrates into the human genome near the EGFR gene and other
genes responsible for regulating cell growth, thus affecting
intracellular signal transduction (45). EGFR is a membrane surface receptor
with tyrosine kinase activity that is widely distributed on the
surface of mammalian cells. It can stimulate different signal
responses in various cells and activates a variety of downstream
signal transduction pathways that are known to stimulate cell
proliferation and enhance cell mobility and organ repair, similar
to PI3K/p-AKT/Bad and Ras/Raf/MEK/ERK pathways (46). p-EGFR protein is robustly
upregulated in HBx-infected human placental tissues and trophoblast
cells, HBx reduces human placental trophoblast cell apoptosis by
activating the EGFR/AKT pathway (47). However, the site and mechanism
underlying EGFR activation by HBx remain to be elucidated and
require further study.
Co-transfection of HBx and EGFR promoter plasmids in
JEG-3 and HTR-8 cells significantly elevated EGFR promoter driven
luciferase expression relative to the control group. In the present
study, it was revealed that HBx protein acts on the EGFR promoter
and activates EGFR promoter driven expression. When EGFR was
overexpressed in cells, PI3K and p-AKT were localized to the
cytoplasm and PI3K and p-Akt levels significantly increased,
whereas AKT expression was not affected. Additionally, the
percentage of cells undergoing apoptosis was also significantly
decreased. However, when EGFR expression was knocked down in
EGFR-overexpressing cells, PI3K and p-AKT levels were significantly
decreased, AKT expression was unaffected and the percentage of
cells undergoing apoptosis was significantly increased.
Collectively, the present results suggest that EGFR promotes AKT
phosphorylation in trophoblast cells by increasing expression of
its upstream effector, PI3K, which inhibits apoptosis. The present
study demonstrated that HBx protein activates EGFR expression by
acting on the EGFR promoter in placental trophoblast cells and
inhibits trophoblast cell apoptosis via the downstream PI3K/p-AKT
signalling pathway (Fig. S2).
In conclusion, HBx protein expression increased in
trophoblast cells with increasing titres of HBV DNA in maternal
blood. The findings of the present study indicate that HBx protein
promotes Smc5/6 degradation to enhance HBV replication in placental
trophoblast cells. Furthermore, HBx protein expression is
upregulated by increasing HBV replication. HBx protein also
activates EGFR expression by acting on the EGFR promoter and
inhibits trophoblast cell apoptosis via the downstream PI3K/p-AKT
signalling pathway. HBx prolongs the life of trophoblast cells
infected with HBV and provides a latent place for viruses to escape
(48). It was hypothesised that the
HBx protein, via its functions described above, is involved in the
mechanism underlying viral infection of placental trophoblast cells
and foetuses, thereby promoting the development of HBV intrauterine
infection. Therefore, HBx protein plays a crucial role in HBV
infection of placental trophoblasts and increases intrauterine
infection risk.
Supplementary Material
Western blot analysis of EGFR
expression. Protein fold change is displayed as the ratio of the
EGFR protein band to the β-actin protein band.
*P<0.05 vs. mock. sh, short hairpin; mock, shRNA
control vector.
HBx protein promotes degradation of
Smc5/6 to enhance HBV replication in placental trophoblast cells.
HBx protein expression is upregulated by increasing HBV
replication. HBx protein also activates EGFR expression by acting
on the EGFR promoter and inhibits trophoblast cell apoptosis
through the downstream signalling pathway PI3K/p-AKT. HBx,
hepatitis B virus X; Smc5, structural maintenance of chromosomes 5;
HBV, hepatitis B virus; pg, pregenomic; ccc, covalently closed
circular.
Acknowledgements
The authors would like to acknowledge the technical
assistance provided by Mrs. Rongjuan Chen (Institute of Infectious
Diseases, 5th Medical Center of Chinese PLA General Hospital,
Beijing); Mrs. Lanlan Si (Institute of Infectious Diseases, 5th
Medical Center of Chinese PLA General Hospital, Beijing); Dr Kai
Zhang (Institute of Infectious Diseases, Xiang'an Hospital of
Xiamen University); and Mr. Qi Li (Institute of Infectious
Diseases, 5th Medical Center of Chinese PLA General Hospital,
Beijing).
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81370721) and the National
Natural Science Foundation of China (grant no. 81771615).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GB conceived and designed the experiments. YL and DX
provided the resources and designed the experiments. YYL, WZ and FG
performed the experiments. YYL and YZ analyzed the data. YYL and GB
wrote the paper. YYL and GB confirmed the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Institutional
Review Board of the First Affiliated Hospital, Xi'an Jiaotong
University. All experiments were performed in accordance with
applicable guidelines and regulations. Informed written consent was
obtained from all study participants after a detailed explanation
of the study at the time of blood and serum collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dandri M: Epigenetic modulation in chronic
hepatitis B virus infection. Semin Immunopathol. 42:173–185.
2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Liu CJ and Kao JH: NOhep: Toward Global
Control of Hepatitis B virus infection-an introduction. J Infect
Dis. 216 (Suppl 8)(S749)2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Maini MK and Bertoletti A: HBV in 2016:
Global and immunotherapeutic insights into hepatitis B. Nat Rev
Gastroenterol Hepatol. 14:71–72. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wu Q, Xu C, Li J, Li L, Yan G, Yue L, Zeng
Y, Huang H, Deng G and Wang Y: Evolution and mutations of hepatitis
B virus quasispecies in genotype B and C during vertical
transmission. J Med Virol. 88:1018–1026. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Eke AC, Eleje GU, Eke UA, Xia Y and Liu J:
Hepatitis B immunoglobulin during pregnancy for prevention of
mother-to-child transmission of hepatitis B virus. The Cochrane
Database Syst Rev. 2(CD008545)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li Y, Shen C, Yang L, Yang Y, Wang M, Li
S, Chen F, Yang M, Peng L, Ma J, et al: Intra-host diversity of
hepatitis B virus during mother-to-child transmission: The X gene
may play a key role in virus survival in children after
transmission. Arch Virol. 165:1279–1288. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gong J and Liu X: Effect of HBIG combined
with hepatitis B vaccine on blocking HBV transmission between
mother and infant and its effect on immune cells. Exp Ther Med.
15:919–923. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Su WJ, Chen HL and Chang MH: Breakthrough
hepatitis B Virus (HBV) infection from mother-to-infant
transmission is the key problem hindering HBV eradication. J Infect
Dis. 208:1036–1037. 2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liu J, Feng Y, Wang J, Li X, Lei C, Jin D,
Feng W, Yang Y, He Y, Li Y, et al: An ‘immune barrier’ is formed in
the placenta by hepatitis B immunoglobulin to protect the fetus
from hepatitis B virus infection from the mother. Hum Vaccin
Immunother. 11:2068–2076. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang DD, Yi LZ, Wu LN, Yang ZQ, Hao HY,
Shi XH, Wang B, Feng SY, Feng YL and Wang SP: Relationship between
maternal PBMC HBV cccDNA and HBV serological markers and its effect
on HBV intrauterine transmission. Biomed Environ Sci. 32:315–323.
2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bai GQ, Li SH, Yue YF and Shi L: The study
on role of peripheral blood mononuclear cell in HBV intrauterine
infection. Arch Gynecol Obstet. 283:317–321. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ke C, Xiao X, Gan L and Zhou Y:
Preliminary study of hepatitis B virus replication in primary
placental trophoblastic cells. Prog Obstet Gynecol. 24:1–5.
2015.
|
|
13
|
Chen Y, Wang L, Xu Y, Liu X, Li S, Qian Q,
Hu B, Zhou A, Chen T and Zhao Y: Role of maternal viremia and
placental infection in hepatitis B virus intrauterine transmission.
Microbes Infect. 15:409–415. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhao Y, Di F and Zhong Y: Progress in
research on mechanism of HBV intrauterine transmission. Chin J Clin
(Electronic Edition). 9:1433–1436. 2015.
|
|
15
|
Zhang SL, Yue YF, Bai GQ, Shi L and Jiang
H: Mechanism of intrauterine infection of hepatitis B virus. World
J Gastroenterol. 10:437–438. 2004.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zuo Y, Li D, Xu D, Chen C and Wang X:
Meta-analysis of intrauterine infection rate of HBV in different
periods of pregnancy. J Fourth Military Med Univ. 9:853–855.
2002.(In Chinese).
|
|
17
|
Feitelson MA, Bonamassa B and Arzumanyan
A: The roles of hepatitis B virus-encoded X protein in virus
replication and the pathogenesis of chronic liver disease. Expert
Opin Ther Targets. 18:293–306. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lee HR, Cho YY, Lee GY, You DG, Yoo YD and
Kim YJ: A direct role for hepatitis B virus X protein in inducing
mitochondrial membrane permeabilization. J Viral Hepat. 25:412–420.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zoulim F, Saputelli J and Seeger C:
Woodchuck hepatitis virus X protein is required for viral infection
in vivo. J Virol. 68:2026–2030. 1994.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lucifora J, Arzberger S, Durantel D,
Belloni L, Strubin M, Levrero M, Zoulim F, Hantz O and Protzer U:
Hepatitis B virus X protein is essential to initiate and maintain
virus replication after infection. J Hepatol. 55:996–1003.
2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Decorsiere A, Mueller H, van Breugel PC,
Abdul F, Gerossier L, Beran RK, Livingston CM, Niu C, Fletcher SP,
Hantz O and Strubin M: Hepatitis B virus X protein identifies the
Smc5/6 complex as a host restriction factor. Nature. 531:386–389.
2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mitra B and Guo H: Hepatitis B virus X
protein crosses out Smc5/6 complex to maintain covalently closed
circular DNA transcription. Hepatology. 64:2246–2249.
2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Murphy CM, Xu Y, Li F, Nio K,
Reszka-Blanco N, Li X, Wu Y, Yu Y, Xiong Y and Su L: Hepatitis B
virus X protein promotes degradation of SMC5/6 to enhance HBV
replication. Cell Rep. 16:2846–2854. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bai G, Wang Y, Zhang L, Tang Y and Fu F:
The study on the role of hepatitis B virus X protein and apoptosis
in HBV intrauterine infection. Arch Gynecol Obstet. 285:943–949.
2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bai G, Fu F, Tang Y and Wang Y: Effect of
hepatitis B virus infection on apoptosis of a human choriocarcinoma
cell line in vitro. J Obstet Gynaecol Res. 39:1200–1211.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hung CM, Huang WC, Pan HL, Chien PH, Lin
CW, Chen LC, Chien YF, Lin CC, Leow KH, Chen WS, et al: Hepatitis B
Virus X upregulates HuR protein level to stabilize HER2 expression
in hepatocellular carcinoma cells. Biomed Res Int.
2014(827415)2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Chen JY, Chen YJ, Yen CJ, Chen WS and
Huang WC: HBx sensitizes hepatocellular carcinoma cells to
lapatinib by up-regulating ErbB3. Oncotarget. 7:473–489.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Graham CH, Hawley TS, Hawley RG,
MacDougall JR, Kerbel RS, Khoo N and Lala PK: Establishment and
characterization of first trimester human trophoblast cells with
extended lifespan. Exp Cell Res. 206:204–211. 1993.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Abou-Kheir W, Barrak J, Hadadeh O and
Daoud G: HTR-8/SVneo cell line contains a mixed population of
cells. Placenta. 50:1–7. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang J, Shen T, Huang X, Kumar GR, Chen X,
Zeng Z, Zhang R, Chen R, Li T, Zhang T, et al: Serum hepatitis B
virus RNA is encapsidated pregenome RNA that may be associated with
persistence of viral infection and rebound. J Hepatol. 65:700–710.
2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Huang H, Wang J, Li W, Chen R, Chen X,
Zhang F, Xu D and Lu F: Serum HBV DNA plus RNA shows superiority in
reflecting the activity of intrahepatic cccDNA in treatment-naïve
HBV-infected individuals. J Clin Virol. 99-100:71–78.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ma L, Alla NR, Li X, Mynbaev OA and Shi Z:
Mother-to-child transmission of HBV: Review of current clinical
management and prevention strategies. Rev Med Virol. 24:396–406.
2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mavilia MG and Wu GY: Mechanisms and
prevention of vertical transmission in chronic viral hepatitis. J
Clin Transl Hepatol. 5:119–129. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Peng S, Wan Z, Liu T, Zhu H and Du Y:
Incidence and risk factors of intrauterine transmission among
pregnant women with chronic hepatitis B virus infection. J Clin
Gastroenterol. 53:51–57. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Xiao Y, Sun K, Duan Z, Liu Z, Li Y, Yan L,
Song Y, Zou H, Zhuang H, Wang J and Li J: Quasispecies
characteristic in ‘a’ determinant region is a potential predictor
for the risk of immunoprophylaxis failure of
mother-to-child-transmission of sub-genotype C2 hepatitis B virus:
A prospective nested case-control study. Gut. 69:933–941.
2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shih YF and Liu CJ: Mother-to-infant
transmission of hepatitis B virus: Challenges and perspectives.
Hepatol Int. 11:481–484. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Allweiss L and Dandri M: The role of
cccDNA in HBV maintenance. Viruses. 9(156)2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Giersch K, Allweiss L, Volz T, Dandri M
and Lutgehetmann M: Serum HBV pgRNA as a clinical marker for cccDNA
activity. J Hepatol. 66:460–462. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Abdul F, Filleton F, Gerossier L, Paturel
A, Hall J, Strubin M and Etienne L: Smc5/6 antagonism by HBx is an
evolutionarily conserved function of Hepatitis B virus infection in
mammals. J Virol. 92:e00769–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Liang TJ: Virology: The X-Files of
hepatitis B. Nature. 531:313–314. 2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Jeppsson K, Carlborg KK, Nakato R, Berta
DG, Lilienthal I, Kanno T, Lindqvist A, Brink MC, Dantuma NP, Katou
Y, et al: The chromosomal association of the Smc5/6 complex depends
on cohesion and predicts the level of sister chromatid
entanglement. PLoS Genet. 10(e1004680)2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Jeppsson K, Kanno T, Shirahige K and
Sjogren C: The maintenance of chromosome structure: Positioning and
functioning of SMC complexes. Nature reviews. Molecular cell
biology. 15:601–614. 2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Van Breugel PC, Robert EI, Mueller H,
Decorsière A, Zoulim F, Hantz O and Strubin M: Hepatitis B virus X
protein stimulates gene expression selectively from
extrachromosomal DNA templates. Hepatology. 56:2116–2124.
2012.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wang W, Shi Y, Bai G, Tang Y, Yuan Y,
Zhang T and Li C: HBxAg suppresses apoptosis of human placental
trophoblastic cell lines via activation of the PI3K/Akt pathway.
Cell Biol Int. 40:708–715. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen YJ, Chien PH, Chen WS, Chien YF, Hsu
YY, Wang LY, Chen JY, Lin CW, Huang TC, Yu YL and Huang WC:
Hepatitis B virus-encoded X protein downregulates EGFR expression
via inducing MicroRNA-7 in hepatocellular carcinoma cells. Evid
Based Complement Alternat Med. 2013(682380)2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wee P and Wang Z: Epidermal growth factor
receptor cell proliferation signaling pathways. Cancers (Basel).
9(52)2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wang W, Bai G, Zhang Y, Zhang T, Li C,
Yuan Y, Liu S and Wang C: HBxAg suppresses cell apoptosis and
promotes the secretion of placental hormones in human placental
trophoblasts via activation of the EGFR/Akt pathway. Cell Biol Int.
42:237–247. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Rawat S and Bouchard MJ: The hepatitis B
virus (HBV) HBx protein activates AKT to simultaneously regulate
HBV replication and hepatocyte survival. J Virol. 89:999–1012.
2015.PubMed/NCBI View Article : Google Scholar
|