1. Introduction
Lower back pain (LBP) is one of the most common
symptoms and/or disorders globally, affecting people of all ages,
and is now the number one cause of disability worldwide (1,2). It
has been reported that ~84% of the general population suffers from
LBP during their lifetime (3), and
roughly 10% of them become chronically disabled (4). Due to the extremely high prevalence
of LBP, it seriously harms society and the economy, placing a heavy
burden on health care systems globally (5).
Intervertebral disc degeneration (IDD) is a major
pathological contributor to LBP. The intervertebral disc is a
complex fibrocartilaginous tissue that connects adjacent vertebral
bodies and confers spinal mobility; it is also the largest
avascular structure in the human body, consisting of the nucleus
pulposus (NP), annulus fibrosus (AF) and cartilage endplate (CEP).
Lacking a blood supply, disc cells retrieve oxygen and nutrients
mainly by diffusion through CEPs. Thus, they have a limited
capacity for self-repair after damage or degeneration (6). The extracellular matrix (ECM) is
mainly synthesized and secreted by NP cells (NPCs) and is composed
of type II collagen (collagen II) and aggrecan, helping to resist
compression and maintain disc height. According to previous
studies, IDD is mainly characterized by dysregulation of NPC
survival, an imbalance between ECM anabolism and catabolism, and
aberrant activation of the inflammatory response (7,8).
As the molecular mechanisms of IDD are complicated
and remain unclear, circular RNAs (circRNAs) have attracted growing
attention in the past few years. circRNAs are a novel class of
covalently closed, single-stranded and endogenous non-coding RNAs,
unlike linear RNAs, without 5'-3' polarities and polyadenylation.
Most circRNAs are produced from exons by a backsplicing mechanism
in eukaryotes with cell type- and developmental stage-specific
expression patterns (9). Compared
with linear RNAs, circRNAs are generally more stable and resistant
to RNase R digestion owing to their loop structure. circRNAs are
abundant and evolutionarily conserved, and mainly localize to the
cytoplasm. They exert important biological functions by acting as
microRNA (miRNA/miR) sponges, which are also known as competing
endogenous RNA (ceRNA) mechanisms, in the pathological process of
various diseases (10).
Previous studies have shown that circRNAs play
critical roles in various musculoskeletal diseases, including
osteoarthritis, rheumatoid arthritis, osteoporosis, osteosarcoma,
osteonecrosis, scoliosis and spinal cord injury (11-17).
Recently, accumulating evidence has uncovered the significant
differential expression of circRNAs between IDD and control groups.
Gain-of-function, loss-of-function and rescue experiments were
generally performed to investigate the biological functions and
regulatory pathways of circRNAs. Luciferase reporter, pulldown and
immunoprecipitation assays were performed to confirm the direct
interactions among the molecules of signaling pathways of circRNAs
(18,19).
The present review summarizes the current evidence
on the roles of circRNAs in the pathogenesis of IDD, including
proliferation, apoptosis, senescence, mitophagy, inflammation and
ECM metabolism (Table I).
 | Table IRoles of specific circRNAs in
IDD. |
Table I
Roles of specific circRNAs in
IDD.
| First author/s,
year | circRNA | Samples | Cell type | Treatment | Expression | miRNA | Targets | Functions | (Refs.) |
|---|
| Cheng et al,
2018 | circVMA21 | IDD vs.
thoracolumbar fracture or scoliosis | NPC | TNF-α and
IL-1β | ↓ | miR-200c | XIAP | ECM synthesis ↑,
cell apoptosis ↓ | (23) |
| Guo et al,
2018 | circ-GRB10 | Lumbar IDD vs.
traumatic lumbar fracture | NPC | Nutrition
deprivation | ↓ | miR-328-5p | ERBB2 | Cell apoptosis
↓ | (26) |
| Wang et al,
2018 | circ-4099 | IDD vs. vertebral
fracture or scoliosis | NPC | TNF-α | ↑ | miR-616-5p | Sox9 | ECM synthesis ↑,
inflammation ↓ | (30) |
| Song et al,
2018 | circRNA_104670 | Cervical
spondylotic myelopathy vs. Hirayama disease | NPC | NA | ↑ | miR-17-3p | MMP-2 | ECM synthesis ↓,
cell proliferation ↓, cell apoptosis ↑ | (31) |
| Wang et al,
2018 | circSEMA4B | Severe vs. mild
lumbar IDD | NPC | IL-1β | ↓ | miR-431 | Wnt pathway (SFRP1
and GSK-3β) | ECM synthesis ↑,
cell proliferation ↑, cell senescence ↓ | (24) |
| Xie et al,
2019 | circERCC2 | Cervical
spondylotic myelopathy vs. Hirayama disease | NPC | TBHP | ↓ | miR-182-5p | SIRT1 | ECM synthesis ↑,
cell apoptosis ↓, cell senescence ↓, cell mitophagy ↑ | (25) |
| Cui and Zhang,
2020 | circ_001653 | IDD vs. spinal cord
injury | NPC | NA | ↑ | miR-486-3p | CEMIP | ECM synthesis ↓,
cell proliferation ↓, cell apoptosis ↑ | (32) |
| Xiang et al,
2020 | circRNA-CIDN | IDD vs. idiopathic
scoliosis | NPC | Compression | ↓ | miR-34a-5p | SIRT1 | ECM synthesis ↑,
cell apoptosis ↓ | (33) |
| Guo et al,
2020 | circ-FAM169A | IDD vs. traumatic
vertebral fracture | NPC | NA | ↑ | miR-583 | BTRC, NF-κB
pathway | ECM synthesis ↓,
inflammation ↑ | (34) |
| Guo et al,
2020 | circ-TIMP2 | Lumbar IDD vs.
traumatic lumbar fracture | NPC | TNF-α and
IL-1β | ↑ | miR-185-5p | MMP-2 | ECM synthesis
↓ | (35) |
| Xiao et al,
2020 |
circRNA_0058097 | Severe vs. mild
IDD | CEP cell | Tension | ↑ | miR-365a-5p | HDAC4 | Cell morphology ECM
synthesis ↓ | (36) |
| Song et al,
2020 |
circRNA_0000253 | Severe vs. mild
IDD | NPC | NA | ↑ | miR-141-5p | SIRT1 | ECM synthesis ↓,
cell proliferation ↓, cell apoptosis ↑ | (37) |
| Li et al,
2020 | circ-FAM169A | Lumbar IDD vs.
thoracolumbar fracture or scoliosis | NPC | NA | ↑ | miR-583 | Sox9 | NA | (42) |
| Guo et al,
2020 | circ-GRB10 | Lumbar IDD vs.
traumatic lumbar fracture | NPC | NA | ↓ | miR-328-5p | Erk1/2, miR-141-3p,
FUS (upstream targets) | ECM synthesis
↑ | (27) |
| Chen et al,
2020 | circGLCE | IDD vs. traumatic
lumbar fracture | NPC | IL-1β | ↓ | miR-587 | STAP1 | ECM synthesis ↑,
cell apoptosis ↓ | (28) |
| Kong et al,
2020 | circ_0059955 | IDD vs. normal
discs from cadavers | NPC | NA | ↓ | NA | ITCH | Cell proliferation
↑, cell apoptosis ↓, cell cycle arrest ↓, inflammation ↓ | (44) |
| Huang et al,
2021 | circPKNOX1 | IDD vs. lumbar
vertebrae trauma | NPC | NA | ↓ | miR-370-3p | KIAA0355 | ECM synthesis
↑ | (29) |
2. Roles of circRNA in ECM metabolism
ECM degradation is a key contributor to the
progression of IDD, leading to loss of compression resistance and
disc height. Under pathological conditions, the balance between the
synthesis and decomposition of the ECM is disturbed, which is
characterized by decreased structural components (collagen II and
aggrecan) and increased matrix-degrading enzymes [matrix
metalloproteinase (MMP)-2, MMP-3, MMP-13, A disintegrin-like and
metalloproteinase with thrombospondin type-1 motifs (ADAMTS)-4 and
ADAMTS-5] (20-22).
Downregulated expression of circRNAs
in ECM metabolism
Cheng et al (23) first elucidated the role of a
specific circRNA in the pathological process of IDD. circVMA21 was
significantly downregulated in IDD tissues compared with control
tissues. Overexpression of circVMA21 reversed the imbalance between
ECM anabolism and catabolism in NPCs after treatment with tumor
necrosis factor-α (TNF-α) and interleukin-1β (IL-1β) in
vitro. Luciferase reporter, pulldown, RNA immunoprecipitation
and fluorescence in situ hybridization assays confirmed that
circVMA21 directly binds with cytoplasmic miR-200c and subsequently
upregulates the expression of the target mRNA, X linked
inhibitor-of-apoptosis protein (XIAP).
Wang et al (24) found that circSEMA4B overexpression
in NPCs attenuated the adverse effects on ECM synthesis induced by
IL-1β treatment in vitro. It was further verified that
circSEMA4B exerted positive functions by sponging miR-431 and
subsequently upregulating the expression of secreted frizzled
related protein 1 and glycogen synthase kinase-3β, which are known
as Wnt signaling-related factors. Similarly, it was reported that
circERCC2 facilitated ECM anabolism in NPCs by sponging miR-182-5p
and elevating the expression level of silent mating type
information regulation 2 homolog 1 (SIRT1) (25).
Guo et al (26) suggested that circ-GRB10 enhanced
NPC survival through the miR-328-5p-Erb-b2 receptor tyrosine kinase
2 (ERBB2) axis. Notably, another study by Guo et al
(27) focused on circ-GRB10 to
elucidate the upstream mechanisms. Forced expression of circ-GRB10
promoted phosphorylated-Erk1/2 expression and ameliorated ECM
synthesis. RNA-binding protein FUS (FUS) showed strongly
downregulated expression, while significant upregulation of
miR-141-3p expression was observed in IDD samples. The positive
effects induced by FUS overexpression could be counteracted by
silencing circ-GRB10. In addition, RNA immunoprecipitation
demonstrated the direct interaction between FUS and circ-GRB10.
Moreover, combined with further investigations and the results of
the previous study (26), Guo
et al (27) established the
signaling circuitry of the circ-GBR10-miR-328-5p-ERBB2-Erk1/2
phosphorylation-miR-141-3p-FUS loop in the pathogenesis of IDD
(Fig. 1).
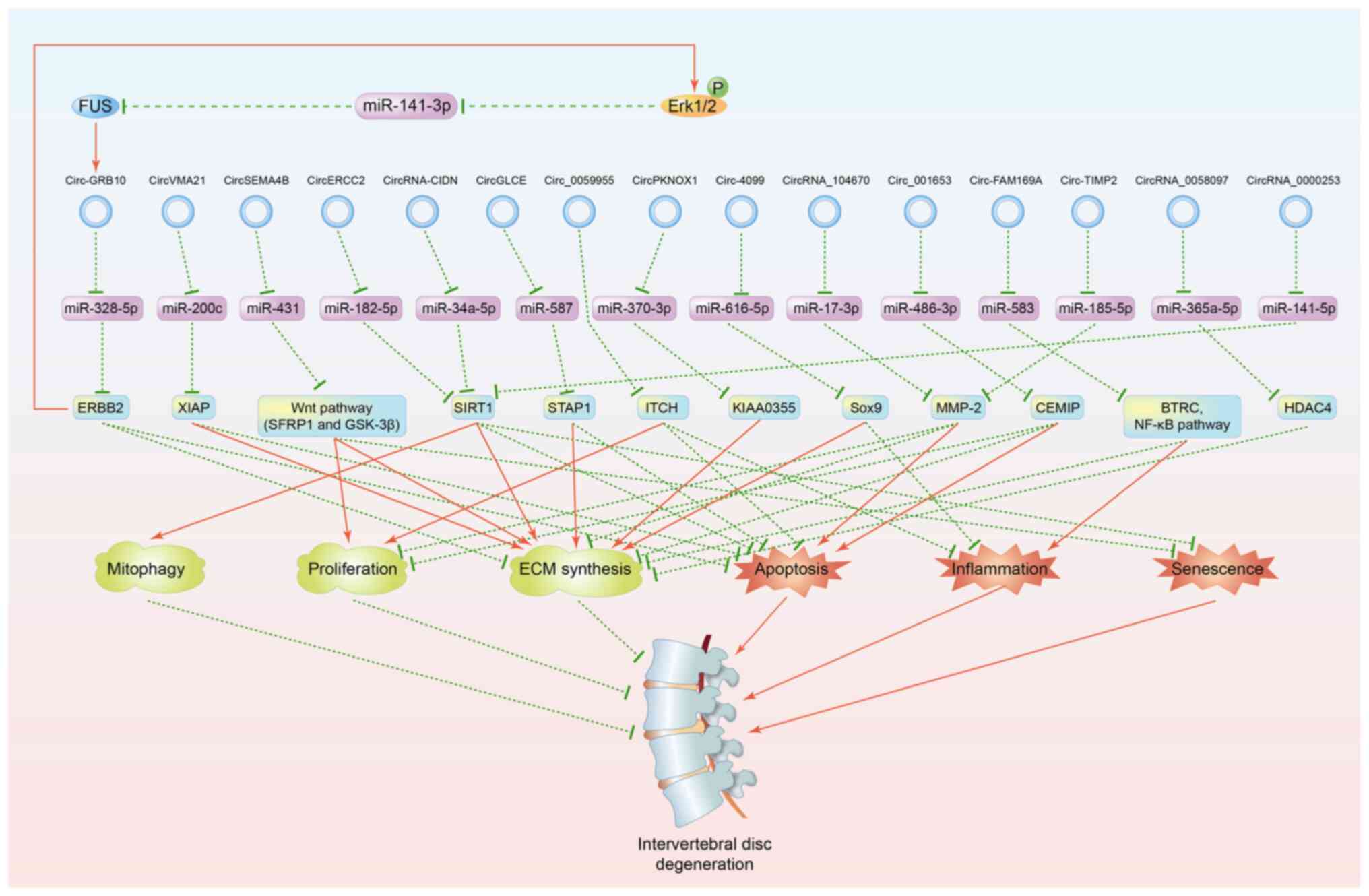 | Figure 1circRNAs involved in the regulation
of IDD. Red solid lines indicate upregulation and green dashed
lines represent downregulation. circRNA, circular RNA; miR,
microRNA; P, phosphorylation; ECM, extracellular matrix; IDD,
intervertebral disc degeneration; FUS, RNA-binding protein FUS;
ERBB2, Erb-b2 receptor tyrosine kinase 2; XIAP, X linked
inhibitor-of-apoptosis protein; SFRP1, secreted frizzled related
protein 1; GSK-3β, glycogen synthase kinase-3β; SIRT1, silent
mating type information regulation 2 homolog 1; STAP1, signal
transducing adaptor family member 1; ITCH, itchy E3 ubiquitin
protein ligase; Sox9, SRY-related high mobility group-box gene 9;
MMP-2, matrix metalloproteinase 2; CEMIP, cell migration-inducing
hyaluronan binding protein; BTRC, β-transducin repeat containing;
NF-κB, nuclear factor-κB; HDAC4, histone deacetylase 4. |
Chen et al (28) confirmed that circGLCE attenuates
ECM degradation by competitively absorbing miR-587 to upregulate
signal transducing adaptor family member 1 expression. Huang et
al (29) reported that
circPKNOX1 facilitated ECM synthesis and restrained ECM
decomposition. Dual luciferase assays indicated that circPKNOX1
directly interacts with miR-370-3p and subsequently upregulates
KIAA0355 expression.
Upregulated expression of circRNAs in
ECM metabolism
Wang et al (30) identified the significant
upregulation of circ-4099 expression in NPCs after treatment with
TNF-α in vitro. Overexpression of circ-4099 promoted ECM
anabolism, which was rescued by miR-616-5p mimics. Further
experiments revealed that circ-4099 ameliorates ECM synthesis
through the miR-616-5p-SRY-related high mobility group-box gene 9
(Sox9) axis. It was reported that knockdown of circRNA_104670
increased ECM synthesis, which was counteracted by miR-17-3p
inhibition (31). Luciferase
reporter and enhanced green fluorescent protein/red fluorescent
reporter assays demonstrated that circRNA_104670 aggravates ECM
degradation by sponging miR-17-3p and upregulating MMP-2
expression. Cui and Zhang (32)
showed that silencing circ_001653 in NPCs ameliorated ECM synthesis
and inhibited ECM decomposition. It was further determined that
circ_001653 sponges miR-486-3p, which targets cell
migration-inducing hyaluronan binding protein (CEMIP) to regulate
ECM metabolism.
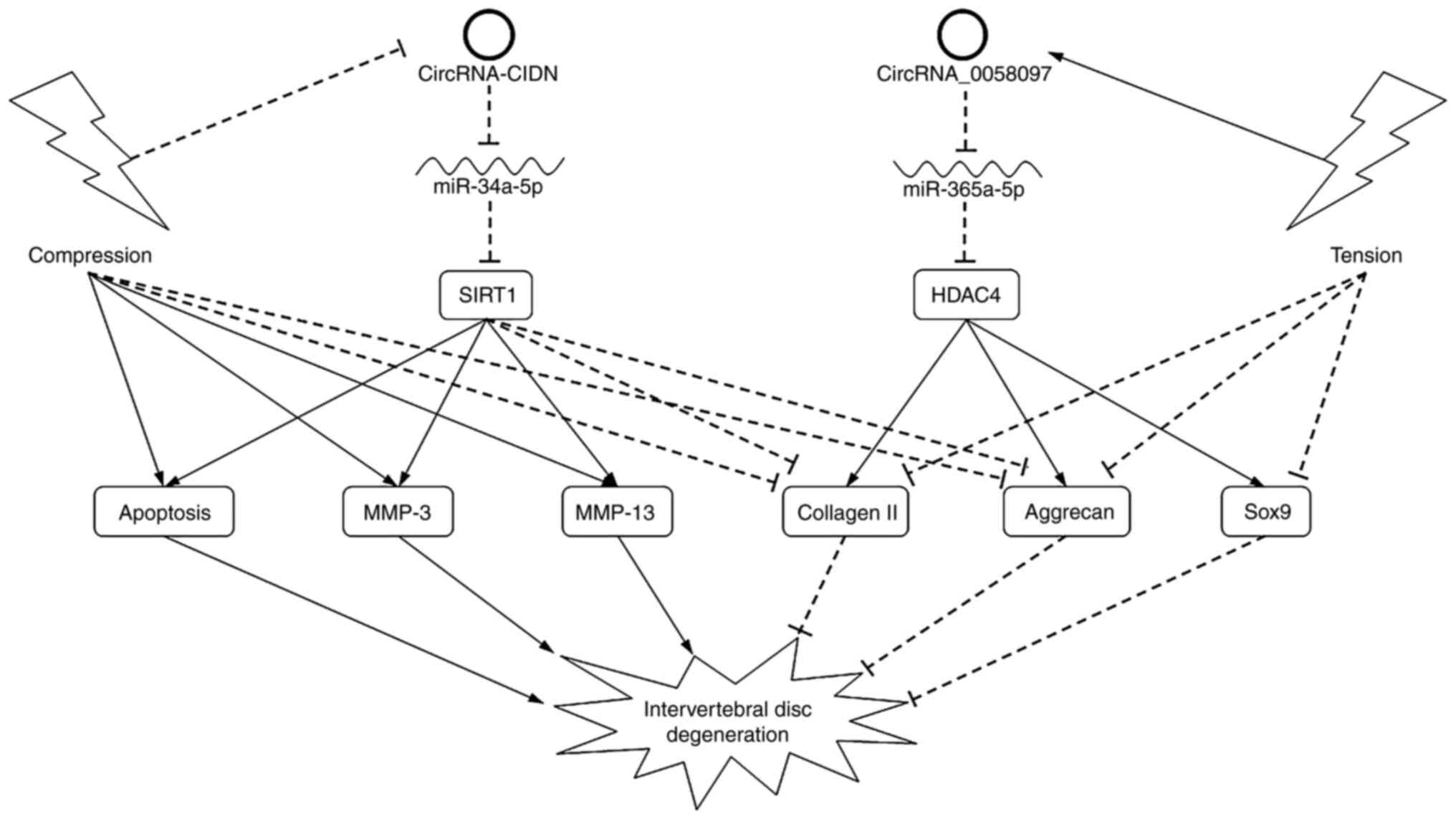
In the study by Xiang et al (33), compression treatment rather than
inflammatory cytokines (ICs) was applied to treated NPCs and
induced IDD. Highly expressed in IDD samples and
compression-treated NPCs, circRNA-CIDN could ameliorate the
pro-catabolic effects in NPCs induced by compression in
vitro and ex vivo. Luciferase reporter and RNA
immunoprecipitation assays verified the direct binding between
circRNA-CIDN and miR-34a-5p, and between miR-34a-5p and SIRT1.
Rescue experiments indicated that circRNA-CIDN overexpression
blocked the pro-catabolic effects of the miR-34a-5p mimic, while
knockdown of SIRT1 impaired the protective effects of circRNA-CIDN
in compression-treated NPCs. circRNA-CIDN therefore alleviates ECM
catabolism induced by compression through the miR-34a-5p-SIRT1
pathway. Guo et al (34,35)
reported that both circ-FAM169A and circ-TIMP2 shift ECM
homeostasis towards catabolism in NPCs by functioning as ceRNAs
through miR-583-β-transducin repeat containing (BTRC) and
miR-185-5p-MMP-2 signaling, respectively.
Xiao et al (36) focused on endplate chondrocytes
instead of NPCs and found that circRNA_0058097 expression was
upregulated in endplate chondrocytes after treatment with
intermittent cyclic tension in vitro. Tension treatment
decreased collagen II, aggrecan and Sox9 expression, and changed
the cell shape in endplate chondrocytes. Further assays showed that
circRNA_0058097 negatively regulated ECM metabolism by sponging
miR-365a-5p and activating the expression of histone deacetylase 4
(HDAC4) in endplate chondrocytes induced by tension loading
(Fig. 2).
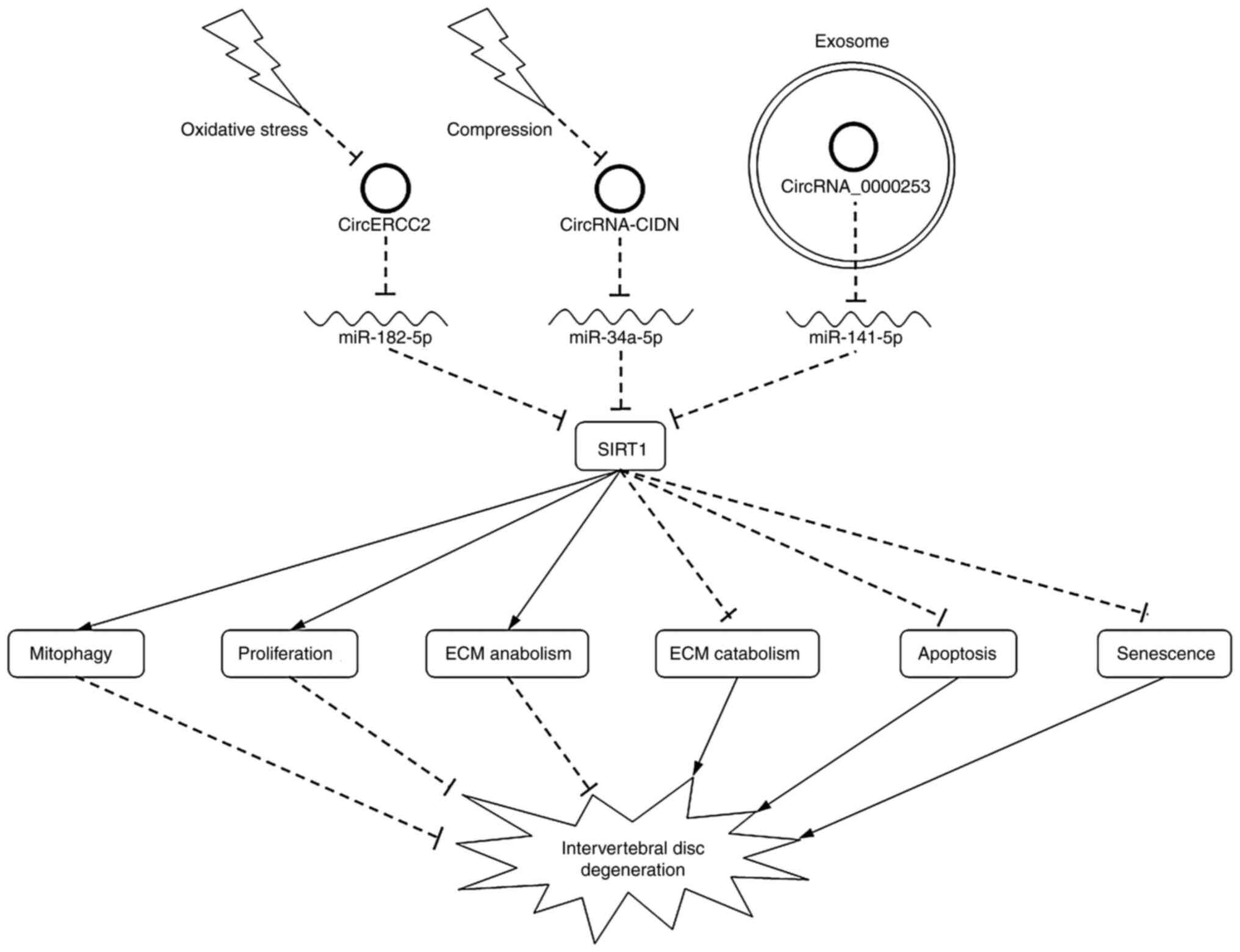
We previously focused on the roles of
exosome-transported circRNAs in IDD (37) and found that degenerative NPCs
secreted more exosomes than the controls in NPC culture medium.
Silencing circRNA_0000253, which was strongly upregulated in
degenerative NPC exosomes, promoted ECM synthesis. Further
investigations were performed both in vitro and in
vivo, and indicated that exosome-transported circRNA_0000253
aggravates IDD via the miRNA-141-5p-SIRT1 axis (Fig. 3).
Collectively, circRNA-miRNA-mRNA axes play crucial
roles in regulating ECM homeostasis.
3. Roles of circRNA in cell proliferation
and apoptosis
The intervertebral disc, as the largest avascular
tissue in the human body, is a cartilaginous structure with low
cell density, despite the cells in the intervertebral disc being
responsible for ECM synthesis and maintenance. IDD is characterized
by a diminution in cell number and viability. Thus, cell
proliferation and apoptosis are crucial to the pathogenesis of IDD
(38,39). For example, it was found that
circVMA21 overexpression attenuated cell apoptosis in NPCs after
treatment with TNF-α and IL-1β by acting as an miR-200c sponge and
upregulating XIAP (23). Urban
et al (40) clarified the
strong association between nutrition and IDD. It has been
demonstrated that loss of nutrient supply can lead to cell death
and an increase in ECM degradation, and hence to IDD (41). Guo et al (26) reported that circ-GRB10 expression
was significantly downregulated and miR-328-5p expression was
significantly upregulated in IDD samples compared with normal
controls. Upregulation of circ-GRB10 expression significantly
suppressed cell apoptosis in NPCs after nutrient deprivation in
vitro. Further experiments demonstrated that circ-GRB10 serves
as a regulator of NPC survival by sequestering miR-328-5p activity
and subsequently promoting the expression of ERBB2. Li et al
(42) found that circ-FAM169A
expression was strongly upregulated in IDD tissues. Dual luciferase
reporter assays corroborated the direct binding among circ-FAM169A,
miR-583 and Sox9. Although functional and rescue experiments were
lacking, functional enrichment analyses suggested that the
circ-FAM169A-miR-583 axis might be involved in ECM metabolism and
NPC survival.
In our previous study, interfering with
circRNA_104670 facilitated the proliferation and suppressed the
apoptosis of NPCs, as evaluated by MTT assays and cell flow
cytometry (31). Wnt/β-catenin
signaling is involved in a multitude of biological processes,
including cell proliferation, apoptosis and differentiation,
embryonic development and tissue organization (43). It was reported that upregulation of
circSEMA4B expression decreased proliferation and activated
senescence through the miR-431-Wnt pathway in NPCs after treatment
with IL-1β (24). circ_001653
inhibition promoted proliferation and suppressed apoptosis in NPCs
via miR-486-3p-CEMIP (32).
circRNA-CIDN overexpression inhibited apoptosis via the
miR-34a-5p-SIRT1 axis in NPCs after compression loading (33). Our previous study showed that
exosome-transported circRNA_0000253 could activate apoptosis while
decreasing the proliferation of NPCs by competitively absorbing
miRNA-141-5p (37). It was found
that circGLCE inhibition increased the rate of apoptosis in NPCs
after IL-1β treatment by acting as a sponge for miR-587(28).
Notably, Kong et al (44) focused on the interaction between
circRNA and target protein rather than the extensively studied
classic ceRNA mechanisms. This study found that the expression
level of hsa_circ_0059955 was significantly lower in IDD specimens
than that in control specimens. Knockdown of hsa_circ_0059955
inhibited proliferation and induced apoptosis and
G0/G1 phase arrest in NPCs. The expression of
itchy E3 ubiquitin protein ligase (ITCH) was negatively correlated
with hsa_circ_0059955. Forced expression of ITCH counteracted the
suppression of NPC proliferation induced by hsa_circ_0059955
inhibition. These results suggested the existence of the
hsa_circ_0059955-ITCH axis. However, further evidence verifying the
direct binding between hsa_circ_0059955 and the ITCH protein is
needed to make the hypothesis more convincing.
Thus, the aforementioned studies suggest that
different circRNAs could exert opposing effects on NPC
proliferation and apoptosis through ceRNA mechanisms.
4. Roles of circRNA in inflammation
Inflammatory mediators and signaling pathways are
recognized as major contributors to the onset and development of
IDD. Elevated levels of inflammatory molecules have been detected
in IDD tissues compared with those in healthy controls (45). Furthermore, ICs, particularly TNF-α
and IL-1β, have been demonstrated to stimulate and deteriorate the
progression of IDD (46). Thus,
treatment of NPCs with ICs has been widely applied to construct
in vitro IDD models (23,24,28,30,35).
The nuclear factor-κB (NF-κB) signaling pathway is closely
implicated in numerous complex biological processes, such as
immunity and inflammatory responses. There are two distinct
pathways leading to the activation of the NF-κB signaling cascades.
The canonical pathway is mediated by nuclear translocation of RelA
(p65), c-Rel and p50, while the non-canonical pathway is mediated
by nuclear translocation of p52 and RelB (47). It has been identified that the
NF-κB signaling pathway is a master regulator of inflammation and
catabolism in IDD (48).
In the study by Wang et al (30), an IDD NPC model was generated by
TNF-α treatment, and a higher expression level of circ-4099 was
observed. When pathway inhibitors of the mitogen-activated protein
kinase (MAPK) and NF-κB pathways were applied, the upregulation of
circ-4099 expression induced by TNF-α was reversed, indicating that
TNF-α enhanced circ-4099 expression via the MAPK and NF-κB
pathways. Moreover, circ-4099 overexpression significantly
decreased the expression level of IL-1β, TNF-α and prostaglandin E2
in NPCs, while miR-616-5p mimics could impair the anti-inflammatory
effects of circ-4099.
As a component of the Skp-Cullin-F-box-containing E3
ubiquitin ligase complex, BTRC recognizes the NF-κB inhibitor IκBα
and precursor p100 for proteasomal degradation and processing,
respectively. Therefore, BTRC plays an important role in both the
canonical and non-canonical NF-κB activation pathways (49). Guo et al (34) reported that circ-FAM169A was
upregulated in IDD tissues. Knockdown of circ-FAM169A not only
alleviated ECM degradation but also inhibited IL-1β, TNF-α and IKBα
expression, while circ-FAM169A overexpression exhibited the
opposite effects. Silencing BTRC could reverse the adverse effects
of circ-FAM169A. In addition, luciferase reporter assays confirmed
the direct binding among circ-FAM169A, miR-583 and BTRC.
5. Roles of circRNA in senescence
Cellular senescence is characterized by irreversible
proliferative arrest in response to multiple stresses, such as
telomere dysfunction, DNA damage, organelle stress and oncogene
activation (50). A growing body
of evidence has indicated that cellular senescence participates in
the pathological process of a number of chronic age-associated
diseases, including IDD (51-54).
To date, the detection of senescent cells depends on a combination
of various markers, including senescence-associated-β-galactosidase
(SA-β-gal), aberrant cell morphology, the nucleoside analogs p21
and p16, nuclear senescence-associated heterochromatin foci and
components of the senescence-associated secretory phenotype
(55). Previous studies identified
that circSEMA4B and circERCC2 both inhibited cellular senescence,
as determined using SA-β-gal staining, in NPCs stimulated by IL-1β
and tert-butyl hydroperoxide (TBHP), respectively (24,25).
Hence, circRNAs could affect NPC senescence through miRNA-mRNA
pathways.
6. Roles of circRNA in mitophagy
Mitochondria are double membrane-enclosed
organelles, playing pivotal roles in energy production, reactive
oxygen species (ROS) generation and cell death (56). Mitochondrial dysfunction is
associated with the occurrence and development of various diseases.
Therefore, dynamic and relatively stable control of mitochondrial
quantity and quality is crucial for preserving the physiological
functions of cells (57).
Mitophagy, a special form of autophagy, is a conserved and
essential process to maintain mitochondrial homeostasis by
selectively eliminating dysfunctional mitochondria (56).
The PTEN induced kinase 1 (PINK1)/Parkin-mediated
mitophagy pathway is a classical and well-studied pathway in
degenerative diseases (58). Wang
et al (59) discovered that
PINK1 expression was upregulated in IDD tissues. NPCs were treated
with H2O2 to induce oxidative stress, which
aggravated mitochondrial dysfunction and cellular senescence, and
activated mitophagy in NPCs. Silencing PINK1 enhanced oxidative
stress-induced NPC senescence by inhibiting PINK1/Parkin-mediated
mitophagy. Zhang et al (60) reported that Parkin expression was
elevated both in IDD specimens and NPCs stimulated by TNF-α.
Knockdown of Parkin increased mitochondrial damage, ROS production
and apoptosis in NPCs, while upregulating Parkin expression
alleviated the progression of IDD via Parkin-mediated mitophagy.
Wang et al (61) revealed
that SIRT1 attenuated ROS accumulation, mitochondrial injury and
cellular senescence through PINK1-dependent mitophagy in NPCs
treated with high-magnitude compression. Therefore, mitophagy
appears to be closely correlated with IDD development.
Hirayama disease is a special neurological disorder,
predominantly affecting male adolescents. Thus, non-degenerative NP
tissues could be obtained from patients undergoing anterior
cervical discectomy and fusion due to this disease (62). We previously showed that circERCC2
expression was significantly downregulated in IDD tissues compared
with that in Hirayama disease samples (25). Forced expression of circERCC2
decreased the rate of apoptosis and activated mitophagy, which was
evaluated by detecting the key markers for mitophagy (PINK1,
Parkin, p62 and LC3II/I ratio), in NPCs induced by TBHP in
vitro. The direct binding between circERCC2 and miR-1825p, and
between miR182-5p and SIRT1 was confirmed by dual-luciferase
assays. Further analysis was performed and determined that
circERCC2 alleviates IDD by regulating NPC apoptosis and mitophagy
through the miR-182-5p-SIRT1 axis.
7. Limitations and future directions
Over the past few years, a growing number of studies
have shown that numerous circRNAs can contribute to the development
and progression of IDD, particularly NPC phenotypic modulation
(Fig. 1). circRNAs are abundant,
highly conserved and stable, with tissue- and development-specific
expression patterns. Therefore, the in-depth and reliable
elucidation of the association between circRNAs and IDD will
eventually facilitate the establishment of novel diagnostic and
treatment methods.
However, there are still some limitations in the
available studies. First, to the best of our knowledge, 17 studies
have been published that investigated the potential roles of
circRNAs in IDD. Of these, 15 mainly focused on ceRNA mechanisms,
indicating that circRNAs exert biological functions by acting as
miRNA sponges, and established circRNA-miRNA-mRNA axes. Notably,
Kong et al (44) attempted
to study the interaction between circRNA and target protein instead
of classic ceRNA mechanisms. The results suggested that
hsa_circ_0059955 inhibition negatively regulated NPC survival via
the ITCH protein. However, this finding was not confirmed, as
experiments verifying the direct binding between hsa_circ_0059955
and ITCH protein were not conducted. Notably, circRNAs not only
sponge and sequester proteins, but also act as protein scaffolds to
mediate the formation of circRNA-protein complexes and the cellular
localization of specific proteins (9). Notably, although circRNAs are
generally considered to be non-coding, as they lack 5' caps and
polyadenylated tails that are essential for translation initiation,
some circRNAs can function as templates for protein synthesis
through cap-independent translation driven by internal ribosome
entry sites or N6-methyladenosine (9). Accordingly, future studies are
required to discover other functional mechanisms of circRNAs in IDD
in addition to their serving as miRNA sponges.
Second, most of the present studies concentrated on
the downstream mechanisms of circRNAs, but the question of what
causes the aberrantly altered expression of circRNAs in IDD has
been less discussed. Notably, studies by Guo et al (26,27)
established a complex signaling circuitry of circ-GBR10 rather than
a linear pathway in IDD. It was found that an RNA-binding protein,
FUS, was an upstream regulator of circ-GRB10. RNA
immunoprecipitation further confirmed the direct interaction
between circ-GRB10 and the FUS protein. Combining the results of
two published studies by Guo et al (26,27),
a circ-GBR10-miR-328-5p-ERBB2-Erk1/2 phosphorylation-miR-141-3p-FUS
loop was constructed. In the future, it will be worthwhile to fully
investigate circRNAs with their upstream regulators and downstream
targets to establish circRNA networks. Further elucidation of the
crosstalk among circRNAs can help us to recognize the key nodes to
improve diagnosis and treatment.
Third, IDD is an extremely complex process. Studies
tend to focus on NPCs, as they are the main type of cells in the NP
and are responsible for ECM production. However, IDD also includes
the deterioration of both AF and CEPs. Xiao et al (36) focused on endplate chondrocytes
instead of NPCs and demonstrated that inhibition of circRNA_0058097
could exert positive effects on endplate chondrocyte degeneration
via the miR-365a-5p-HDAC4 axis. More studies are needed to reveal
how circRNAs affect the IDD process by regulating the functions of
cells in AF and CEPs. Furthermore, numerous studies simulated IDD
by culturing NPCs with ICs, such as IL-1β and TNF-α. In addition to
ICs, oxidative stress, nutrient deprivation and mechanical loading
are crucial factors inducing IDD (46,63-65).
Xiang et al (33) indicated
that circRNA-CIDN alleviated IDD induced by compression through the
miR-34a-5p-SIRT1 axis. Thus, more research should be performed in
different IDD models. In addition, proliferation, apoptosis,
inflammation and ECM metabolism have been extensively studied. Only
two studies described senescence (24,25),
and one study discussed mitophagy in IDD (25). Therefore, the important phenotypes
mediated by circRNAs, including cellular senescence, autophagy,
mitophagy and ferroptosis, in IDD have not been sufficiently
illuminated (50,57,66,67).
Recently, exosome-transported circRNAs have
attracted increasing attention from scientists. Exosomes are small
membrane vesicles that originate from multivesicular endosomes by
inverse budding. Secreted by most eukaryotes, exosomes can carry
various molecules, including lipids, proteins and nucleic acids,
from one cell to another (68). It
was previously noted that exosomal circRNA_0000253 can aggravate
the progression of IDD via the miRNA-141-5p-SIRT1 axis (37), and the roles of exosome-transported
circRNAs have become a hot topic in the research field of cancer
(69). Therefore, it is essential
to further determine whether these novel circRNAs are involved in
the pathological process of IDD.
8. Conclusion
In conclusion, the latest evidence has confirmed
that circRNAs play crucial roles in ECM synthesis and
decomposition, inflammation, cellular proliferation, apoptosis,
senescence and mitophagy, mainly by functioning as sponges for
miRNAs in the pathogenesis of IDD.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by the General Program of
National Natural Science Foundation of China (grant nos. 81871552,
81972093 and 82072488).
Availability of data and materials
Not applicable.
Authors' contributions
ZL, FZ and JJ were responsible for the
conceptualization of the study. FZ, JJ and FL were responsible for
the study design, and ZL, FL, XM and XX for the search strategy,
article inclusion and interpretation. All authors were involved in
original draft preparation. ZL, FZ and JJ were responsible for
reviewing and editing the manuscript, and FZ and JJ supervised the
project. All authors have read and approved the manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hartvigsen J, Hancock MJ, Kongsted A, Louw
Q, Ferreira ML, Genevay S, Hoy D, Karppinen J, Pransky G, Sieper J,
et al: What low back pain is and why we need to pay attention.
Lancet. 391:2356–2367. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cieza A, Causey K, Kamenov K, Hanson SW,
Chatterji S and Vos T: Global estimates of the need for
rehabilitation based on the Global Burden of Disease study 2019: A
systematic analysis for the Global Burden of Disease Study 2019.
Lancet. 396:2006–2017. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Walker BF: The prevalence of low back
pain: A systematic review of the literature from 1966 to 1998. J
Spinal Disord. 13:205–217. 2000.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Walker BF, Muller R and Grant WD: Low back
pain in Australian adults: Prevalence and associated disability. J
Manipulative Physiol Ther. 27:238–244. 2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Vlaeyen JW, Maher CG, Wiech K, Van Zundert
J, Meloto CB, Diatchenko L, Battie MC, Goossens M, Koes B and
Linton SJ: Low back pain. Nat Rev Dis Primers. 4(52)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wise CA, Sepich D, Ushiki A, Khanshour AM,
Kidane YH, Makki N, Gurnett CA, Gray RS, Rios JJ, Ahituv N and
Solnica-Krezel L: The cartilage matrisome in adolescent idiopathic
scoliosis. Bone Res. 8(13)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Adams MA and Roughley PJ: What is
intervertebral disc degeneration, and what causes it? Spine (Phila
Pa 1976). 31:2151–2161. 2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shen S, Wu Y, Chen J, Xie Z, Huang K, Wang
G, Yang Y, Ni W, Chen Z, Shi P, et al: CircSERPINE2 protects
against osteoarthritis by targeting miR-1271 and ETS-related gene.
Ann Rheum Dis. 78:826–836. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Cai Y, Liang R, Xiao S, Huang Q, Zhu D,
Shi GP, Ouyang Q and Yang M: Circ_0088194 promotes the invasion and
migration of rheumatoid arthritis fibroblast-like synoviocytes via
the miR-766-3p/MMP2 axis. Front Immunol. 12(628654)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Han S, Kuang M, Sun C, Wang H, Wang D and
Liu Q: Circular RNA hsa_circ_0076690 acts as a prognostic biomarker
in osteoporosis and regulates osteogenic differentiation of hBMSCs
via sponging miR-152. Aging (Albany NY). 12:15011–15020.
2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wu Y, Xie Z, Chen J, Chen J, Ni W, Ma Y,
Huang K, Wang G, Wang J, Ma J, et al: Circular RNA circTADA2A
promotes osteosarcoma progression and metastasis by sponging
miR-203a-3p and regulating CREB3 expression. Mol Cancer.
18(73)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hao Y, Lu C, Zhang B, Xu Z, Guo H and
Zhang G: CircPVT1 up-regulation attenuates steroid-induced
osteonecrosis of the femoral head through regulating
miR-21-5p-mediated Smad7/TGFβ signalling pathway. J Cell Mol Med.
25:4608–4622. 2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chen C, Tan H, Bi J, Li Z, Rong T, Lin Y,
Sun L, Li X and Shen J: Identification of competing endogenous RNA
regulatory networks in vitamin A deficiency-induced congenital
scoliosis by transcriptome sequencing analysis. Cell Physiol
Biochem. 48:2134–2146. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Peng P, Zhang B, Huang J, Xing C, Liu W,
Sun C, Guo W, Yao S, Ruan W, Ning G, et al: Identification of a
circRNA-miRNA-mRNA network to explore the effects of circRNAs on
pathogenesis and treatment of spinal cord injury. Life Sci.
257(118039)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li Z, Chen X, Xu D, Li S, Chan MT and Wu
WK: Circular RNAs in nucleus pulposus cell function and
intervertebral disc degeneration. Cell Prolif.
52(e12704)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li Y, Zhou S, Peng P, Wang X, Du L, Huo Z
and Xu B: Emerging role of circular RNA in intervertebral disc
degeneration: Knowns and unknowns (Review). Mol Med Rep.
22:3057–3065. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Le Maitre CL, Pockert A, Buttle DJ,
Freemont AJ and Hoyland JA: Matrix synthesis and degradation in
human intervertebral disc degeneration. Biochem Soc Trans.
35:652–655. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Binch ALA, Shapiro IM and Risbud MV:
Syndecan-4 in intervertebral disc and cartilage: Saint or synner?
Matrix Biol. 52-54:355–362. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang SZ, Rui YF, Lu J and Wang C: Cell and
molecular biology of intervertebral disc degeneration: Current
understanding and implications for potential therapeutic
strategies. Cell Prolif. 47:381–390. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cheng X, Zhang L, Zhang K, Zhang G, Hu Y,
Sun X, Zhao C, Li H, Li YM and Zhao J: Circular RNA VMA21 protects
against intervertebral disc degeneration through targeting miR-200c
and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis.
77:770–779. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang X, Wang B, Zou M, Li J, Lü G, Zhang
Q, Liu F and Lu C: CircSEMA4B targets miR-431 modulating
IL-1β-induced degradative changes in nucleus pulposus cells in
intervertebral disc degeneration via Wnt pathway. Biochim Biophys
Acta Mol Basis Dis. 1864:3754–3768. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Xie L, Huang W, Fang Z, Ding F, Zou F, Ma
X, Tao J, Guo J, Xia X, Wang H, et al: CircERCC2 ameliorated
intervertebral disc degeneration by regulating mitophagy and
apoptosis through miR-182-5p/SIRT1 axis. Cell Death Dis.
10(751)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Guo W, Zhang B, Mu K, Feng SQ, Dong ZY,
Ning GZ, Li HR, Liu S, Zhao L, Li Y, et al: Circular RNA GRB10 as a
competitive endogenous RNA regulating nucleus pulposus cells death
in degenerative intervertebral disk. Cell Death Dis.
9(319)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Guo W, Mu K, Zhang B, Sun C, Zhao L, Li
HR, Dong ZY and Cui Q: The circular RNA circ-GRB10 participates in
the molecular circuitry inhibiting human intervertebral disc
degeneration. Cell Death Dis. 11(612)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen Z, Zhang W, Deng M, Li Y and Zhou Y:
CircGLCE alleviates intervertebral disc degeneration by regulating
apoptosis and matrix degradation through the targeting of
miR-587/STAP1. Aging (Albany NY.). 12:21971–21991. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Huang Y, Gao J, Wang J, Ye H, Yao T, Xu Y,
Chen Z, Shen S and Ma J: Inhibition of intervertebral disc disease
progression via the circPKNOX1-miR-370-3p-KIAA0355 axis. Cell Death
Discov. 7(39)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang H, He P, Pan H, Long J, Wang J, Li Z,
Liu H, Jiang W and Zheng Z: Circular RNA circ-4099 is induced by
TNF-α and regulates ECM synthesis by blocking miR-616-5p inhibition
of Sox9 in intervertebral disc degeneration. Exp Mol Med. 50:1–14.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Song J, Wang HL, Song KH, Ding ZW, Wang
HL, Ma XS, Lu FZ, Xia XL, Wang YW, Fei-Zou and Jiang JY:
CircularRNA_104670 plays a critical role in intervertebral disc
degeneration by functioning as a ceRNA. Exp Mol Med. 50:1–12.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Cui S and Zhang L: Circ_001653 silencing
promotes the proliferation and ECM synthesis of NPCs in IDD by
downregulating miR-486-3p-mediated CEMIP. Mol Ther Nucleic Acids.
20:385–399. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Xiang Q, Kang L, Wang J, Liao Z, Song Y,
Zhao K, Wang K, Yang C and Zhang Y: CircRNA-CIDN mitigated
compression loading-induced damage in human nucleus pulposus cells
via miR-34a-5p/SIRT1 axis. EBioMedicine. 53(102679)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Guo W, Mu K, Zhang B, Sun C, Zhao L, Dong
ZY and Cui Q: The circular RNA FAM169A functions as a competitive
endogenous RNA and regulates intervertebral disc degeneration by
targeting miR-583 and BTRC. Cell Death Dis. 11(315)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Guo W, Zhang B, Sun C, Duan HQ, Liu WX, Mu
K, Zhao L, Li HR, Dong ZY and Cui Q: Circular RNA derived from
TIMP2 functions as a competitive endogenous RNA and regulates
intervertebral disc degeneration by targeting miR-185-5p and matrix
metalloproteinase 2. Int J Mol Med. 46:621–632. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Xiao L, Ding B, Xu S, Gao J, Yang B, Wang
J and Xu H: CircRNA_0058097 promotes tension-induced degeneration
of endplate chondrocytes by regulating HDAC4 expression through
sponge adsorption of miR-365a-5p. J Cell Biochem. 121:418–429.
2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Song J, Chen ZH, Zheng CJ, Song KH, Xu GY,
Xu S, Zou F, Ma XS, Wang HL and Jiang JY: Exosome-Transported
circRNA_0000253 competitively adsorbs MicroRNA-141-5p and increases
IDD. Mol Ther Nucleic Acids. 21:1087–1099. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ding F, Shao ZW and Xiong LM: Cell death
in intervertebral disc degeneration. Apoptosis. 18:777–785.
2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhao CQ, Jiang LS and Dai LY: Programmed
cell death in intervertebral disc degeneration. Apoptosis.
11:2079–2088. 2006.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Urban JP, Smith S and Fairbank JC:
Nutrition of the intervertebral disc. Spine (Phila Pa 1976).
29:2700–2709. 2004.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Grunhagen T, Wilde G, Soukane DM,
Shirazi-Adl SA and Urban JP: Nutrient supply and intervertebral
disc metabolism. J Bone Joint Surg Am. 88 (Suppl 2):S30–S35.
2006.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li Y, Pan D, Liu S, Xing X, Zhou H, Zhang
B, Zhang D, Li B, Li G, Tao B, et al: Identification of
circ-FAM169A sponges miR-583 involved in the regulation of
intervertebral disc degeneration. J Orthop Translat. 26:121–131.
2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kahn M: Can we safely target the WNT
pathway? Nat Rev Drug Discov. 13:513–532. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kong D, Gu R, Zhang C and Yin R: Knockdown
of hsa_circ_0059955 induces apoptosis and cell cycle arrest in
nucleus pulposus cells via inhibiting itchy E3 ubiquitin protein
ligase. Drug Des Devel Ther. 14:3951–3963. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lyu FJ, Cui H, Pan H, Mc Cheung K, Cao X,
Iatridis JC and Zheng Z: Painful intervertebral disc degeneration
and inflammation: From laboratory evidence to clinical
interventions. Bone Res. 9(7)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wang Y, Che M, Xin J, Zheng Z, Li J and
Zhang S: The role of IL-1beta and TNF-alpha in intervertebral disc
degeneration. Biomed Pharmacother. 131(110660)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Miraghazadeh B and Cook MC: Nuclear
factor-kappaB in autoimmunity: Man and Mouse. Front Immunol.
9(613)2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhang GZ, Liu MQ, Chen HW, Wu ZL, Gao YC,
Ma ZJ, He XG and Kang XW: NF-κB signalling pathways in nucleus
pulposus cell function and intervertebral disc degeneration. Cell
Prolif. 54(e13057)2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Shi M, Cho H, Inn KS, Yang A, Zhao Z,
Liang Q, Versteeg GA, Amini-Bavil-Olyaee S, Wong LY, Zlokovic BV,
et al: Negative regulation of NF-κB activity by brain-specific
TRIpartite Motif protein 9. Nat Commun. 5(4820)2014.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Zhang Y, Yang B, Wang J, Cheng F, Shi K,
Ying L, Wang C, Xia K, Huang X, Gong Z, et al: Cell senescence: A
nonnegligible cell state under survival stress in pathology of
intervertebral disc degeneration. Oxid Med Cell Longev.
2020(9503562)2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Baker DJ and Petersen RC: Cellular
senescence in brain aging and neurodegenerative diseases: Evidence
and perspectives. J Clin Invest. 128:1208–1216. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Grootaert MOJ, Moulis M, Roth L, Martinet
W, Vindis C, Bennett MR and De Meyer GRY: Vascular smooth muscle
cell death, autophagy and senescence in atherosclerosis. Cardiovasc
Res. 114:622–634. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Lee S and Schmitt CA: The dynamic nature
of senescence in cancer. Nat Cell Biol. 21:94–101. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wang F, Cai F, Shi R, Wang XH and Wu XT:
Aging and age related stresses: A senescence mechanism of
intervertebral disc degeneration. Osteoarthritis Cartilage.
24:398–408. 2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Di Micco R, Krizhanovsky V, Baker D and
d'Adda di Fagagna F: Cellular senescence in ageing: From mechanisms
to therapeutic opportunities. Nat Rev Mol Cell Biol. 22:75–95.
2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Youle RJ and Narendra DP: Mechanisms of
mitophagy. Nat Rev Mol Cell Biol. 12:9–14. 2011.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Sun K, Jing X, Guo J, Yao X and Guo F:
Mitophagy in degenerative joint diseases. Autophagy: Sep 24, 2020
(Epub ahead of print). doi: 10.1080/15548627.2020.1822097.
|
|
58
|
Doblado L, Lueck C, Rey C, Samhan-Arias
AK, Prieto I, Stacchiotti A and Monsalve M: Mitophagy in human
diseases. Int J Mol Sci. 22(3903)2021.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Wang Y, Shen J, Chen Y, Liu H, Zhou H, Bai
Z, Hu Z and Guo X: PINK1 protects against oxidative stress induced
senescence of human nucleus pulposus cells via regulating
mitophagy. Biochem Biophys Res Commun. 504:406–414. 2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zhang Z, Xu T, Chen J, Shao Z, Wang K, Yan
Y, Wu C, Lin J, Wang H, Gao W, et al: Parkin-mediated mitophagy as
a potential therapeutic target for intervertebral disc
degeneration. Cell Death Dis. 9(980)2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Wang Y, Wang H, Zhuo Y, Hu Y, Zhang Z, Ye
J, Liu L, Luo L, Zhao C, Zhou Q and Li P: SIRT1 alleviates
high-magnitude compression-induced senescence in nucleus pulposus
cells via PINK1-dependent mitophagy. Aging (Albany NY).
12:16126–16141. 2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Lyu F, Zheng C, Wang H, Nie C, Ma X, Xia
X, Zhu W, Jin X, Hu Y, Sun Y, et al: Establishment of a
clinician-led guideline on the diagnosis and treatment of Hirayama
disease using a modified Delphi technique. Clin Neurophysiol.
131:1311–1319. 2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Wang XB, Wang H, Long HQ, Li DY and Zheng
X: LINC00641 regulates autophagy and intervertebral disc
degeneration by acting as a competitive endogenous RNA of
miR-153-3p under nutrition deprivation stress. J Cell Physiol.
234:7115–7127. 2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Dai S, Liang T, Shi X, Luo Z and Yang H:
Salvianolic acid B protects intervertebral discs from oxidative
stress-induced degeneration via activation of the JAK2/STAT3
signaling pathway. Oxid Med Cell Longev.
2021(6672978)2021.PubMed/NCBI View Article : Google Scholar
|
|
65
|
He R, Wang Z, Cui M, Liu S, Wu W, Chen M,
Wu Y, Qu Y, Lin H, Chen S, et al: HIF1A Alleviates
compression-induced apoptosis of nucleus pulposus derived stem
cells via upregulating autophagy. Autophagy: Jan 18, 2021 (Epub
ahead of print). doi: 10.1080/15548627.2021.1872227.
|
|
66
|
Lan T, Shiyu-Hu Shen Z, Yan B and Chen J:
New insights into the interplay between miRNAs and autophagy in the
aging of intervertebral discs. Ageing Res Rev.
65(101227)2021.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Yang RZ, Xu WN, Zheng HL, Zheng XF, Li B,
Jiang LS and Jiang SD: Involvement of oxidative stress-induced
annulus fibrosus cell and nucleus pulposus cell ferroptosis in
intervertebral disc degeneration pathogenesis. J Cell Physiol.
236:2725–2739. 2021.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang
W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs:
Biogenesis, effect and application in human diseases. Mol Cancer.
18(116)2019.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Seimiya T, Otsuka M, Iwata T, Shibata C,
Tanaka E, Suzuki T and Koike K: Emerging roles of exosomal circular
RNAs in cancer. Front Cell Dev Biol. 8(568366)2020.PubMed/NCBI View Article : Google Scholar
|