Introduction
Ovarian cancer (OC) is one of the leading causes of
tumor-associated deaths and most common gynecological malignancies
(1). Current standard treatments,
including surgery and adjuvant chemotherapy or radiotherapy, have
made significant improvements in diagnosing and treating OC
(2). However, the prognosis is
still unfavorable, especially for patients with OC at advanced
stage when first diagnosed. It has been widely accepted that OC
pathogenesis is accompanied by malignant uncontrolled
proliferation, extensive invasion and lymphatic metastasis, which
is regulated by aberrantly expressed genes (3,4).
Consequently, there is a great need for research to elucidate the
molecular mechanisms associated with OC pathogenesis to help
improve the patient outcomes.
The family of discs large-associated protein (DLGAP)
composed of five members (DLGAP1-5) were originally found in rats,
which have three key domains, including a 14-amino-acid repeat
domain (5), a dynein light chain
domain (6) and a guanylate
kinase-associated protein homology domain (7). DLGAP1-4 have been reported to
participate in a variety of neurological disorders, such as
cerebellar ataxia, obsessive compulsive disorder and autism
spectrum disease (8-10).
In the present study, DLGAP5 (also named as HURP or KIAA0008) is of
great interest for it is mainly associated with to various types of
cancer (11-13).
For instance, DLGAP5 was identified as a cycle cycle-related gene
associated with long-term in vitro proliferation in patients
with acute myeloid leukemia (14).
Bioinformatics analysis by different investigators indicated that
DLGAP5 was correlated with poor prognosis in anaplastic thyroid
carcinoma (15), non-small cell
lung cancer (16,17), pancreatic carcinoma (18) and glioblastoma (19). Loss-of-functional experiments showed
that DLGAP5 could promote cell growth, proliferation and migration
in hepatocellular carcinoma (20,21).
Notably, Liu et al (22) and
Shen et al (23)
consistently showed that DLGAP5 was aberrantly expressed and
correlated with prognosis in OC. Nevertheless, the biological
function of DLGAP5 in OC still has not been reported yet.
Therefore, the present study first searched the
Oncomine microarray database (http://www.oncomine.org) and Kaplan-Meier Plotter
(http://kmplot.com/analysis/) to
investigate the expression pattern and prognostic value of DLGAP5
in OC. By performing loss-of-functional assays, the effects of
DLGAP5 on cell proliferation, cell cycle progression and apoptosis
was further explored in OC cells. The findings of the present study
might provide evidence that targeting DLGAP5 could be a promising
therapeutic strategy in OC.
Materials and methods
Oncomine gene expression analysis
The expression of DLGAP5 in OC was analyzed using
publicly available Oncomine microarray database (http://www.oncomine.org) by searching the following
key words: Gene name, DLGAP5; primary filter, ‘Cancer vs. Normal
Analysis’; and cancer type ‘Ovarian cancer’. A total of six
datasets, including Yoshihara Ovarian (24), Welsh Ovarian (25), Bonome Ovarian (26), The Cancer Genome Atlas (TCGA,
http://tcga-data.nci.nih.gov/tcga/),
Adib Ovarian (27) and Lu Ovarian
(28) were screened to compare the
expression pattern of DLGAP5 between OC tissues with normal
tissues. The DLGAP5 expression data were log-transformed and
median-centered per array with normalized standard deviation
(SD).
Kaplan-Meier plotter analysis
The prognostic value of DLGAP5 was further evaluated
in OC using Kaplan-Meier Plotter (http://kmplot.com/analysis/), a database that contains
information on 22,277 genes and their effect on survival in 2,977
breast, 1,464 ovarian and 1,715 patients with lung cancer. In the
present study, patients with OC and complete overall survival
information were classified into high and low DLGAP5 expression
groups. The effects of DLGAP5 on overall survival rate were
displayed using a Kaplan-Meier survival plot. Meanwhile, the hazard
ratio was calculated with 95% confidence intervals, log rank
P-value and median survival time.
Cell culture and transfection
Two human OC cell lines, including SKOV3 and CAOV3
were purchased from the American Type Culture Collection and
routinely cultured in RPMI-1640 medium (Sigma-Aldrich; Merck KGaA)
supplemented with 10% FBS (Sigma-Aldrich; Merck KGaA) at 37˚C in a
humidified incubator containing 5% CO2. For DLGAP5
knockdown, SKOV3 and CAOV3 cell were seeded into six-well plates at
a density of 2x105 cells per well and transfected with
50 nM small interfering (si)RNA targeting DLGAP5 (si-DLGAP5-1:
5'-ATTCAAACCACATCAGCCTGC-3' or si-DLGAP5-2:
5'-TTGACCGGAGAAGATAGAGGA-3') or negative control (si-NC:
5'-GAAACACAGCCTGTACGCCTG-3') (all chemically synthesized from
Shanghai GenePharma Co., Ltd.) using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Subsequent experiments were performed
48 h post transfection.
Cell counting kit-8 (CCK-8) assay
Transfected cells were seeded in a 96-well plate at
a density of 3,000 cells per and cultured overnight. At 24, 48 and
72 h, cells in each well were incubated with 10 µl CCK-8 solution
(Biosharp Life Sciences) for 2 h at 37˚C. Absorbance at 450 nm was
recorded using a microplate reader. Each sample was performed in
triplicate.
Colony-formation assay
Transfected cells were plated in six-well culture
plates at 500 cells per well and cultured to attach for 24 h. After
incubation for 14 consecutive days at 37˚C, naturally formed
colonies were fixed for 30 min on ice with 70% ethanol and stained
with 0.1% crystal violet for 15 min at room temperature. Images of
the colonies (each colony contains at least 50 cells) were captured
and counted in five randomly selected fields under a light
microscope (magnification, x25). Each sample was performed in
triplicate.
Flow cytometry analysis
Transfected cells were harvested by trypsinization,
washed with cold PBS and fixed with 70% cold ethanol for 30 min at
ice. For cell cycle analysis, fixed cells were washed and stained
with propidium iodide (PI)/RNase A solution (BD Biosciences) in
darkness for 30 min at 37˚C. Stained cells were applied for cell
cycle distribution analysis using a FACSCalibur flow cytometer (BD
Biosciences). For apoptosis analysis, fixed cells were stained with
Annexin V-fluorescein isothiocyanate/PI apoptosis detection (BD
Biosciences), followed by flow cytometry analysis according to the
manufacturer's instructions. Data were analyzed using
FlowJo™ v.10.8 software (FlowJo, LLC).
Western blot analysis
All protein samples were extracted and the
corresponding protein concentration were determined according to
the instructions of RIPA lysis buffer and BCA protein assay
provided by Beyotime Institute of Biotechnology, respectively.
Next, protein samples (30 µg/lane) were separated on 12% SDS-PAGE
gel and transferred onto polyvinylidene fluoride membranes. After
conventional blocking with 5% skimmed non-fat dry milk (Cell
Signaling Technology, Inc.) for 2 h at room temperature, the
membranes were incubated overnight at 4˚C with primary
antibodies against CDK1 (1:1,000; cat. no. ab131450; Abcam), Cyclin
B1 (1:1,000; cat. no. 4138; Cell Signaling Technology, Inc.), Bax
(1:1,000; cat. no. 2772; Cell Signaling Technology, Inc.), Bcl-2
(1:1,000; cat no. sc-7382; Santa Cruz Biotechnology Inc.) and GAPDH
(1:1,000; cat. no. G9545; Sigma-Aldrich; Merck KGaA) at 4˚C,
followed by incubation with secondary antibody conjugated to
horseradish peroxidase (1:5,000; cat. no. sc-2357; Santa Cruz
Biotechnology, Inc.) for 2 h at room temperature. The protein bands
were visualized with an enhanced chemiluminescence substrate
(Beyotime Institute of Biotechnology). Results of western blots
were analyzed using ImageLab software version 4.1 (Bio-Rad
Laboratories, Inc.).
Statistical analysis
Data were analyzed using GraphPad Prism 6.0
(GraphPad Software, Inc.) and expressed as mean ± SD of three
independent experiments. One way ANOVA followed by the post hoc
Tukey's HSD test was used to assess the differences among different
groups. The Renyi test was used for analyzing survival curves.
P<0.05 was used to indicate a statistically significant
difference.
Results
DLGAP5 expression in human OC
In order to obtain the expression pattern of DLGAP5
in OC, multiple cancer microarray data sets available from Oncomine
database were downloaded to determine the differential mRNA
expression of DLGAP5 between OC tissues and normal tissues. As
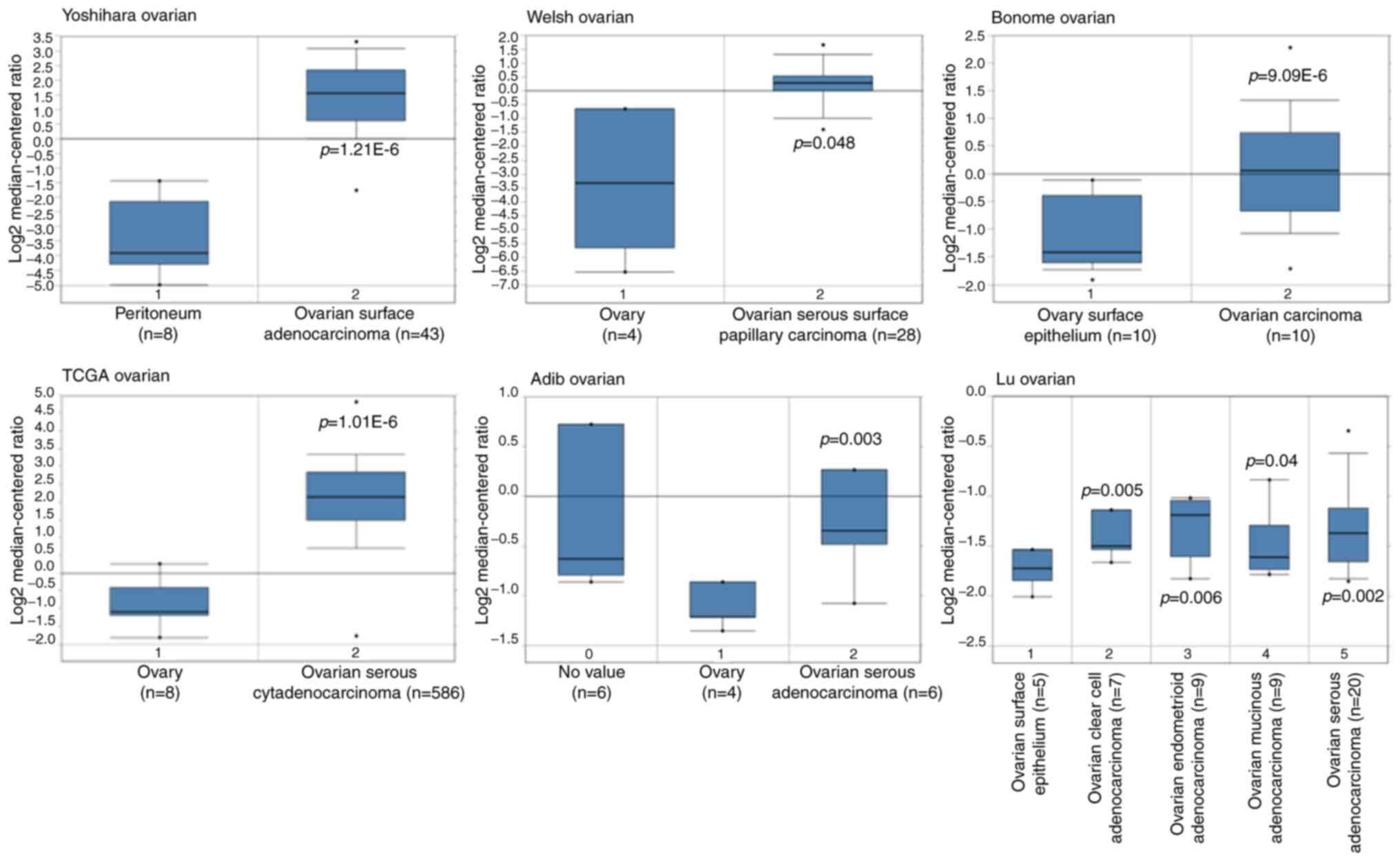
shown in Fig. 1, the mRNA
expression of DLGAP5 was markedly increased in OC tissues,
including epithelium OC (for example, serous, endometrioid,
mucinous and clear cell subtypes) vs. normal tissues in the
Yoshihara Ovarian dataset (P=1.21x106), Welsh Ovarian
dataset (P=0.048), Bonome Ovarian dataset (P=9.09E-6), TCGA
database, (P=1.01E-6), Adib Ovarian dataset (P=0.003) and Lu
Ovarian dataset (P<0.05). The aforementioned observations
highlighted the overexpression of DLGAP5 in the development of
OC.
Prognostic value of DLGAP5 in OC
Next, the focus was on the survival information of
patients with OC to analyze the prognostic value of DLGAP5 using
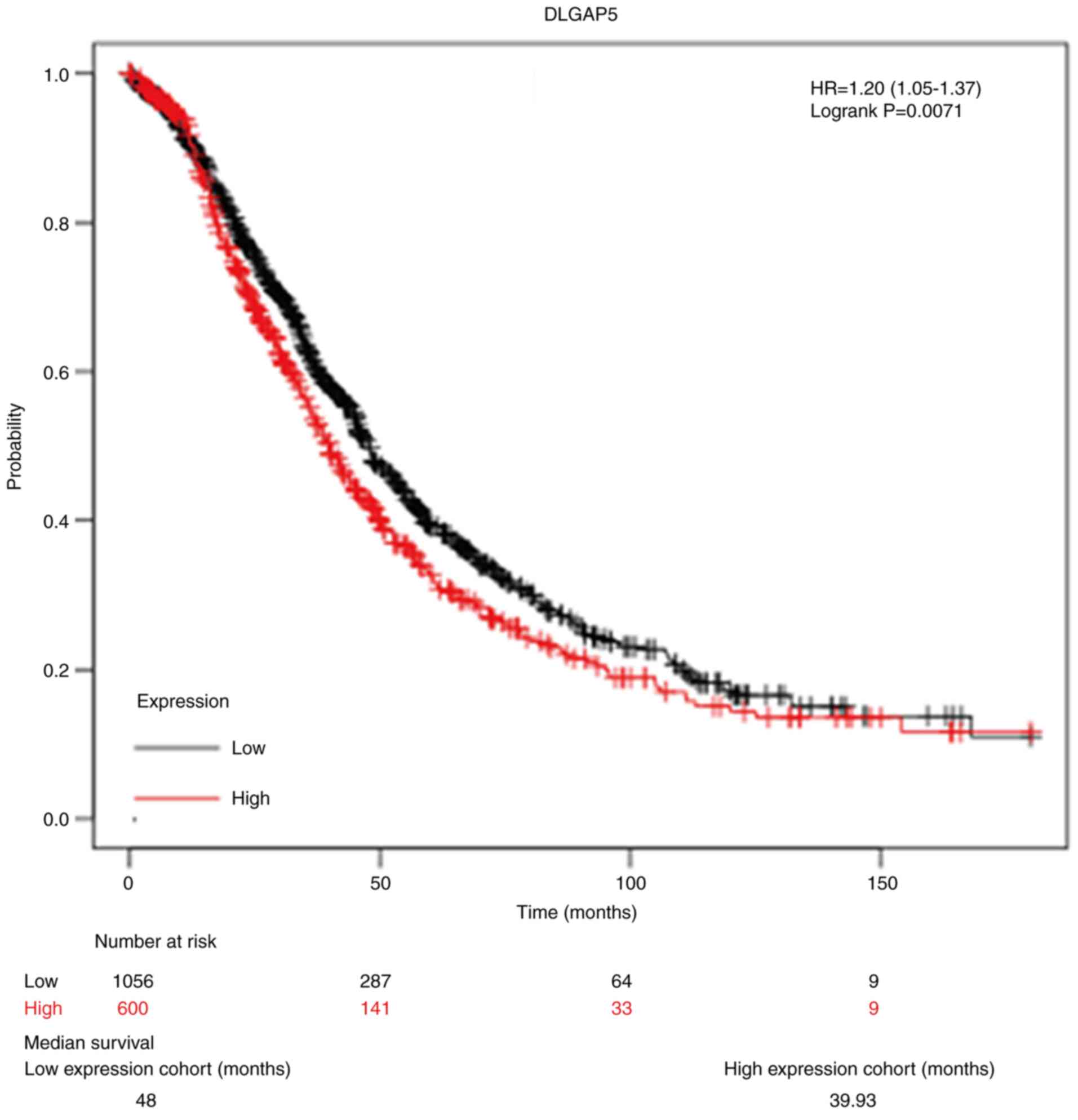
online Kaplan-Meier analysis. As depicted in Fig. 2, 1,656 patients with OC were divided
into high DLGAP5 expression group (n=600) and low DLGAP5 expression
group (n=1,056) according to the median value of DLGAP5 expression.
Kaplan-Meier survival curve analysis showed that high expression of
DLGAP5 was negatively associated with overall survival (HR=1.20,
95% CI: 1.05-1.37, P=0.0071). The median survival in the low
expression group and high expression group was 48 and 39.93 months,
respectively.
Knockdown of DLGAP5 significantly
suppresses cell proliferation in OC cells
Considering the overexpression of DLGAP5 in OC
tissues, loss-of-functional experiments were subsequently performed
to investigate the function of DLGAP5 in OC cells. Firstly, two OC
cell lines (SKOV3 and CAOV3) were transfected with two different
siRNAs targeting DLGAP5. As demonstrated by western blot analysis,
both si-DLGAP5-1 and si-DLGAP5-2 caused a notable downregulation in
DLGAP5 expression in SKOV3 and CAOV3 cells compared with si-NC
transfection, while there was no significant change in DLGAP5
expression between control and si-NC group (Fig. 3A). Furthermore, si-DLGAP5-2
transfection caused stronger suppressive effects on DLGAP5 protein
expression compared with si-DLGAP5-1 transfection. CCK-8 assay
showed that significant inhibition of cell proliferation was
observed in SKOV3 (at 48 and 72 h) and CAOV3 (at 24, 48 and 72 h)
cells after transfection with either si-DLGAP5-1 or si-DLGAP5-2
compared with si-NC transfection, while there was no significant
change in cell proliferation ability between control and si-NC
group (Fig. 3B). si-DLGAP5-2
transfection had stronger suppressive effects on the proliferation
ability of SKOV3 and CAOV3 cells compared with the effects of
si-DLGAP5-1. Thus, si-DLGAP5-2 transfection was selected for the
subsequent experiments. Consistent with CCK-8 assay, the number of
colonies was significantly decreased in the si-DLGAP5-2
transfection group compared with the si-NC group in both SKOV3 and
CAOV3 cells (Fig. 3C and D). The aforementioned data indicated that
DLGAP5 might be an oncogene by promoting cell proliferation in
OC.
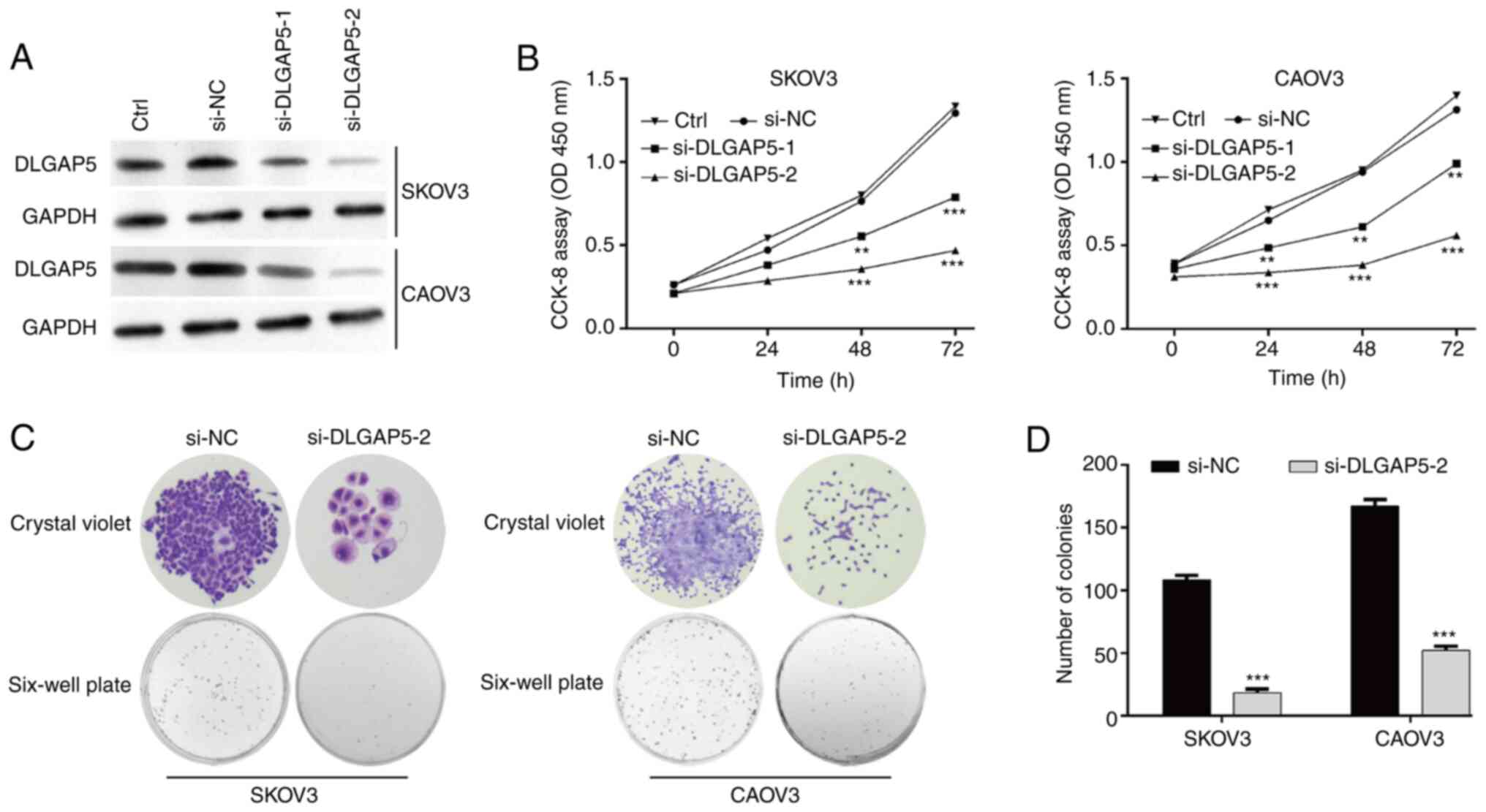 | Figure 3Knockdown of DLGAP5 significantly
suppresses cell proliferation in ovarian cancer cells. (A) The
expression of DLGAP5 protein was measured in SKOV3 and CAOV3 cells
in control (Ctrl), si-NC, si-DLGAP5-1 or si-DLGAP5-2 group. (B)
Cell Counting Kit-8 assay was performed to analyze cell
proliferation in SKOV3 and CAOV3 cells in Ctrl, si-NC, si-DLGAP5-1
or si-DLGAP5-2 group. (C) Representative images of colonies in
SKOV3 and CAOV3 cells after transfection with si-DLGAP5-2 or si-NC
are displayed in left panel (magnification, x50) and (D)
quantification of colonies is shown in right panel. Data are
expressed as mean ± SD of three independent experiments.
**P<0.01, ***P<0.001, vs. si-NC. si-,
small interfering RNA; Ctrl, control; NC, negative control. |
Knockdown of DLGAP5 causes
G2/M phase arrest and apoptosis in OC cells
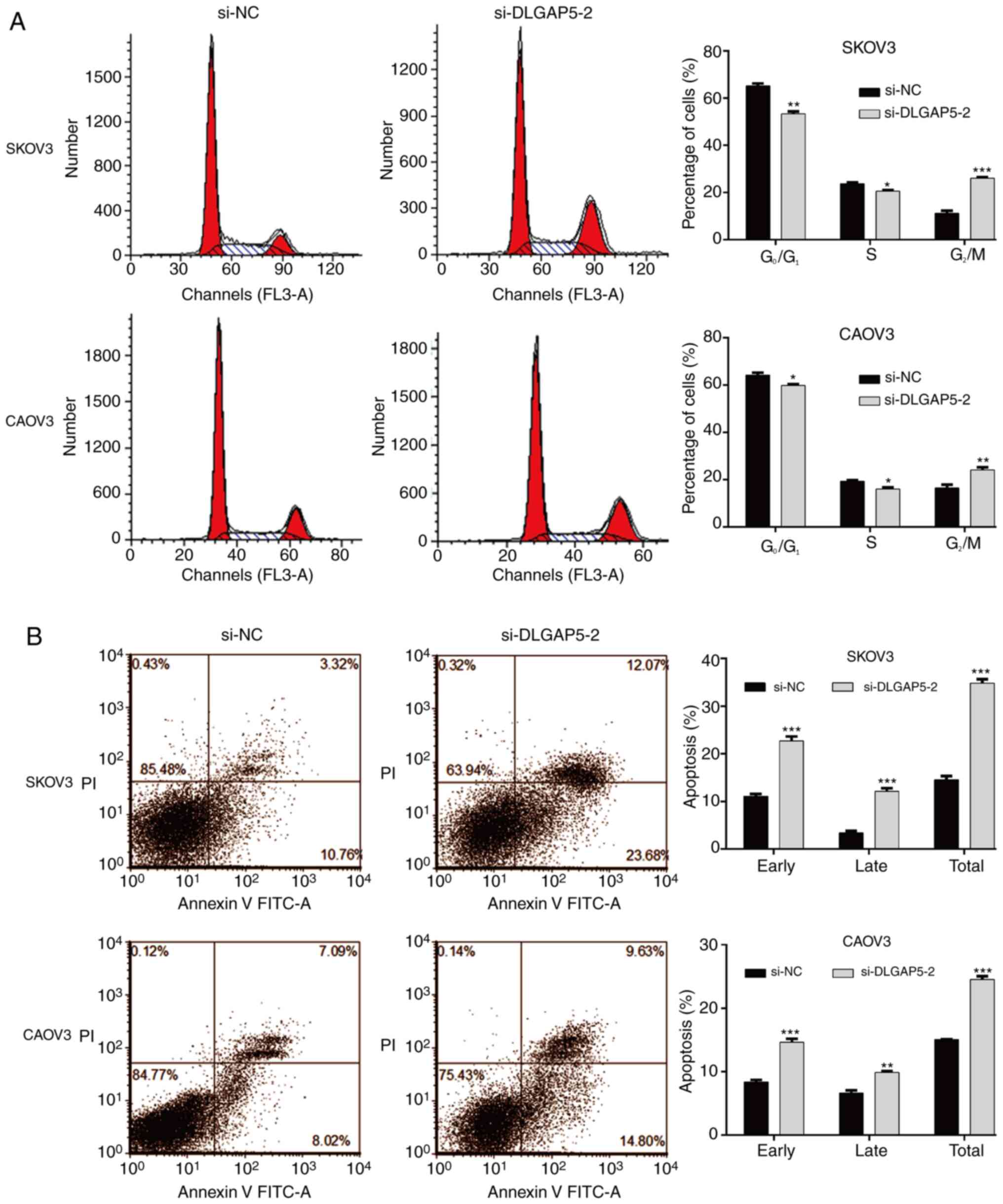
Subsequently, the effects of DLGAP5 knockdown on
cell cycle progression and apoptosis was studied by flow cytometry
analysis. As shown in Fig. 4A, the
percentage of cells at G0/G1 and S phases was
significantly decreased, accordingly accompanied with increased
cell proportion at G2/M phase in si-DLGAP5-2 group
compared with the si-NC group in SKOV3 and CAOV3 cells, which
indicated that knockdown of DLGAP5 caused cell cycle
G2/M phase arrest. Moreover, it was found that knockdown
of DLGAP5 remarkedly increased the early, late and overall
apoptotic cells in both SKOV3 and CAOV3 cells (Fig. 4B).
Knockdown of DLGAP5 regulates the
protein expression levels associated with G2/M
transition and apoptosis
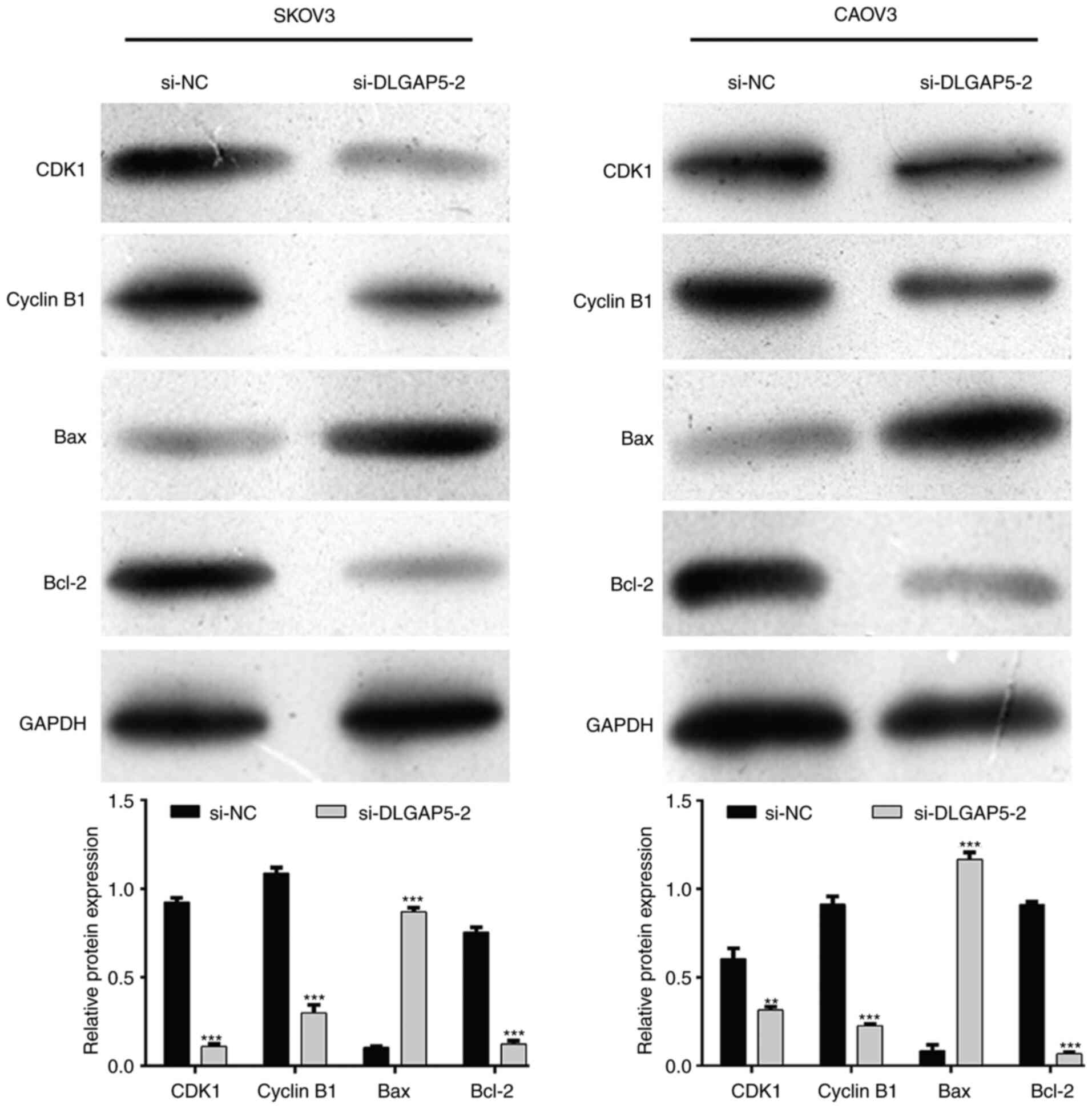
To explore the mechanisms underlying DLGAP5
regulating cell cycle progression and apoptosis, the expression
levels of major cell cycle-promoting factors and apoptosis
regulatory factors were analyzed. As shown in Fig. 5, the expression of CDK1 and Cyclin
B1, associated with the G2/M checkpoint, was notably
downregulated after DLGAP5 knockdown in both SKOV3 and CAOV3 cells.
Meanwhile, DLGAP5 knockdown activated the pro-apoptotic Bax, while
suppressing the anti-apoptotic Bcl-2 expression in both SKOV3 and
CAOV3 cells.
Discussion
In the present study, DLGAP5 expression was found to
be significantly overexpressed in OC tissues and negatively
associated with overall survival in patients with OC by
bioinformatics analysis. The Lu Ovarian dataset in Fig. 1 demonstrated that the mRNA
expression of DLGAP5 was different between mucinous OC and serous,
endometrioid and clear cell OC, of which endometrioid OC presented
higher DLGAP5 mRNA expression. The present study focused on the
expression of DLGAP5 or prognosis in tumor tissues; however, more
clinical samples should be collected to confirm this. In fact,
DLGAP5 has been reported to be upregulated in hepatocellular cancer
(13,29), urinary bladder transitional cell
carcinoma (30) and transitional
cell carcinoma (31). Consistent
with the bioinformatic analysis, DLGAP5 was identified and
validated as one of the most upregulated genes regulating cell
division and mitotic spindle formation in ovarian granulosa cells
with cancer-like characteristics (32). DLGAP5 was also screened as a hub
gene and was associated with the prognosis of endometrial carcinoma
(33). Interestingly, the Gene
Expression Omnibus database showed that DLGAP5 was one of the core
gene with more than ten degrees in epithelial OC (22). GEPIA database analysis indicated
that DLGAP5 was highly expressed in OC specimens and was associated
with worse prognosis (23).
Furthermore, DLGAP5 was overexpressed and associated with worse
overall survival in distinct molecular colorectal cancer subtypes
(34). The aforementioned evidences
suggested that DLGAP5 might be candidate target for the diagnosis
and treatment of OC.
Next, two OC cell lines (SKOV3 and CAOV3) were
selected to investigate the biological function of DLGAP5, by
successfully constructing DLGAP5-silenced OC cell lines. Two
different siRNAs targeting DLGAP5 (si-DLGAP5-1 and si-DLGAP5) were
designed to knockdown DLGAP5 expression and it was found that
si-DLGAP5-2 transfection caused stronger suppressive effects on
DLGAP5 protein expression compared with si-DLGAP5-1 transfection.
Notably, si-DLGAP5-2 transfection had stronger suppressive effects
on the proliferation ability of SKOV3 and CAOV3 cells compared with
si-DLGAP5-1 transfection. The differences in knockdown efficiency
between si-DLGAP5-2 and si-DLGAP5-1 are ascribed to the different
sequences designed for targeting DLGAP5 and different response of
OC cells to the designed sequences. It was further found that
knockdown of DLGAP5 significantly suppressed colony formation,
induced cell cycle G2/M phase arrest and apoptosis in OC
cells. At the molecular level, it was further demonstrated that
knockdown of DLGAP5 downregulated the expression of CDK1, Cyclin B1
and Bcl-2, while upregulating Bax expression in both SKOV3 and
CAOV3 cells. To the best of our knowledge, normal cell cycle
regulation comprised of G0/G1, S,
G2/M stages requires the balance of growth factors and
growth inhibitor factors, while its deregulation often causes
uncontrolled malignant proliferation involved in the pathogenesis
of tumors (35,36). DLGAP5, as a microtubule-associated
protein, promotes spindle formation (37), novel tubulin sheet formation
(38) and the capture of spindle by
kinetochore (39). Associated
studies indicated that DLGAP5 is periodically changed as the cells
progress from M to G0/G phase, whose degradation could
be modulated by CDK1/Cyclin B (40,41).
Through GSEA analysis, Zhang et al (42) found that DLGAP5 is involved in the
G1/S phase transition and M/G1 transition. In
addition, DLGAP5 participated in the NUSAP1 knockdown
downregulating the expression of genes associated with
G2/M phase, including CDK1 expression in invasive breast
cancer cells (42). Liao et
al (20) also reported that the
silencing of DLGAP5 gene expression by RNA interference
significantly suppressed cell growth and colony formation in
vitro in hepatocellular carcinoma cells. Based on the
aforementioned facts, one might speculate that knockdown of DLGAP5
inhibited cell proliferation by inducing G2/M phase
arrest via downregulating CDK1/Cyclin B1 expression in OC cells.
Meanwhile, it was found that the knockdown of DLGAP5 promoted
apoptotic cells by regulating Bax and Bcl-2 expression levels.
Similarly, Kuo et al (21)
indicated that knockdown of DLGAP5 sensitized SK-Hep-1 cells to
cisplatin and resulted in smaller tumors in nude mice via promoting
apoptosis in hepatocellular carcinoma. The overexpression of DLGAP5
suppressed γ-irradiation-induced apoptosis in prostate cancer cells
(43).
In conclusion, it was revealed that DLGAP5 was
overexpressed in OC tissues and predicted poor prognosis in
patients with OC. The potential mechanisms of DLGAP5 in
facilitating OC cell proliferation via regulating G2/M
phase and apoptosis was also demonstrated, suggesting that DLGAP5
might be a potential therapeutic target of OC. In addition, some
limitations of the present study were as follows: A non-cancerous
cell line as a normal/negative control was not used in the present
study. DLGAP5 expression will be analyzed in clinical samples by
immunohistochemistry, and the association between DLGAP5 expression
and the prognosis of ovarian cancer will be determined in
subsequent investigations.
Acknowledgements
Not applicable.
Funding
Funding: This study was sponsored by the Hubei Natural Science
Foundation (grant no. 2017 CFB335).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ designed the study, performed the experiments and
made substantial contributions to the conception of this work. YL
performed the experiments and assessed the literature. ST and XQ
collected the data and were involved in drafting the manuscript or
revising it critically for important intellectual content. LL and
JTZ analyzed the data. JZ and BL performed the experiments and gave
suggestions for the revision of this manuscript. HZ and JZ
confirmed the authenticity of all the raw data and designed this
work. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lheureux S, Braunstein M and Oza AM:
Epithelial ovarian cancer: Evolution of management in the era of
precision medicine. CA Cancer J Clin. 69:280–304. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Reid BM, Permuth JB and Sellers TA:
Epidemiology of ovarian cancer: A review. Cancer Biol Med. 14:9–32.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Smith RA, Andrews KS, Brooks D, Fedewa SA,
Manassaram-Baptiste D, Saslow D, Brawley OW and Wender RC: Cancer
screening in the United States, 2018: A review of current American
cancer society guidelines and current issues in cancer screening.
CA Cancer J Clin. 68:297–316. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Han CY, Patten DA, Richardson RB, Harper
ME and Tsang BK: Tumor metabolism regulating chemosensitivity in
ovarian cancer. Genes Cancer. 9:155–175. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sabio G, Arthur JS, Kuma Y, Peggie M, Carr
J, Murray-Tait V, Centeno F, Goedert M, Morrice NA and Cuenda A:
p38gamma regulates the localisation of SAP97 in the cytoskeleton by
modulating its interaction with GKAP. EMBO J. 24:1134–1145.
2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Naisbitt S, Valtschanoff J, Allison DW,
Sala C, Kim E, Craig AM, Weinberg RJ and Sheng M: Interaction of
the postsynaptic density-95/guanylate kinase domain-associated
protein complex with a light chain of myosin-V and dynein. J
Neurosci. 20:4524–4534. 2000.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tong J, Yang H, Eom SH, Chun C and Im YJ:
Structure of the GH1 domain of guanylate kinase-associated protein
from Rattus norvegicus. Biochem Biophys Res Commun. 452:130–135.
2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Minocherhomji S, Hansen C, Kim HG, Mang Y,
Bak M, Guldberg P, Papadopoulos N, Eiberg H, Doh GD, Møllgård K, et
al: Epigenetic remodelling and dysregulation of DLGAP4 is linked
with early-onset cerebellar ataxia. Hum Mol. 23:6163–6176.
2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ryu S, Oh S, Cho EY, Nam HJ, Yoo JH, Park
T, Joo YH, Kwon JS and Hong KS: Interaction between genetic
variants of DLGAP3 and SLC1A1 affecting the risk of atypical
antipsychotics-induced obsessive-compulsive symptoms. Am J Med
Genet B Neuropsychiatr Genet. 156B:949–959. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kajimoto Y, Shirakawa O, Lin XH, Hashimoto
T, Kitamura N, Murakami N, Takumi T and Maeda K: Synapse-associated
protein 90/postsynaptic density-95-associated protein (SAPAP) is
expressed differentially in phencyclidine-treated Rats and is
increased in the nucleus accumbens of patients with schizophrenia.
Neuropsychopharmacology. 28:1831–1839. 2003.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye
QH, Wang L, Zhou J, Qiu SJ, Li Y, et al: A decade's studies on
metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol.
130:187–196. 2004.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Stangeland B, Mughal AA, Grieg Z, Sandberg
CJ, Joel M, Nygård S, Meling T, Murrell W, Vik Mo EO and Langmoen
IA: Combined expressional analysis, bioinformatics and targeted
proteomics identify new potential therapeutic targets in
glioblastoma stem cells. Oncotarget. 6:26192–26215. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tsou AP, Yang CW, Huang CY, Yu RC, Lee YC,
Chang CW, Chen BR, Chung YF, Fann MJ, Chi CW, et al: Identification
of a novel cell cycle regulated gene, HURP, overexpressed in human
hepatocellular carcinoma. Oncogene. 22:298–307. 2003.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hatfield KJ, Reikvam H and Bruserud O:
Identification of a subset of patients with acute myeloid leukemia
characterized by long-term in vitro proliferation and altered cell
cycle regulation of the leukemic cells. Expert Opin Ther Targets.
18:1237–1251. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Weinberger P, Ponny SR, Xu H, Bai S,
Smallridge R, Copland J and Sharma A: Cell cycle M-phase genes are
highly upregulated in anaplastic thyroid carcinoma. Thyroid.
27:236–252. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Schneider MA, Christopoulos P, Muley T,
Warth A, Klingmueller U, Thomas M, Herth FJ, Dienemann H, Mueller
NS, Theis F and Meister M: AURKA, DLGAP5, TPX2, KIF11 and CKAP5:
Five specific mitosis-associated genes correlate with poor
prognosis for non-small cell lung cancer patients. Int J Oncol.
50:365–372. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Shi YX, Yin JY, Shen Y, Zhang W, Zhou HH
and Liu ZQ: Genome-scale analysis identifies NEK2, DLGAP5 and ECT2
as promising diagnostic and prognostic biomarkers in human lung
cancer. Sci Rep. 7(8072)2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhou Z, Cheng Y, Jiang Y, Liu S, Zhang M,
Liu J and Zhao Q: Ten hub genes associated with progression and
prognosis of pancreatic carcinoma identified by co-expression
analysis. Int J Biol Sci. 14:124–136. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhou Y, Yang L, Zhang X, Chen R, Chen X,
Tang W and Zhang M: Identification of potential biomarkers in
glioblastoma through bioinformatic analysis and evaluating their
prognostic value. Biomed Res Int. 2019(6581576)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liao W, Liu W, Yuan Q, Liu X, Ou Y, He S,
Yuan S, Qin L, Chen Q, Nong K, et al: Silencing of DLGAP5 by siRNA
significantly inhibits the proliferation and invasion of
hepatocellular carcinoma cells. PLoS One. 8(e80789)2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kuo TC, Chang PY, Huang SF, Chou CK and
Chao CC: Knockdown of HURP inhibits the proliferation of
hepacellular carcinoma cells via downregulation of gankyrin and
accumulation of p53. Biochem Pharmacol. 83:758–768. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu J, Meng H, Li S, Shen Y, Wang H, Shan
W, Qiu J, Zhang J and Cheng W: Identification of potential
biomarkers in association with progression and prognosis in
epithelial ovarian cancer by integrated bioinformatics analysis.
Front Genet. 10(1031)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shen J, Yu S, Sun X, Yin M, Fei J and Zhou
J: Identification of key biomarkers associated with development and
prognosis in patients with ovarian carcinoma: Evidence from
bioinformatic analysis. J Ovarian Res. 12(110)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yoshihara K, Tajima A, Komata D, Yamamoto
T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K,
et al: Gene expression profiling of advanced-stage serous ovarian
cancers distinguishes novel subclasses and implicates ZEB2 in tumor
progression and prognosis. Cancer Sci. 100:1421–1428.
2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Welsh JB, Zarrinkar PP, Sapinoso LM, Kern
SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA and Hampton GM:
Analysis of gene expression profiles in normal and neoplastic
ovarian tissue samples identifies candidate molecular markers of
epithelial ovarian cancer. Proc Natl Acad Sci USA. 98:1176–1181.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bonome T, Levine DA, Shih J, Randonovich
M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC,
Boyd J and Birrer MJ: A gene signature predicting for survival in
suboptimally debulked patients with ovarian cancer. Cancer Res.
68:5478–5486. 2008.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Adib TR, Henderson S, Perrett C, Hewitt D,
Bourmpoulia D, Ledermann J and Boshoff C: Predicting biomarkers for
ovarian cancer using gene-expression microarrays. Br J Cancer.
90:686–692. 2004.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lu KH, Patterson AP, Wang L, Marquez RT,
Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I,
et al: Selection of potential markers for epithelial ovarian cancer
with gene expression arrays and recursive descent partition
analysis. Clin Cancer Res. 10:3291–3300. 2004.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chang ML, Lin SM and Yeh CT: HURP
expression-assisted risk scores identify prognosis distinguishable
subgroups in early stage liver cancer. PLoS One.
6(e26323)2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Huang YL, Chiu AW, Huan SK, Wang YC, Ju JP
and Lu CL: Prognostic significance of hepatoma-up-regulated protein
expression in patients with urinary bladder transitional cell
carcinoma. Anticancer Res. 23:2729–2733. 2003.PubMed/NCBI
|
|
31
|
Chiu AW, Huang YL, Huan SK, Wang YC, Ju
JP, Chen MF and Chou CK: Potential molecular marker for detecting
transitional cell carcinoma. Urology. 60:181–185. 2002.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Brązert M, Kranc W, Chermuła B, Kowalska
K, Jankowski M, Celichowski P, Jeseta M, Piotrowska-Kempisty H,
Pawelczyk L, Zabel M, et al: Human ovarian granulosa cells isolated
during an IVF procedure exhibit differential expression of genes
regulating cell division and mitotic spindle formation. J Clin Med.
8(2026)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang W, Gao L, Wang C, Wang S, Sun D, Li
X, Liu M, Qi Y, Liu J and Lin B: Combining bioinformatics and
experiments to identify and verify key genes with prognostic values
in endometrial carcinoma. J Cancer. 11:716–732. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Branchi V, Garcia SA, Radhakrishnan P,
Győrffy B, Hissa B, Schneider M, Reißfelder C and Schölch S:
Prognostic value of DLGAP5 in colorectal cancer. Int J Colorectal
Dis. 34:1455–1465. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Sterlacci W, Tzankov A, Veits L, Zelger B,
Bihl MP, Foerster A, Augustin F, Fiegl M and Savic S: A
comprehensive analysis of p16 expression, gene status, and promoter
hypermethylation in surgically resected non-small cell lung
carcinomas. J Thorac Oncol. 6:1649–1657. 2011.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Singhal S, Amin KM, Kruklitis R, DeLong P,
Friscia ME, Litzky LA, Putt ME, Kaiser LR and Albelda SM:
Alterations in cell cycle genes in early stage lung adenocarcinoma
identified by expression profiling. Cancer Biol Ther. 2:291–298.
2003.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Santarella RA, Koffa MD, Tittmann P, Gross
H and Hoenger A: HURP wraps microtubule ends with an additional
tubulin sheet that has a novel conformation of tubulin. J Mol Biol.
365:1587–1595. 2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Koffa MD, Casanova CM, Santarella R,
Köcher T, Wilm M and Mattaj IW: HURP is part of a Ran-dependent
complex involved in spindle formation. Curr Biol. 16:743–754.
2006.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Casanova CM, Rybina S, Yokoyama H,
Karsenti E and Mattaj IW: Hepatoma up-regulated protein is required
for chromatin-induced microtubule assembly independently of TPX2.
Mol Biol Cell. 19:4900–4908. 2008.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yu CT, Hsu JM, Lee YC, Tsou AP, Chou CK
and Huang CY: Phosphorylation and stabilization of HURP by
Aurora-A: Implication of HURP as a transforming target of Aurora-A.
Mol Cell Biol. 25:5789–5800. 2005.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hsu JM, Lee YC, Yu CT and Huang CY: Fbx7
functions in the SCF complex regulating Cdk1-cyclin
B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis
by a proline-rich region. J Biol Chem. 279:32592–32602.
2004.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhang X, Pan Y, Fu H and Zhang J:
Nucleolar and spindle associated protein 1 (NUSAP1) inhibits cell
proliferation and enhances susceptibility to epirubicin in invasive
breast cancer cells by regulating cyclin D kinase (CDK1) and DLGAP5
expression. Med Sci Monit. 24:8553–8564. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Hassan M, El Khattouti A, Ejaeidi A, Ma T,
Day WA, Espinoza I, Vijayakumar S and Gomez CR: Elevated expression
of hepatoma up-regulated protein inhibits γ-irradiation-induced
apoptosis of prostate cancer cells. J Cell Biochem. 117:1308–1318.
2016.PubMed/NCBI View Article : Google Scholar
|